Abstract
The nucleotide sequences of CAP59 genes from five serotypes of Cryptococcus neoformans were analyzed for their phylogenetic relationships. Approximately 600-bp genomic DNA fragments of the CAP59 gene were amplified from each isolate by PCR and sequenced. The CAP59 nucleotide sequences of C. neoformans showed more than 90% similarity among the five serotypes. By phylogenetic analysis, their sequences were divided into three clusters: serotypes A and AD, serotypes B and C, and serotype D. In addition, the results of reduced amino acid sequences were similar to the nucleotide sequence data. These data revealed that serotype AD was genetically close to serotype A rather than serotype D, although it had been considered to be a mixed type of serotype A and D by serological analysis. Furthermore, the nucleotide sequences of the serotype B and C isolates of C. neoformans were very similar to each other. These results indicated that serotype B and C isolates belonging to C. neoformans var. gattii were genetically homogeneous and closely related. The molecular analysis of the CAP59 gene will provide useful information for the differentiation of serotypes of C. neoformans and for an understanding of their phylogenetic relationships.
Isolates of Cryptococcus neoformans from human cryptococcosis have been identified by morphological, biochemical, and serological analyses. C. neoformans was divided into five serotypes: A, B, C, D, and AD (1, 2). The serotyping of C. neoformans has been performed immunologically with antisera against the mucopolysaccharide capsule component of the yeast. However, there are some limitations in the serotyping, since noncapsulated mutants and nontypeable isolates have been reported (5, 6). Furthermore, we previously reported that an isolate of C. neoformans var. neoformans (serotype A, D, or AD) showed atypical biochemical characteristics (7). Thus, a new, easier, and more reliable method is required to confirm the serotype.
There have been many investigations of methods for molecular analysis of C. neoformans. A CAP59 gene deletion by homologous integration resulted in an acapsular phenotype, as indicated by Chang and Kwon-Chung (3). Moreover, they demonstrated that capsule-related virulence was accounted for molecularly and that the CAP59 gene is required for capsule formation.
Therefore, this study attempted to differentiate five serotypes of C. neoformans by molecular analysis of CAP59 genes and to investigate their phylogenetic relationships.
MATERIALS AND METHODS
Fungal strains and culture.
The strains of each of the serotypes A, B, C, and D, including the reference strains, and the three strains of serotype AD were used in this study (Table 1). These isolates were inoculated on sunflower seed agar (8) at 25°C, allowed to grow for 5 or 6 days, and then cultured in Sabouraud broth (peptone, 1%; glucose, 4%; yeast extract, 0.05%) at 25°C for 10 days.
TABLE 1.
Isolates of C. neoformans used in this studya
| Strain | Serotype | Origin |
|---|---|---|
| TLD-ui005b | A | Pigeon droppings |
| TLD-ui006 | A | Pigeon droppings |
| TLD-ui014 | A | Pigeon droppings |
| TLD-ui021 | A | Pigeon droppings |
| TLD-ui031 | A | Pigeon droppings |
| TLD-ui045 | A | Pigeon droppings |
| TLD-0261c | D | ATCC34873g |
| TLD-0262 | D | ATCC34874g |
| TLD-0383 | D | HE37 |
| TLD-0384 | D | HE47 |
| TLD-0386 | D | HE151 |
| TLD-0387 | D | HE152 |
| TLD-0343d | AD | CBS 132h |
| TLD-0344 | AD | TIMM 0377 |
| TLD-0345 | AD | TIMM 0384 |
| TLD-0263e | B | ATCC32609i |
| TLD-0341 | B | ATCC24065 |
| TLD-0367 | B | M 9228 |
| TLD-0368 | B | M 9243 |
| TLD-0369 | B | M 9245 |
| TLD-0377 | B | ATCC 76110 |
| TLD-0264f | C | ATCC32608i |
| TLD-0342 | C | NIH 18 |
| TLD-0371 | C | M 9230 |
| TLD-0372 | C | M 9246 |
| TLD-0373 | C | M 9247 |
| TLD-0374 | C | M 9248 |
TLD, Teikyo University School of Medicine Laboratory of Dermatology; ATCC, American Type Culture Collection; NIH, National Institutes of Health; HE, Federal Institute for Health Protection of Consumers and Veterinary Medicine; CBS, Centraalbureau voor Schimmelcultures; TIMM, Teikyo Institute of Medical Mycology; M, Meiji College of Pharmacy.
CAP59 accession no., AB019367.
CAP59 accession no., L26508.
CAP59 accession no., AB019368.
CAP59 accession no., AB019369.
CAP59 accession no., AB019370.
Cross between type strains of F. neoformans; NIH430 × NIH12.
Type strain of C. neoformans.
Type strain of Filobasidiella bacillispora.
Isolation of genomic DNA.
The yeasts were collected by centrifugation at 3,800 × g for 5 min and then homogenized in liquid nitrogen. The samples were lysed with 1 mg of Zymolyase-100T (Takara, Kyoto, Japan) per ml in a lysis buffer containing 0.1 mM EDTA, 1% sodium dodecyl sulfate, 10 mM Tris hydrochloride (pH 8.0), and 0.3% 2-mercaptoethanol at 37°C for 16 h. High-molecular-weight DNAs were obtained from these samples by phenol and chloroform extraction. These DNA samples dissolved in TE buffer (10 mM Tris hydrochloride [pH 8.0]–1 mM EDTA) were used for PCR amplification.
PCR amplification.
The genomic DNA samples (200 ng) of the yeast were amplified by PCR in a reaction mixture (20 μl) containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 200 mM (each) deoxynucleoside triphosphate, 1.0 U of Taq polymerase (Takara), and 0.5 μg each of a pair of primers. The PCR primers were prepared based on the sequences conserved in the C. neoformans CAP59 gene (3). The primer sequences used for amplification of the C. neoformans CAP59 gene were 5′-GAG TGT CTC CGC AAC CCG CA-3′ (primer CAP59 S-2; nucleotides 590 to 598 in the C. neoformans serotype D CAP59 gene [DDBJ-EMBL-GenBank accession no. L26508]) and 5′-CCT ACT CTG CCA AAT CAA CTC-3′ (primer CAP59 R-2; nucleotides 1176 to 1155 in the C. neoformans serotype D CAP59 gene). With these primers, a 597-bp fragment containing about 30% of the coding sequence of the CAP59 gene was amplified (Fig. 1). The PCR amplification was carried out for 35 cycles consisting of template denaturation (1 min at 94°C), primer annealing (1 min at 53°C), and polymerization (3 min at 72°C). The PCR products were electrophoresed through 2% agarose gel and then stained with ethidium bromide and UV irradiation.
FIG. 1.
The CAP59 gene has five exons. The longest exon, a 597-bp fragment containing about 30% of the coding sequence of the gene, was amplified.
Cloning and sequencing of PCR products.
The PCR products from each isolate were gel purified and cloned into pCRII vector (Invitrogen, San Diego, Calif.). The plasmid DNAs from more than three clones of each species were extracted with a Qiagen (Studio City, Calif.) plasmid kit and sequenced by the dideoxy chain termination method using an ABI PRISM 310 genetic analyzer (Perkin-Elmer, Foster City, Calif.).
Phylogenetic analysis.
To examine the phylogenetic relationships, the nucleotide and reduced amino acid sequences were analyzed by Clustal W multiple sequence alignment programs (10) and a phylogenetic tree was constructed by the TREEVIEW phylogeny display program (9). Bootstrap analysis was performed on 1,000 random samples taken from multiple alignments as described by Felsenstein (4).
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the DDBJ-EMBL-GenBank database under the following accession numbers: Filobasidiella neoformans serotype A, AB019367; F. neoformans serotype AD, AB019368; F. neoformans serotype B, AB019369; F. neoformans serotype C, AB019370. Serotype AD was genetically close to serotype A rather than serotype D.
RESULTS
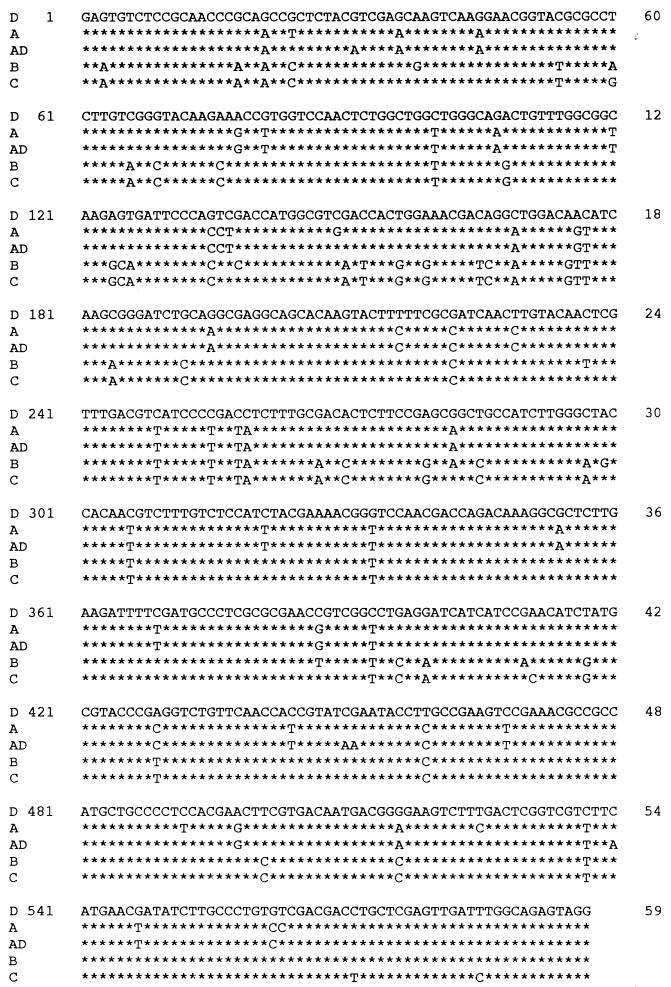
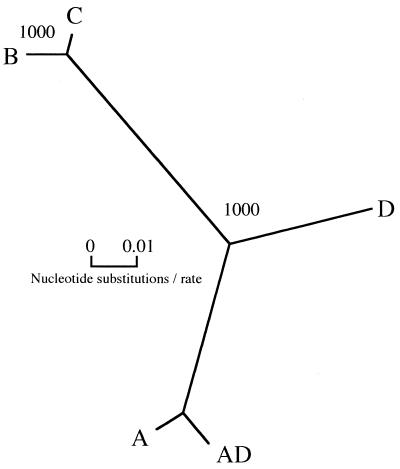
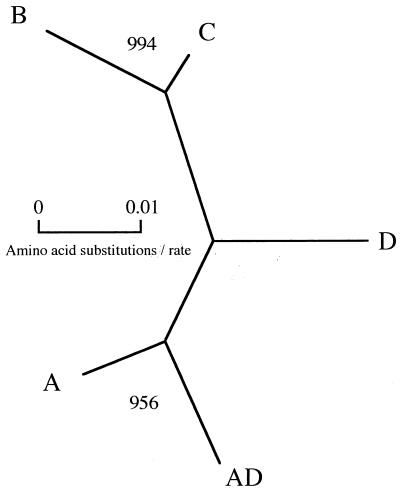
Amplification of the yeast DNAs with degenerate CAP59 primers yielded fragments of approximately 600 bp, consistent with the sizes of CAP59 gene fragments reported previously (3). Nucleotide sequence analysis of the CAP59 gene fragments indicated more than 90% sequence similarity among the isolates of five serotypes (Fig. 2). In addition, reduced amino acid sequence analysis of the CAP59 gene fragments produced the same results. An especially high degree (>92%) of nucleotide and reduced amino acid sequence similarity was noted among the CAP59 gene fragments of serotype A, serotype AD, and serotype D. The phylogenetic analysis revealed that the nucleotide sequences of CAP59 gene fragments from five serotypes of C. neoformans were divided into three clusters: serotypes A and AD, serotypes B and C, and serotype D (Fig. 3). Furthermore, the phylogenetic analysis indicated that reduced amino acid sequences of CAP59 gene fragments were similar to the nucleotide sequences and genetically distinct from each other in serotype (Fig. 4).
FIG. 2.
Alignment of CAP59 sequences of C. neoformans. The sequences of five serotypes of the yeast were aligned by using the Clustal W computer programs. The asterisks indicate conserved serotype D sequence (DDBJ-EMBL-GenBank accession no. L26508).
FIG. 3.
Phylogenetic tree showing relationships of CAP59 gene fragments from five serotypes of C. neoformans based on DNA sequences. The DNA sequences were compared by Clustal W multiple sequence alignment programs, and a phylogenetic tree was constructed by TREEVIEW. Bootstrap analysis was performed on 1,000 random samples. The numbers at the branches were determined by bootstrap analysis, indicating the times in 1,000 repeat samples in a monophylogenic grouping.
FIG. 4.
A phylogenetic tree showing relationships of CAP59 gene fragments from five serotypes of C. neoformans based on reduced amino acid sequences. The reduced amino acid sequences were compared by Clustal W multiple sequence alignment programs, and a phylogenetic tree was constructed by TREEVIEW. Bootstrap analysis was performed on 1,000 random samples. The numbers at the branches were determined by bootstrap analysis, indicating the times in 1,000 repeat samples in a monophylogenic grouping.
DISCUSSION
The isolates of C. neoformans were analyzed using nucleotide sequences of CAP59 genes from five serotypes of the yeast. The phylogenetic relationships based on the CAP59 gene sequence alignment from the isolates of five serotypes agreed with the biochemical and serological analyses, and CAP59 gene fragments showed more than 90% sequence similarity to each other. An especially high degree (>98%) of nucleotide sequence similarity was noted in the CAP59 gene fragments between serotypes A and AD, and also between serotypes B and C. There were no differences in the sequences of the CAP59 gene between isolates of mating types a and α in the same serotype. Although serotype AD has been considered to be a mixed type of serotype A and serotype D by serological analysis, our study revealed that the nucleotide sequence similarity between serotype A and serotype AD was very close and that these two serotypes were included in the same cluster.
Furthermore, the band patterns of serotypes B and C were shown to be similar to each other as in our previous report, by PCR fingerprinting with an FM1 random primer (5′-AGC CGC CTC CAT GGC CCC AGG-3′) (7). The serotypes B and C were genetically homogeneous and closely related in both studies.
The sequence analysis of the CAP59 gene in this study should be very useful for understanding the evolution of C. neoformans as well as for the differentiation of these five serotypes.
ACKNOWLEDGMENTS
We express our deepest gratitude to K. J. Kwon-Chung (National Institute of Health), P. Kielstein (Federal Institute for Health Protection of Consumers and Veterinary Medicine), Katsuhisa Uchida (Teikyo University Research Center for Medical Mycology), and Takako Shinoda (Meiji College of Pharmacy) for providing the isolates.
REFERENCES
- 1.Ansheng L, Nishimura K, Taguchi H, Tanaka R, Shaoxi W, Miyaji M. The isolation of Cryptococcus neoformans from pigeon droppings and serotyping of naturally and clinically sourced isolates in China. Mycopathologia. 1993;124:1–5. doi: 10.1007/BF01103049. [DOI] [PubMed] [Google Scholar]
- 2.Bennett J E, Kwon-Chung K J, Howard D H. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein J. Confidence and phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 5.Fromtling R A, Shadomy H J, Jacobson E S. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia. 1982;79:23–29. doi: 10.1007/BF00636177. [DOI] [PubMed] [Google Scholar]
- 6.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura Y, Kano R, Sato H, Watanabe S, Takahashi H, Hasegawa A. Isolates of Cryptococcus neoformans serotype A and D developed on canavanine-glycine-bromthymol blue medium. Mycoses. 1997;41:35–40. doi: 10.1111/j.1439-0507.1998.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 8.Pal M, Onda C, Hasegawa A. Sexual compatibility of clinical and environmental isolates of Cryptococcus neoformans. Jpn J Med Mycol. 1991;32:101–106. [Google Scholar]
- 9.Page R D M. TREEVIEW: an application to display phyologenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 10.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]