ABSTRACT
Selective autophagy is a specific elimination of certain intracellular substrates by autophagic pathways. The most studied macroautophagy pathway involves tagging and recognition of a specific cargo by the autophagic membrane (phagophore) followed by the complete sequestration of targeted cargo from the cytosol by the double-membrane vesicle, autophagosome. Until recently, the knowledge about selective macroautophagy was minimal, but now there is a panoply of links elucidating how phagophores engulf their substrates selectively. The studies of selective autophagy processes have further stressed the importance of using the in vivo models to validate new in vitro findings and discover the physiologically relevant mechanisms. However, dissecting how the selective autophagy occurs yet remains difficult in living organisms, because most of the organelles are relatively inaccessible to observation and experimental manipulation in mammals. In recent years, zebrafish (Danio rerio) is widely recognized as an excellent model for studying autophagic processes in vivo because of its optical accessibility, genetic manipulability and translational potential. Several selective autophagy pathways, such as mitophagy, xenophagy, lipophagy and aggrephagy, have been investigated using zebrafish and still need to be studied further, while other selective autophagy pathways, such as pexophagy or reticulophagy, could also benefit from the use of the zebrafish model. In this review, we shed light on how zebrafish contributed to our understanding of these selective autophagy processes by providing the in vivo platform to study them at the organismal level and highlighted the versatility of zebrafish model in the selective autophagy field.
Abbreviations: AD: Alzheimer disease; ALS: amyotrophic lateral sclerosis; Atg: autophagy-related; CMA: chaperone-mediated autophagy; CQ: chloroquine; HsAMBRA1: human AMBRA1; KD: knockdown; KO: knockout; LD: lipid droplet; MMA: methylmalonic acidemia; PD: Parkinson disease; Tg: transgenic.
KEYWORDS: Aggrephagy, lipophagy, mitophagy, selective autophagy, xenophagy, zebrafish
Debut of zebrafish as a new autophagy model
Autophagy describes a set of evolutionarily conserved lysosomal degradation pathways of cytoplasmic components ranging from single proteins to large organelles [1]. The process involves adaptation to nutrient starvation or elimination of pathogens and is fundamental during development to maintain normal cell homeostasis [2]. There are several variations that are defined as macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). Macroautophagy is characterized by formation of autophagosomes, the double-membrane vesicles that enclose a portion of the cytosol or specific cargo and fuse with the lysosomes or late endosomes (Figure 1, top panel), whereas microautophagy involves direct acquisition of cytoplasm or specific substrates by invaginating lysosomal or endosomal membranes; and in CMA, soluble proteins containing the KFERQ-like pentapeptide are recognized by the HSPA8/HSC70 (heat shock protein family A (Hsp70) member 8) chaperone and translocated across the lysosomal membrane. CMA is in principle a selective process where single polypeptides are recognized as unfolded/misfolded when exposing their KFERQ-like binding sites to HSPA8 [3]. For a long time, autophagy was considered to be nonselective and deliver a bulk cytosol to the lysosomes for degradation and recycling. It is now known that in many instances it is a selective delivery of specific cargo to the lytic compartment, e.g. targeted elimination of protein aggregates by aggrephagy, pathogens by xenophagy, degradation of organelles, such as mitochondria, endoplasmic reticulum, peroxisomes and lipid droplets by mitophagy, reticulophagy, pexophagy and lipophagy, respectively, as recently reviewed [4,5].
Figure 1.
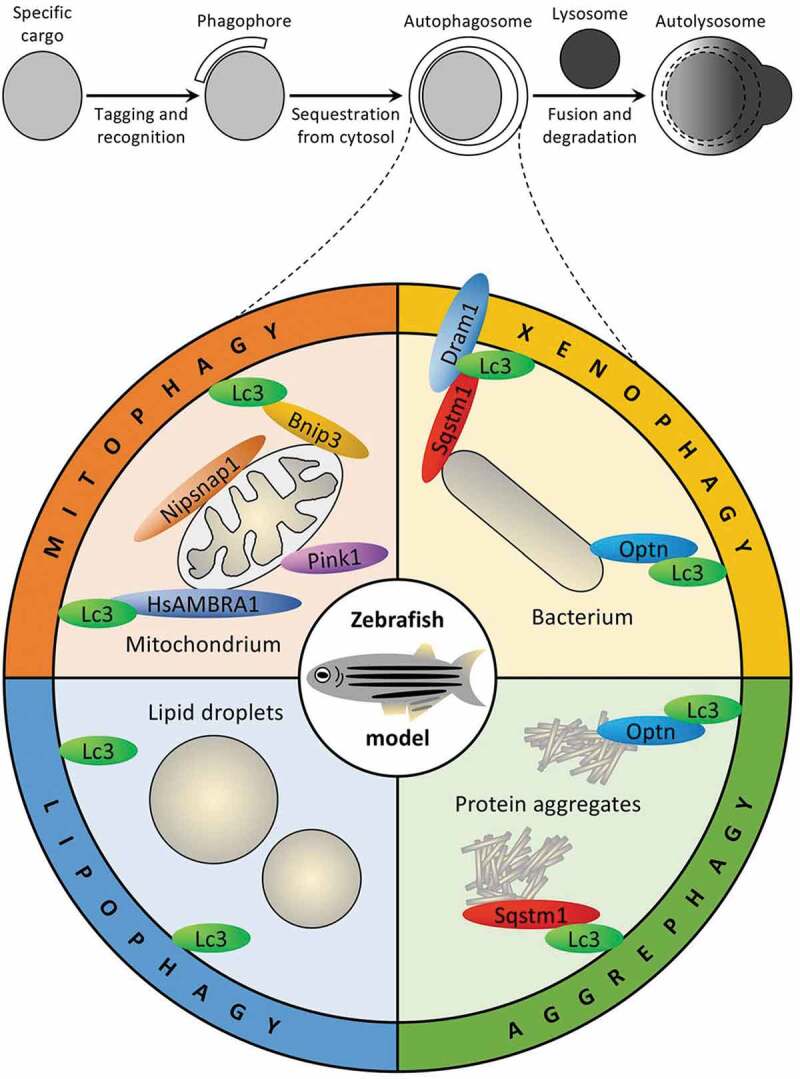
Selective autophagy pathways and selectivity factors studied in zebrafish. Top panel displays main steps shared by all selective macroautophagy pathways. Bottom panel shows selective macroautophagy pathways (mitophagy, xenophagy, lipophagy and aggrephagy) and their selectivity factors studied in zebrafish to date. See text for details
Zebrafish is a well-established model to study autophagy now. However, the field of zebrafish autophagy was initiated only in 2009 when Daniel Klionsky and colleagues identified two zebrafish Atg8 homologs, Map1lc3b/Lc3 (microtubule-associated protein 1 light chain 3 beta) and Gabarapa/Gabarap (GABA(A) receptor-associated protein a), which are involved in autophagosome formation, cargo recognition and recruitment to the autophagic membrane [6]. The group created transgenic (Tg) zebrafish lines, Tg(CMV:eGFP-map1lc3b) and Tg(CMV:eGFP-gabarapa), and described temporal expression patterns of Map1lc3b and Gabarapa at the early embryonic stage, which can be affected by autophagy inhibitors. The authors discussed that several autophagy-related (atg) genes, such as ulk1/Atg1, atg2, atg3, atg4, atg5, becn1/Vps30/Atg6, atg7, lc3/Atg8, atg9, atg12 and atg16 are present in the zebrafish genome, which suggested that this organism may be useful as a model system for studying the mechanism and function of autophagy. These findings provide the first instance of monitoring autophagy in zebrafish. The transparency of zebrafish embryos further attracted the attention of cell biologists using zebrafish as a model to study autophagic processes. Since the initial publication by He et al. in 2009, the interest in zebrafish autophagy models has grown substantially. A dual Tg(CMV:eGFP-map1lc3b; eef1a1l1:mCherry-lamp1) reporter line was used to further elucidate the role of lysosome acidification in senescence in larval zebrafish [7]. The development of this new generation of dual fluorescent probes further strengthened the use of zebrafish model in autophagy field, as reviewed in [8].
There are many features that make zebrafish the organism of choice for autophagy studies (see Table 1). Zebrafish model has received a lot of attention to close the gap between in vitro and in vivo assays as its embryogenesis is a rapid ex utero process amenable to noninvasive intravital imaging and longitudinal analysis. Moreover, embryo transparency during the early life stages makes observation of the whole body and organ formation possible. It allows researchers to label specific proteins and cell types for high-resolution fluorescence microscopy. In addition, antibodies that can be used to analyze autophagy in zebrafish have been listed elsewhere [9]. Using microinjection techniques at 1–4 cell stage of the embryo, zebrafish transient gene knockdowns (KDs) can be generated by application of antisense morpholino oligonucleotides what allows for a rapid test of gene involvement in autophagy. However, the off-target and less durable effects of morpholinos should be considered. The effects can be evaluated by using multiple morpholinos or by using alternative approaches, e.g. injection of four-guide clustered regularly interspaced short palindromic repeats-Cas9 ribonucleoprotein complexes that can cause biallelic gene disruption in G0 [10]. However, the potential drawback of gene disruption is genetic compensation due to mutant mRNA decay that can be minimized by creating mutant alleles that do not transcribe [11]. The same is also important for gene knockout (KO) strategies with transcription activator-like effector nucleases or clustered regularly interspaced short palindromic repeats that are often used to generate stable lines and might explain why the morpholino gene KD and the KO of the same gene with other approaches not always yield the same outcomes. Finally, exploiting the Gal4-UAS and Cre-lox systems, it is possible to define the functional roles of atg-genes in the cell- or tissue-specific manner [12–15].
Table 1.
Comparison of different animal models widely used in autophagy research (adapted from [87])
| Model | Caenorhabditis elegans | Drosophila melanogaster | Danio rerio | Mus musculus |
|---|---|---|---|---|
| Life cycle | Short (3.5 days) | Short (12 days) | Long (10–12 weeks) | Long (50–60 days) |
| Gene homology for human diseases | 65% | 75% | 84% | >90% |
| Anatomy | Relatively simple | Relatively simple | Vertebrate | Vertebrate |
| Husbandry and animal costs | Very low | Low | High | Very high |
| Screening assays | Simple | Simple | High-throughput and high-content | Low throughput |
The zebrafish model has several advantages over other animal model systems used in autophagy field (Table 1). It has a high fecundity rate when a pair of adult zebrafish can breed rapidly (approximately every 7 days) and produce as many as 100 to 200 eggs at a time. This is helpful where large number of animals is needed for phenotypic screening to produce more accurate and reproducible results. Zebrafish is becoming a popular and useful model in the biomedical field also because its genome has a high degree of conservation with human genome. The zebrafish genome study highlighted that 70% of human protein-coding genes are related to zebrafish genes and 82% of disease-causing human proteins have a zebrafish ortholog making zebrafish an attractive model organism for human disease research [16]. The ability to study molecular and cell biology of autophagy in the context of specific diseases, especially those for which mouse is not a good model of human processes, is another advantage of zebrafish. Here, we review the advances made using zebrafish for studying the selective autophagy processes and associated diseases.
Zebrafish as a tool for studying selective autophagy processes
While CMA was recently identified in fish (medaka) extending the availability of this important pathway beyond mammals and birds [17], most of the zebrafish research has been focused on the selective macroautophagy pathways, such as mitophagy, xenophagy, lipophagy and aggrephagy, to date (Figure 1). The common theme for these 4 pathways is tagging of the specific cargo to be degraded with ubiquitin by the specific E3 ubiquitin-protein ligase followed by recognition of the ubiquitinated substrate by at least one ubiquitin-binding selective autophagy receptor. The autophagic receptors are the proteins that bring the core autophagic machinery to the cargos and bridge them with the phagophore. It is important to note that some receptors can recognize their substrate directly, bypassing the need for its ubiquitination. Usually, such receptors have narrower substrate specificity and are normal constituents of cargo surface where they are turned on when needed by phosphorylation of the key protein binding sites. To close the gap on the other end and connect their substrates with a growing phagophore, autophagic receptors bind the phagophore/autophagosome membrane protein that belongs to the Atg8 family using their Atg8-family interacting motif, which is also known as the LC3-interacting region. Recently, both the selective autophagy receptors and Atg8-family proteins were extensively reviewed in the excellent publications [5,18].
Because most of the early selective autophagy studies in mammals were done in vitro using cell lines often overexpressing the proteins of interest and using non-physiological stresses, such as drug exposure, many findings require further validation in vivo. For example, understanding of how protein members of two Atg8 subfamilies – LC3 and GABARAP – play their unique roles in the selective autophagy is incomplete. Thus, it is now vital to contextualize how selective autophagy occurs in the multicellular, multiorgan environment of the entire organism under physiological conditions. To showcase zebrafish selective autophagy tools (Table 2), we provide recent examples of mitophagy, xenophagy, lipophagy and aggrephagy studies done in zebrafish and summarize their outcomes for autophagy field.
Table 2.
Tools to study selective autophagy in zebrafish
| Line/Reporter/Construct | Relevant selective autophagy pathway | Reference |
|---|---|---|
| Tg(CMV:eGFP-map1lc3b) | Many | [6] |
| Tg(CMV:eGFP-gabarapa) | Many | [6] |
| Tg(CMV:eGFP-map1lc3b; eef1a1l1:mCherry-lamp1) | Many | [7] |
| Tg(lyz:RFP-GFP-map1lc3b) | Many | [53] |
| Tg(elavl3:eGFP-map1lc3b) | Many | [85] |
| Tg(lyz:GFP-sqstm1) | Many | [52] |
| Cox8A-GFP-mCherry | Mitophagy | [26] |
| pCS2 + Venus-iLID-ActA | Mitophagy | [30] |
| pCS2 + HsAMBRA1-RFP-SspB | Mitophagy | [30] |
| Tg(5kbneurod:mito-mEos) | Mitophagy | [38] |
| Tg(ubi:mito-Keima) | Mitophagy | [39] |
| Tg(ubi:mito-GR) | Mitophagy | [39] |
| apoBb.1NLuc/Nluc | Lipophagy | [69] |
| Tg(rho:eGFP-tau) | Aggrephagy | [82] |
| Dendra-MAPT/tau (in pDendra2) | Aggrephagy | [82] |
| Dendra-MAPT/tauWT (in pDestTol2CG2) | Aggrephagy | [83] |
| Dendra-MAPT/tauA152T (in pDestTol2CG2) | Aggrephagy | [83] |
| C99-GFP-P2A-GFP | Aggrephagy | [84] |
Mitophagy
Mitophagy is known to be one of the best studied selective autophagy processes, which involves the removal of damaged mitochondria via autophagy, prevents cellular damage and apoptosis [19]. Disruptions in mitophagy are linked to muscular dysfunction [20], neurodegenerative diseases, such as Parkinson disease (PD) [21], and other diseases (see below). Zebrafish model has been widely used in understanding the role of mitophagy in neurodegeneration, as reviewed [22]. Despite there are multiple mechanisms observed in vitro by which mitochondria are sequestered for autophagic clearance, the mitophagy in vivo is understudied.
Early evidence linking mitophagy to PD arose when mutations in both PINK1 (PTEN induced kinase 1) and PRKN (parkin RBR E3 ubiquitin protein ligase) were found to be associated with Parkinsonism in humans [23]. Several independent studies have demonstrated that PINK1 detects mitochondrial dysfunction and then, signals PRKN to ubiquitinate specifically the damaged mitochondria for their removal by autophagy [24]. Recently, the zebrafish PD model with pink1 gene deletion was developed and characterized by mitochondria defects and loss of neurons [25]. This model was further exploited for drug screening. By using this tool, the drugs such as piperazine phenothiazines (e.g. trifluoperazine) were found to ameliorate the pink1 defect by activating autophagy. Later, the first zebrafish mitophagy reporter line expressing Cox8a (cytochrome c oxidase subunit 8A)-GFP-mCherry, the GFP-mCherry tandem-tagged mitochondrial protein, Cox8a, was constructed [26]. This reporter was used to measure mitophagic flux, the ratio of red (lysosomal) to yellow (cytosolic) mitochondrial puncta, and made it possible to confirm the role of another PD-related gene, NIPSNAP1, in mitophagy in vivo using the zebrafish model (Figure 1).
Many human disorders have been linked to imbalances in mitochondrial quality control process [27]. For example, BDH2 (3-hydroxy butyrate dehydrogenase 2) catalyzes a rate-limiting step in the formation of small iron-binding compounds, siderophores, that facilitate the import of iron into mitochondria. It has been shown that inactivation of the bdh2 gene in zebrafish embryos leads to heme deficiency and slows down erythroid maturation [28]. Interestingly, bdh2 inactivation in zebrafish results in dysfunction of mitochondria, their increased degradation by mitophagy and premature loss of these organelles that can be rescued by reintroduction of the bdh2 gene [29]. In another study, the in vivo induction of mitophagy through the optogenetic bimodular system was used in zebrafish ambra1a KD larvae [30]. Zebrafish Ambra1a (autophagy/beclin-1 regulator 1a) is an ortholog of human AMBRA1 (HsAMBRA1) and protects zebrafish from myopathy and accumulation of swollen, and aberrant mitochondria in skeletal muscles [31]. It has been previously reported that HsAMBRA1 can bind LC3 to enhance autophagosome production following mitochondrial damage [32]. In the aforementioned optogenetic system, the authors used Venus-iLID-ActA, which is a more stable version of GFP (Venus) combined with the improved light-induced dimer (iLID) blue light sensor and mitochondrial anchor (ActA) [30]. Despite the fact that Venus-iLID-ActA fully colocalized with Tomm20b (translocase of outer mitochondrial membrane 20b), murine TOMM20 is rapidly degraded by the proteasome under mitophagy-inducing conditions [33]. Nevertheless, Venus-iLID-ActA was co-injected with HsAMBRA1-RFP-SspB-expressing plasmid in zebrafish ambra1a embryos and the question of whether mitophagy could be induced by blue light was addressed [30]. In the absence of light, HsAMBRA1 remains in the cytosol, but after short bursts of blue light iLID changes its conformation what allows it to bind with a high affinity to SspB forcing HsAMBRA1 to the mitochondria’s outer membrane. In double-positive Venus-iLID-ActA/HsAMBRA1-RFP-SspB embryos, mitochondria appeared rounded, while the Venus-iLID-ActA intensity per single fiber was significantly decreased, suggesting an ongoing mitochondrial clearance i.e. mitophagy. However, no changes were detected in Venus-iLID-ActA/RFP-SspB embryos without HsAMBRA1 [30]. These studies showcase that both lack and overabundance of mitophagy leads to pathology in zebrafish that can be efficiently ameliorated by various pharmacological, genetic and optogenetic approaches. One of the biggest advantages of using zebrafish model for research is its amenability to drug exposure. Many drugs were used previously to induce or inhibit mitophagy in the in vitro studies and could potentially be used in the zebrafish mitophagy research. They have been comprehensively reviewed recently [34,35].
Interestingly, zebrafish model was also used to recapitulate methylmalonic acidemia (MMA), which is a common inherited metabolic disease caused by the deficiency of an enzyme found in mitochondria and known as MMUT (methylmalonyl-CoA mutase) [36]. Using cell culture model, deficiency of MMUT was shown to lead to mitophagy dysfunction and accumulation of damaged mitochondria. Because the Mmut KO mice developed neither structural changes, nor significant kidney failure, they could not recapitulate the kidney disease associated with human MMA patients. In contrast, the mmut-deficient zebrafish exhibits the MMA disease-relevant phenotype, including liver/kidney mitochondriopathy, behavioral changes and excessive mortality. This study shows a potential of this zebrafish model to study the involvement of mitophagy in MMA in vivo. In another study, zebrafish model system was explored to see the effects of resveratrol on aging-related alterations in retinas, including mitochondrial DNA integrity and copy number, mitochondrial fusion and fission, mitophagy and autophagy [37]. Because resveratrol increased Pink1 expression in aging zebrafish retina among other effects that increase the quality and function of mitochondria, it holds a potential for prevention of the aging-induced oculopathy in zebrafish and other vertebrates. Together, these studies illustrate a high utility of zebrafish model in exploring various human mitophagy-related diseases and their potential therapeutic interventions.
Recently, Mandal et al. developed Tg(5kbneurod:mito-mEos) to interrogate the behavior of mitochondria in neurons in vivo in the embryonic and larval zebrafish posterior lateral line mechanosensory system [38]. This reporter could also be used to study mitophagy, because it allows to distinguish the preexisting mitochondria from new mitochondria. The mito-mEos fusion protein can be stably photoconverted from “green” to “red” using 405 nm illumination. Therefore, it allows a pulse-chase analysis of “red” mitochondria turnover and is especially useful in the regions of neurons with high turnover, such as axon terminals. Finally, the group of Goessling generated the zebrafish mitophagy reporter lines with the pH-sensitive biosensors, Tg(ubi:mito-Keima) and Tg(ubi:mito-GR) [39]. These tools enable quantitative in vivo light microscopy analysis of mitophagy in real-time during embryonic development or in response to stress. They have helped to detect frequent mitophagy events during development in the heart, vasculature, liver, kidney, brain and spinal cord. This is the first intravital time-lapse mitophagy analysis tracking simultaneously the mitochondrial fission, pH change and trafficking of mitochondria to autolysosomes in a vertebrate. Moreover, it has already provided a very useful mechanistic insight that Bnip3, but not other mitophagy receptors, such as Bnip3la/Nix or Fundc1, is required for hypoxia-induced mitophagy in vivo [39] (Figure 1). Taken together, a plethora of mitophagy studies using zebrafish as a model contributed a lot of in vivo knowledge to the selective autophagy field. Because mitochondrial dynamics and its molecular mechanisms were primarily investigated in cell culture in the past, even more in vivo studies are needed to provide novel mechanistic insights on mitophagy in vertebrates under physiological and pathophysiological conditions.
Xenophagy
Xenophagy is a selective autophagy that is activated during host-pathogen interactions. It is the ability to capture invasive viruses, bacteria, fungi and parasites, thus safeguarding the health of host cells. Most of the xenophagy studies carried out in zebrafish model focused on bacterial infection, as reviewed [40]. Using zebrafish, a wide variety of pathogenic bacteria, including Salmonella enterica serovar Typhimurium [41,42], Shigella flexneri [43,44], Pseudomonas aeruginosa [45,46], Burkholderia cenocepacia [47,48], Listeria monocytogenes [49], Staphylococcus aureus [50–53], Mycobacterium marinum [54], Mycobacterium abscessus [55], Mycobacterium leprae [56] and Vibrio cholerae [57] have been investigated providing new insights about the cellular response to infection in vivo. For example, it was shown that Salmonella plasmid virulence gene spvB from S. enterica serovar Typhimurium that causes bacterial gastroenteritis can inhibit autophagic activity and enhance bacterial virulence [58]. Taking advantage of a zebrafish model for tuberculosis, it has been shown that Dram1 (DNA damage regulated autophagy modulator 1) acts downstream of the pathogen recognition and activates the selective autophagy of mycobacteria [59] (Figure 1). It was shown that dram1 expression was significantly reduced during M. marinum infection in the absence of Myd88 (MYD88 innate immune signal transduction adaptor). Morpholino KD of myd88 or dram1 in eGFP-Map1lc3b embryos infected with M. marinum resulted in reduced number of eGFP-Map1lc3b puncta. On the other hand, overexpression of dram1 increased the autophagy response in infected cells promoting a colocalization between Map1lc3b and bacteria. The authors concluded that this Dram1-mediated mechanism downstream of Myd88 has a protective function during mycobacterial infection using zebrafish model [59]. Further research in the stable dram1 mutant of zebrafish has validated this conclusion [60]. In addition, it has also been shown that Sting1 (stimulator of interferon response cGAMP interactor 1) and autophagic receptors, Sqstm1/p62 (sequestosome 1) and Optn (optineurin), are essential for the selective autophagy defense during mycobacterial infection [59,61] (Figure 1). It would be also interesting to study the role of other ubiquitin-binding receptors, such as CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2) in the context of bacterial infection in zebrafish model as it has been shown to be an important hub for the anti-bacterial autophagy [62]. Zebrafish also served as a new model for the in vivo study of xenophagy of S. flexneri that causes shigellosis in human. It has been shown in zebrafish that the intracellular S. flexneri can escape from the phagosomes to the cytosol where it induces septin caging formation and becomes a target of the Sqstm1-mediated autophagy [44]. Collectively, the use of zebrafish to study the host-pathogen interactions has contributed to our understanding of xenophagy. It can be further used for the development of new therapeutic strategies against pathogens and zebrafish will continue to serve as in vivo platform for investigation of the mechanisms underlying microbial pathogenesis.
Lipophagy
Lipophagy is the selective autophagic degradation of the intracellular lipid droplets (LDs) [63]. Human lipid metabolism genes with corresponding zebrafish paralogs have been reviewed and the results indicated that lipid metabolism is conserved between the two species [64]. On the organismal level, LD degradation in white adipose tissue is critical to increase circulating fatty acids that provide fuel in the non-adipose tissues during nutrient insufficiency. Although LDs have historically been recognized for their importance in energy storage, a growing body of literature has more recently identified important roles for LDs in cell signaling and other functions that link LD accumulation to the etiology of numerous diseases [65,66]. A major challenge in the lipophagy field is to construct markers of the LDs that accurately measure the engulfment and degradation of these organelles through the autophagy pathway with high sensitivity. The zebrafish larva is a powerful tool for the study of lipid biology, because it allows the visualization of digestive processes at both the organ and subcellular levels [67,68]. Ideally, a fluorescent neutral lipid stain that is resistant to the lysosomal quenching and degradation would allow an optimal measurement of lipophagic flux via co-localization with the LysoTracker family of dyes. Recently, the group of Farber created a zebrafish reporter line that has been genetically engineered to produce glowing lipoproteins and named it “LipoGlo” [69]. This line gives the zebrafish model an added advantage for the lipophagy-related studies. Recent lipidomics studies in zebrafish have further enhanced its attractiveness as a model for lipophagy and lipid-related diseases [70,71]. In many lipid-related diseases, lipid biomarkers, such as ceramides, which play a tremendous role in disease diagnosis, have been identified. Recently, Pant et al., found novel variants in a gene (DEGS1), which is responsible for leukodystrophy in human patients. Using targeted lipidomics studies in a zebrafish degs1 KD model, the group was able to identify specific long chain sphingolipids that are altered in both patients and zebrafish [71]. In addition, the group of Miller using high cholesterol diet-fed larvae and adult zebrafish created a useful model of hypercholesterolemia and atherogenesis, which can be further exploited for lipophagy studies [72,73]. Recently, the first evidence emerged that lipophagy in zebrafish might indeed exist, as 36 h of starvation decreases the abundance of LDs stained with BODIPY 493/503 in zebrafish liver [74]. Researchers have shown that when zebrafish were on a high-fat diet with 13% of lipid for one week and, then, fasted for 24 hours with or without chloroquine (CQ), the use of CQ leads to higher LD and triacylglycerol accumulation in the liver than fasting without CQ. The protein levels of lipidated Map1lc3b that represents autophagosomes are also higher in the CQ-treated fish consistent with the autophagic degradation of LDs [74]. These results in zebrafish are in agreement with the results in a zebrafish liver cell line where inhibition of lipophagy with CQ decreases the lipid catabolism and anabolism indicating that lipophagy is an important cellular process in zebrafish [75]. Together, the use of zebrafish to study lipophagy can provide novel insights for our understanding of this selective autophagy process. These studies might be useful for the development of new therapeutic approaches against many human lipid accumulation diseases where lipophagy could serve as a therapeutic target.
Aggrephagy
Aggrephagy, which means the selective autophagic degradation of protein aggregates [76], has emerged as an important mechanism for understanding the TARDBP/TDP-43 (TAR DNA binding protein) proteinopathies, tauopathies, synucleinopathies and polyglutamine disorders. It is known that approximately 30% of newly synthesized proteins are misfolded [77]. Therefore, the protein quality control must operate continuously to manage the influx of misfolded proteins. The selectivity of aggrephagy toward specific cargos is mediated largely by autophagic receptors, such as SQSTM1 and OPTN. Indeed, the zebrafish genetic model with sqstm1 KD suggests preferential targeting of motor neurons by sqstm1 loss-of-function, the phenotype consistent with the amyotrophic lateral sclerosis (ALS) that supports the role of aggrephagy in ALS [78]. The morpholino KD of OPTN ortholog in zebrafish also causes the motor axonopathy phenotype, as in the zebrafish sqstm1 ALS model [79]. Despite further studies failed to confirm this phenotype in stable optn mutant [80], overexpression of human OPTN disease variants in zebrafish did cause axonal defects that could be efficiently ameliorated by the morpholino KD of map2k5 (mitogen-activated protein kinase kinase 5), most probably, via augmenting autophagy [81].
Several studies also used zebrafish to model Alzheimer disease (AD). To analyze whether PICALM (phosphatidylinositol binding clathrin assembly protein) affects the clearance of MAPT (microtubule associated protein tau) in vivo, the expression construct consisting of a green-to-red photoconvertible fluorescent protein, Dendra, fused to human MAPT (Dendra-tau) was generated and co-expressed with the human PICALM in zebrafish [82]. The human PICALM reduced autophagosome formation and slowed down the clearance of red Dendra-tau after photoconversion in zebrafish suggesting that PICALM indeed modulates autophagy and MAPT clearance. In a different study, expression of a rare variant of the human MAPT, A152T, which is a risk factor for the frontotemporal dementia and AD, in zebrafish was found to be associated with the disruption of proteasome but not autophagy function and delayed MAPT-A152T clearance [83]. Interestingly, upregulation of autophagy improved the clearance of MAPT-A152T in zebrafish and decreased its toxicity suggesting that autophagy upregulation could be a viable therapeutic strategy for these tauopathies. It has also been shown that ERBB2 (erb-b2 receptor tyrosine kinase 2) is at higher levels in the hippocampal regions of AD patients [84]. By using the HEK293 cells overexpressing the C-terminal 99 residues of APP (amyloid beta precursor protein), it was showed that the treatment with ERBB2 inhibitor decreased the levels of both ERBB2 and the C-terminal 99 residues of APP. This result was validated in zebrafish model of amyloidopathy. A discovery of the non-canonical function of Erbb2 in modulating autophagy and establishment of Erbb2 as a therapeutic target for AD have been accomplished using zebrafish model [84]. These three studies in zebrafish showcase the paramount role of aggrephagy in AD and important role of zebrafish model in validating the in vitro findings with human disease-related proteins.
Recently, a novel stable transgenic zebrafish line that expresses eGFP-Map1lc3b under the neuron-specific elavl3 promoter has been developed [85]. This transgenic line allowed the quantification of autophagosome number, as well as measuring autophagic flux, in neurons. Using lysosome protease inhibitors, the authors were able to show an increase in eGFP-Map1lc3b positive puncta in this transgenic line, which might be useful in determining how the aggregate-prone proteins, such as MAPT, are regulated by aggrephagy in relation to neurodegenerative diseases. However, after discovery of the single-membrane MAP1LC3B conjugation events in the endolysosomal compartments during a non-canonical autophagy, care must be taken in the interpretation of eGFP-Map1lc3b dots [86].
Notably, mounting evidence in zebrafish models demonstrated the selectivity of autophagy in degradation of the aggregate-prone proteins via autophagic receptors, which link autophagic cargos to Map1lc3b at the autophagic membrane (Figure 1). These findings will serve as a proof of principle that selective autophagy processes, such as aggrephagy, can be easily studied in vivo using zebrafish. In summary, zebrafish models of neurological diseases provided fundamental insights into aggrephagy. These insights will have broad implications for the design of novel therapeutic strategies that can control these diseases.
Conclusions and prospects
We have illustrated how zebrafish models can reveal key aspects of autophagy and provide fundamental advances in understanding the biology of selective autophagy processes. Despite major recent advances, the exact relevance of many selective autophagy processes in vivo remains obscure. It is expected that the study of selective autophagy pathways using zebrafish model will continue to illuminate the complexity that underlies these processes in vertebrates, including human. The full potential of zebrafish as a tool in the selective autophagy field has yet to be realized and employing advanced gene editing, and high spatiotemporal resolution in vivo microscopy techniques will further promote this model for unlocking fundamental aspects of selective autophagy processes. Clearly, the key strengths of the zebrafish are its versatility and possibility of high-resolution dynamic observations of autophagic flux that combined can enable rapid drug discovery. Therefore, to fully harness the power of this model, the research community will need to embrace the extensive but often low cost (when compared to rodents) functional testing.
Most studies by now focused on understanding the selective autophagy mechanisms in yeast and cell culture models. However, many researchers have already developed the reliable zebrafish models to understand the selective autophagy processes and their relevance in pathophysiology. For example, our laboratory is working on lipophagy and looking forward to making a contribution to this new and exciting area of autophagy research using zebrafish models of atherosclerosis. In zebrafish literature, there is no mention of other important selective autophagy pathways, such as pexophagy or reticulophagy, and they are yet to be explored. We predict that unique properties of zebrafish model will be valuable to investigate the molecular and cell biology of the remaining selective autophagy processes in vivo and to discover the unforeseen aspects of their mechanisms. Indeed, for the in-depth understanding of the selective autophagy pathways in human, it will be critical to complement the existing in vitro models with the in vivo studies in vertebrates. Together with advances in RNA sequencing and proteomics approaches, high-throughput studies of the selective autophagy processes in zebrafish will provide new insights to autophagy field. Finally, another challenge will be to integrate the molecular and cellular data on the selective autophagy in zebrafish with such data in other vertebrate models, including mice. Together, the improved understanding of the selective autophagy in live vertebrates will provide vital clues for the development of new therapeutic strategies against the wide spectrum of human diseases.
Funding Statement
This work was supported by the NIH grant, GM119571, to Taras Y. Nazarko.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Lahiri V, Hawkins WD, Klionsky DJ.. Watch what you (Self-) eat: autophagic mechanisms that modulate metabolism. Cell Metab. 2019. Apr 2;29(4):803–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mizushima N, Komatsu M.. Autophagy: renovation of cells and tissues. Cell. 2011. Nov 11;147(4):728–741. [DOI] [PubMed] [Google Scholar]
- [3].Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013. Jul;15(7):713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kirkin V. History of the selective autophagy research: how did it begin and where does it stand today? J Mol Biol. 2020. Jan 3;432(1):3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019. Oct 17;76(2):268–285. [DOI] [PubMed] [Google Scholar]
- [6].He C, Bartholomew CR, Zhou W, et al. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-gabarap zebrafish embryos. Autophagy. 2009. May;5(4):520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sasaki T, Lian S, Khan A, et al. Autolysosome biogenesis and developmental senescence are regulated by both Spns1 and v-ATPase. Autophagy. 2017. Feb;13(2):386–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lopez A, Fleming A, Rubinsztein DC. Seeing is believing: methods to monitor vertebrate autophagy in vivo. Open Biol. 2018. Oct 24;8(10):180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mathai BJ, Meijer AH, Simonsen A. Studying autophagy in zebrafish. Cells. 2017. Jul 9;6(3):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu RS, Lam II, Clay H, et al. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev Cell. 2018. Jul 2;46(1):112–125 e4. [DOI] [PubMed] [Google Scholar]
- [11].El-Brolosy MA, Kontarakis Z, Rossi A, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019. Apr;568(7751):193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009. Oct 16;10:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Soroldoni D, Hogan BM, Oates AC. Simple and efficient transgenesis with meganuclease constructs in zebrafish. Methods Mol Biol. 2009;546:117–130. [DOI] [PubMed] [Google Scholar]
- [14].Linney E, Hardison NL, Lonze BE, et al. Transgene expression in zebrafish: A comparison of retroviral-vector and DNA-injection approaches. Dev Biol. 1999. Sep 1;213(1):207–216. [DOI] [PubMed] [Google Scholar]
- [15].Asakawa K, Suster ML, Mizusawa K, et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008. Jan 29;105(4):1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013. Apr 25;496(7446):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lescat L, Veron V, Mourot B, et al. Chaperone-mediated autophagy in the light of evolution: insight from fish. Mol Biol Evol. 2020 Oct 1;37(10):2887–2899. [DOI] [PubMed] [Google Scholar]
- [18].Wesch N, Kirkin V, Rogov VV. Atg8-family proteins-structural features and molecular interactions in autophagy and beyond. Cells. 2020. Sep 1;9(9):2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011. Jan;12(1):9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Drake JC, Yan Z. Mitophagy in maintaining skeletal muscle mitochondrial proteostasis and metabolic health with ageing. J Physiol. 2017. Oct 15;595(20):6391–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].de Vries RL, Przedborski S. Mitophagy and Parkinson’s disease: be eaten to stay healthy. Mol Cell Neurosci. 2013. Jul;55:37–43. [DOI] [PubMed] [Google Scholar]
- [22].Wager K, Russell C. Mitophagy and neurodegeneration: the zebrafish model system. Autophagy. 2013. Nov 1;9(11):1693–1709. [DOI] [PubMed] [Google Scholar]
- [23].Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson’s disease: the PINK1-parkin link. Biochim Biophys Acta. 2011. Apr;1813(4):623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015. Jan 21;85(2):257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Y, Nguyen DT, Olzomer EM, et al. Rescue of Pink1 deficiency by stress-dependent activation of autophagy. Cell Chem Biol. 2017. Apr 20;24(4):471–480 e4. [DOI] [PubMed] [Google Scholar]
- [26].Princely Abudu Y, Pankiv S, Mathai BJ, et al. NIPSNAP1 and NIPSNAP2 act as “Eat me” signals for mitophagy. Dev Cell. 2019. May 20;49(4):509–525 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chinnery PF, Turnbull DM. Mitochondrial DNA and disease. Lancet. 1999. Jul;354(Suppl 1):SI17–21. [DOI] [PubMed] [Google Scholar]
- [28].Devireddy LR, Hart DO, Goetz DH, et al. A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell. 2010. Jun 11;141(6):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davuluri G, Song P, Liu Z, et al. Inactivation of 3-hydroxybutyrate dehydrogenase 2 delays zebrafish erythroid maturation by conferring premature mitophagy. Proc Natl Acad Sci U S A. 2016. Mar 15;113(11):E1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].D’Acunzo P, Strappazzon F, Caruana I, et al. Reversible induction of mitophagy by an optogenetic bimodular system. Nat Commun. 2019. Apr 4;10(1):1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Skobo T, Benato F, Grumati P, et al. Zebrafish ambra1a and ambra1b knockdown impairs skeletal muscle development. PLoS One. 2014;9(6):e99210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Strappazzon F, Nazio F, Corrado M, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015. Mar;22(3):419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yoshii SR, Kishi C, Ishihara N, et al. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011. Jun 3;286(22):19630–19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Georgakopoulos ND, Wells G, Campanella M. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017. Jan 19;13(2):136–146. [DOI] [PubMed] [Google Scholar]
- [35].Palikaras K, Princz A, Tavernarakis N. Mitophagy modulators. Encycl Biomed Gerontol. 2020;2:433–446. [Google Scholar]
- [36].Luciani A, Schumann A, Berquez M, et al. Impaired mitophagy links mitochondrial disease to epithelial stress in methylmalonyl-CoA mutase deficiency. Nat Commun. 2020. Feb 20;11(1):970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang N, Luo Z, Jin M, et al. Exploration of age-related mitochondrial dysfunction and the anti-aging effects of resveratrol in zebrafish retina. Aging (Albany NY). 2019. May 19;11(10):3117–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mandal A, Pinter K, Drerup CM. Analyzing neuronal mitochondria in vivo using fluorescent reporters in zebrafish. Front Cell Dev Biol. 2018;6:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wrighton PJ, Shwartz A, Heo J-M, et al. Live imaging defines the dynamics and molecular basis of in vivo mitophagy. bioRxiv. 2020. Mar 26;010405. .biorxiv.org/content/ 10.1101/2020.03.26.010405v4 [DOI] [Google Scholar]
- [40].Torraca V, Mostowy S. Zebrafish infection: from pathogenesis to cell biology. Trends Cell Biol. 2018. Feb;28(2):143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tyrkalska SD, Candel S, Angosto D, et al. Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat Commun. 2016. Jul 1;7:12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hall CJ, Flores MV, Oehlers SH, et al. Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell. 2012. Feb 3;10(2):198–209. [DOI] [PubMed] [Google Scholar]
- [43].Willis AR, Moore C, Mazon-Moya M, et al. Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr Biol. 2016. Dec 19;26(24):3343–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mostowy S, Boucontet L, Mazon Moya MJ, et al. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 2013;9(9):e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Clatworthy AE, Lee JS, Leibman M, et al. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect Immun. 2009. Apr;77(4):1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Brannon MK, Davis JM, Mathias JR, et al. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell Microbiol. 2009. May;11(5):755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vergunst AC, Meijer AH, Renshaw SA, et al. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect Immun. 2010. Apr;78(4):1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mesureur J, Feliciano JR, Wagner N, et al. Macrophages, but not neutrophils, are critical for proliferation of Burkholderia cenocepacia and ensuing host-damaging inflammation. PLoS Pathog. 2017. Jun;13(6):e1006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Levraud JP, Disson O, Kissa K, et al. Real-time observation of Listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect Immun. 2009. Sep;77(9):3651–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Prajsnar TK, Hamilton R, Garcia-Lara J, et al. A privileged intraphagocyte niche is responsible for disseminated infection of Staphylococcus aureus in a zebrafish model. Cell Microbiol. 2012. Oct;14(10):1600–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Prajsnar TK, Cunliffe VT, Foster SJ, et al. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 2008. Nov;10(11):2312–2325. [DOI] [PubMed] [Google Scholar]
- [52].Gibson JF, Prajsnar TK, Hill CJ, et al. Neutrophils use selective autophagy receptor Sqstm1/p62 to target Staphylococcus aureus for degradation in vivo in zebrafish. Autophagy. 2020. Jun 19:1–10. pubmed.ncbi.nlm.nih.gov/32559122/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Prajsnar TK, Serba JJ, Dekker BM, et al. The autophagic response to Staphylococcus aureus provides an intracellular niche in neutrophils. Autophagy. 2020. Mar 15:1–15. pubmed.ncbi.nlm.nih.gov/32174246/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cronan MR, Tobin DM. Fit for consumption: zebrafish as a model for tuberculosis. Dis Model Mech. 2014. Jul;7(7):777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bernut A, Herrmann JL, Kissa K, et al. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A. 2014. Mar 11;111(10):E943–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Madigan CA, Cambier CJ, Kelly-Scumpia KM, et al. A macrophage response to Mycobacterium leprae phenolic glycolipid initiates nerve damage in leprosy. Cell. 2017. Aug 24;170(5):973–985 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Runft DL, Mitchell KC, Abuaita BH, et al. Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl Environ Microbiol. 2014. Mar;80(5):1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li YY, Wang T, Gao S, et al. Salmonella plasmid virulence gene spvB enhances bacterial virulence by inhibiting autophagy in a zebrafish infection model. Fish Shellfish Immunol. 2016. Feb;49:252–259. [DOI] [PubMed] [Google Scholar]
- [59].van der Vaart M, Korbee CJ, Lamers GE, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected]. Cell Host Microbe. 2014. Jun 11;15(6):753–767. [DOI] [PubMed] [Google Scholar]
- [60].Zhang R, Varela M, Forn-Cuni G, et al. Deficiency in the autophagy modulator Dram1 exacerbates pyroptotic cell death of Mycobacteria-infected macrophages. Cell Death Dis. 2020. Apr 24;11(4):277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang R, Varela M, Vallentgoed W, et al. The selective autophagy receptors Optineurin and p62 are both required for zebrafish host resistance to mycobacterial infection. PLoS Pathog. 2019. Feb;15(2):e1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ravenhill BJ, Boyle KB, von Muhlinen N, et al. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol Cell. 2019. Apr 18;74(2):320–329 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Quinlivan VH, Farber SA. Lipid uptake, metabolism, and transport in the larval zebrafish. Front Endocrinol (Lausanne). 2017;8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Welte MA. Expanding roles for lipid droplets. Curr Biol. 2015. Jun 1;25(11):R470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Welte MA, Gould AP. Lipid droplet functions beyond energy storage. Biochim Biophys Acta Mol Cell Biol Lipids. 2017. Oct;1862(10Pt B):1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Miyares RL, de Rezende VB, Farber SA. Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis Model Mech. 2014. Jul;7(7):915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zeituni EM, Farber SA. Studying lipid metabolism and transport during zebrafish development. Methods Mol Biol. 2016;1451:237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Thierer JH, Ekker SC, Farber SA. The LipoGlo reporter system for sensitive and specific monitoring of atherogenic lipoproteins. Nat Commun. 2019. Jul 31;10(1):3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fraher D, Sanigorski A, Mellett NA, et al. Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Rep. 2016. Feb 16;14(6):1317–1329. [DOI] [PubMed] [Google Scholar]
- [71].Pant DC, Dorboz I, Schluter A, et al. Loss of the sphingolipid desaturase DEGS1 causes hypomyelinating leukodystrophy. J Clin Invest. 2019. Mar 1;129(3):1240–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fang L, Green SR, Baek JS, et al. In vivo visualization and attenuation of oxidized lipid accumulation in hypercholesterolemic zebrafish. J Clin Invest. 2011. Dec;121(12):4861–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Stoletov K, Fang L, Choi SH, et al. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res. 2009. Apr 24;104(8):952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang J, Han S-L, Li L-Y, et al. Lipophagy is essential for lipid metabolism in fish. Sci Bull. 2018. Jul 30;63(14):879–882. [DOI] [PubMed] [Google Scholar]
- [75].Wang J, Han SL, Lu DL, et al. Inhibited lipophagy suppresses lipid metabolism in zebrafish liver cells. Front Physiol. 2019;10:1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007. Jul-Aug;3(4):300–322. [DOI] [PubMed] [Google Scholar]
- [77].Princiotta MF, Finzi D, Qian SB, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003. Mar;18(3):343–354. [DOI] [PubMed] [Google Scholar]
- [78].Lattante S, de Calbiac H, Le Ber I, et al. Sqstm1 knock-down causes a locomotor phenotype ameliorated by rapamycin in a zebrafish model of ALS/FTLD. Hum Mol Genet. 2015. Mar 15;24(6):1682–1690. [DOI] [PubMed] [Google Scholar]
- [79].Korac J, Schaeffer V, Kovacevic I, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013. Jan 15;126(Pt 2):580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Paulus JD, Link BA. Loss of optineurin in vivo results in elevated cell death and alters axonal trafficking dynamics. PLoS One. 2014;9(10):e109922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Jo M, Chung AY, Yachie N, et al. Yeast genetic interaction screen of human genes associated with amyotrophic lateral sclerosis: identification of MAP2K5 kinase as a potential drug target. Genome Res. 2017. Sep;27(9):1487–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Moreau K, Fleming A, Imarisio S, et al. PICALM modulates autophagy activity and tau accumulation. Nat Commun. 2014. Sep 22;5:4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lopez A, Lee SE, Wojta K, et al. A152T tau allele causes neurodegeneration that can be ameliorated in a zebrafish model by autophagy induction. Brain. 2017. Apr 1;140(4):1128–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang BJ, Her GM, Hu MK, et al. ErbB2 regulates autophagic flux to modulate the proteostasis of APP-CTFs in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2017. Apr 11;114(15):E3129–E3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Khuansuwan S, Barnhill LM, Cheng S, et al. A novel transgenic zebrafish line allows for in vivo quantification of autophagic activity in neurons. Autophagy. 2019. Aug;15(8):1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Florey O, Kim SE, Sandoval CP, et al. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011. Oct 16;13(11):1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Strange K. Drug discovery in fish, flies, and worms. Ilar J. 2016. Dec;57(2):133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]


