ABSTRACT
Under adverse conditions, tRNAs are processed into fragments called tRNA-derived stress-induced RNAs (tiRNAs) by stress-responsive ribonucleases (RNases) such as angiogenin (ANG). Recent studies have reported several biological functions of synthetic tiRNAs lacking post-transcriptional modifications found on endogenous tiRNAs. Here we describe a simple and reproducible method to efficiently isolate ANG-cleaved tiRNAs from endogenous tRNAs. Using this in vitro method, more than 50% of mature tRNAs are cleaved into tiRNAs which can be enriched using complementary oligonucleotides. Using this method, the yield of isolated endogenous 5ʹ-tiRNAGly-GCC was increased about fivefold compared to when tiRNAs were obtained by cellular treatment of ANG. Although the non-specific ribonuclease activity of ANG is much lower than that of RNase A, we show that ANG cleaves physiologically folded tRNAs as efficiently as bovine RNase A. These results suggest that ANG is highly specialized to cleave physiologically folded tRNAs. Our method will greatly facilitate the analysis of endogenous tiRNAs to elucidate the physiological functions of ANG.
KEYWORDS: tRNA, tiRNAs, tRNA-derived fragments, stress
Introduction
Transfer RNAs (tRNAs) are traditionally considered as adapter molecules that assist the ribosome to produce proteins. Beyond this canonical function, tRNAs also participate in non-canonical functions ranging from cell signalling, survival and apoptosis, and stress responses (reviewed in [1,2]). In addition to the functions of mature full-length tRNAs, a growing number of studies have shown that diverse biological functions are triggered by small non-coding RNAs (ncRNAs) derived from both precursor and mature forms of tRNAs. These ncRNAs are often named tRNA-derived RNAs (tdRs) or tRNA-derived fragments (tRFs) (reviewed in refs [3–6]). As various classes of tdRs/tRFs have been identified thanks to the development of high-throughput sequencing technologies [7], this field is currently gaining significant attention and growing rapidly.
One of the major and best-studied class of tRFs generated by specific cleavage around anticodon loops in response to stress are known as tRNA-derived stress-induced RNAs (tiRNAs) [8], or tRNA halves [9]. Specific ribonucleases (RNases) are responsible for tiRNA production [10–12]. One such RNase is angiogenin (ANG), a secreted RNase that is a member of the RNase A superfamily (reviewed in [13,14]). In optimal growth conditions, ANG is found in both the nucleus, where it promotes the synthesis of ribosomal RNAs, and the cytoplasm, where it complexes with the endogenous RNase inhibitor, RNH1. However, under various stress conditions such as oxidative stress or nutrient starvation, ANG becomes activated by dissociation from RNH1 or by translocation from the nucleus to the cytoplasm [15]. Activated ANG targets mature tRNAs in their anticodon loops, thus producing two subclasses of tiRNAs: 5ʹ- and 3ʹ-tiRNAs [8] (or 5ʹ- and 3ʹ-halves of tRNAs [9], respectively).
tiRNAs have been implicated in several biological functions. It has been reported that both 5ʹ- and 3ʹ-tiRNAs promote cell survival under stress through their interaction with cytochrome C [16]. In addition, specific 5ʹ-tiRNAs, derived from tRNAAla and tRNACys, inhibit translation in vitro and in cellulo [17]. In cells, 5ʹ-tiRNAAla/Cys-mediated inhibition of mRNA translation promotes the formation of stress granules (SGs) [17,18], prosurvival RNA granules that help cells to cope with stress [19–21]. It should be noted that most of these experiments were performed using synthetic tiRNA molecules, although gel-purified endogenous 5ʹ-tiRNAs also promoted translation arrest and SG formation [8,17,18].
The most striking difference between endogenous RNAs and synthetic counterparts is that endogenous RNAs are post-transcriptionally modified during maturation. Post-transcriptional RNA modifications, found in both mRNAs and ncRNAs, play various roles in cell physiology (reviewed in [22]). Extensive RNA modifications are a characteristic feature of tRNAs [23,24]. Many tRNA modifications are evolutionarily conserved reflecting their critical roles in tRNA folding and in codon-anticodon interactions (reviewed in [25]). Because synthetic RNAs do not have such modifications, studies using synthetic RNAs may not reflect the physiological roles of their endogenous counterparts. Recent data have suggested that specific RNA modifications modulate the efficiency of ANG-mediated tRNA cleavage (reviewed in [26]). Loss of 5-methylcytosine (m5C) modifications through deletion of Dnmt2 or NSun2 enhances ANG-mediated tRNA cleavage [27–29]. Dnmt2/NSun2 double knockout mice completely lack m5C modifications in tRNAs and show reduced overall protein synthesis, suggesting that m5C modifications are crucial for the regulation of protein synthesis [27]. Moreover, it was shown that modulation of Nsun2 activity contributes to stem cell fate by fine-tuning ANG-mediated tiRNA production [30]. Furthermore, it has been recently reported that the presence of a pseudouridine modification at a specific position of a tRF regulates differentiation of stem cells through inhibition of protein synthesis [31]. These reports reemphasize the importance of studying tRFs in their physiological context.
Recently, we have developed a simple and reproducible method for isolating endogenous tiRNAs using biotinylated antisense oligos [32]. Using this method, we demonstrated that endogenous tiRNAs can behave similarly to their synthetic counterparts. First, we showed that endogenous 5ʹ-tiRNAAla-AGC assembles G-quadruplexes (G4) [32], that are required for translation inhibition, SG formation and cytoprotection [33,34]. In addition, endogenous 5ʹ-tiRNAGly-GCC was shown to be significantly more potent than its synthetic counterpart at inhibiting translation of mRNA reporters [32]. Although these data using endogenous tiRNAs have started to elucidate the more physiological roles of tiRNAs in the cell, there remain some problems for efficient isolation of endogenous tiRNAs. The most serious problem is very low yield of target tiRNA due to low efficiency of ANG-mediated tiRNA production towards some specific tRNA species. The percentage of stress-induced tRNA cleavage (typically by oxidative stress [35]) in cells is only 1–2% [8], and this efficiency of tiRNA generation is quite comparable with approach when cells are treated with recombinant ANG. Because recombinant ANG added to cell culture media is rapidly internalized and cleaves mature cytoplasmic tRNAs [8,36], the easiest way to generate tiRNAs is to treat cells with recombinant ANG. However, cells should be seeded relatively sparsely when treating with ANG because confluent cells can inhibit the uptake of ANG through down regulation of its receptor, PLXNB2 [37,38], which limits the yield of endogenous tiRNA. Moreover, ANG overexpression does not efficiently induce tiRNA production, as a possible consequence of its active secretion from the cell [39]. Therefore, a new method for tiRNA production was needed to prepare specific endogenous tiRNAs for functional analysis.
Here we report an efficient and cost-effective method of in lysate ANG digestion of cytoplasmic tRNAs for the production of endogenous tiRNAs. In our method, more than 50% of mature tRNAs are cleaved into tiRNAs. By comparing our method with conventional in vitro ANG digestion, we also show that the high specificity of ANG for tRNA substrates depends on their physiological structural conformation in the cell. Intriguingly, the exquisite specificity of ANG for tRNAs suggests that its major physiologic role is to cleave tRNAs in the cell. We also show that the enzymatic activity of ANG on physiologically folded tRNAs is comparable to that of bovine RNase A, despite the fact that its non-specific RNase activity is significantly lower than that of RNase A [40–42].
Results
It was previously reported that the enzymatic activity of ANG towards purified tRNAs is up to 105-fold lower than that of RNase A in vitro [41], which is apparently inconsistent with the evidence that ANG plays important roles in the stress response by actively cleaving tRNAs into tiRNAs in the cell (reviewed in [14]). Traditionally, the enzymatic activity of ANG has been assessed in vitro [40–42], using purified RNAs. We hypothesized that the efficiency of ANG-induced tRNA cleavage is more efficient within cells due to certain factors such as different conformation of tRNAs in cells when compared to purified counterparts. To test this hypothesis, we examined the tRNA-cleaving efficiency of ANG in the cell lysate according to the flowchart shown in Figure 1. Because magnesium ion (Mg2+) plays an important role in RNA folding in the cell (reviewed in [43]), we used a lysis buffer (named ‘ANG Digestion Buffer’) lacking EDTA which chelates Mg2+.
Figure 1.
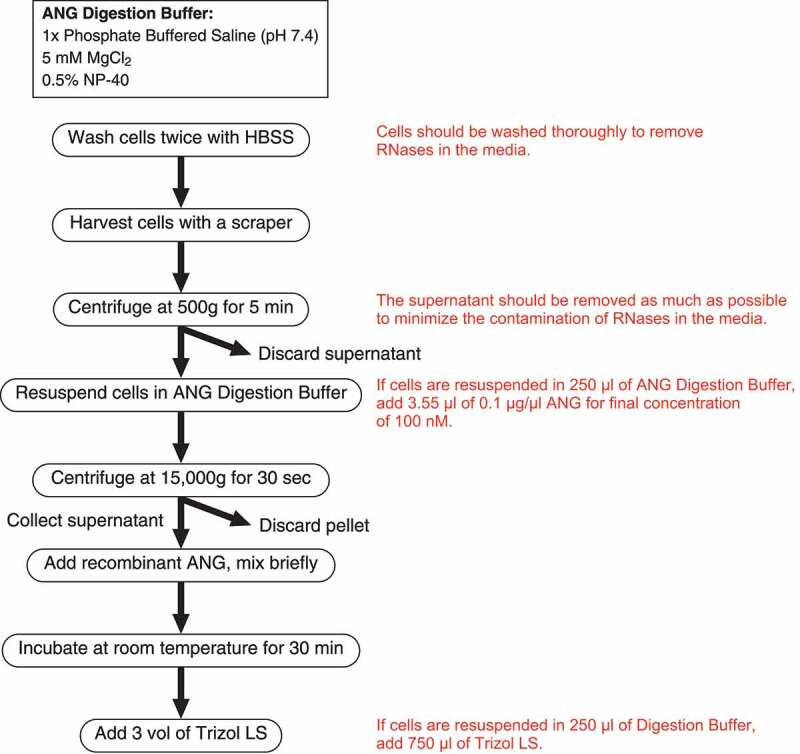
Flowchart of in lysate ANG digestion
First, we conducted dose-response and time-course experiments to investigate the characteristics of this method (Figure 2(a–d)). As demonstrated in Figure 2(a), ANG cleaved tRNAs into tiRNAs in a dose-dependent manner. Even the highest concentration of ANG (100 nM) only tRNA was cleaved (Figure 2(a)), suggesting that ANG is specific for tRNAs under these conditions. Northern blotting for tRNAGly-GCC also showed the dose-dependent increase of 5ʹ-tiRNAGly-GCC (Figure 2(b)). It should be noted that the amount of mature tRNAGly-GCC was significantly decreased by higher concentrations (>10 nM) of ANG (Figure 2(b)). As in vivo ANG treatment, typically performed at 0.5 µg/ml (35.7 nM), does not induce a significant decrease of mature tRNAs [8], this method was estimated to be a more efficient way to generate tiRNAs than in vivo ANG treatment. Mean reduction of mature tRNAGly-GCC after treatment of 100 nM ANG was 77.6 ± 5.2% (n = 3, data not shown). In a time-course experiment, mature tRNAs and tRNAGly-GCC were rapidly cleaved into tiRNAs in the presence of ANG (Figure 2(c and d), respectively). Two kinds of 5ʹ-tiRNAGly-GCC were observed by Northern blotting as previously reported [10,12,44] and, interestingly, the shorter form of 5ʹ-tiRNAGly-GCC (the lower band) seemed to be generated after the production of the longer form (the upper band) (Figure 2(d)), suggesting that the shorter form may be generated by additional cleavage of the longer form. In contrast, when the lysate was treated with bovine RNase A (bRNase A) (Figure 2(e,f)), RNAs were non-specifically degraded at higher concentrations (>10 nM), resulting in a smear of bands in SYBR Gold staining (Figure 2(e)). Not only tRNAGly-GCC but also 28S rRNA were cleaved in a dose-dependent manner (Figure 2(f)), suggesting that the smear of bands observed by SYBR Gold staining was mainly due to the fragmentation of ribosomal RNAs. Interestingly, the RNA-cleaving activity of bRNase A seemed to increase abruptly at 10 nM (Figure 2(e,f)), which is likely because of the inactivation of bRNase A by the endogenous RNase inhibitor, RNH1 [45]. In RNH1-null U2OS cells (ΔRNH1 cells), both bRNase and ANG cleaved target RNAs at lower concentrations (as low as 1 nM) compared to wild-type (RNH1-intact) U2OS cells (Supplementary Fig. 1), which suggests that bRNase A was strongly inhibited by RNH1 at below 10 nM. As RNH1 forms a one-to-one complex with RNase A family members including ANG (reviewed in [46]), RNH1 concentration in the lysate in Figure 2(e,f) was estimated to be approximately 3 nM. It should be noted that tiRNA-generating activity of ANG was comparable with that of bRNase A even in ΔRNH1 cells, whereas 28S rRNA was actively degraded only by bRNase A (Supplementary Fig. 1B). Interestingly, it seemed that ANG-mediated tiRNA production was not efficiently inhibited by RNH1 compared to bRNase A (Figure 2) because ANG cleaved tRNAs into tiRNAs dose-dependently from the lowest concentration (1 nM). However, pre-treatment of ANG with RNase inhibitor significantly inhibited tRNA cleavage similarly to bRNase A (Supplementary Fig. 2). One possibility is that RNH1 may inactivate ANG more slowly than bRNase A, allowing ANG to cleave tRNAs until it is inactivated. Further study is necessary to clarify the difference between ANG and bRNase A in RNH1-mediated inactivation.
Figure 2.
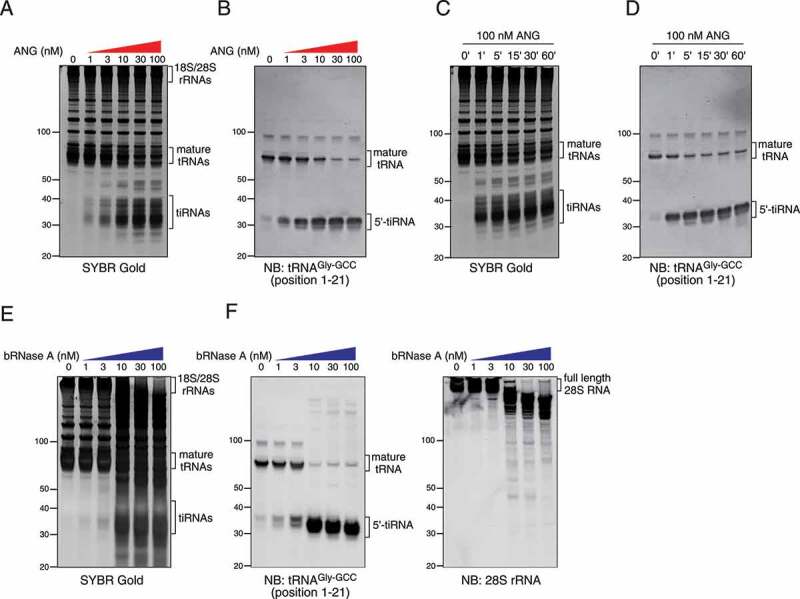
Dose-dependency and time course of in lysate digestion method. (a-b) Dose-dependency of in lysate ANG digestion. (a) SYBR Gold staining and (b) Northern blotting for 5ʹ-tiRNAGly-GCC. (c-d) Time/response of in lysate ANG digestion. (c) SYBR Gold staining and (d) Northern blotting for 5ʹ-tiRNAGly-GCC. (d–e) Dose-dependency of in lysate digestion with bovine RNase A (bRNase A). (d) SYBR Gold staining and (e) Northern blotting for 5ʹ-tiRNAGly-GCC and 28S rRNA are shown
Next, we compared the efficiency and specificity of RNA cleavage among three kinds of tiRNA-producing methods, namely in vivo ANG treatment, in vitro ANG digestion and in lysate ANG digestion (Figure 3). As shown in Figure 3(a), mature tRNAs were more efficiently cleaved into tiRNAs by in lysate ANG digestion compared to in vivo ANG treatment. Specific 5ʹ-tiRNAs (5ʹ-tiRNAGly-GCC and 5ʹ-tiRNAAla-AGC) were also more efficiently generated by in lysate ANG digestion than in vivo ANG treatment (Figure 3(b)). In contrast, RNA smears were produced by in vitro ANG digestion as revealed by SYBR Gold staining (Figure 3(a)), suggesting non-specific RNA degradation. Although 5ʹ-tiRNAGly-GCC was slightly increased, mature tRNAAla-AGC did not seem to be cleaved by in vitro ANG digestion. We also examined the cleavage of other RNA molecules such as 7SK RNA and 28S rRNA by these methods (Figure 3(b)). In lysate ANG digestion did not induce the cleavage of 7SK RNA or 28S rRNA similarly to in vivo ANG treatment (Figure 3(b)); however, when purified total RNA was incubated with ANG (in vitro ANG digestion), both 7SK RNA and 28S rRNA were cleaved into various length fragments (Figure 3(b)), suggesting that ANG may significantly decrease both activity and specificity of tRNA cleavage under this non-physiological condition. In addition, we also confirmed that in lysate ANG digestion produced both 5ʹ- and 3ʹ-tiRNAs from tRNAGly-GCC and tRNAiMet-CAT more efficiently than in vivo ANG treatment, without loss of specificity (Supplementary Fig. 3). These data show that the in lysate ANG digestion method can generate tiRNAs more efficiently than in vivo ANG treatment without loss of specificity for tRNA targets.
Figure 3.
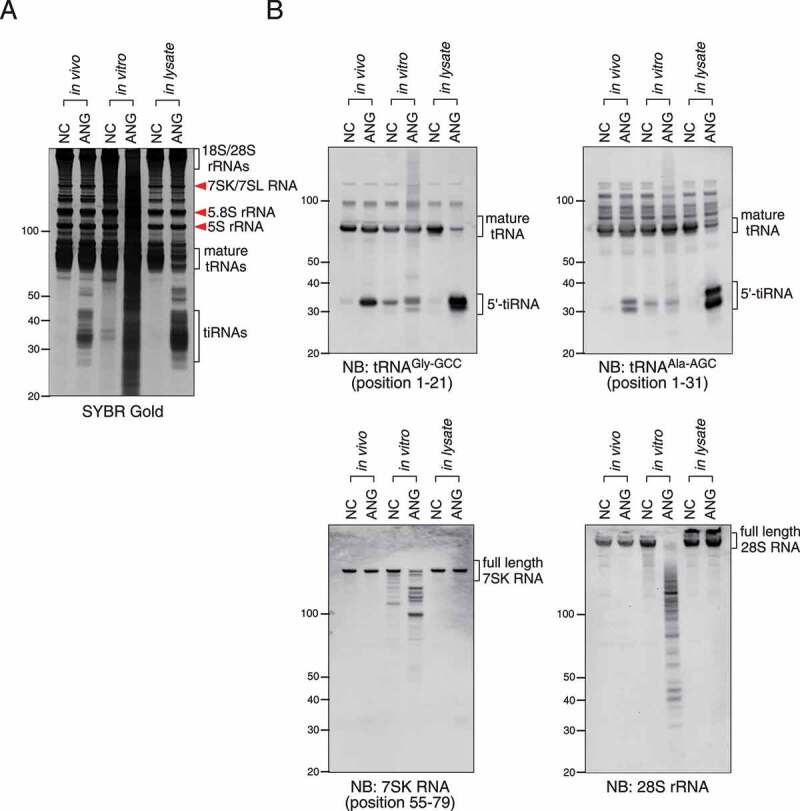
Comparison of efficiency and specificity among in vivo ANG treatment, in vitro ANG digestion and in lysate ANG digestion. (a) SYBR Gold staining. (b) Northern blotting for 5ʹ-tiRNAGly-GCC, 5ʹ-tiRNAAla-AGC, 7SK RNA and 28S rRNA
We hypothesized that the extremely high specificity and efficiency of ANG-mediated tRNA cleavage observed using in lysate ANG digestion was mainly due to the physiological conformation of tRNAs. It is known that Mg2+ ion is essential to maintain RNAs in a physiologically folded state [43], and EDTA chelates Mg2+ and decreases the stability of RNAs [47]. To clarify the importance of RNA conformation, we examined the effect of EDTA-induced conformational disruption of RNAs on in lysate ANG digestion method (Figure 4(a,b)). As RNase A family members, including ANG, are metal-independent RNases [48], their enzymatic activity is not inhibited by chelation of metal ions. Surprisingly, EDTA pre-treatment (20 mM) of lysates almost completely abolished ANG-mediated tRNA cleavage (Figure 4(a)). The production of 5ʹ-tiRNAGly-GCC was also almost completely inhibited by EDTA (Figure 4(b)). Interestingly, in contrast to tRNAs, ANG-mediated 28S rRNA cleavage was increased by EDTA pre-treatment (Figure 4(b)). We also examined the effect of EDTA pre-treatment on bRNase A-mediated RNA cleavage. In clear contrast to ANG, bRNase A-mediated RNA cleavage was increased by EDTA pre-treatment, as 28S and 18S rRNAs were quantitatively cleaved into shorter fragments (Figure 4(c)). Northern blotting showed that full-length 28S rRNA was completely lost by bRNase with EDTA pre-treatment (Figure 4(d)). However, interestingly, bRNase A-mediated cleavage of tRNAGly-GCC was not decreased by EDTA (Figure 4(d)), which was a striking contrast to the ANG-mediated tRNAGly-GCC cleavage (Figure 4(b)). To exclude the possibility that the ANG-mediated tRNA cleavage was inhibited by the chelation of other divalent metal ions, especially Ca2+, we also examined the effect of EGTA, which specifically chelates Ca2+ ion, on ANG-mediated tRNA cleavage (Figure 4(e,f)). EGTA pre-treatment (10 mM) did not affect ANG-mediated tiRNA production as revealed by SYBR Gold staining (Figure 4(e)). Northern blotting also showed no effect of EGTA on the production of specific 5ʹ-tiRNAs (5ʹ-tiRNAGly-GCC and 5ʹ-tiRNAAla-AGC) (Figure 4(f)), supporting the conclusion that the inhibitory effect of EDTA was due to conformational disruption of RNAs by chelating Mg2+, but not Ca2+ . In summary, these data suggest that ANG is highly specialized to cleave physiologically folded tRNAs inside the cell.
Figure 4.
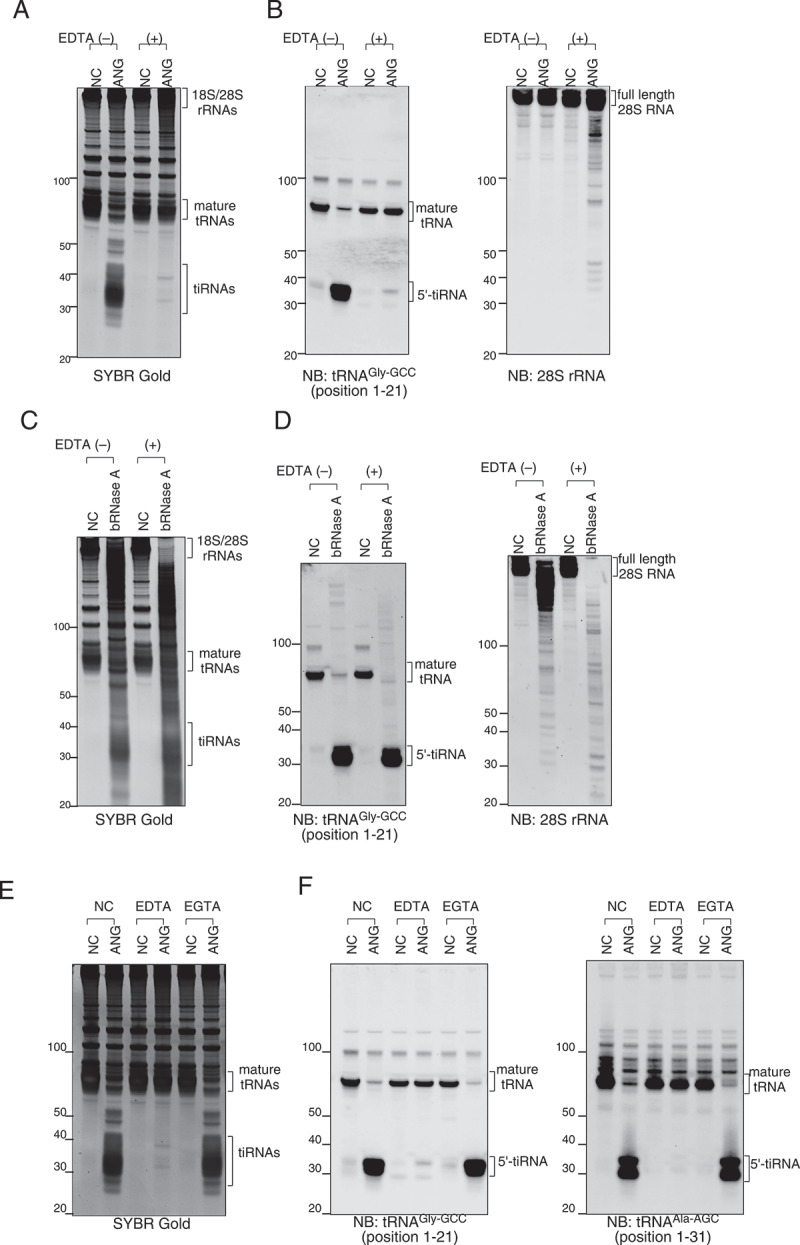
Effect of RNA conformation on ANG-mediated cleavage of tRNA and 28S rRNA. (a-b) EDTA pre-treatment (20 mM) of the lysate abolishes ANG-mediated tRNA cleavage and slightly promotes 28S rRNA cleavage. (a) SYBR Gold staining and (b) Northern blotting for 5ʹ-tiRNAGly-GCC and 28S rRNA. (c-d) bRNase A-mediated digestion (10 nM) is not affected by EDTA pre-treatment. (c) SYBR Gold staining and (d) Northern blotting for 5ʹ-tiRNAGly-GCC and 28S rRNA. (e-f) Calcium chelation with EGTA (10 mM) does not inhibit ANG-mediated tRNA cleavage. (e) SYBR Gold staining and (f) Northern blotting for 5ʹ-tiRNAGly-GCC and 5ʹ-tiRNAAla-AGC
Finally, as another purpose of this study was to improve the yield of endogenous tiRNAs obtained by pulldown methods, we compared the yields of endogenous 5ʹ-tiRNAGly-GCC pulldown between in vivo ANG treatment and in lysate ANG digestion (Figure 5). Using RNAs obtained by each method from one 15-cm dish per sample, we performed 5ʹ-tiRNAGly-GCC pulldown as reported previously [32]. As shown in Figure 5(a and b), the image of SYBR Gold staining (Figure 5(a)) was almost identical to the Northern blotting image (Figure 5(b)), suggesting that 5ʹ-tiRNAGly-GCC was specifically pulled down using this method. Densitometry analysis showed that following in lysate ANG digestion, the yield of 5ʹ-tiRNAGly-GCC was increased about fivefold (5.13 ± 0.52, p < 0.0001) compared with in vivo ANG digestion (Figure 5(b)). Unfortunately, the yields of endogenous 5ʹ-tiRNAGly-GCC were too low to measure the concentrations even when in lysate ANG digestion was performed using confluent cells in one 15-cm dish. When 5ʹ-tiRNAAla-AGC pulldown was performed combined with in lysate ANG digestion using RNAs obtained from twenty 15-cm dishes with confluent U2OS cells (Supplementary Fig. 4), the final yield was about 1.72 µg (169 pmol), which is sufficient for some functional analyses [32].
Figure 5.
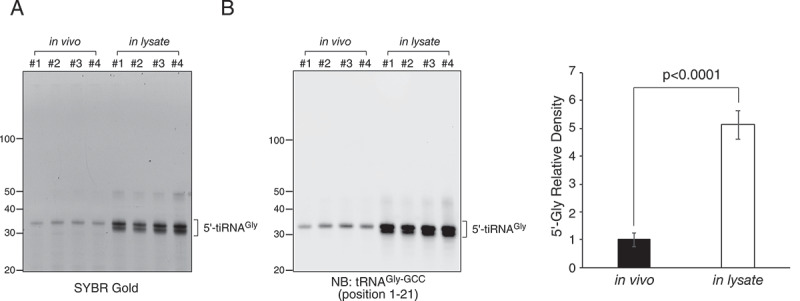
Comparison of yield of 5ʹ-tiRNAGly-GCC pulldown between in vivo ANG treatment and in lysate ANG digestion. (a) SYBR Gold staining of purified RNA obtained by 5ʹ-tiRNAGly-GCC pulldown method. (b) Northern blotting of purified RNA against tRNAGly-GCC. Densitometry analysis of 5ʹ-tiRNAGly-GCC signals is also shown. Data are presented as the mean ± SD (n = 4)
Discussion
The field of tRNA-derived fragments (tRFs) has gained significant attention in recent years [5–7]. Among the various subclasses of tRFs, tiRNAs [8] (also called tRNA halves [9]), are one of the best-studied. Although several biological functions of tiRNAs have been reported (reviewed in [14]), most studies have used synthetic mimics of tRFs. Researchers have no other choice than ignoring the fact that synthetic RNA molecules have significantly different chemical compositions than natural ones because of the difficulty in isolating endogenous tiRNAs. We and others recently developed a simple and reproducible method to pull down and purify endogenous tiRNAs [32,49]; however, the yield of endogenous tiRNAs is low mainly because of the low efficiency of ANG-mediated tRNA cleavage. Therefore, we are in need of developing methods to generate tiRNAs more efficiently compared with the conventional methods such as in vivo ANG treatment of the cell, and in vitro ANG digestion.
Here, we developed a novel, highly efficient method to generate ANG-mediated tiRNAs, called in lysate ANG digestion (Figure 1). In contrast to cellular treatment with ANG [8], this method improved ANG-mediated tRNA cleavage from 1% to 2% to over 70% (Figure 2(a–d)). At the same time, ANG-mediated cleavage remained highly specific to tRNAs similarly to in vivo ANG treatment (Figure 3), which was in striking contrast to in vitro ANG digestion that showed the fragmentation of 7SK RNA and 28S rRNA (Figure 3(b)). Using this method combined with the pulldown method reported previously [32], the yield of endogenous 5ʹ-tiRNAGly-GCC was increased about fivefold compared with when ANG was generated by cellular treatment with ANG (Figure 5). As tRNAs can be cleaved by in lysate ANG digestion over 30-fold compared to in vivo ANG treatment (over 70% vs 1–2%, respectively), it is possible that the yield may further increase by optimizing the condition of the pulldown method.
In addition to high specificity and efficiency, our method also has some other advantages. First, it is highly reproducible. When performing in vivo ANG treatment, tiRNA production is affected by cell density, because high cell density can inhibit ANG uptake into the cells through down-regulation of PLXNB2 [37,38], which decreases reproducibility of the amount of tiRNA production. On the other hand, in in lysate ANG digestion, we can seed cells confluently and can easily treat the lysate with a fixed concentration of ANG. Second, it is cost effective, which is a big advantage because recombinant ANG is difficult to isolate and/or it is commercially expensive, although available. For in vivo ANG treatment, relatively large amount of ANG is required because it should be dissolved in cell culture media [8]. The amount of recombinant ANG required for in vivo ANG treatment (0.5 µg/ml) and our new method (100 nM) per 15-cm dish is 4 µg and 0.355 µg, respectively. Third, our method will enable us to examine the physiological enzymatic activity of ANG in the cell more deeply than before. For example, we can analyse dose-dependency experiments like in Figure 2(a,b) and examine the effect of some compounds such as EDTA and EGTA like in Figure 4, which will further elucidate physiological roles of ANG in the cell. Furthermore, this method can be applied to studies about other RNases that work inside the cell such as RNase L [50] because our method should reflect more physiological cleavage of RNAs in the cell compared with the in vitro digestion method.
As RNase A family enzymes including RNase A and ANG cleave the 3ʹ-side of pyrimidine residues in single-strand RNAs [48], conformational disruption of RNAs is theoretically predicted to facilitate cleavage. When the conformation of RNAs was disrupted by EDTA (Figure 4), or when purified total RNA was incubated with ANG (Figure 3), RNAs such as 7SK RNA and 28S rRNAs were more actively cleaved into various length fragments by both ANG and bRNase A. On the other hand, ANG-mediated tRNA cleavage was paradoxically abolished by EDTA (Figure 4(a,b)), which suggests that ANG-mediated RNA cleavage is regulated by two factors: i) specificity for physiologically folded tRNAs (tRNA-specific cleavage) and ii) sequence-dependent non-specific cleavage (non-specific cleavage). It has been reported that the enzymatic activity of ANG towards tRNAs is up to 105-fold lower than that of RNase A [41]. However, because the report was based on in vitro analysis, it is unlikely that the result reflects the tRNA-specific cleavage of ANG in the cell. Our data showed that the in lysate tRNA-cleaving activity of ANG is comparable to that of bRNase A (Figure 2(a,b,e,f) and Supplementary Fig. 1), suggesting that tRNA-cleaving activity of ANG in cells is much higher than estimated from in vitro studies. Therefore, under physiological conditions in the cell, it is likely that ANG dominantly cleaves tRNAs even at low concentrations because of 1) high tRNA-cleaving activity, 2) very low activity of non-specific cleavage, and 3) inhibition of non-specific cleavage by folding of RNAs. On the other hand, under in vitro conditions (Figure 3) or in EDTA-containing lysates (Figure 4), non-specific cleavage is likely to become dominant because the conformational disruption of RNAs causes both 1) inhibition of tRNA-specific cleavage and 2) facilitation of non-specific cleavage by increasing unfolded RNAs. Interestingly, EDTA pre-treatment of lysate did not decrease bRNase A-mediated tRNA cleavage (Figure 4(c,d)), suggesting that the specificity for physiologically folded tRNAs is a unique feature of ANG compared to bRNase A. Taken together, our results demonstrate that ANG is highly specialized to cleave tRNAs inside the cell.
Which cells are suitable for in lysate ANG digestion, ΔRNH1 cells or RNH1-intact cells? From the viewpoint of cost-effectiveness, ΔRNH1 cells should be better because they induce ANG-mediated tiRNA production at lower concentrations (Figure 2 and Supplementary Fig. 1). However, in ΔRNH1 cells, a substantial amount of tiRNAs are constitutively generated even in the absence of stress because the endogenous RNase A family enzymes are not inactivated (Supplementary Fig. 1 and [12]). Therefore, it is inevitable that tiRNAs generated by other RNase A family enzymes are mixed with ANG-induced tiRNAs, which prevents obtaining pure ANG-induced tiRNAs. For this reason, we think that RNH1-intact cells are more suitable for in lysate ANG digestion.
In summary, we describe a novel method to efficiently and specifically generate ANG-cleaved tiRNAs. In addition to the improvement of the yield of endogenous tiRNAs, these studies have revealed a unique feature of ANG in the cell, namely an exquisite specificity for physiologically folded tRNAs. Our in lysate ANG digestion method will not only facilitate studies using endogenous tiRNAs but also clarifies the physiological functions of ANG in the cell.
Material and methods
Cell culture and treatment
U2OS cells were purchased from the American Type Culture Collection (ATCC). RNH1-null U2OS cells (ΔRNH1 cells) were generated as previously reported [12]. Cells were cultured at 37°C in a CO2 incubator in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum (FBS) (Sigma-Aldrich) and 1% of penicillin/streptomycin (Sigma-Aldrich).
For in vivo angiogenin (ANG) treatment, U2OS cells were seeded at 6 × 106 on a 15 cm dish the day before ANG treatment. The cells were incubated with DMEM supplemented with 0.5 μg/ml recombinant human ANG (R&D systems) for 1 h. After washing with Hank’s balanced salt solution (HBSS), total RNA was extracted with Trizol reagent (ThermoFisher Scientific).
In vitro ANG digestion was performed according to the procedure described in [51] with slight modification. Briefly, total RNA in 30 mM HEPES (pH 7.0), 30 mM NaCl was heated at 90°C for 2 min and cooled down to room temperature. MgCl2 and BSA were added to final concentrations of 2 mM and 0.01%, respectively, and further incubated at 37°C for 5 min. Recombinant human ANG was added to a final concentration of 0.2 µM and incubated at 37°C for 4 h.
In lysate ANG digestion
For in lysate ANG digestion, U2OS cells were seeded at 1 × 107 cells on a 15 cm dish the day before ANG digestion. The cells were washed with HBSS twice, scraped with using a cell lifter, then collected into a 1.5 mL microcentrifuge tube. After centrifugation at 500 g for 5 min, the supernatant was carefully removed and the cells were resuspended in 250 µl of ANG digestion buffer (1x phosphate-buffered saline (pH 7.4) containing 5 mM MgCl2 and 0.5% NP-40), mixed well with pipetting and brief vortex mixing. The cells were centrifuged at 15,000 g for 30 sec and the supernatant was collected into a new microcentrifuge tube. Recombinant human ANG was added to the lysate to a final concentration of 100 nM, unless otherwise indicated. After mixing briefly, the lysate was incubated at room temperature for 30 min, then mixed with 750 µl of Trizol LS reagent (ThermoFisher Scientific). The flowchart of this method is shown in Figure 1.
Pulldown of endogenous tiRNAs
Pulldown of endogenous tiRNAs was performed as previously described [32]. Biotinylated oligonucleotide DNA probes were synthesized by IDT. The sequences of the probes were shown as follows:
probe for 5ʹ-tiRNAGly-GCC pulldown: 5ʹ-GCAGGCGAGAATTCTACCACTGAACCACCCATGCT-biotin-3ʹ
probe for 5ʹ-tiRNAAla-AGC pulldown: 5ʹ-AAGCACGCGCTCTACCACTGAGCTA-biotin-3ʹ
Northern blotting
Northern blotting was performed using digoxigenin (DIG)-tailed oligonucleotide probes as previously described [32]. Oligonucleotide probes were synthesized by IDT. DIG-labelled probes were prepared using the DIG Oligonucleotide tailing kit (2nd generation; Roche) according to the manufacturer’s instructions. The sequences of the probes were as follows:
probe for 5ʹ-tiRNAGly-GCC (position 1–21): 5ʹ-CTACCACTGAACCACCCATGC-3ʹ
probe for 5ʹ-tiRNAGly-GCC (position 37–64): 5ʹ-GCCGGGAATCGAACCCGGGCCTCCCGCG-3ʹ
probe for 5ʹ-tiRNAiMet-CAT (position 1–21): 5ʹ-CTTCCGCTGCGCCACTCTGCT-3ʹ
probe for 3ʹ-tiRNAiMet-CAT (position 43–72): 5ʹ-TAGCAGAGGATGGTTTCGATCCATCGACCT-3ʹ
probe for 5ʹ-tiRNAAla-AGC (position 1–31): 5ʹ-AAGCACGCGCTCTACCACTGAGCTACACCCCC-3ʹ
probe for 28S rRNA: 5ʹ-GGGTGAACAATCCAACGCTTGGTG-3ʹ
probe for 7SK RNA: 5ʹ-CCTAGCCAGCCAGATCAGCCGAATC-3ʹ
Densitometry and statistical analyses
The amount of isolated 5ʹ-tiRNA-Gly was quantified by densitometry using Image J software from the National Institute of Health (NIH). Data are shown as mean ± standard deviation. Student’s t-test was performed to compare the two groups. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Victoria Ivanova for assistance with preliminary experiments and Anderson and Ivanov lab members for helpful critiques.
Funding Statement
This work was supported by the National Institutes of Health [R35 GM126901 to P.A., RO1 GM126150 to P.I.], and by the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (PS KAKENHI, grant number 26860094 to Y.A.). Funding for open access charge: National Institutes of Health.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Raina M, Ibba M.. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Phizicky EM, Alfonzo JD.. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sun C, Fu Z, Wang S, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16–25. [DOI] [PubMed] [Google Scholar]
- [4].Polacek N, Ivanov P. The regulatory world of tRNA fragments beyond canonical tRNA biology. RNA Biol. 2020;17:1057–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Keam SP, Hutvagner G. tRNA-derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel). 2015;5:1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. 2016;41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yamasaki S, Ivanov P, Hu GF, et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. [DOI] [PubMed] [Google Scholar]
- [10].Tosar JP, Segovia M, Castellano M, et al. Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res. 2020;48:12874–12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nechooshtan G, Yunusov D, Chang K, et al. Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res. 2020;48:8035–8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Akiyama Y, Lyons SM, Fay MM, et al. Multiple ribonuclease A family members cleave transfer RNAs in response to stress. bioRxiv. 2019;811174. [Google Scholar]
- [13].Sheng J, Xu Z. Three decades of research on angiogenin: a review and perspective. Acta Biochim Biophys Sin (Shanghai). 2016;48:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lyons SM, Fay MM, Akiyama Y, et al. RNA biology of angiogenin: current state and perspectives. RNA Biol. 2017;14:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pizzo E, Sarcinelli C, Sheng J, et al. Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J Cell Sci. 2013;126:4308–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Saikia M, Jobava R, Parisien M, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol. 2016;215:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol. 2019;11. DOI: 10.1101/cshperspect.a032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nachtergaele S, He C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017;14:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, et al. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lorenz C, Lunse CE, Morl M. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017;7. DOI: 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lyons SM, Fay MM, Ivanov P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018;592:2828–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tuorto F, Liebers R, Musch T, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. [DOI] [PubMed] [Google Scholar]
- [28].Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Blanco S, Bandiera R, Popis M, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guzzi N, Ciesla M, Ngoc PCT, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–16 e26. [DOI] [PubMed] [Google Scholar]
- [32].Akiyama Y, Kharel P, Abe T, et al. Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs. RNA Biol. 2020;17:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lyons SM, Gudanis D, Coyne SM, et al. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci USA. 2014;111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Saikia M, Krokowski D, Guan BJ, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hu G, Xu C, Riordan JF. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J Cell Biochem. 2000;76:452–462. [DOI] [PubMed] [Google Scholar]
- [37].Yu W, Goncalves KA, Li S, et al. Plexin-B2 mediates physiologic and pathologic functions of angiogenin. Cell. 2017;171:849–64 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci USA. 1997;94:2204–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Su Z, Kuscu C, Malik A, et al. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem. 2019;294:16930–16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shapiro R, Riordan JF, Vallee BL. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry. 1986;25:3527–3532. [DOI] [PubMed] [Google Scholar]
- [41].Lee FS, Vallee BL. Characterization of ribonucleolytic activity of angiogenin towards tRNA. Biochem Biophys Res Commun. 1989;161:121–126. [DOI] [PubMed] [Google Scholar]
- [42].Harper JW, Vallee BL. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry. 1989;28:1875–1884. [DOI] [PubMed] [Google Scholar]
- [43].Bowman JC, Lenz TK, Hud NV, et al. Cations in charge: magnesium ions in RNA folding and catalysis. Curr Opin Struct Biol. 2012;22(3):262–272. [DOI] [PubMed] [Google Scholar]
- [44].Tosar JP, Gambaro F, Darre L, et al. Dimerization confers increased stability to nucleases in 5ʹ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 2018;46:9081–9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shapiro R, Vallee BL. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc Natl Acad Sci USA. 1987;84:2238–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Darzynkiewicz Z, Traganos F, Sharpless T, et al. Conformation of RNA in situ as studied by acridine orange staining and automated cytofluorometry. Exp Cell Res. 1975;95:143–153. [DOI] [PubMed] [Google Scholar]
- [48].Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Drino A, Oberbauer V, Troger C, et al. Production and purification of endogenously modified tRNA-derived small RNAs. RNA Biol. 2020;17:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liang SL, Quirk D, Zhou A. RNase L: its biological roles and regulation. IUBMB Life. 2006;58:508–514. [DOI] [PubMed] [Google Scholar]
- [51].Czech A, Wende S, Morl M, et al. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 2013;9:e1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


