ABSTRACT
The Dynein motor is responsible for the localization of numerous mRNAs within Drosophila oocytes and embryos. The RNA binding protein, Egalitarian (Egl), is thought to link these various RNA cargoes with Dynein. Although numerous studies have shown that Egl is able to specifically associate with these RNAs, the nature of these interactions has remained elusive. Egl contains a central RNA binding domain that shares limited homology with an exonuclease, yet Egl binds to RNA without degrading it. Mutations have been identified within Egl that disrupt its association with its protein interaction partners, BicaudalD (BicD) and Dynein light chain (Dlc), but no mutants have been described that are specifically defective for RNA binding. In this report, we identified a series of positively charged residues within Egl that are required for RNA binding. Using corresponding RNA binding mutants, we demonstrate that specific RNA cargoes are more reliant on maximal Egl RNA biding activity for their correct localization in comparison to others. We also demonstrate that specification and maintenance of oocyte fate requires maximal Egl RNA binding activity. Even a subtle reduction in Egl’s RNA binding activity completely disrupts this process. Our results show that efficient RNA localization at the earliest stages of oogenesis is required for specification of the oocyte and restriction of meiosis to a single cell.
KEYWORDS: RNA localization, molecular motor, RNA binding domain, cell polarity, cargo adaptor
Introduction
Localization of mRNAs is a conserved mechanism for the spatial and temporal sorting of proteins [1]. Numerous mRNAs are localized throughout oogenesis and embryogenesis in Drosophila melanogaster, making this a powerful model system in which to explore mechanisms of localization [2]. In most instances, the specific sorting of these mRNAs requires motor-based transport on cytoskeletal filaments [3]. For example, in the Drosophila egg chamber, the plus-end microtubule motor, Kinesin-1, is required for the posterior localization of oskar (osk) mRNA [4]. The microtubule minus-end motor, cytoplasmic Dynein (hereafter Dynein), is responsible for the localization of numerous mRNAs including gurken (grk) at the dorsal-anterior corner and bicoid (bcd) along the anterior margin of the oocyte [5–9].
A central question in this field concerns the mechanism by which these diverse mRNA cargoes are linked to appropriate motors for transport. In the case of osk, an atypical Tropomyosin links this transcript with Kinesin [10,11]. Most mRNAs that are transported within the oocyte and embryo by Dynein are thought to be linked to this motor via the RNA binding protein, Egalitarian (Egl) [12–14]. Recent studies have revealed the details of this process. Dynein light chain (Dlc/LC8) promotes the dimerization of Egl. Once Egl dimerizes, it is able to bind to mRNA cargo more efficiently [15]. This enables the Egl/RNA complex to associate with BicaudalD (BicD), which in turn links the complex with Dynein [16,17].
Egl interacts with numerous mRNA cargoes in oocytes and embryos [12–14]. However, the nature of this interaction has remained elusive. For instance, it is generally accepted that Egl recognizes a common structural motif in its various cargoes [18]. The putative RNA binding region within Egl contains features of a 3ʹ-5ʹ exonuclease, and bears similarity to the E. coli protein, RNaseD [19,20]. Despite these similarities, Egl is not an exonuclease. Mutating the conserved residues required for exonuclease activity does not compromise the function of Egl [21]. In addition, binding studies indicate that Egl binds to mRNAs without degrading them [12]. The key residues within this unique RNA binding domain required for Egl binding to its diverse set of cargoes are not known.
To identify amino acids that are critical for Egl’s RNA binding activity, we undertook a mutational approach. Our analysis revealed a stretch of positively charged residues that are essential for RNA binding. Characterization of these mutants in vivo revealed that some cargoes rely critically on optimal Egl RNA binding activity for their correct localization, while others are less sensitive. We also determined that specification and maintenance of oocyte fate requires maximal Egl RNA binding activity. Even a subtle disruption of Egl’s RNA binding activity interferes with these processes. Defective oocyte specification likely arises due to inefficient or incomplete localization of mRNA cargo to the presumptive oocyte and early-stage egg chambers.
Results
Generation of Egl RNA binding mutants
As a starting point for identifying residues within Egl that are critical for RNA binding, we compared the sequence of Egl between Drosophila species that had diverged over 40 million years ago. In particular, we examined the RNA binding domain for positively charged residues that are highly conserved. This analysis revealed 15 conserved Lysine or Arginine residues. For our initial experiments, we generated three mutants; egl_rbd1, egl_rbd2, and egl_rbd3. In egl_rbd1, all 15 positively charged residues were converted to Alanine (Figure 1(a)). egl_rbd2 mutants contained Alanine substitutions at 11 conserved positions, and egl_rbd3 contained Alanine substations at 8 positions that clustered towards the 3ʹend of the RNA binding domain (Figure 1(a)).
Figure 1.
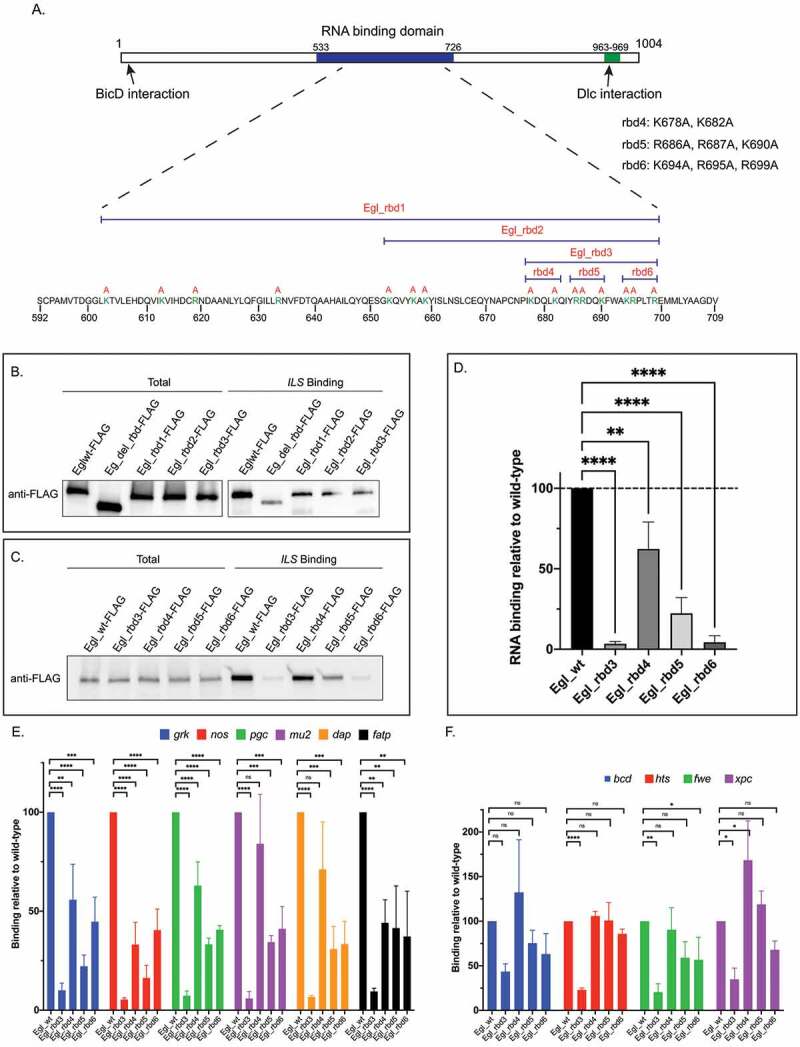
Identification of Egl RNA binding mutants. (a) Schematic showing the domain structure of Egl. The RNA binding domain, and the BicD and Dlc interaction sites are shown. The positively charged residues that were mutated are shown in green. The specific residues mutated in Egl_rbd4, Egl_4bd5 and Egl_rbd6 are also shown. (b) The indicated Flag-tagged Egl constructs were expressed in S2 cells. Cell lysates were incubated with Streptavidin beads coated with the ILS localization element. After binding and wash steps, the bounds proteins were eluted and analysed by western blotting using the FLAG antibody. The total fraction was also analysed. (c) A similar RNA binding experiment was performed using ovarian lysates from flies expressing the indicated Egl constructs. Bound proteins were eluted and analysed by blotting using the FLAG antibody. A total fraction is also shown. (d) Quantification of the experiment shown in C. The experiment was repeated three times and the band intensity in the bound fractions was determined. Values are shown relative to the wild-type construct. (e-f) Ovarian lysates were prepared from flies expressing the indicated Egl constructs. The FLAG-tagged proteins were immunoprecipitated using FLAG beads. The co-precipitating RNAs were extracted, reverse transcribed and analysed using quantitative PCR. RNA enrichment was normalized to the level of gamma tubulin mRNA that co-precipitated in each pellet. The values are shown relative to binding observed with wild-type Egl. ****p ≤ 0.0001, ***p ≤ 0.001, ** p ≤ 0.01, *p ≤ 0.05, ns = not significant; a one-way ANOVA was used to calculate statistical significance
We next compared the RNA binding activity between wild-type Egl and the mutants. An Egl construct lacking the entire RNA binding domain was used as a control (egl_del_rbd). Wild-type and mutant constructs were expressed in Drosophila S2 cells and lysates were subjected to binding experiments using a well-characterized localization element from the I factor retro-transposon (ILS) [12]. As expected, in comparison to wild-type Egl, Egl_del_rbd retained minimal RNA binding activity (Figure 1(b)). Interestingly, Egl_rbd1, Egl_rbd2, and Egl_rbd3 were as defective for binding the localization element as the deletion construct (Figure 1(b)). Because Egl_rbd3 contains the fewest amino acid substitutions, yet was as defective as the deletion mutant, residues within this region must be critical for mediating RNA binding.
In order to further narrow down the amino acids required for RNA binding, three mutants in addition to egl_rbd3 were generated and introduced into flies; egl_rbd4, egl_rbd5, and egl_rbd6 (Figure 1(a)). These constructs also contained silent mutations that render them refractory to short hairpin (sh)RNA-mediated depletion of endogenous Egl. The constructs were introduced into the background of the egl shRNA strain and were crossed to an early-stage maternal Gal4 driver. This driver results in depletion of Egl from stage 5 onwards [13]. Thus, mid and late-stage egg chambers from these flies were depleted of endogenous Egl, but expressed transgenic FLAG-tagged wild-type Egl, Egl_rbd3, Egl_rbd4, Egl_rbd5, or Egl_rbd6. Ovarian lysates were prepared and tested for RNA binding activity using ILS (Figure 1(c)). The RNA binding activity of Egl_rbd4 was only modestly reduced (Figure 1(c,d)). However, Egl_rbd5 and Egl_rbd6 showed significantly compromised RNA binding, with Egl_rbd6 displaying the more severe binding deficit (Figure1(c,d)). Similar results were obtained using the TLS localization element (Supplemental Figure 1(a)).
We next determined how effectively these mutants bound native mRNAs in Drosophila ovaries. Wild-type or mutant versions of Egl containing a C-terminal FLAG tag were expressed in the female germline. Ovarian lysates were prepared and subjected to immunoprecipitation using FLAG antibody beads. The co-precipitating RNAs were extracted and analysed using reverse transcription followed by quantitative PCR. gamma-tubulin mRNA, which co-precipitates non-specifically, was used to normalize the RNA in the bound fractions. Vazquez-Pianzola and colleagues identified numerous mRNAs bound by Egl in embryos [14]. We tested several of these mRNAs to determine their association with Egl in ovaries, and chose the top 10 candidates that reliably co-precipitated with wild-type Egl for further experiments
Our analysis revealed that the RNA cargoes can be classified into two groups based on their association with Egl mutants. The first group consists of gurken (grk), nanos (nos), polar granule component (pgc), mutator2 (mu2), dacapo (dap) and fatty acid transport protein1 (fatp) (Figure 1(e)). Egl_rbd3 displayed a five-fold reduction in binding to these mRNAs (Figure 1(e)). Egl_rbd5 and Egl_rbd6 displayed a two-fold reduction in binding, whereas Egl_rbd4 was only minimally affected (Figure 1(e)). The second group of RNAs included bicoid (bcd), hu li tai shao (hts), flower (fwe) and xeroderma pigmentosum complementation group C (xpc). Egl_rbd3 displayed a three-fold reduction in binding to these cargoes, whereas Egl_rbd4, Egl_rbd5, and Egl_rbd6 were either unaffected or only minimally affected (Figure 1(f)). Thus, RNAs in group one are much more compromised than those in group two for binding to the Egl mutants identified in this study. This suggests that there might exist differences in the way in which Egl interacts with its various cargoes.
Egl RNA binding mutants exhibit compromised binding to BicD
The Dynein adaptor, BicaudalD (BicD), interacts directly with Egl [12]. However, in vivo, the Egl-BicD interaction is regulated by cargo binding to Egl. Once Egl binds to RNA cargo, it has a much higher affinity for BicD [16,17]. We therefore examined the ability of our Egl mutants to associate with BicD in ovarian lysates. As expected, Egl_rbd3 and Egl_rbd6 were compromised for binding BicD. By contrast, Egl_rbd4 and Egl_rbd5 were largely unaffected (Figure 2(a,b)).
Figure 2.
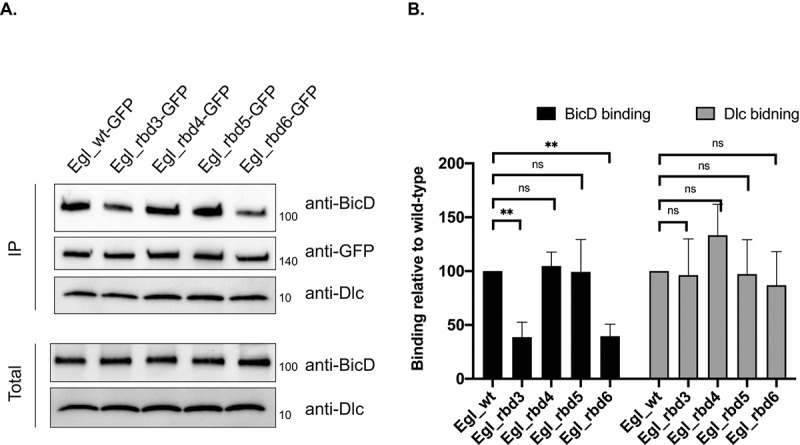
Egl RNA binding mutants are defective for binding to BicD. (a) A co-immunoprecipitation experiment was performed using ovarian lysates expressing the indicated Egl constructs. The lysates were incubated with GFP-Trap beads. After binding and wash steps, the bound proteins were eluted in SDS buffer and analysed by western blotting using the indicated antibodies. The total and IP fraction are shown. (b) Quantification of the binding experiment shown in (a). The experiment was repeated three times and the band intensities were for BicD and Dlc were determined. Binding is shown relatively to the BicD and Dlc value obtained with wild-type Egl. **p ≤ 0.01, ns = not significant; a one-way ANOVA was used to calculate statistical significance
The BicD-binding difference between Egl_rbd5 and Egl_rbd6 was somewhat surprising because both mutants were similarly defective for binding RNA cargoes such as grk, nos and pgc (Figure 1(e)). However, Egl_rbd6 was more defective than Egl_rbd5 for binding the ILS and TLS localization elements (Figure 1(c) and Supplemental Figure 1(a)). Thus, there might exist numerous uncharacterized cargoes for which Egl_rbd6 is more defective in its binding activity in comparison to Egl_rbd5. As a result, the aggregate interaction between Egl_rbd6 and BicD is more severely compromised.
In addition to binding BicD, Egl also interacts directly with Dynein light chain (Dlc) [21]. The interaction with Dlc is required for dimerization of Egl, a prerequisite for optimal RNA binding activity [15]. We therefore examined the ability of these mutants to interact with Dlc. Consistent with the notion that the Egl-Dlc interaction occurs prior to RNA binding, all four mutants retained the ability to associate with Dlc (Figure 2(a,b)).
Localization of Egl RNA binding mutants
Within early and mid-stage egg chambers, Egl localizes preferentially within the oocyte [19]. The same localization pattern is observed for BicD, the Dynein motor, and the Dynactin complex, a critical regulator of Dynein activity [19,22,23]. Thus, this localization pattern is assumed to be indicative of cargo transport into the oocyte. Egl also localizes to the nuage, an electron-dense area that surrounds nurse cell nuclei [24]. In contrast to wild-type Egl, Egl_rbd3 was neither enriched within the oocyte, nor was it localized to the nuage (Figure 3(a,a’,b,b’,f)). A similar, but slightly milder phenotype was observed for Egl_rbd6 (Figure 3(e,e’,f)). Egl_rbd5 was enriched within the oocyte, though to a slightly lesser extent than the wild-type (Figure 3(d,f)). The localization of Egl_rbd5 to the nuage was unaffected (Figure 3(d’)). Lastly, Egl_rbd4 displayed essentially the same localization pattern as wild-type Egl within the oocyte and at the nuage (Figure 3(c,c’,f)).
Figure 3.
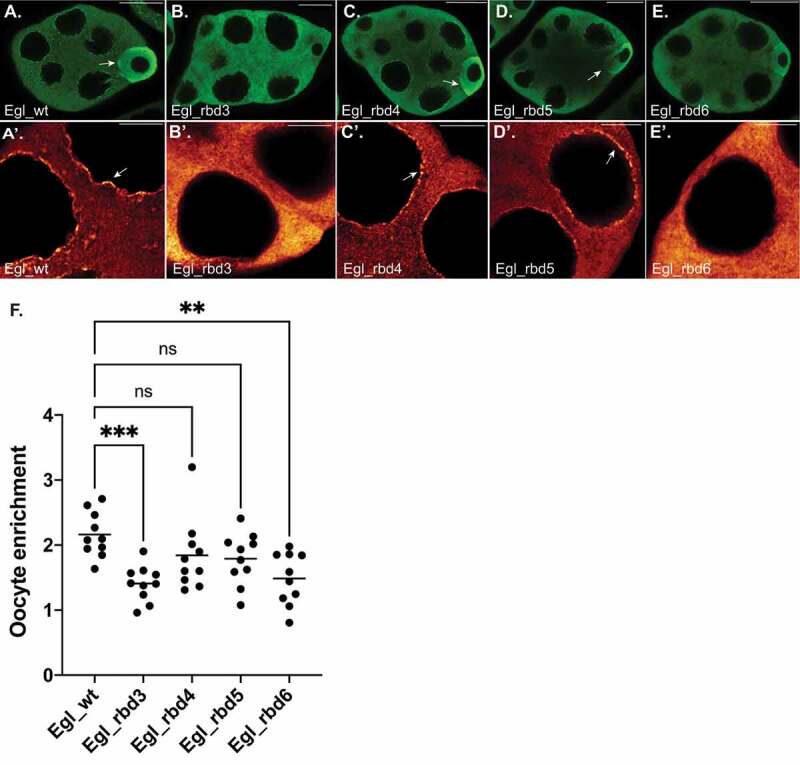
Localization of Egl RNA binding mutants. (a–e) Ovaries were dissected from flies expressing GFP tagged versions of the indicated constructs. The ovaries were fixed and processed for immunofluorescence using a poly-clonal GFP antibody. The relative enrichment of the Egl to the oocyte (arrows) is shown in A-E and the localization to the nuage is shown in A’-E’ (arrows). (f) The enrichment of Egl within the oocyte relative to the entire egg chamber was quantified for the indicated genotypes. n = 10 egg chambers for each genotype. ***p ≤ 0.001, ** p ≤ 0.01, ns = not significant; a one-way ANOVA was used to calculate statistical significance. The scale bar in a-e is 20 microns; the scale bar in a’-e’ is 5 microns
Based on the localization of these mutants, and their ability to associate with RNA and BicD, we conclude that they represent an allelic series with the strongest to weakest mutants in the following order; Egl_rbd3, Egl_rbd6, Egl_rbd5 and finally Egl_rbd4.
Microtubule organization in Egl RNA binding mutants
Depletion of Egl in mid and late-stage egg chambers results in profound disorganization of oocyte microtubules [13]. In wild-type egg chambers, microtubule minus-ends are enriched at the anterior of the oocyte and along the cortex. In contrast, depletion of Egl results in microtubule minus-ends being diffusely localized within the oocyte [13]. At present, the mechanism by which Egl regulates the organization of oocyte microtubules is unknown. In order to determine whether the RNA binding activity of Egl is required for this function, we examined the localization of microtubules in Egl RNA binding mutants. Egg chambers were first stained using an antibody against alpha-Tubulin to reveal the microtubule lattice. In wild-type oocytes, alpha-Tubulin is detected primarily at the anterior margin of the oocyte and along the oocyte cortex (Figure 4(a,k)). A similar pattern was also observed in Egl_rbd4, Egl_rbd5, and Egl_rbd6 egg chambers (Figure 4(c-e,k)). By contrast, alpha-Tubulin signal was detected throughout the oocyte in Egl_rbd3 mutants (Figure 4(b,k)).
Figure 4.
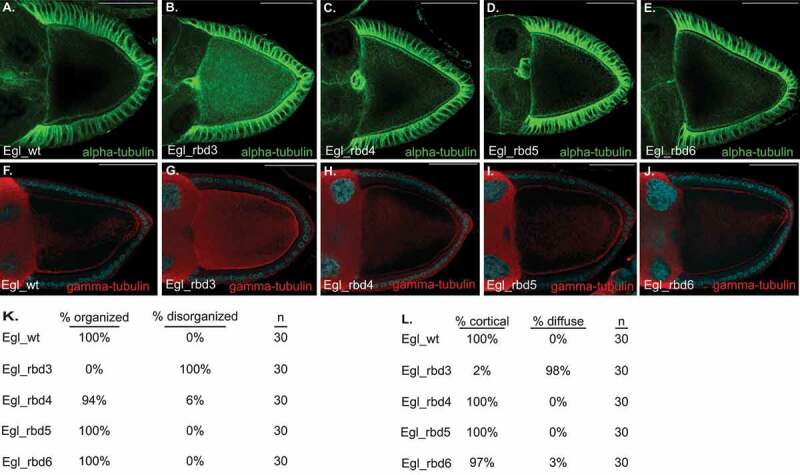
Microtubule organization requires Egl RNA binding activity. (a–e) Ovaries were dissected and fixed from flies expressing FLAG tagged versions of the indicated constructs. Ovaries were processed for immunofluorescence using a FITC-conjugated alpha-Tubulin antibody. Quantification of these phenotypes is shown in panel K. (f–j) Ovaries from the same strains were also processed for immunofluorescence using a gamma-Tubulin antibody. Quantification of the gamma-Tubulin phenotype is shown in panel l. The scale bar is 50 microns
Egg chambers were next stained using an antibody against gamma-Tubulin, a marker for microtubule minus ends. As expected, gamma-Tubulin was detected around the oocyte cortex of egg chambers expressing wild-type Egl, Egl_rbd4, Egl_rbd5, and Egl_rbd6, with minimal signal observed within the interior of the oocyte (Figure 4(F,h-j,l)). By contrast, gamma-Tubulin signal was diffusely distributed throughout the oocyte in Egl_rbd3 mutants (Figure 4(g,l)). This phenotype is similar to that observed upon depletion of Egl [13]. These results suggest that although microtubules are organized in the milder RNA binding mutants, they are disorganized in Egl_rbd3 oocytes. Consistent with this notion, the oocyte nucleus was correctly localized at the dorsal anterior corner of egg chambers expressing wild-type Egl, Egl_rbd4, Egl_rbd5, and Egl_rbd6, but was often delocalized in Egl_rbd3 mutants (Supplemental Figure 1(b-g)). The localization of the oocyte nucleus at the dorsal-anterior corner requires a correctly organized microtubule cytoskeleton [25].
Egl_rbd3 is compromised for binding to RNA as well as BicD (Figure 1 and Figure 2). Thus, RNA binding, BicD binding, or both are required for organization of oocyte microtubules. However, Egl_rbd6 is also compromised for binding BicD (Figure 2), yet microtubule organization is relatively unaffected in this milder RNA binding mutant. Based on these results, we conclude that organization of oocyte microtubules requires at least some level of Egl RNA binding activity. A complete loss of RNA binding, as is observed in Egl_rbd3 mutants, disrupts this process. Alternatively, a structural defect in Egl_rbd3 that is not present in Egl_rbd6 might also account for their different microtubule organization phenotypes. If such a defect exists, however, it is likely to be subtle, as Egl_rbd3 retained its ability to dimerize and to interact with Dlc (Figure 2 and Supplemental Figure 1(h)).
RNA localization in mid-stage egg chambers
Numerous mRNAs exhibit specific localization patterns in mid and late stage Drosophila egg chambers. Many of these mRNAs are localized via a Dynein-based mechanism, and the cargo adaptor for these mRNAs is thought to be Egl. Consistent with this notion, depletion or mutation of Egl results in delocalization of grk and bcd mRNAs [13,21]. For this analysis, we examined the localization of grk, mu2, bcd and hts mRNAs in mid and late-stage egg chambers.
grk mRNA localizes to the dorsal-anterior corner of stage10 egg chambers [26] (Figure 5(a)). A similar pattern was observed in Egl_rbd4, Egl_rbd5, and Egl_rbd6 mutants (Figure 5(b-d)). However, quantification of localized signal revealed that the enrichment of grk mRNA at this site was reduced in Egl_rbd5 mutants (Figure 5(m)). A similar reduction was observed in Egl_rbd4 and Egl_rbd6 mutants. However, the difference was not statistically significant (Figure 5(m)).
Figure 5.
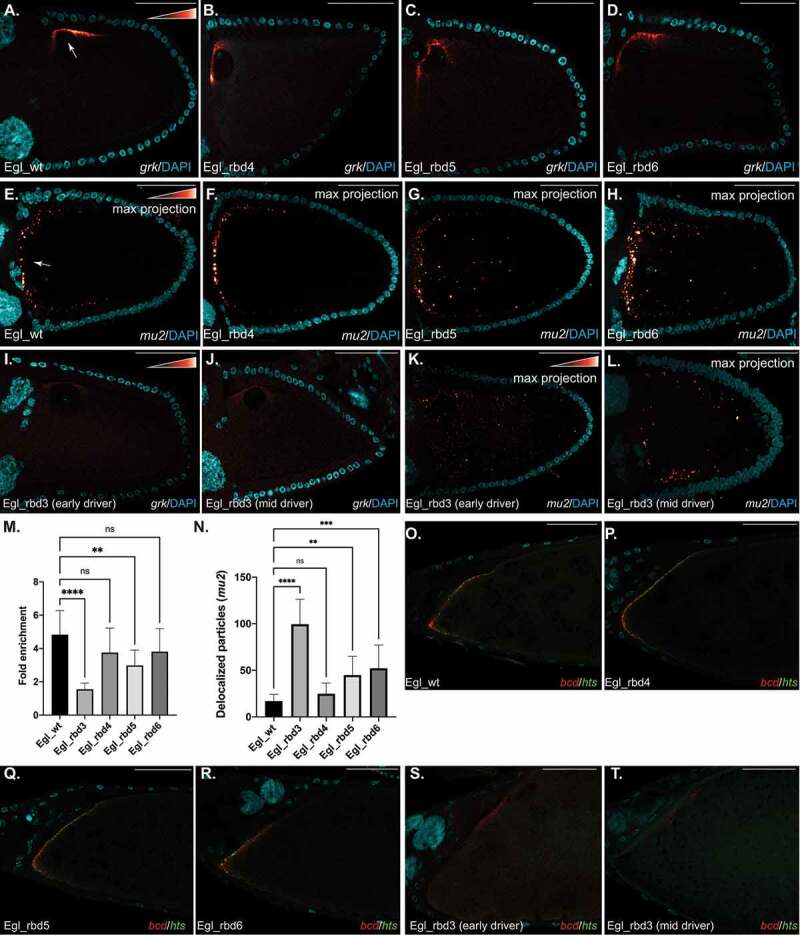
mRNA localization in stage 10 egg chambers. (a–d) Ovaries from flies expressing the indicated Egl constructs were dissected and processed for in situ hybridization using probes against grk mRNA. The egg chambers were counter stained with DAPI to reveal nuclei (Cyan). The in situ signal for this and subsequent experiments is shown using a red to white look up table (LUT). Red pixels indicate low-intensity signal and white pixels indicate high-intensity signal. The arrow in panel A indicates the normal localization of grk to the dorsal-anterior corner of wild-type egg chambers. Quantification of the dorsal-anterior enrichment of grk is shown in panel M. (e–h) The same strains were processed using in situ hybridization using probes against mu2 mRNA. The arrow in panel E indicates the normal localization of mu2 mRNA to distinct particles that cluster at the anterior margin of the oocyte. Quantification of the delocalized particles in each of the indicated genotypes is shown in panel N. (i–l) Ovaries from flies expressing Egl_rbd3 and depleted of endogenous Egl using either the early-stage driver (i, k) or the mid-stage driver (j, l) were processed for in situ hybridization using probes against grk mRNA (i, j) or mu2 mRNA (k, l). All mu2 in situs shown in this figure represent maximum projection images. (o–r) Ovaries from flies expressing the indicated Egl constructs were dissected and processed for in situ hybridization using probes against bcd (red) and hts (green) mRNAs. The egg chambers were counter stained with DAPI (Cyan). The anterior margin of stage14 egg chambers are shown. Ovaries from flies expressing Egl_rbd3 and depleted of endogenous Egl using either the early-stage driver (s) or the mid-stage driver (t) were also processed for in situ hybridization using probes against bcd (red) and hts (green) mRNAs. ****p ≤ 0.0001, ***p ≤ 0.001, ** p ≤ 0.01, ns = not significant; a one-way ANOVA was used to calculate statistical significance. The scale bar is 50 microns
mu2 mRNA has been shown to localize to the anterior margin of the oocyte [27]. Using single molecule fluorescent in situ hybridization (smFISH), we detected numerous discrete puncta of mu2 mRNA along the anterior margin of the oocyte in egg chambers expressing wild-type Egl (Figure 5(e)). A similar pattern was observed in Egl_rbd4 mutants (Figure 5(f)). Interestingly, although some anterior enrichment of mu2 could be detected in egg chambers expressing Egl_rbd5 and Egl_rbd6, many puncta were delocalized away from the anterior of the oocyte (Figure 5(g,h,n)).
Both grk and mu2 mRNAs were significantly delocalized in egg chambers expressing Egl_rbd3 (Figure 5(I,k)). However, as noted, microtubules are disorganized in these oocytes (Figure 4). Thus, using this early-stage driver, we were not able to conclude whether this defect results from loss of RNA binding activity or from microtubule disorganization. In order to distinguish between these possibilities, we depleted endogenous Egl using a mid-stage driver. Depletion of Egl with this driver does not compromise the organization of oocyte microtubules [13]. Consistent with the notion that Egl RNA binding activity is required for the localization of grk and mu2 mRNAs, both transcripts were also delocalized in egg chambers that were depleted of endogenous Egl using the mid-stage driver (Figure 5(j,l)).
We next examined the localization of bcd and hts mRNAs, both of which localize along the anterior margin of mid and late-stage egg chambers (Figure 5(o)) [28,29,30]. Both mRNAs were delocalized in egg chambers expressing Egl_rbd3, regardless of whether endogenous Egl was depleted using the early or mid-stage driver (Figure 5(s,t)). Thus, at least some level of Egl RNA binding activity is required for the localization of these mRNAs. However, in contrast to Egl_rbd3, bcd and hts mRNAs remained localized in egg chambers expressing Egl_rbd4, Egl_rbd5 and Egl_rbd6 (Figure 5(p-r)). As indicated in a previous section, milder RNA binding mutants such as Egl_rbd5 and Egl_rbd6 are more compromised for binding to grk and mu2 than they are for binding to bcd and hts (Figure 1(e,f)). Thus, although grk and mu2 are not efficiently localized in the milder RNA binding mutants, bcd and hts remain unaffected in these same mutants.
Oocyte specification in Egl RNA binding mutants
Specification of the oocyte occurs in the germarium. A germline stem cell divides to produce a daughter cell referred to as a cystoblast. The cystoblast undergoes four rounds of mitosis to produce a cyst that contains sixteen interconnected cells. Two cells within the cyst will begin to accumulate oocytes specific factors. Formation of the synaptonemal complex is also evident in these two cells, indicating initiation of a meiotic program (Figure 6(a)) [31]. One of these cells is eventually specified as the oocyte while the other exits meiosis and reverts to nurse cell fate. Null mutants in egl fail to specify an oocyte [32,33]. Paradoxically, numerous cells within the cyst of egl nulls enter into meiosis and form a synaptonemal complex. However, by region 2B of the germanium, meiosis is terminated in all cells of the cyst, an oocyte is never specified, and the egg chamber eventually degenerates [32,34].
Figure 6.
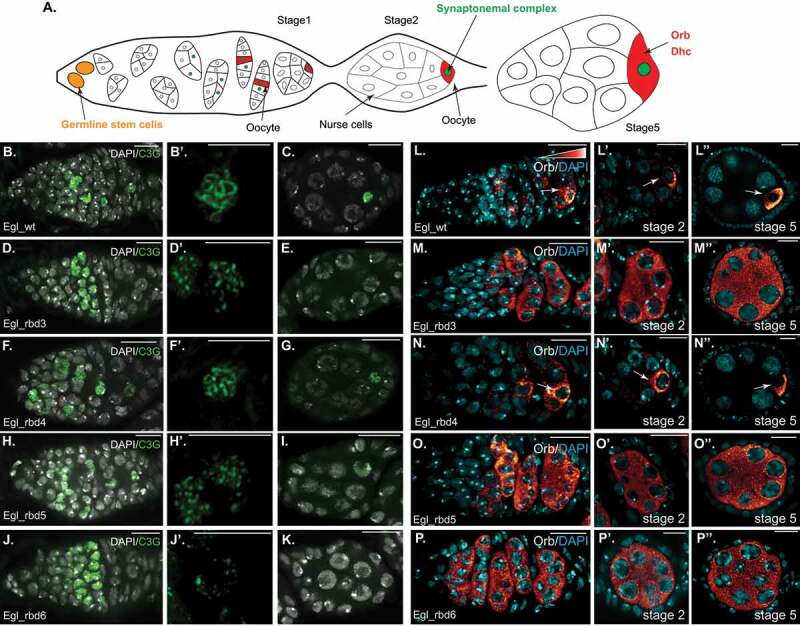
Oocyte specification requires optimal Egl RNA binding activity. (a) Schematic illustrating how the oocyte is specified, and the enrichment of factors such as Orb and Dhc within the oocyte. (b–k) Ovaries from the indicated genotypes were processed for immunofluorescence using the c(3)g antibody (green), a marker for the synaptonemal complex. The egg chambers were also counter-stained with DAPI to reveal nuclei (greyscale). The germarium is shown in b, d, f, h and j (scale bar is 10 microns), a magnified view of the synaptonemal complex from a cell in region 2B is shown in b’, d’, f’, h’ and j’ (scale bar is 5 microns) and a stage2 egg chamber is shown in c, e, g, i and k (scale bar is 10 microns). (l–p) The same flies were also processed for immunofluorescence using an antibody against Orb. Egg chambers were counter-stained for DAPI (cyan). The germarium is shown in L-P, representative stage2 egg chambers are shown in l’-p’ and representative stage5 egg chambers are shown in l”-p” (scale bar is 10 microns). Arrows indicate the oocyte enrichment of Orb
The mechanism by which Egl functions in oocyte specification and restriction of meiosis to a single cell is unknown. Given the known role of Egl in Dynein-mediated transport, the RNA binding activity of Egl might be required for this function. In order to test this, endogenous Egl was depleted in the germanium using the nanos-Gal4 driver. This produced a phenotype that is identical to egl nulls (data not shown). We then co-expressed the egl shRNA along with wild-type Egl or the RNA binding mutants. In order to visualize the synaptonemal complex, germaria were stained using the C(3)G antibody [35]. Expression of Egl_wt completely rescued this phenotype; meiosis was restricted to a single cell, the oocyte was specified, and egg chamber maturation proceeded as normal (Figure 6(b-c)). This process was disrupted in flies expressing Egl_rbd3, producing a phenotype that was identical to egl nulls (Figure 6(d-e)). Interestingly, even though numerous cells formed a synaptonemal complex in these mutants, the C(3)G staining pattern was unusual. In wild-type, the synaptonemal complex forms along the length of homologous chromosomes [35]. By contrast, in Egl_rbd3 mutants, the C(3)G staining was punctuate rather than the more uniform appearance in wild-type germaria (Figure 6(b’,d’)).
A similar phenotype, resembling egl nulls, was observed in Egl_rbd6 mutants (Figure 6(j-k), while a slightly milder phenotype was observed in Egl_rbd5 mutants (Figure 6(h-i)). In the latter germaria, the synaptonemal complex persisted in stage1 egg chambers but was lost by stage2 (Figure 6(I)). In addition, the appearance of C(3)G staining was intermediate between the punctuate appearance of Egl_rbd3 and Egl_rbd6 mutants and the more uniform staining seen in wild-type (Figure 6(h’)). As expected, Egl_rbd4 mutants produced the mildest phenotype (Figure (f-g)). However, these mutants were not normal either. Oocyte fate was lost in approximately 80% of ovarioles resulting in their eventual degeneration (Supplemental Figure 1(I-k)).
We next examined the localization of Orb protein in these mutants. Orb has been shown to be enriched in the two cells that initially form the synaptonemal complex and eventually becomes highly enriched within the oocyte (Figure 6(a)) [34]. Like Egl, Orb is also required for oocyte specification [36]. Orb displayed the expected localization pattern in wild-type germaria and early-stage egg chambers (Figure 6(l)). A similar pattern was observed in the 20% of ovarioles from Egl_rbd4 mutants that developed normally (Figure 6(n)). However, Orb was delocalized in egg chambers which eventually lost oocyte fate (Supplemental Figure 1(h,I)). Interestingly, in the context of Orb localization, Egl_rbd3, Egl_rbd5 and Egl_rbd6 produced an identical phenotype. In these mutants, Orb was never localized to a single cell (Figure 6(m,o,p)). Thus, in contrast to synaptonemal complex formation, which occurs at least partially in Egl_rbd5 mutants, Orb localization depends critically on Egl RNA binding activity. Based on these results, we conclude that specification and maintenance of oocyte fate requires maximal Egl RNA binding activity.
Early-stage cargo transport in Egl RNA binding mutants
Consistent with the notion that oocyte specificity factors are actively transported into the presumptive oocyte, Dynein heavy chain (Dhc), the motor subunit of Dynein, is enriched within the oocyte [37]. The oocyte enrichment pattern is clearly observed in egg chambers expressing Egl_wt and Egl_rbd4 but is disrupted in Egl_rbd3 mutants (Figure 7(a-c)). Interestingly, oocyte enrichment is still observed in stage1 egg chambers of Egl_rbd6 mutants but is lost by stage2 (Figure 7(e)). Egl_rbd5 mutants produce a slightly milder phenotype and Dhc is enriched in oocytes until stage2 (Figure 7(d)). This suggests that in the milder RNA binding mutants, at least some transport of cargo into the oocyte might still occur.
Figure 7.
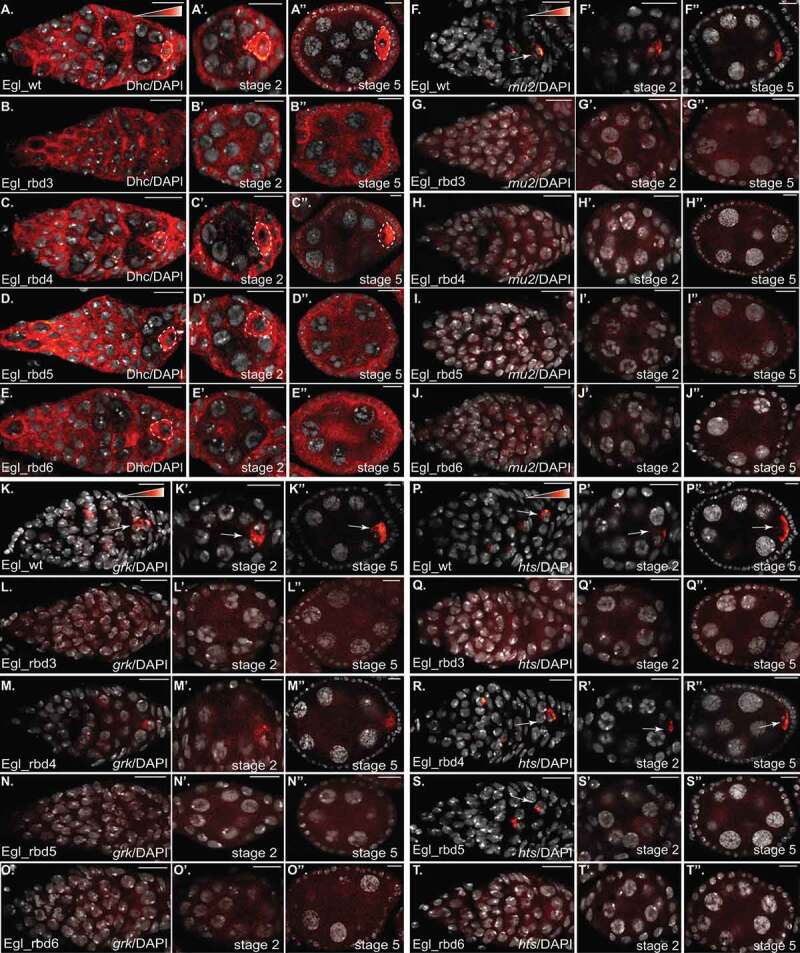
Dynein mediated transport of mRNA cargo in the germarium and early-stage egg chambers. (a–e) The indicated genotypes were processed or immunofluorescence using an antibody against Dhc (red to white LUT). The oocyte enrichment of Dynein is indicated by dashed lines. The egg chambers were also counter-stained with DAPI to reveal nuclei (greyscale). The germarium is shown in a-e, representative stage2 egg chambers are shown in A’-E’ and representative stage5 egg chambers are shown in a”-e”. The same flies were also processed for in situ hybridization using probes against mu2 (f-j), grk (k-o) and hts (p-t). The egg chambers were also counter-stained with DAPI to reveal nuclei (greyscale). The layout is the same as shown for Dhc. Arrows indicate the oocyte enrichment of the respective mRNAs. The scale bar is 10 microns
Among the known cargoes of Egl, mu2, grk, and hts mRNAs specifically localize to the oocyte within the germarium [27,30,38]. We therefore examined the localization of these cargoes in our RNA binding mutants. mu2 mRNA displayed the expected localization pattern in egg chambers expressing Egl_wt (Figure 7(f)). However, this mRNA was delocalized in all of the RNA binding mutants, including the mildest mutant, Egl_rbd4 (Figure 7(g-j), Supplemental Figure 1(l)). grk mRNA was slightly less affected. Although grk was correctly localized in the germaria and early-stage egg chambers of Egl_rbd4 mutants, the level of oocyte enrichment was reduced in comparison to the wild-type (Figure 7(K,M), Supplemental Figure 1(l)). Similar to mu2, grk was completely delocalized in Egl_rbd3, Egl_rbd5, and Egl_rbd6 mutants (Figure 7(l,n,o)). Consistent with the RNA binding results (Figure 1(e,f)), hts mRNA displayed the mildest defect. hts mRNA was correctly localized in the germaria and early-stage egg chambers of Egl_rbd4 mutants. However, the level of oocyte enrichment was reduced (Figure 7(p,r), Supplemental Figure 1(l)). hts mRNA was completely delocalized in Egl_rbd3 and Egl_rbd6 mutants (Figure 7(q,t)). Interestingly, in Egl_rbd5 mutants, hts mRNA was correctly localized in the germanium, but was delocalized in stage2 and later egg chambers (Figure 7(s)).
We can make two conclusions based on these results. The first is that Egl mediated mRNA transport in the germarium and early-stage egg chambers requires optimal RNA binding activity. Even a mild defect in RNA binding, which can be tolerated in later stage egg chambers, results in defective localization of the same transcripts at early stages. Second, some cargoes such as mu2 and grk are much more reliant on optimal Egl RNA binding activity in comparison to cargoes such as hts. Defective cargo transport into the presumptive oocyte likely results in a failure to specify and maintain the fate of the oocyte.
Discussion
Most of the mRNA cargoes that are known to be transported by Dynein in Drosophila oocytes and embryos are likely linked to this motor via Egl [12,14]. Thus, Egl must have the capacity to simultaneously associate with a diverse set of cargoes, yet have sufficient specificity to ensure that non-localized cargoes are not accidentally linked to Dynein. The binding affinity of Egl for its various cargoes must also not be excessively high. Tight binding would preclude unloading of cargoes once they are delivered to their site of localization. Perhaps due to these functional constraints, the RNA binding domain of Egl differs extensively from many other previously characterized RNA binding proteins.
The most recognizable feature within the RNA binding domain of Egl is a motif typically found in exonucleases, enzymes that degrade RNA. In fact, residues associated with catalysis are conserved in Egl. However, mutation of these amino acids does not disrupt the function of Egl [21]. Thus, it is unlikely that Egl possesses exonuclease activity. In this regard, Egl is similar to Exuperantia (Exu) and Maelstrom (Mael). Both Exu and Mael contain an exonuclease-like RNA binding domain, yet like Egl, neither protein appears to possess catalytic activity [39,40]. It is likely that these proteins have evolved from ancient RNA degrading enzymes but have been coopted to perform a different function in Drosophila. Another similarity between Egl and Exu is that both proteins bind RNA as dimers [40]. However, while Egl associates with numerous Dynein-localized cargoes [12,14], Exu is much more specific and seems to be primarily involved in the binding and localization of bcd mRNA [28,29,40].
In this study, we have identified residues within a small stretch of Egl that are required for RNA binding (Figure 1(a)). The two most compromised mutants are Egl_rbd3, in which eight positively charged residues have been mutated, and Egl_rbd6, in which three positively charged residues have been mutated. Our analysis revealed unexpected differences in the sensitivity of cargo localization to Egl’s RNA binding activity. For instance, binding of Egl to cargoes such as grk and mu2 was almost completely eliminated in the Egl_rbd3 background, and reduced approximately two-fold in the Egl_rbd6 background (Figure 1(e)). In contrast, cargoes such as bcd and hts displayed about three-fold reduced binding to Egl_rbd3 and were only modestly affected in the Egl_rbd6 background (Figure 1(f)). One possible reason for this difference is that additional amino acids within the RNA binding domain of Egl, apart from the ones identified in our study, might interact with cargoes such as bcd and hts. This might result in a more stable association. Another possibility is that these cargoes might contain multiple localization elements, each of which is capable of associating with Egl. Thus, the additive nature of the Egl-cargo interaction ensures that at least some binding occurs even in the Egl_rbd3 background. It is important to note, however, that this level of binding is insufficient for the proper localization of bcd and hts mRNAs (Figure 5(s,t)). Further studies are required to distinguish between these scenarios.
In addition to its role in mRNA localization in mid-stage egg chambers and embryos, Egl is also required for oocyte specification and restricting meiosis to a single cell. In egl nulls, all 16 cells of the developing cyst enter meiosis. However, this process is not sustained, and by region 2B of the germarium, no meiotic markers are observed in any cell. In addition, oocyte-specific markers such as Orb fail to concentrate in a single cell. Consequently, oocyte specification is aborted, and all cells of the cyst adopt a nurse cell fate [32–34].
Our findings indicate that the RNA binding activity of Egl is required for restriction of meiosis to single cells. Not surprisingly, the strongest RNA binding mutant, Egl_rbd3, displays a phenotype that is identical to egl nulls (Figure 6(d)). However, even Egl_rbd6 mutants, which produce a milder phenotype in mid-stage egg chambers, resembled egl nulls in this respect (Figure 6(j)). A slightly less severe phenotype was observed in Egl_rbd5 mutants. In these mutants, all sixteen cells initially entered meiosis before this process became restricted to a single cell. However, by stage2, the meiotic marker was no longer detected, indicating that oocyte fate was lost (Figure 6(h)). Orb, an oocyte marker, becomes enriched to a single cell within region 2B of the germanium [41]. This pattern was observed in ovarioles expressing Egl_wt and in 54% of ovarioles expressing Egl_rbd4. However, this failed to occur in Egl_rbd3, Egl_rbd5, and Egl_rbd6 mutants (Figure 6(m-p)). Based on these results, we conclude that enactment of the proper meiotic program, and specification of oocyte fate, requires optimal Egl RNA binding activity.
How might Egl function in meiosis? As noted previously, loss of Egl causes all cells of the cyst to enter meiosis. However, in bicD nulls, the opposite phenotype is observed; meiosis does not initiate in any cells within the cyst [34]. Thus, this meiotic function of Egl might not require the Dynein motor. By contrast, specification and maintenance of oocyte fate is likely the result of Dynein-mediated transport of mRNAs into the presumptive oocyte. Dynein becomes enriched within the presumptive oocyte within the germarium and remains enriched in the oocyte during early oogenesis [23]. Disruption of the Dynein motor, or mutation of Dynein activators such as Lis1 also compromises oocyte specification [37,42]. We therefore examined the localization of grk, mu2, and hts in the germarium and early-stage egg chambers. All three cargoes have been shown to localize to the presumptive oocyte in the germarium and to the oocyte in stage1 egg chambers [27,30,38]. Interestingly, the localization of all three cargoes was significantly affected at these stages, even in Egl_rbd4, the mildest mRNA binding mutant.
These findings raise an important question. Why is mRNA localization so much more compromised in early-stage egg chambers whereas the same cargoes are only modestly delocalized when endogenous Egl is depleted in mid and late-stage egg chambers? Although we do not have a definitive answer to this question, it is known that the localization of cargoes such as grk and bcd in mid-stage egg chambers involves an anchoring mechanism in addition to active transport [9,43]. Thus, even though these mRNAs may not be efficiently transported in the background of the milder Egl RNA binding mutants, as long as their anchoring is efficient, their overall localization would not be drastically affected. By contrast, in the germarium and early-stage egg chambers, anchoring mechanisms might not yet be fully active. Thus, at these stages, continuous active transport by Dynein may be required to correctly localize these transcripts. Consequently, even subtle defects in Egl’s ability to bind these cargoes would result in their pronounced delocalization.
Our results suggest that Dynein-mediated localization of mRNA is essential for specification of oocyte fate. However, the identity of this cargo, or even whether it represents single mRNA species, is not known. A possible candidate for this function is Orb. Both orb mRNA and protein are highly enriched in the oocyte from very early stages [41]. The localization of orb mRNA was recently shown to require sequences in its 3ʹUTR. Mutations within this region result in the mRNA and protein being delocalized from the oocyte and a defect in oocyte specification [41]. However, in this background Egl and Dynein were also delocalized [41]. Thus, cargoes localized by Egl and Dynein are likely to be delocalized as well. In addition, Orb is an RNA binding protein that has been shown to associate with, and possibly regulate the translation, of thousands of mRNAs [44]. As such, although Orb is clearly required for oocyte specification, it probably mediates this function by regulating one or more of its target mRNAs.
In conclusion, by identifying critical residues within Egl that are required for RNA binding, we have gained mechanistic insight into the process of mRNA localization. Our findings revealed that certain cargoes are much more reliant on Egl’s RNA binding activity for their correct localization in comparison to others. Second, maximal Egl RNA binding activity is most critical at the earliest stages of oogenesis. Even a minor disruption in Egl’s ability to bind to RNA cargo at these stages results in defective specification and maintenance of oocyte fate.
Supplementary Material
Acknowledgments
We thank Simon Bullock for providing the ILS and TLS aptamer constructs and Scott Hawley for providing the C(3)G antibody. We are grateful to the Bloomington Stock Center, Developmental Studies Hybridoma Bank, and the Drosophila Genomics Resource Center for providing fly strains, antibodies, cell lines, and DNA constructs. This work was supported by a grant from the National Institutes of Health to G.B.G (R01GM100088).
Funding Statement
This work was supported by the National Institute of General Medical Sciences [R01GM100088].
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Materials and methods
Fly stocks
The following shRNA stocks were used:
eb1 shRNA (Bloomington stock center; #36,680, donor TRiP)
egl shRNA-1 (Bloomington stock center; #43,550, donor TRiP).
shRNA expression was driven using:
P{w[+mC]=matalpha4-GAL-VP16}V37 (Bloomington Stock Center, #7063; donor Andrea Brand) for early-stage expression.
w[*]; P{w[+mC]=matalpha4-GAL-VP16}V2H (Bloomington Stock Center, #7062; donor Andrea Brand) for mid-stage expression.
P{w[+mC]=GAL4::VP16-nos.UTR}CG6325[MVD1] (Bloomington Stock center, #4937; donor Ruth Lehmann) for expression in the germanium.
Egl_wt, Egl_rbd3, Egl_rbd4, Egl_rbd5, and Egl_rbd6 tagged on the C-terminal end with either 3xFLAG or GFP were constructed by cloning the respective coding regions into the pUASp-attB-K10 vector [45]. The GFP tagged transgenic constructs were inserted at ZH-86Fb site (Bloomington stock center; #24,749, donor Konrad Basler & Johannes Bischof) and the FLAG tagged constructs were inserted at su(Hw)attP1 site (Bloomington stock center; #34,760, donor Norbert Perrimon). The strains were injected by BestGene Inc. Fly stocks and crosses used for these experiments were maintained at 25°C.
Antibodies
The following antibodies were used: anti-FLAG (Sigma Aldrich, F1804, 1:5000 for western, 1:500 for immunofluorescence), mouse anti-BicD (Clones 1B11 and 4C2, Developmental Studies Hybridoma Bank, 1:30 for immunofluorescence; 1:300 for western, donor R. Steward), rabbit anti-Ctp (Abcam, ab51603, 1:5000 for western), GFP nanobody booster (Chromotek; 1:500 for immunofluorescence), mouse anti-GFP (Clontech, JL-8, 1:5000 for western), rabbit anti-GFP (Chromotek; 1:3000 for immunofluorescence), alpha-tubulin FITC (Sigma Aldrich, F2168, 1:150 for immunofluorescence), mouse anti-γ-tubulin (Sigma, T5326, 1:100 for immunofluorescence), C3G (generous gift from S. Hawley, mouse anti-C(3)G, 1:500), Orb (mouse anti-Orb, clone 4H8, 1:40 dilution), mouse anti-Dhc (Developmental Studies Hybridoma Bank, clone 2C11-2, 1:250; for immunofluorescence, donor J. Scholey), mouse anti-Lamin DmO (Developmental Studies Hybridoma Bank, clone ADL84.12; 1:200; for immunofluorescence, donor P. A. Fisher). The following secondary antibodies were used: goat anti-rabbit Alexa 594, 555 and 488 (Life Technologies, 1:400, 1:400 and 1:200, respectively); goat anti-mouse Alexa 594, 555 and 488 (Life Technologies, 1:400, 1:400 and 1: 200, respectively) goat anti-mouse HRP (Pierce, 1:5000); and goat anti-rabbit HRP (Pierce, 1:5000).
DNA constructs
The transgenes for expressing Egl_wt, Egl_rbd3, Egl_rbd4, Egl_rbd5, and Egl_rbd6 were cloned into the pUASp-attB-K10 vector [45]. A construct containing the wild-type Egl coding region fused to either a C terminal 3xFLAG tag or a GFP tag was used as the template for generating the mutants. The mutants were constructed by replacing the wild-type sequence with a gene synthesized double stranded DNA fragment using Gibson assembly (NEB). The gene synthesized fragments were constructed by Genewiz. The final constructs were verified by sequencing prior to injection into flies. Constructs were expressed in S2 cells by co-transfecting along with an Act5c-Gal4 plasmid (gift from Jocelyn McDonald).
Cell culture
The S2 cells used in this study were obtained from the Drosophila Genomics Resource Center (S2-DRSC line, stock number 181). Cells were grown in Schneider’s medium containing 10% heat inactivated fetal calf serum and were transfected using Effectene (Qiagen) according to instructions provided by the manufacturer.
In vitro protein/RNA interaction
The binding of Egl constructs to the ILS and TLS localization elements was performed as previously described [12,15]. Briefly, 5μg of RNA was first refolded in 10μL of Drosophila extraction buffer (DXB: 25mM HEPES pH 6.5, 50mM KCl, 1mM MgCl2, 250mM sucrose, 0.1% NP40, supplemented with 10μM MgATP and 1mM DTT at the time of use). Refolded RNA was then incubated with High Capacity Streptavidin Agarose Beads (Pierce) in 90μL DXB for 1.5 hours at 4°C while nutating. Extracts from either S2 cells or frozen Drosophila ovaries were prepared using RIPA lysis buffer (50mM Tris pH 7.5, 200mM NaCl, 0.2mM EDTA, 0.05% NP40, Halt protease inhibitor cocktail, Pierce). 800ug of total protein was used in each binding experiment. The extracts were incubated with the RNA-bound streptavidin beads for 15 min at room temperature followed by 30 min at 4°C with nutation. The beads were then rinsed 4 times in lysis buffer, the bound proteins were eluted by boiling in Laemmli buffer and examined by western blotting.
Analysing in vivo protein-mRNA association
The binding of Egl constructs to native mRNAs from Drosophila ovaries was analysed as previously described [15]. In brief, 600ug of total lysate was used in each immunoprecipitation. The tagged proteins were immunoprecipitated using FLAG M2 antibody beads (Sigma). Co-precipitating RNAs were reverse transcribed using random hexamers and Superscript III (Life Technologies) according to directions provided by the manufacturer. Quantitative (qPCR) was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). A Bio-Rad CFX96 Real-Time PCR System was used for this experiment. Fold enrichment was calculated by comparing ct values for each RNA to that obtained for γ-tubulin.
Protein–protein interaction
A co-immunoprecipitation experiment was used to analyse protein–protein interaction in vivo using a previously described procedure [15]. In brief, ovaries were homogenized into lysis buffer (50mM Tris pH 7.5, 150mM NaCl, 0.2mM EDTA, 0.05% NP40 and Halt protease inhibitor cocktail, Pierce) and cleared by centrifugation at 10,000g at 4°C for 5 min. 800ug of total protein was used per immunoprecipitation. Immunoprecipitation was performed by incubating lysates at 4°C for 1 hour with GFP-trap beads (Chromotek). The beads were then washed four times with lysis buffer. Co-precipitating proteins were eluted in Laemmli buffer, run on a gel, and analysed by western blotting.
Immunofluorescence and in situ hybridization
Immunofluorescence and in situ hybridization were performed as described [15]. Flies were fattened on yeast pellet for 2 to 3 days prior to dissection. Ovaries were dissected and fixed in 4% formaldehyde (Pierce) for 20 min at room temperature. For immunofluorescence experiments, primary antibody was incubated in blocking buffer (PBS + 0.1% Triton X-100 + 0.2% BSA, Promega) overnight at 4°C. Next, the samples were washed three times in PBST (PBS + 0.1% Triton X-100) and incubated overnight with the fluorescent secondary antibody in the same blocking buffer. The next day, the samples were washed four times with PBST, stained with DAPI and mounted onto slides using Prolong Diamond (Life technologies). For in situ hybridization, the ovaries were dissected and fixed as above. After fixation, ovaries were stored in 100% methanol at −20°C for 1 hour. Next, the samples were re-hydrated with three 10 min washes using a solution of PBST and 100% methanol (3:7, 1:1, 7:3) and rinsed four times with PBST. The hydrated samples were then washed for 10 minutes in Wash Buffer (4xSSC, 35% deionized formamide, 0.1% Tween-20). Fluorescent oligo probes (Stellaris probes) were obtained from Biosearch technologies. Fluorescent probes diluted in Hybridization Buffer (10% dextran sulphate, 0.1 mg/ml salmon sperm ssDNA, 100 µl vanadyl ribonucleoside (NEB biolabs), 20ug/ml RNAse-free BSA, 4x SSC, 0.1% Tween-20, 35% deionized formamide) were applied to the ovaries and incubated overnight at 37°C. The next day, the samples were washed twice with pre-warmed Wash Buffer for 30 min. After two rinses with PBST and two rinses with PBS, the samples were stained with DAPI, and mounted onto slides using Prolong Diamond.
Microscopy
Images were captured on either a Zeiss LSM 780 inverted confocal microscope or an inverted Leica Stellaris confocal microscope. Images were processed for presentation using Fiji, Adobe Photoshop, and Adobe Illustrator. All imaging experiments were performed at the Augusta University Cell Imaging Core.
Quantification
All western images were acquired on a Bio Rad ChemiDoc MP. Band intensities were quantified using the Bio Rad Image lab software. RNA and protein enrichment was quantified by measuring the average pixel intensity of the localized signal and dividing by the average pixel intensity of the delocalized signal. For oocyte enrichment of Egl constructs depicted in Figure 4, and for the oocyte enrichment of mu2, hts and grk mRNAs in stage 2 egg chambers (Supplemental Figure 1(l)), the mean localized signal in the oocyte was compared to the mean signal in the rest of the egg chamber. In order to quantify the dorsal-anterior enrichment for grk mRNA in stage 10 egg chambers (Figure 5), the mean signal at the dorsal-anterior of the oocyte was compared to the mean signal in the rest of the oocyte. Signal intensity was quantified using the Zeiss Zen Black software. For quantification of mu2 localization, the particle analysis tool in Fiji was used. Localized particles were considered to be within 20 microns from the anterior margin of the oocyte. The remainder of the particles were considered to be delocalized. For quantification of microtubule organization, egg chambers were scored for the localization of alpha-tubulin and gamma-tubulin. For alpha tubulin quantification, egg chambers with a microtubule density at the anterior margin and around the oocyte cortex were considered ‘organized’ as this reflected the staining pattern of wild-type egg chambers. If significant alpha-tubulin staining was observed in the middle of the oocyte, this was scored as ‘disorganized’. For gamma-tubulin quantification, egg chambers were scored as ‘cortical’ if the signal for gamma-tubulin was detected primarily around the cortex of the egg chamber. By contrast, if signal was detected within the entire egg chamber, it was scored as ‘diffuse’.
References
- [1].Suter B. RNA localization and transport. Biochim Biophys Acta Gene Regul Mech. 2018;1861(23):938–951. [DOI] [PubMed] [Google Scholar]
- [2].Lasko P. mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb Perspect Biol. 2012;4(10):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Goldman CH, Gonsalvez GB. The role of microtubule motors in mRNA localization and patterning within the Drosophila Oocyte. Results Probl Cell Differ. 2017;63:149–168. [DOI] [PubMed] [Google Scholar]
- [4].Brendza RP, Serbus LR, Duffy JB, et al. A function for kinesin I in the posterior transport of oskar mRNA and staufen protein. Science. 2000;289(5487):2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Clark A, Meignin C, Davis I. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development. 2007;134(10):1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jaramillo AM, Weil TT, Goodhouse J, et al. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci. 2008;121(6):887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].MacDougall N, Clark A, MacDougall E, et al. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell. 2003;4(3):307–319. [DOI] [PubMed] [Google Scholar]
- [8].Mische S, Li M, Serr M, et al. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol Biol Cell. 2007;18(6):2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Trovisco V, Belaya K, Nashchekin D, et al. bicoid mRNA localises to the Drosophila oocyte anterior by random Dynein-mediated transport and anchoring. Elife. 2016;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gaspar I, Sysoev V, Komissarov A, et al. An RNA-binding atypical tropomyosin recruits kinesin-1 dynamically to oskar mRNP s. EMBO J. 2017;36(3):319–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Veeranan-Karmegam R, Boggupalli DP, Liu G, et al. A new isoform of Drosophila non-muscle Tropomyosin 1 interacts with Kinesin-1 and functions in oskar mRNA localization. J Cell Sci. 2016;129(22):4252–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dienstbier M, Boehl F, Li X, et al. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23(13):1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sanghavi P, Liu G, Veeranan-Karmegam R, et al. Multiple roles for Egalitarian in polarization of the Drosophila egg chamber. Genetics. 2016;203(1):415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vazquez-Pianzola P, Schaller B, Colombo M, et al. The mRNA transportome of the BicD/Egl transport machinery. RNA Biol. 2017;14(1):73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goldman CH, Neiswender H, Veeranan-Karmegam R, et al. The Egalitarian binding partners Dynein light chain and Bicaudal-D act sequentially to link mRNA to the Dynein motor. Development. 2019;146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McClintock MA, Dix CI, Johnson CM, et al. RNA-directed activation of cytoplasmic dynein-1 in reconstituted transport RNPs. Elife. 2018;;7:e36312. doi: 10.7554/eLife.36312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sladewski TE, Billington N, Ali MY, et al. Recruitment of two dyneins to an mRNA-dependent Bicaudal D transport complex. Elife. 2018;77:e36306. doi: 10.7554/eLife.36306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bullock SL, Ringel I, Ish-Horowicz D, et al. A’-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol. 2010;17(6):703–709. doi: 10.1038/nsmb.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 1997;11(4):423–435. [DOI] [PubMed] [Google Scholar]
- [20].Moser MJ, Holley WR, Chatterjee A, et al. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25(24):5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Navarro C, Puthalakath H, Adams JM, et al. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6(5):427–435. [DOI] [PubMed] [Google Scholar]
- [22].Januschke J, Gervais L, Dass S, et al. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr Biol. 2002;12(23):1971–1981. [DOI] [PubMed] [Google Scholar]
- [23].Li M, McGrail M, Serr M, et al. Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. J Cell Biol. 1994;126(6):1475–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kordyukova M, Morgunova V, Olovnikov I, et al. Subcellular localization and Egl-mediated transport of telomeric retrotransposon HeT-A ribonucleoprotein particles in the Drosophila germline and early embryogenesis. PLoS One. 2018;13(8):e0201787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhao T, Graham OS, Raposo A, et al. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science. 2012;336(6084):999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75(1):165–174. [PubMed] [Google Scholar]
- [27].Kasravi A, Walter MF, Brand S, et al. Molecular cloning and tissue-specific expression of the mutator2 gene (mu2) in Drosophila melanogaster. Genetics. 1999;152(3):1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Berleth T, Burri M, Thoma G, et al. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7(6):1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].St Johnston D, Driever W, Berleth T, et al. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development. 1989;107 Suppl(Suppl):13–19. [DOI] [PubMed] [Google Scholar]
- [30].Yue L, Spradling AC. hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev. 1992;6(12b):2443–2454. [DOI] [PubMed] [Google Scholar]
- [31].Huynh JR, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol. 2004;14(11):R438–449. [DOI] [PubMed] [Google Scholar]
- [32].Carpenter AT. Egalitarian and the choice of cell fates in Drosophila melanogaster oogenesis. Ciba Found Symp. 1994;182: 223–246. discussion 246-254. [PubMed] [Google Scholar]
- [33].Theurkauf WE, Alberts BM, Jan YN, et al. A central role for microtubules in the differentiation of Drosophila oocytes. Development. 1993;118(4):1169–1180. [DOI] [PubMed] [Google Scholar]
- [34].Huynh JR, St Johnston D. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development. 2000;127(13):2785–2794. [DOI] [PubMed] [Google Scholar]
- [35].Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15(23):3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lantz V, Chang JS, Horabin JI, et al. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 1994;8(5):598–613. [DOI] [PubMed] [Google Scholar]
- [37].McGrail M, Hays TS. The microtubule motor cytoplasmic dynein is required for spindle orientation during germline cell divisions and oocyte differentiation in Drosophila. Development. 1997;124(12):2409–2419. [DOI] [PubMed] [Google Scholar]
- [38].Roth S, Neuman-Silberberg FS, Barcelo G, et al. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81(6):967–978. [DOI] [PubMed] [Google Scholar]
- [39].Chen KM, Campbell E, Pandey RR, et al. Metazoan Maelstrom is an RNA-binding protein that has evolved from an ancient nuclease active in protists. RNA. 2015;21(5):833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lazzaretti D, Veith K, Kramer K, et al. The bicoid mRNA localization factor Exuperantia is an RNA-binding pseudonuclease. Nat Struct Mol Biol. 2016;23(8):705–713. [DOI] [PubMed] [Google Scholar]
- [41].Barr J, Gilmutdinov R, Wang L, et al. The Drosophila CPEB protein Orb specifies Oocyte Fate by a 3′UTR-dependent autoregulatory loop. Genetics. 2019;213(4):1431–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu Z, Xie T, Steward R. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development. 1999;126(20):4477–4488. [DOI] [PubMed] [Google Scholar]
- [43].Delanoue R, Herpers B, Soetaert J, et al. Drosophila squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13(4):523–538. [DOI] [PubMed] [Google Scholar]
- [44].Stepien BK, Oppitz C, Gerlach D, et al. RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc Natl Acad Sci U S A. 2016;113(45):E7030–E7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Koch R, Ledermann R, Urwyler O, et al. Systematic functional analysis of Bicaudal-D serine phosphorylation and intragenic suppression of a female sterile allele of BicD. PLoS One. 2009;4(2):e4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


