ABSTRACT
MicroRNAs (miRNAs) are essential non-coding riboregulators of gene expression in plants and animals. In plants, miRNAs guide their effector protein named ARGONAUTE (AGO) to find target RNAs for gene silencing through target RNA cleavage or translational inhibition. miRNAs are derived from primary miRNA transcripts (pri-miRNAs), most of which are transcribed by the DNA-dependent RNA polymerase II. In plants, an RNase III enzyme DICER-LIKE1-containing complex processes pri-miRNAs in the nucleus into miRNAs. To ensure proper function of miRNAs, plants use multiple mechanisms to control miRNA accumulation. On one hand, pri-miRNA levels are controlled through transcription and stability. On the other hand, the activities of the DCL1 complex are regulated by many protein factors at transcriptional, post-transcriptional and post-translational levels. Notably, recent studies reveal that pri-miRNA structure/sequence features and modifications also play important roles in miRNA biogenesis. In this review, we summarize recent progresses on the mechanisms regulating miRNA biogenesis.
KEYWORDS: Plant miRNA biogenesis, pri-miRNA transcription, pri-miRNA stability, pri-miRNA processing, dcl1
1. Introduction
Small non-coding RNAs (sRNAs) are riboregulators of gene expression and play crucial roles in various biological processes [1–5]. In eukaryotes, small RNAs can be classified into four groups based on their origins: microRNA (miRNA), small interfering RNA(siRNA), PIWI-interacting RNA (piRNA) and transfer RNA-derived small RNA (tsRNA) [1,2,6–8]. These small RNAs can repress gene expression at the transcriptional (TGS) and/or post-transcriptional (PTGS) levels [3,9]. miRNAs, ~20-24 nucleotides (nt) in size, which are encoded by plants, animals and some viruses, are distinguished from other small RNAs by their highly precise excision from imperfect stem-loop residing in the primary miRNA transcripts (pri-miRNAs) [10]. In plants, miRNAs mainly repress gene expression through mediating target RNA cleavage or translational inhibition [1,2].
Most plant miRNAs are encoded by genes (MIRs) residing in the intergenic regions, while some miRNAs are derived from introns of protein-coding genes and other non-coding RNAs [1,11,12]. The majority of plant MIRs are transcribed by the DNA-dependent RNA polymerase II (Pol II) to generate pri-miRNAs [1,2]. The imperfect stem-loop structure of pri-miRNAs is recognized and processed by the Dicer-like RNase III endonuclease 1 (DCL1) into short precursors (pre-miRNAs) with the help of the dsRNA-binding protein HYPONASTIC LEAVES 1 (HYL1) and the zinc finger protein SERRATE (SE). Pre-miRNAs are further processed by DCL1 to generate imperfect miRNA/miRNA* duplexes [2,13], which consist of a duplex region and 2-nt 3ʹ overhangs at each end, in the nucleus [13]. After processing, each strand of the duplex is 2ʹ-O-methalated at the 3ʹ end by the small RNA methyltransferase HUA ENHANCER 1 (HEN1), which protects miRNAs from degradation [14]. Then, one strand (miRNA) of the miRNA/miRNA* duplex is selected as the guide strand and loaded into Argonaute-1 (AGO1) to assemble miRNA-induced silencing complex (miRISC), while the passenger strand (miRNA*) is eliminated [13]. It should be noted that some miRNAs are loaded into AGO1 homologs instead of AGO1 in plants [15–17]. Recent studies suggest that miRISC assembles in the nucleus and is exported to the cytosol in a CRM1(EXPO1)/NES-dependent manner [18,19]. However, another study shows that some unloaded miRNAs are present in the cytoplasm, indicating the miRISC assembly may also occur in the cytoplasm [20].
To date, hundreds of miRNAs have been identified in plants. These miRNAs modulate development and physiology of plants such as the development of seed, root, shoot and flower, phase transition, and responses to biotic and abiotic stresses [1,3,21,22]. Misregulation of miRNA accumulation has been shown to cause developmental defects or diseases, suggesting that miRNA biogenesis is a precisely controlled process. Indeed, studies have found that miRNA biogenesis is modulated at both transcriptional and post-transcriptional levels. In this review, we seek to summarize recent progresses in the regulation of miRNA biogenesis.
2. Regulation of pri-miRNA accumulation
2.1. Pri-miRNA transcription
Like mRNAs, the Pol II-dependent pri-miRNAs undergo capping, splicing and polyadenylation [23–25]. Their transcription and processing are regulated by general and specific transcription factors (Fig. 1) [25,26]. In addition, the transcription of some pri-miRNAs is spatiotemporally modulated to ensure proper function of corresponding miRNAs.
Figure 1.
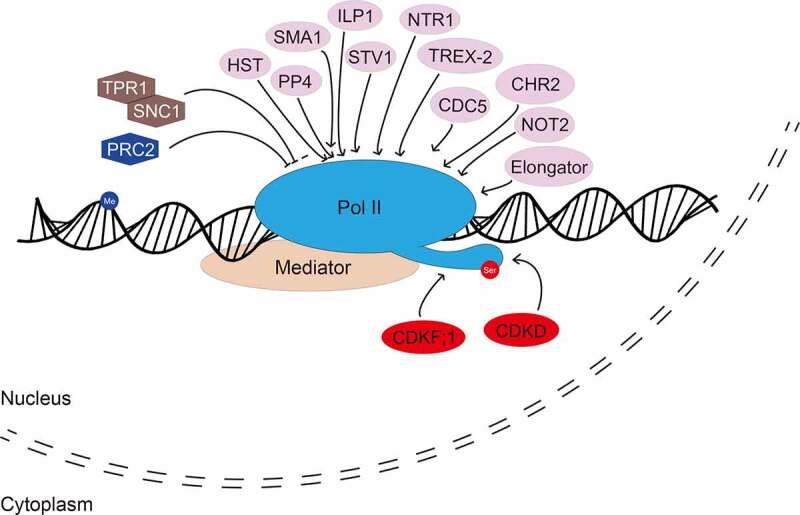
Regulation of MIR transcription
Multiple transcription factors are required for the regulation of RNA polymerase II (Pol II)-mediated MIR transcription. The activity of Pol II is also regulated by phosphorylation at the C-terminal domain (CTD) of its largest subunit. Positive and negative regulators are marked with ellipse and hexagon, respectively. Regulators involved in phosphorylation of CTD and histone methylation of MIR promoters are marked with red colour and blue colour, respectively. Me: methylated DNA; ser: serine amino acid within the CTD.
The Mediator complex is a general transcriptional coactivator facilitating the recruitment of RNA Pol II to MIR promoters (Fig. 1) [27]. Many additional proteins have been shown to stimulate MIR transcription. These factors include ELP2 and ELP5 of the Elongator complex [28], the transcription factor NEGATIVE ON TATA LESS 2 (NOT2) [29], the DNA-binding protein CELL DIVISION CYCLE 5 (CDC5) [30], the ribosomal protein SHORT VALVE 1 (STV1) [31], PROTEIN PHOSPHATASE 4 (PP4) [32], the splicing factor SMALL1(SMA1) [33], the ATPase CHROMATIN REMODELLING FACTOR 2 (CHR2) [34], the exportin 5 homolog HASTY (HST) [35], the pore-associated proteins THO/HRP1 PHENOTYPE 1 (THP1) and SUPPRESSOR OF ACTIN 3A (SAC3A) of the TREX-2 complex (Fig. 1) [19]. In the loss-of-function mutants of these proteins, the levels of pri-miRNAs, MIR promoter activities and/or the Pol II occupancy at MIR promoters are reduced, suggesting that these proteins modulate MIR transcription by directly or indirectly promoting the recruitment of Pol II to MIR promoters [19,28–35]. In addition, some proteins negatively regulate miRNA transcription (Fig. 1). Disruption of the F-Box protein CONSTITUTIVE EXPRESSER OF PR GENE 1 (CPR1) increases the transcript levels of the disease resistance protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 (SNC1), resulting in reduced pri-miRNA transcription. Moreover, overexpression of SNC1 and the transcriptional corepressor TOPLESS RELATED 1 (TPR1) represses the transcription of MIRs (Fig. 1) [36]. These results suggest that SNC1 is a repressor of MIR transcription. However, some protein factors can act as both a transcriptional activator and repressor. For instance, mutations in the CYCLING DOF TRANSCRIPTION FACTORS 2 (CDF2) increases the transcription of some MIRs, while represses that of others [37].
Besides affecting MIR promoter activity, some protein factors, such as INCREASED LEVEL OF POLYPLOIDY1-1D (ILP1) and NTC-RELATED PROTEIN 1 (NTR1), facilitate transcriptional elongation of MIR genes (Fig. 1) [38]. Notably, the RNA silencing effector protein AGO1 can negatively affect the transcription of MIR161 and MIR173 via disassembling Pol II under salt stress conditions [39]. Moreover, phosphorylation of the C-terminal domain (CTD) of the largest subunit of Pol II is also required for efficient MIR transcription (Fig. 1). Disruption of CYCLIN-DEPENDENT KINASE F;1 (CDKF;1) and CDKD kinases, which catalyse Ser phosphorylation of CTD, impairs MIR transcription, 5ʹ-capping, 3ʹ-processing and splicing [40]. Chromatin features such as histone acetylation and methylation can also influence MIR transcription. For example, Polycomb Repressive Complex 2 (PRC2) represses MIR156A/C transcription through increasing H3K27me3 deposition at their promoters [41].
Some specific transcription factors regulate the transcription of individual MIR genes in a spatiotemporal manner. For instance, flower development can be coordinately regulated by several protein factors that affect the transcription of MIR172. The SANT-domain-containing protein POWERDRESS (PWR) can facilitate Pol II occupancy at the promoters of MIR172a, b and c but not MIR172d or e [42]. The transcription factor APETALA 2 (AP2)-dependent recruitment of the transcription co-repressors LEUNIG (LUG) and SEUSS (SEU) represses MIR172 expression [43]. During shade avoidance, PHYTOCHROMOE INTERACTING FACTORS (PIFs) bind five MIR156 promoters and repress their expression [44]. MIR156 can also be positively regulated by the B3 domain transcription factor FUSCA3 (FUS3) during the transition from embryo to seedling development [45]. Interestingly, another B3 transcription factor ABSCISIC ACID INSENSETIVE3 (ABI3) promotes the expression of MIR156 during the early stages of seed development while repressing their expression during late development [46]. In addition, ABI3 negatively regulates the transcription of MIR160B [46]. During cell wall remodelling, the transcription factor for photomorphogenesis HYPOCOTYL5 (HY5) negatively regulates MIR775A in aerial organs whereas positively in roots of Arabidopsis [47,48].
2.2. Pri-miRNA stability
Eukaryotes possess nuclear RNA surveillance machineries that cooperate with RNA processing to control maturation and degradation of RNAs [49,50]. Previous studies demonstrated that the 5ʹ-to-3ʹ exoribonucleases (XRNs) and the 3ʹ-to-5ʹ exoribonuclease complex exosome are responsible for the elimination of pri-miRNA processing intermediates [51–53]. Notably, most pri-miRNAs are non-coding RNAs that contain premature stop codon, and are potential targets of the nuclear RNA machineries. Thus, protection of pri-miRNAs from degradation during co-transcriptional processing is likely crucial for maintaining the amounts of pri-miRNAs required for miRNA biogenesis. Indeed, accelerated degradation of pri-miRNAs has been shown to reduce miRNA accumulation [54–59].
DAWDLE (a forkhead-associated domain-containing protein, DDL) has been shown to stabilize pri-miRNAs in plants (Fig. 2) [54]. DDL is an RNA-binding protein and interacts with both DCL1 and pri-miRNAs. Lack of DDL reduces the accumulation of pr-miRNAs and miRNAs without affecting MIR promoter activity and transcription, suggesting that DDL may protect pri-miRNAs from degradation. Besides DDL, several components of the MOS4-associated complex (MAC), including PRL1 (PLEIOTROPIC REGULATORY LOCUS1, a conserved WD-40 protein), MAC3 (a U-box type E3 ubiquitin ligase), also play a role in stabilizing pri-miRNAs (Fig. 2). MAC is a conserved complex. Its homolog complexes include the CDC5-SNEVprp [19]-pso [4] complex of human and the Nineteen complex (NTC) of yeast are associated with the spliceosome and participate in splicing [60,61]. Like DDL, these MAC proteins associate with the DCL1 complex and are required for the accumulation of pri-miRNAs but not for MIR transcription [55,56]. However, how these proteins stabilize pri-miRNAs are still poorly understood.
Figure 2.
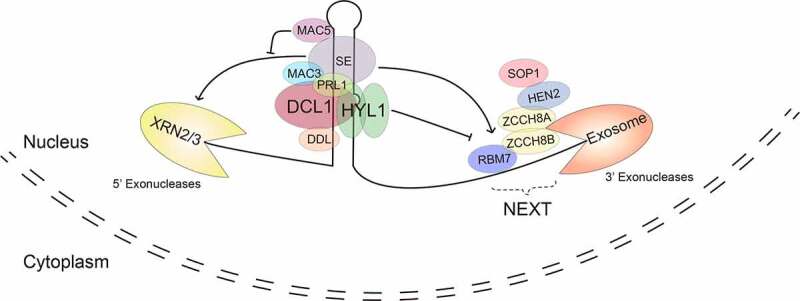
Regulation of pri-miRNA stability
Multiple protein factors are required for the regulation of pri-miRNA stability. Unprocessed pri-miRNA can be degraded by 5ʹ-3ʹ exoribonucleases (XRNs) and 3ʹ-5ʹ exosomes in an SE-dependent manner. MAC5 and HYL1 protect pri-miRNAs from degradation by XRNs and exosomes, respectively. The functional mechanisms of DDL, PRL1 and MAC3 in the regulation of pri-miRNA stability are still unknown.
Recently, several enzymes that degrades pri-miRNAs in the nucleus have been identified. Over accumulation of 3ʹ-phosphadenosine 5ʹ-phosphate (PAP), an inhibitor of the XRNs, increases pri-miRNA levels and stability [57]. Moreover, the introduction of loss-of-function xrn2 or xrn3 mutation into mac5a, in which lack of MAC5A, another component of MAC, reduces pri-miRNA stability, increases pri-miRNA half-lives, and partially recovers the accumulation of pri-miRNAs [58]. These results show that XRN2/XRN3 catalyzes the degradation of pri-miRNAs in the nucleus (Fig. 2). In addition, the levels of some pri-miRNAs are increased in the hen2 mutant, which lacks HUA1 EHANCHER2 (HEN2), a component of the nuclear exosome targeting complex (NEXT) that promotes RNA degradation by nuclear exosome, suggesting that exosome is also responsible for the degradation of pri-miRNAs (Fig. 2) [59,62].
Evidences also reveal that the interplay between protein involved in miRNA biogenesis and ribonucleases leads to pri-miRNA degradation and stabilization. SE, a key factor in the DCL1 complex, appears to have a role in recruiting ribonucleases to pri-miRNAs (Fig. 2), given the fact that SE interacts with both NEXT and XRN2 [58,59,62]. Supporting this notion, in mac5, the degradation of pri-miRNAs by XRN2 or XRN3 requires a functional SE [58]. This result also suggests that the function of MAC5 is to protect pri-miRNAs from SE-dependent 5ʹ-to-3ʹ ribonucleases (Fig. 2). Interestingly, hen2 and sop1 (mutation in another component of NEXT) increase stability and accumulation of some pri-miRNAs in hyl1, resulting in elevated levels of some miRNAs and partial recovery of developmental defects of hyl1 [62]. In contrast, these two mutations do not have a similar impact on dcl1 [62]. These results suggest that HYL1 is able to protect some pri-miRNAs from SE-dependent exosome activities (Fig. 2), in addition to its role in pri-miRNA processing [62]. Taken together, these recent findings suggest that plants may use SE to recruit enzymes to degrade pri-miRNA processing intermediates. However, at the same time, plants also employ other components of the DCL1 complex such as DDL, HYL1 and MAC to coordinately protect unprocessed pri-miRNAs from degradation by these ribonucleases.
3. Regulation of miRNA biogenesis by pri-miRNA structure features, splicing and modification
As the substrates of the DCL1 complex, pri-miRNAs themselves also affect the processing efficiency in various ways. It has been shown that pri-miRNA structure is an import factor for the cleavage site selection and processing efficiency of DCL1. Moreover, splicing and alternative splicing of pri-miRNAs also contribute to the processing efficiency of some pri-miRNAs. Interestingly, nucleotide modifications within the pri-miRNAs can also modulate the production of some miRNAs.
3.1. Sequence and structure features of pri-miRNAs
The stem-loop of pri-miRNAs varies in length, structure and positioning of the miRNA/miRNA* duplex. These features play crucial roles in determining processing efficiency and manner. Most pri-miRNAs have an imperfect lower stem of ~15 bp, which is often followed by a large bulge, below the miRNA/miRNA* duplex [63–65]. This structure is required for the accurate initial cleavage distal to loop (base-to-loop cleavage). In contrast, some pri-miRNAs such as pri-miR159a and pri-miR319a possess long upper stem, resulting in loop-to-base cleavage [66–68].
Interestingly, a few of pri-miRNAs contain multibranched terminal loops [69]. This structure heterogeneity results in bi-directional processing of pri-miRNAs, although the base-to-loop processing more efficiently produces miRNAs [69]. However, the efficient processing pri-miR157c depends on its branched terminal loop, suggesting that the DCL1 complex can recognize alternative structures of pri-miRNAs under some circumstances [69,70]. In addition, the presence of specific GC-rich sequence in the miRNA:miRNA* region has been observed and may be required for efficient and accurate processing of pri-miRNAs [71]. Moreover, certain nucleotide pairs at pri-miRNAs’ cleavage site are preferred for efficient processing pri-miRNAs [72]. Interestingly, the identity of the nucleotides at mismatched positions of the stem is also important for efficient pri-miRNA processing [72]. Furthermore, some conserved sequence and structures of pri-miRNAs proximal to the miRNA/miRNA* duplex have been identified from various plant species and likely are important for miRNA biogenesis [73].
3.2. Splicing of pri-miRNAs
Like protein-coding genes, many MIRs contain introns [74]. The splicing of introns has important roles in the processing of some pri-miRNAs. Splicing of introns can change the structure of some pri-miRNAs, which in turn can affect processing. In rice, the presence of intron disrupts the formation of stem-loop structure of pri-miR842 and pri-miR846, and therefore, splicing of these introns is required for their processing [75]. In contrast, the stem-loop of pri-miR162a is composed of unspliced intron and exon [76]. As a result, splicing of intron inhibits the production of miR162 [76]. In addition, the introns downstream of stem loop from pri‐miR161, pri‐miR163 and pri‐miR172b are required for efficient production of corresponding miRNAs [77,78], although the introns can be replaced by unrelated ones. Further study shows that the 5ʹ splicing site, but not the 3ʹ splicing site, of these introns, stimulates processing efficiency by affecting the selection of polyadenylation site [77–79].
Alternative splicing also plays a role in regulating miRNA biogenesis. In rice, MIR528 contains two alternative splicing sites, resulting in two forms of pri-miR528 [80]. One has a long 3ʹ end and less efficiently processed, while the other one has a short 3ʹ end and efficiently processed [80]. The levels of two forms change during development, resulting in differential accumulation of miR528 [80]. In Arabidopsis, pri-miR400 localizes in the intron of a host gene and needs to be spliced out for efficient processing [81]. However, heat stress can cause retention of pri-miR400 containing intron in the host gene transcript, resulting in less accumulation of miR400 [81].
3.3. Pri miRNA modifications
RNA modifications have recently emerged as important post-transcriptional regulators [82]. Particularly, the N [6]-methyadenosine (m6A) modification is crucial to regulate gene expression in mammals and plants [83,84]. A recent study links m6A to miRNA biogenesis [85,86]. mRNA adenosine methylase (MTA) is responsible for the deposition of m6A into RNA molecules [87]. MTA is able to interact with Pol II and Tough (TGH), a known miRNA processing regulator [85,86]. In the mta mutant, the abundances of m6A and miRNAs are significantly reduced. Further evidences show that a subset of pri-miRNAs is bound and methylated by MTA, which in turn ultimately promotes pri-miRNA processing [85]. Low level of m6A may affect stem-loop region of pri-miRNAs, and thus reduce the association of pri-miRNAs with HYL1 [85]. Intriguingly, untemplated cytidine or uridine addition on the trimmed pre-miRNAs can restore the intact length pre-miRNAs, and therefore, promote their processing [88].
4. Regulation of the DCL1 complex
The levels and activities of the DCL1 complex are modulated at multiple levels to control miRNA biogenesis [1,4,5]. On one hand, the recognition and processing of pri-miRNAs by the DCL1 complex require the assistance of many protein factors. On the other hand, DCL1, HYL1 and SE themselves are regulated at transcriptional, post-transcriptional and post-translational levels.
4.1. The recognition and processing of pri-miRNAs by DCL1
Besides DCL1, HYL1 and SE are two core components for miRNA biogenesis. HYL1 is a dsRNA-bind protein that recognizes the stem region of pri-miRNA. Studies show that HYL1 plays important roles in positioning of pri-miRNAs into DCL1. Lack of HYL1 causes miscleavage of pri-miRNAs by DCL1 and reduces the accumulation of miRNAs at global levels. SE is a C2H2 zinc finger domain and binds pri-miRNAs. Disruption of SE also causes reduced miRNA levels at global levels. It has been proposed that DCL1, HYL1 and SE form a subnuclear loci called the D-body [2,89–91]. A recent study shows that SE-mediated phase separation is crucial for D-body assembly [92]. SE contains three intrinsically disordered regions (IDRs), which are required for phase separation and efficient miRNA processing [92]. D-body co-transcriptionally processes pri-miRNAs [2,89–91]. The co-transcriptional recruitment of DCL1 to pri-miRNAs requires the Elongator complex [28], NOT2 [29], MAC3 [55] and MAC7 [93]. Mutations in these proteins reduce the amount of the DCL1 complex without affecting the protein levels of DCL1, HYL1 and SE, suggesting that they facilitate the assembly of the DCL1 complex [28,29,55,93]. It should be noted that the ubiquitin ligase activity of MAC3 is required for miRNA biogenesis [55], suggesting that protein ubiquitination may play a role in the assembly of the DCL1 complex. The DEAH-box helicase PHYTOPHTHORA SUPPRESSOR OF RNA SILENCING 1 (PINP1) [94] and the RNA binding protein MODIFIER OF SNC1, 2 (MOS2) [95] also affect the formation of the DCL1 complex. MOS2 [95], STV1 [31] and TGH [86] bind pri-miRNAs and assist the recruitment of the DCL1 complex to pri-miRNAs.
In the past decades, many protein factors modulating DCL1 activities have been identified. NOT2 and Elongator, in addition to their role in pri-miRNA transcription, interact with the DCL1 complex to facilitate pri-miRNA processing (Fig. 3) [28,29]. In addition, CDC5 [30], PRL1 [56], MAC3 [55], MAC5 [58] and MAC7 [93], also interact with the DCL1 complex and promotes its activity (Fig. 3). These five proteins are components of MAC, suggesting that MAC plays a multiple role in pri-miRNA processing. Additional factors include TGH [86], DDL [54] and MOS2 [95], which also interact with DCL1 to enhance its activity (Fig. 3). Moreover, several pre-mRNA processing factors including the CAP-BINDING PROTEIN 20 (CBP20) and CBP80 [96] SMA1 [33], the pre-mRNA processing factor 6 homolog STABILIZED1 (STA1) [97], HIGH OSMOTIC STRESS GENE EXPRESSION 5 (HOS5) [98], ARGININE/SERINE-RICH SPLICING FACTOR 40 (RS40) and RS41 [98], the U1 snRNP Subunit LETHAL UNLESS CBC 7 RL (LUC7rl) [11], the THO2 in the THO/TREX complex [99], GLYCINE-RICH RBP 7 (GPR7) [100], SICKLE (SIC, a proline-rich protein) [101], the PRE-MRNA-PROCESSING PROTEIN (PRP)39b, PRP40a, PRP40b [11], also interact with the DCL1 complex and positively regulate pri-miRNA processing (Fig. 3). In addition, the RECEPTOR FOR ACTIVATED C KINASE 1 (RACK1) interacts with SE to promotes pri-miRNA processing (Fig. 3) [102]. Notably, these protein factors also modulate the splicing. These discoveries suggest that miRNA biogenesis and general RNA processing are interconnected. Some specific factors modulate the activity of DCL in a spatiotemporal manner. For example, the DEAD-BOX RNA HELICASE 27 (RH27) interacts with DDL, HYL1 and SE, and promotes miRNA biogenesis in embryos, shoot apical meristem and root apical meristem [103]. Besides positive factors, negative factors of the DCL1 complex are also identified. Despite of its positive role in promoting MIR transcription, CHR2, an DNA/RNA helicase, is able to unwinds pri-miRNAs, which in turn represses pri-miRNA processing (Fig. 3) [34]. CDF2 decoys the DCL and HYL1 to inhibit the activity of the DCL1 complex [37].
Figure 3.
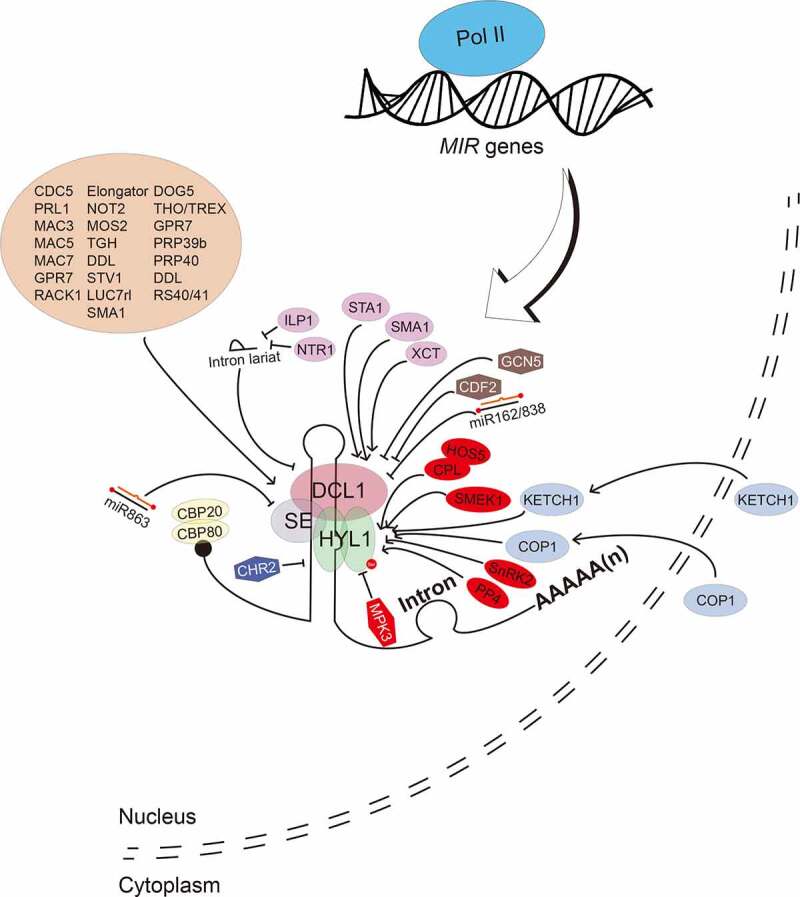
Regulation of pri-miRNA processing
Processing of pri-miRNAs requires the core dicing body (DCL1, SE and HYL1) as well as many other accessory factors (brown ellipse). The abundance and activities of DCL1, SE and HYL1 are regulated by multiple protein factors and miRNAs. Positive and negative regulators are marked with ellipse and hexagon, respectively. Regulators involved in phosphorylation of HYL1 are marked with red colour.
4.2. Regulation of DCL1, HYL1 and SE
Studies also reveal that the expression and activities of DCL1, HYL1 and SE is controlled at transcriptional, post-transcriptional and post-translational levels. These multifaceted regulations appear to be crucial for proper accumulation of miRNAs.
Factors that control the transcript levels of DCL1, HYL1 and/or SE have been identified. XAP5 CIRCADIAN TIMEKEEPER (XCT), a nuclear-localized protein and STA1 have been shown to promote the expression of DCL genes including DCL1 [97,104], while the histone acetyltransferase GENERAL CONTROL NON-REPRESSED PROTEIN 5 (GCN5) appears to repress the transcription of DCL1, HYL1 and SE (Fig. 3) [105]. Moreover, the DELAY OF GERMINATION1 (DOG1) promotes the transcription of DCL1, HYL1, SE, TGH and CDC5 [106]. In addition, proper splicing of the ninth intron of DCL1 requires the splicing factor SMA1 [33], which may affect DCL1 expression. The expression of DCL1 and SE is also subjected to feedback regulation. miR162 and miR863, which are generated by the DCL1 complex, directly target DCL1 and SE for cleavage, respectively, which in turn ensures the correct DCL1 and SE transcript levels [107,108]. Interestingly, the DCL1 transcript levels are also negatively controlled by the DCL1-mediated processing of MIR838, which is derived from the 14th intron of DCL1 pre-mRNAs [109]. Interestingly, the DCL1 protein can be stabilized by light during de-etiolation [110]. However, elevated DCL1 protein levels do not alter miRNA levels in the de-etiolated plants relative to etiolated plants, suggesting the presence of a light-induced suppressor of DCL1 [110]. Indeed, a light-stabilized FORHEAD-ASSOCIATED DOMAIN 2 (FHA2) protein was recently found to inhibit DCL1 activity by limiting its access to pri-miRNAs [111].
Notably, plants also use RNA-based mechanisms to decoy the components of the DCL1 complex. Intron lariat RNAs derived splicing by product, when failing to be disassociated from spliceosome, bind the DCL1 complex and prevent pri-miRNA processing (Fig. 3) [112]. Interestingly, ILP1 and NTR1, two splicing factors, interact with DCL1 and SE, remove intron lariat RNAs from spliceosome, and promotes the accumulation of miRNAs (Fig. 3) [38]. Another example is the transcripts generated from the short-interspersed elements (SINEs). SINE transcripts form a structure similar to pri-miRNAs, which in turn sequesters HYL1 from pri-miRNA processing [113].
Post-translational modifications also play critical roles in modulating the activities of the DCL1 complex. HYL1 is subjected to phosphorylation modification. The MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3) and the SNF1-RELATED PROTEIN KINASE 2 (SnRK2) interact with and phosphorylate HYL1 (Fig. 3) [114,115]. Phosphorylation has been shown to inhibit HYL1 function, because de-phosphorylation of HYL1 by the C-TERMINAL DOMAIN PHOSPHATASE-LIKE 1 (CPL1) and its homolog CPL2, is required for HYL1 localization to D-bodies and pri-miRNA processing (Fig. 3) [116]. CPL1 and CPL2 require the assistance of HOS5 to dephosphorylate HYL1 in young vegetative and reproductive tissues [117]. Moreover, SUPPRESSOR OF MEK 1 (SMEK1), together with the Protein Phosphatase 4 (PP4), also dephosphorylates HYL1, which in turn prevents HYL1 degradation (Fig. 3) [118]. Interestingly, phosphorylation also stabilizes HYL1 in the dark by keeping it in the nucleus so that during dark-to-light transition, light-mediated de-phosphorylation is able to active HYL1 for miRNA biogenesis [119]. In addition, Constitutive Photomorphogenic 1 (COP1), a RING-finger E3 ligase, moves from the nucleus to the cytoplasm and stabilizes HYL1 in the light by repressing unknown protease activity [120]. SE is also phosphorylated by in vitro [29,115], but the functional significance of this modification remains to be identified.
The activity of SE and HYL1 is modulated at additional layers. An importin-beta protein KARYOPHERIN ENABLING THE TRANSPORT OF THE CYTOPLASMIC HYL1 (KETCH1) transports HYL1 from the cytoplasm into the nucleus to promote miRNA production (Fig. 3) [121]. Moreover, a recent study shows that SE interacts with the 20S core proteasome, which is able to degrade dysfunctional SE in a ubiquitin-independent manner [122]. The elimination of disordered SE is required for the protection of the functional DCL1 complex.
Conclusion and perspectives
In the past decades, many protein factors modulating miRNA biogenesis have been identified. Analyses on these proteins and corresponding mutants have led to a better understanding of the process generating miRNAs. However, huge challenges remain to fully understand miRNA biogenesis. The biochemical functions for most of these protein factors are still unknown. Many of these factors have multiple roles in miRNA biogenesis. It is unclear whether a complex or multiple subcomplexes are required for a specific component to perform multiple tasks. Moreover, how the protein factors act coordinately to control the biogenesis of individual miRNAs based on pri-miRNA sequence and structure is still an unexplored field. In addition, it is unknown if miRNA biogenesis is spatiotemporally controlled at single cell levels, if so, how this is achieved. Clearly solving these questions requires novel techniques including single-cell biology and in vitro and in vivo systems to reconstitute miRNA biogenesis pathway. Studies also reveal that pri-miRNA stability is determined by the interplay between proteins involved in miRNA processing and ribonucleases, which provide a new regulatory layer of miRNA biogenesis. Further investigation on these observations will lead to a better understanding of how miRNA processing machinery and RNA decay machinery coordinate during miRNA biogenesis. The contribution of m6A methylation on miRNA biogenesis and the fact that many factors involved in miRNA biogenesis function in transcription and splicing of mRNAs suggest that miRNA biogenesis is interconnected with other cellular processes. It will be interesting to further investigate how miRNA biogenesis is integrated into these processes.
Funding Statement
This work is supported by National Institutes of Health [GM127414] and National Science Foundation [MCB- 1818082].
Disclosure statement
No potential conflicts of interest were disclosed.
References
- [1].Wang J, Mei J, Ren G.. Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci. 2019;10:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yu Y, Jia T, Chen X.. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017;216(4):1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li S, Castillo-Gonzalez C, Yu B, et al. The functions of plant small RNAs in development and in stress responses. Plant J. 2017;90(4):654–670. [DOI] [PubMed] [Google Scholar]
- [4].Song X, Li Y, Cao X, et al. Micrornas and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol. 2019;70(1):489–525. [DOI] [PubMed] [Google Scholar]
- [5].Manavella PA, Yang SW, Palatnik J. Keep calm and carry on: miRNA biogenesis under stress. Plant J. 2019;99(5):832–843. [DOI] [PubMed] [Google Scholar]
- [6].Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel). 2015;5(4):1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Czech B, Munafo M, Ciabrelli F, et al. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. 2018;52(1):131–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16(12):727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ozata DM, Gainetdinov I, Zoch A, et al. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
- [10].Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64(1):137–159. [DOI] [PubMed] [Google Scholar]
- [11].Knop K, Stepien A, Barciszewska-Pacak M, et al. Active 5ʹ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. 2017;45(5):2757–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zielezinski A, Dolata J, Alaba S, et al. mirEX 2.0 - an integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 2015;15(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25(7):2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yu B, Yang ZY, Li JJ, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mi SJ, Cai T, Hu YG, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133(1):116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Montgomery TA, Howell MD, Cuperus JT, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128–141. [DOI] [PubMed] [Google Scholar]
- [17].Zhu H, Hu F, Wang R, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145(2):242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bologna NG, Iselin R, Abriata LA, et al. Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microrna pathway. Mol Cell. 2018;69(4):709–19 e5. [DOI] [PubMed] [Google Scholar]
- [19].Zhang B, You C, Zhang Y, et al. Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis. Nat Plants. 2020;6(8):957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dalmadi Á, Gyula P, Balint J, et al. AGO-unbound cytosolic pool of mature miRNAs in plant cells reveals a novel regulatory step at AGO1 loading. Nucleic Acids Res. 2019;47(18):9803–9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shriram V, Kumar V, Devarumath RM, et al. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci. 2016;7:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brant EJ, Budak H. Plant small non-coding RNAs and their roles in biotic stresses. Front Plant Sci. 2018;9:1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. [DOI] [PubMed] [Google Scholar]
- [24].Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xie Z, Allen E, Fahlgren N, et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138(4):2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Megraw M, Baev V, Rusinov V, et al. MicroRNA promoter element discovery in Arabidopsis. RNA. 2006;12(9):1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim YJ, Zheng B, Yu Y, et al. The role of mediator in small and long noncoding RNA production in Arabidopsis thaliana. Embo J. 2011;30(5):814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fang X, Cui Y, Li Y, et al. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat Plants. 2015;1(6):15075. [DOI] [PubMed] [Google Scholar]
- [29].Wang L, Song X, Gu L, et al. NOT2 proteins promote polymerase II–dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25(2):715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang S, Xie M, Ren G, et al. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc Natl Acad Sci U S A. 2013;110(43):17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li S, Liu K, Zhang S, et al. STV1, a ribosomal protein, binds primary microRNA transcripts to promote their interaction with the processing complex in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114(6):1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang S, Quan L, Li S, et al. The PROTEIN PHOSPHATASE4 complex promotes transcription and processing of primary microRNAs in Arabidopsis. Plant Cell. 2019;31(2):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li S, Xu R, Li A, et al. SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2018;46(17):9148–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang Z, Ma Z, Castillo-Gonzalez C, et al. SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature. 2018;557(7706):516–521. [DOI] [PubMed] [Google Scholar]
- [35].Cambiagno DA, Giudicatti AJ, Arce AL, et al. HASTY modulates miRNA biogenesis by linking pri-miRNA transcription and processing. Mol Plant. 2021;14(3):426–439. In Press, [DOI] [PubMed] [Google Scholar]
- [36].Cai Q, Liang C, Wang S, et al. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat Commun. 2018;9(1):5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sun Z, Guo T, Liu Y, et al. The roles of Arabidopsis CDF2 in transcriptional and posttranscriptional regulation of primary microRNAs. PLoS Genet. 2015;11(10):e1005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang J, Chen S, Jiang N, et al. Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana. Nucleic Acids Res. 2019;47(15):7886–7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dolata J, Bajczyk M, Bielewicz D, et al. Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol. 2016;172(1):297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hajheidari M, Farrona S, Huettel B, et al. CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the c-terminal domain of Arabidopsis RNA polymerase II. Plant Cell. 2012;24(4):1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xu M, Hu T, Smith MR, et al. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell. 2016;28(1):28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yumul RE, Kim YJ, Liu X, et al. POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLoS Genet. 2013;9(1):e1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Grigorova B, Mara C, Hollender C, et al. LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development. 2011;138(12):2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xie Y, Liu Y, Wang H, et al. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat Commun. 2017;8(1):348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wang F, Perry SE. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013;161(3):1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tian R, Wang F, Zheng Q, et al. Direct and indirect targets of the arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. 2020;103(5):1679–1694. [DOI] [PubMed] [Google Scholar]
- [47].Zhang H, Guo Z, Zhuang Y, et al. MicroRNA775 regulates intrinsic leaf size and reduces cell wall pectin levels by targeting a galactosyltransferase gene in Arabidopsis. Plant Cell. 2021; In press, 10.1093/plcell/koaa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang H, He H, Wang X, et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65(3):346–358. [DOI] [PubMed] [Google Scholar]
- [49].Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17(4):227–239. [DOI] [PubMed] [Google Scholar]
- [50].Ogami K, Chen Y, Manley JL. RNA surveillance by the nuclear RNA exosome: mechanisms and significance. Noncoding RNA. 2018;4(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kurihara Y, Schmitz RJ, Nery JR, et al. Surveillance of 3′ noncoding transcripts requires FIERY1 and XRN3 in Arabidopsis. G3 (Bethesda). 2012;2(4):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gy I, Gasciolli V, Lauressergues D, et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19(11):3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chekanova JA, Gregory BD, Reverdatto SV, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131(7):1340–1353. [DOI] [PubMed] [Google Scholar]
- [54].Yu B, Bi L, Zheng B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A. 2008;105(29):10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li S, Liu K, Zhou B, et al. MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 2018;30(2):481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang S, Liu Y, Yu B. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 2014;10(12):e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fang X, Zhao G, Zhang S, et al. Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev Cell. 2019;48(3):371–82 e4. [DOI] [PubMed] [Google Scholar]
- [58].Li S, Li M, Liu K, et al. MAC5, an RNA-binding protein, protects pri-miRNAs from SERRATE-dependent exoribonuclease activities. Proc Natl Acad Sci U S A. 2020;117(38):23982–23990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bajczyk M, Lange H, Bielewicz D, et al. SERRATE interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic Acids Res. 2020;48(12):6839–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Deng X, Lu T, Wang L, et al. Recruitment of the NineTeen complex to the activated spliceosome requires AtPRMT5. Proc Natl Acad Sci U S A. 2016;113(19):5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Palma K, Zhao Q, Cheng YT, et al. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21(12):1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gao S, Wang J, Jiang N, et al. Hyponastic leaves 1 protects pri-miRNAs from nuclear exosome attack. Proc Natl Acad Sci U S A. 2020;117(29):17429–17437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mateos JL, Bologna NG, Chorostecki U, et al. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis miR172a precursor. Curr Biol. 2010;20(1):49–54. [DOI] [PubMed] [Google Scholar]
- [64].Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol. 2010;20(1):37–41. [DOI] [PubMed] [Google Scholar]
- [65].Werner S, Wollmann H, Schneeberger K, et al. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010;20(1):42–48. [DOI] [PubMed] [Google Scholar]
- [66].Bologna NG, Mateos JL, Bresso EG, et al. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. The EMBO Journal. 2009;28(23):3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Cuperus JT, Montgomery TA, Fahlgren N, et al. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci U S A. 2010;107(1):466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bologna NG, Schapire AL, Zhai JX, et al. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013;23(10):1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhu H, Zhou Y, Castillo-Gonzalez C, et al. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol. 2013;20(9):1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Moro B, Chorostecki U, Arikit S, et al. Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res. 2018;46(20):10709–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Narjala A, Nair A, Tirumalai V, et al. A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Res. 2020;48(6):3103–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rojas AML, Drusin SI, Chorostecki U, et al. Identification of key sequence features required for microRNA biogenesis in plants. Nat Commun. 2020;11(1):5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chorostecki U, Moro B, Rojas AML, et al. Evolutionary footprints reveal insights into plant microRNA Biogenesis. Plant Cell. 2017;29(6):1248–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Nozawa M, Miura S, Nei M. Origins and evolution of microRNA genes in plant species. Genome Biol Evol. 2012;4(3):230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Jia F, Rock CD. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol Biol. 2013;81(4–5):447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hirsch J, Lefort V, Vankersschaver M, et al. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006;140(4):1192–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bielewicz D, Kalak M, Kalyna M, et al. Introns of plant pri-miRNAs enhance miRNA biogenesis. Embo Rep. 2013;14(7):622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Schwab R, Speth C, Laubinger S, et al. Enhanced microRNA accumulation through stemloop-adjacent introns. Embo Rep. 2013;14(7):615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Knop K, Stepien A, Barciszewska-Pacak M, et al. Active 5 ‘ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. 2017;45(5):2757–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang RX, Li PC, Mei HL, et al. Fine-Tuning of miR528 accumulation modulates flowering time in Rice. Mol Plant. 2019;12(8):1103–1113. [DOI] [PubMed] [Google Scholar]
- [81].Yan K, Liu P, Wu CA, et al. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell. 2012;48(4):521–531. [DOI] [PubMed] [Google Scholar]
- [82].Frye M, Harada BT, Behm M, et al. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jia GF, Fu Y, Zhao X, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Duan HC, Wei LH, Zhang C, et al. ALKBH10B is an RNA N6-methyladenosine demethylase affecting arabidopsis floral transition. Plant Cell. 2017;29(12):2995–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bhat SS, Bielewicz D, Gulanicz T, et al. mRNA adenosine methylase (MTA) deposits m 6 A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2020;117(35):21785–21795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ren G, Xie M, Dou Y, et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109(31):12817–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zhong S, Li H, Bodi Z, et al. MTA is an arabidopsis messenger RNA Adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20(5):1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Song J, Wang X, Song B, et al. Prevalent cytidylation and uridylation of precursor miRNAs in Arabidopsis. Nat Plants. 2019;5(12):1260–1272. [DOI] [PubMed] [Google Scholar]
- [89].Han MH, Goud S, Song L, et al. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci U S A. 2004;101(4):1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17(9):818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Song L, Han MH, Lesicka J, et al. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A. 2007;104(13):5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Xie D, Chen M, Niu J, et al. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat Cell Biol. 2021;23(1):32–39. [DOI] [PubMed] [Google Scholar]
- [93].Jia T, Zhang B, You C, et al. The Arabidopsis MOS4-associated complex promotes microRNA biogenesis and precursor messenger RNA splicing. Plant Cell. 2017;29(10):2626–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Qiao Y, Shi J, Zhai Y, et al. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci U S A. 2015;112(18):5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wu X, Shi Y, Li J, et al. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013;23(5):645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kim S, Yang JY, Xu J, et al. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008;49(11):1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ben Chaabane S, Liu R, Chinnusamy V, et al. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013;41(3):1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chen T, Cui P, Xiong L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015;43(17):8283–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Francisco-Mangilet AG, Karlsson P, Kim MH, et al. THO 2, a core member of the THO/TREX complex, is required for micro RNA production in Arabidopsis. Plant J. 2015;82(6):1018–1029. [DOI] [PubMed] [Google Scholar]
- [100].Koster T, Meyer K, Weinholdt C, et al. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 2014;42(15):9925–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhan X, Wang B, Li H, et al. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci U S A. 2012;109(44):18198–18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Speth C, Willing EM, Rausch S, et al. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013;76(3):433–445. [DOI] [PubMed] [Google Scholar]
- [103].Hou X-L, Chen W-Q, Hou Y, et al. DEAD-BOX RNA HELICASE 27 regulates microRNA biogenesis, zygote division, and stem cell homeostasis. Plant Cell. 2020; In Press, DOI: 10.1093/plcell/koaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Fang X, Shi Y, Lu X, et al. CMA33/XCT Regulates Small RNA production through modulating the transcription of dicer-like genes in Arabidopsis. Mol Plant. 2015;8(8):1227–1236. [DOI] [PubMed] [Google Scholar]
- [105].Kim W, Benhamed M, Servet C, et al. Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 2009;19(7):899–909. [DOI] [PubMed] [Google Scholar]
- [106].Huo H, Wei S, Bradford KJ. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci U S A. 2016;113(15):E2199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13(9):784–789. [DOI] [PubMed] [Google Scholar]
- [108].Niu D, Lii YE, Chellappan P, et al. miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat Commun. 2016;7(1):11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Rajagopalan R, Vaucheret H, Trejo J, et al. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20(24):3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Choi SW, Ryu MY, Viczian A, et al. Light triggers the miRNA-biogenetic inconsistency for de-etiolated seedling survivability in Arabidopsis thaliana. Mol Plant. 2020;13(3):431–445. [DOI] [PubMed] [Google Scholar]
- [111].Park SJ, Choi SW, Kim GM, et al. Light-stabilized FHA2 suppresses miRNA biogenesis through interactions with DCL1 and HYL1. Mol Plant. 2021; In Press, DOI: 10.1016/j.molp.2021.01.020. [DOI] [PubMed] [Google Scholar]
- [112].Li Z, Wang S, Cheng J, et al. Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet. 2016;12(11):e1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Pouch-Pelissier MN, Pelissier T, Elmayan T, et al. SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet. 2008;4(6):e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Raghuram B, Sheikh AH, Rustagi Y, et al. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. Febs J. 2015;282(3):521–536. [DOI] [PubMed] [Google Scholar]
- [115].Yan J, Wang P, Wang B, et al. The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 2017;13(4):e1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Manavella PA, Hagmann J, Ott F, et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151(4):859–870. [DOI] [PubMed] [Google Scholar]
- [117].Karlsson P, Christie MD, Seymour DK, et al. KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc Natl Acad Sci U S A. 2015;112(45):14096–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Su C, Li Z, Cheng J, et al. The protein phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Dev Cell. 2017;41(5):527–39 e5. [DOI] [PubMed] [Google Scholar]
- [119].Achkar NP, Cho SK, Poulsen C, et al. A quick HYL1-dependent reactivation of microRNA production is required for a proper developmental response after extended periods of light deprivation. Dev Cell. 2018;46(2):236–47 e6. [DOI] [PubMed] [Google Scholar]
- [120].Cho SK, Ben Chaabane S, Shah P, et al. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat Commun. 2014;5(1):5867. [DOI] [PubMed] [Google Scholar]
- [121].Zhang Z, Guo X, Ge C, et al. KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114(15):4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Li Y, Sun D, Ma Z, et al. Degradation of SERRATE via ubiquitin-independent 20S proteasome to survey RNA metabolism. Nat Plants. 2020;6(8):970–982. [DOI] [PMC free article] [PubMed] [Google Scholar]


