ABSTRACT
Ribosome-associated non-coding RNAs (rancRNAs) have been recognized as an emerging class of regulatory molecules capable of fine-tuning translation in all domains of life. RancRNAs are ideally suited for allowing a swift response to changing environments and are therefore considered pivotal during the first wave of stress adaptation. Previously, we identified an mRNA-derived 18 nucleotides long rancRNA (rancRNA_18) in Saccharomyces cerevisiae that rapidly downregulates protein synthesis during hyperosmotic stress. However, the molecular mechanism of action remained enigmatic. Here, we combine biochemical, genetic, transcriptome-wide and structural evidence, thus revealing rancRNA_18 as global translation inhibitor by targeting the E-site region of the large ribosomal subunit. Ribosomes carrying rancRNA_18 possess decreased affinity for A-site tRNA and impaired structural dynamics. Cumulatively, these discoveries reveal the mode of action of a rancRNA involved in modulating protein biosynthesis at a thus far unequalled precision.
KEYWORDS: Non-coding rna, ribosome, translation control, rancRNA, l1 stalk
Introduction
The fast regulation of gene expression during stress situations is important to maintain cellular protein homoeostasis in all kingdoms of life. In the case of harsh environmental conditions, translation has to be tightly controlled, since the production of proteins by ribosomes is a highly energy-demanding process [1]. At the very core of protein biosynthesis is the ribosome, a universally conserved RNA-protein machine responsible for polypeptide synthesis and co-translational folding. Several quite well understood signalling pathways, including the mTOR and the MAPK pathways, modulate the translation machinery by phosphorylation/dephosphorylation of essential protein factors [2]. More recently, non-protein-coding RNAs (ncRNAs) have been recognized as additional players in translation regulation. It has been argued that the main advantage of ncRNA regulators for protein synthesis is their almost immediate cellular disposal. These regulators function at the RNA level and thus are not needed to be translated into proteins to become active. Indeed, it has been noted that under certain stress conditions, the down-regulation of protein production was faster than changes in regulatory signalling pathways [3]. RNA-based translational regulators in eukaryotes such as micro RNAs or small-interfering RNAs bind to mRNAs and trigger their degradation, subsequently leading to repression of translation [4–6]. However, a recently discovered new class of ncRNAs, so-called ribosome-associated non-coding RNAs (rancRNAs), binds directly to ribosomes and alters their translational activity [7]. Experimental data suggest that these rancRNAs constitute a new class of regulators of protein synthesis capable of rapidly fine-tuning the rate of translation in a stress-dependent manner [8–16]. However, although rancRNAs are highly abundant cellular transcripts found in many organisms spanning all three domains of life, they are still functionally poorly explored at the molecular level [7,17].
The so far best studied rancRNA is the yeast rancRNA_18 derived from the TRM10 mRNA transcript [11]. It is important for the adaptation to hyperosmotic stress and has been demonstrated to rapidly inhibit protein synthesis in vivo and in vitro most probably at the step of translation initiation. These so far available data suggest that the fast reduction of protein biosynthesis and consequently of metabolic activities in response to salinity stress allows adaptation processes to be established [3,11,18]. It is known, that translation initiation in S. cerevisiae is rapidly down-regulated as a response to osmotic stress, but so far, the underlying mechanisms are not clear [19]. The TOR pathway that inhibits translation initiation in case of nutritional stress, is most likely not involved [20]. The main pathway that is activated upon salinity stress is the well-studied high-osmolarity glycerol (HOG) pathway [21]. It includes the activation of the MAPK Hog1 that relocates to the nucleus to promote changes in the gene expression programme. However, it was shown that these changes can take up to one hour to be achieved [22,23]. Another adaptation mechanism in the case of high salt concentrations is the elevation of phosphatidylinositol 3,5-bisphosphate levels, which occurs early before the HOG pathway induces changes in gene expression and is suggested to maintain the vacuolar and lysosomal ion homoeostasis [24]. Nevertheless, these two stress adaptation mechanisms are not involved in the inhibition of translation initiation when yeast cells encounter hyperosmotic stress. However, rancRNA_18-mediated down regulation of translation initiation has been demonstrated, and it occurs within minutes, thus likely representing the first wave of osmotic stress response in yeast [11].
Here, we dissected the molecular mechanism of rancRNA_18 function on S. cerevisiae ribosomes. Transcriptome-wide analysis discloses rancRNA_18 as a global inhibitor of protein biosynthesis. Cryo-electron microscopy (cryo-EM) and crosslinking analyses revealed the rancRNA_18 interaction pocket close to the E-site tRNA binding site on the large ribosomal subunit. Occupancy of this rancRNA site affects tRNA binding and restricts ribosome structural dynamics, mainly at the L1 stalk region of the large ribosomal subunit, thus suggesting a molecular mode of action during translation inhibition.
Materials and methods
Yeast strains and growth conditions
S. cerevisiae strain BY4742 (MATalpha, his3-Δ1, leu2-Δ0, lys2-Δ0, ura3-Δ0) and all mutant strains derived from it [11] were grown in either YPD or, when transformed with a plasmid, in Sc-Leu medium (yeast synthetic complete drop out medium -Leu; Sigma Aldrich) at 30°C or at various stress conditions [11]. To monitor growth under more stringent hyperosmotic conditions (the ‘redilution assay’), single colonies were picked and grown in Sc-Leu medium supplemented with NaCl (final concentration 0.7 M) for 72 h. Subsequently the cultures were rediluted to an OD600 of 0.1 into fresh stress medium, and cell growth was monitored over 20 h. For the tetracycline-inducible Rnt1 depletion cells were grown in YPD in the presence of 20 µg/ml doxycycline.
Generation of truncated TRM10 versions
For creating mutants carrying truncated versions of the TRM10 locus, where the ATG start codon was mutated to a TGA stop codon, the pRS415 plasmid, that carries the TRM10 start-to-stop codon mutant (called pTRM10_start-to-stop), was used and ligated with various PCR products of truncated versions of the TRM10 locus (see Supplementary Table S1 for DNA oligomer sequences). One-shot TOP ten electrocompetent E.coli cells (Invitrogen) were chosen as the host organism for transformation. Positive clones were selected by plating 200 μl of the transformation mix on LB + Ampicillin (100 μg/ml) plates. The GenElute™ Plasmid Miniprep System (Sigma Aldrich) was used for plasmid isolation. Preparation was performed according to the manufacturer`s instructions.
In order to replace the AGAA tetraloop within the TRM10 open reading frame by a GAAA tetraloop, a one-step PCR-based method for efficient site-directed substitution mutagenesis was employed [25]. The pTRM10 plasmid was used as template for the PCR reaction (see Supplementary Table S1 for DNA oligomer sequences).
Construction of the Δ6 knock-in strain
For the creation of the Δ6 knock-in, a part of the TRM10 gene including the promoter region and 148 bp of the ORF with CTG instead of ATG start codon (see also truncated TRM10 construct Δ6) was amplified and fused to the URA3 gene of K. lactis including promoter and terminator (pUG72) via a short linker [26]. The fusion PCR was performed with primers containing overhangs specific for a locus on chromosome V described [27] (see Supplementary Table S1 for DNA oligomer sequences). The trm10Δ strain was transformed with the PCR product and cells were selected for the expression of Ura3. Formed colonies were verified with PCR for the correct integration of the Δ6-URA3 construct.
Growth competition experiments
For growth competition experiments, the growth of a 1:1 dilution of two different yeast strains which only differ by the presence or absence of rancRNA_18 was examined under unstressed and high salt stress (0.7 M NaCl) conditions. To establish the identical genetic background for both strains, the TRM10_M2 plasmid (carrying the LEU2 marker and containing synonymous mutations in the rancRNA_18 region of the cloned TRM10 ORF thus yielding active Trm10 but inactive rancRNA_18-M2 [11]) was transformed into the trm10Δ6 knock-in strain (which possesses the URA3 marker) and the trm10Δ strain, respectively. The cultures were grown in Sc-Leu media to stationary phase and plated on either Sc-Leu or Sc-Leu-Ura plates. The stationary culture was subsequently diluted to an OD600 of 0.1 and the described procedure was repeated three times. Colony forming units (cfu) of each plate were determined and used to follow growth competition between cells expressing functional rancRNA_18 (genotype: LEU2, URA3) and cells lacking rancRNA_18 (genotype: LEU2, ura3Δ0).
Isolation of 80S ribosomes
S. cerevisiae cells were grown to exponential growth phase and harvested by centrifugation. The cell pellet was resuspended in buffer A (10 mM MgCl2, 100 mM KCl, 50 mM Tris/HCl pH 7.5, 0.4 mM PMSF) and the cells were opened with the Fast Prep as described before [11]. The cell debris was removed by two centrifugations (30,000 × g, 15 min), resulting in an S30 cell extract. The ribosomes were pelleted by ultracentrifugation at 100,000 × g at 4°C for 18 h in a Ti-60 Beckman rotor resulting in a crude ribosome pellet (P100). The ribosomes were diluted in buffer A and 200 A260 units of the sample was then layered on a linear sucrose gradient (10–40%; Gradient master 108; Biocomp) and ribosomes were purified by centrifugation at 100,000 x g at 4°C for 15 h in an SW41 rotor. The gradient was then pumped out using a peristaltic pump while measuring A254 and fractions containing 80S were collected and ribosomes were again pelleted by ultracentrifugation at 100,000 × g at 4°C for 18 h in a Ti-60 Beckman rotor. To obtain highly pure ribosomal subunits, the above described purification procedure was repeated, and finally purified 80S ribosomes were diluted in buffer A.
Northern blot analysis
Northern blotting was performed as described before [11] using 15 µg total RNA extracted from wt or mutant cells grown under the described stress conditions or utilizing RNA obtained from pooled density gradient fractions. All membranes were hybridized to a 32P-5ʹ-end-labelled LNA probe (Exiqon) complementary to the rancRNA_18 sequence.
Metabolic labelling
For metabolic labelling, yeast spheroplasts [28] were prepared and experiments were performed as previously described [11]. In short, 135 µl yeast spheroplasts were pelleted (4,000 rpm, 5 min, 4°C) and resuspended in 180 µl ice-cold 1 M sorbitol. 1 µl synthetic RNA (100 pmol/µl) was added, the sample transferred to an electroporation cuvette (EP-102-2 mm gap; Cell ProjectsTM) followed by electroporation (Voltage: 1,500 V, Capacitance: 25 μIF, Resistance: 200 Ω). Immediately, 1 ml of cold YPD/1 M Sorbitol was added, and the whole sample was divided into two equal portions of 450 µl and transferred to a fresh 1.5 ml Eppendorf tube containing 1 µl of 35S methionine (1,000 Ci/mmol, 10 mCi/ml). The samples were incubated at 30°C for 1 h, and proteins were precipitated by the addition of 1 ml 20% TCA and incubation at 95°C for 20 min or the spheroplasts were pelleted, resuspended in 2x SDS sample buffer and cooked for 5 min at 95°C. Labelled proteins were separated on a 12% SDS polyacrylamide gel and quantified by phosphorimaging. Alternatively, 35S labelled proteins were also quantified directly after TCA precipitation by filtrating the sample through glass microfibre filters (24 mm in diameter, WhatmanTM) and subsequent liquid scintillation counting.
Filter binding studies and competition experiments
For binding experiments, 5 pmol of yeast 80S, 60S and 40S ribosomal particles were isolated and incubated with 4 pmol 5ʹ 32P-end-labelled synthetic RNA together with binding buffer (20 mM Tris acetate pH 7.6, 0.1 M KOAc, 5 mM Mg(OAc)2) in a total reaction volume of 25 μl. Reactions were incubated for 30 min at room temperature and subsequently filled up with binding buffer to a total volume of 200 μl and filtered through a dot blot-filtering device (Minifold I, 96 well Dot-Blot System, Whatman). On the nitrocellulose membrane, it is expected that only larger particles are bound, while on the positively charged nylon membrane the flow through should be bound. Signals were visualized with a phosphorimager (FLA-3000; Fuji Photo Film or Typhoon FLA 9500; GE Healthcare) and analysed with AIDA Image Analyse software or ImageQuant TL 8.1 (GE Healthcare). For the competition assays, 5 pmol of S. cerevisiae 80S ribosomal particles were incubated in a total volume of 25 μl with 1 μl 5ʹ γ-32P-labelled synthetic RNA (4 pmol/μl) and increasing amounts (5 to 100 pmol) of unlabelled poly(U) or yeast bulk tRNA in binding buffer. Reactions were incubated at room temperature for 30 min and treated as described above. For the P-site tRNA competition, experiments were performed with 5 pmol of S. cerevisiae 80S ribosomal particles programmed with 20 pmol heteropolymeric mRNA (coding for MFKSIRYV [29]) and prebound with 5ʹ γ-32P synthetic RNA (4 pmol/μl) in binding buffer in a total volume of 25 µl and incubated at room-temperature for 15 min. Increasing amounts of deacylated tRNAPhe (5–50 pmol) were added as competitor. Reactions were incubated for another 15 min at room temperature and treated as described above. For the A-site tRNA competition, experiments were performed with 5 pmol of S. cerevisiae 80S ribosomal particles (programmed with 20 pmol poly(U) or 50 pmol heteropolymeric mRNA [29], prebound with 20 pmol of deacylated tRNAPhe (or 50 pmol deacylated tRNAfMet in case of heteropolymeric mRNA) in binding buffer in a total volume of 25 µl and incubated at room-temperature for 15 min in order to block the P-site. Subsequently, 1 pmol of N-acetyl-[3H]Phe-tRNAPhe (15,000 cpm/pmol) was bound to the A-site in the presence of increasing amounts (5–200 pmol) of unlabelled rancRNA_18. The reactions were then incubated for another 15 min at room temperature and finally filtrated and treated as described above.
Reverse-transcription PCR
Total RNA was isolated with TRI Reagent (Zymo Research) and reverse-transcription of 1 µg RNA was performed with Superscript II (Thermo Fisher Scientific) according to the manufacturer’s protocol using random hexamer primers. The produced cDNA was amplified with Taq polymerase using a standard PCR protocol.
Microscale thermophoresis (MST)
The measurement method is based on the directed movement of molecules along a temperature gradient, an effect termed ‘thermophoresis’. A local temperature difference ΔT leads to a local change in molecule concentration (depletion or enrichment), quantified by the Soret coefficient ST: chot/ccold = exp(-STΔT). For the measurement, the Monolith N115 from Nanotemper has been used. The experiments were done by using 10 µM Cy5 labelled rancRNA_18 diluted in MST buffer (provided by Nanotemper) and a serial dilution of S. cerevisiae 80S ribosomes starting at 2.5 µM f.c. The binding was performed in binding buffer (20 mM Tris acetate pH 7.6, 0.1 M KOAc, 5 mM Mg(OAc)2). Measurements were performed according to the manufacturer`s instructions.
Crosslinking and primer extension
For crosslinking experiments, synthetic rancRNA_18 carrying two photo-reactive 4-thio-uridine (4tU) residues was 5ʹ 32P-labelled (see Supplementary Table S1 for RNA and DNA oligomer sequences). 50 pmol of radioactively labelled synthetic RNA was incubated with 5 pmol of yeast 80S ribosomal particles and the reaction was performed in binding buffer at room temperature for 30 min in a final volume of 20 μl. Subsequently, samples were placed on ice and photo-irritated with UV light of 366 nm wavelength for 15 min, followed by PCI extraction and EtOH precipitation. The crosslinked protein samples were analysed by 12% tricine gels or by SDS-PAGE (5%) To detect the site(s) of crosslinking to rRNA, unlabelled 4tU-rancRNA_18 was crosslinked to S. cerevisiae 80S ribosomes as described above. Subsequently, primer extension analysis was employed on purified rRNAs as previously described [9].
Puromycin reaction
Peptide bond formation activity was tested using a peptidyl transferase assay. Puromycin reactions were performed in 25 μl binding buffer (20 mM Tris acetate pH 7.6, 0.1 M KOAc, 5 mM Mg(OAc)2 containing 10 pmol of S. cerevisiae 80S ribosomes, 0.8 pmol N-Acetyl-[3H]Phe- tRNAPhe (15,000 cpm/pmol), 10 pmol poly(U) or heteroplymeric mRNA and, if indicated, 100 pmol of synthetic rancRNA. After a pre-incubation for 15 min at 37°C, the reaction was initiated by the addition of puromycin to a final concentration of 1 mM. The reaction was incubated for 30 minutes at 37°C. Reactions were stopped by the addition of 5 μl 10 M KOH and incubation at 37°C for 20 minutes. After the addition of 100 μl 1 M KH2PO4 the reaction product (N-acetyl-[3H]Phe-puromycin) was extracted with 1 ml cold ethyl acetate by vigorously mixing for 1 min and subsequent centrifugation at 13,000 rpm for 1 minute. After centrifugation, 800 μl of the upper organic phase were measured by liquid scintillation counting.
Spheroplasts, polysome profiling and RNA extraction
In order to generate samples for RNA sequencing, 240 OD600 of exponentially growing cells were harvested by centrifugation and spheroplasts were prepared by incubation with Zymolyase 100 T (Zymo Research) [28]. Equal amounts of spheroplasts (each corresponding to 80 OD600 of cells) were electroporated either with or without 3.6 nmol synthetic rancRNA_18 or rancRNA_18-M2 and a small aliquot was transferred to YPD containing 1 M sorbitol to perform metabolic labelling with 35S-methionine for 80 min (labelled culture) [11]. The remaining spheroplasts were immediately resuspended in 40 ml prewarmed YPD containing 1 M sorbitol and recovered in parallel for 80 min in the absence of 35S-methionine (unlabelled culture). Metabolic labelling reactions were analysed by SDS-PAGE and autoradiography, whereas polysome profiling was performed with the unlabelled culture. To this end, 100 µg/ml cycloheximide was added to the culture and the cells were poured on frozen YPD medium containing 1 M sorbitol and 100 µg/ml cycloheximide, harvested by centrifugation and washed with 1 M sorbitol containing 100 µg/ml cycloheximide. Cell lysis was performed in lysis buffer (20 mM HEPES-KOH pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 0.5 mM DTT, 1 mM PMSF, 100 µg/ml cycloheximide) with glass beads using FastPrep24. Cleared and adjusted lysates were loaded on a 10–40% sucrose gradient prepared in lysis buffer without PMSF and centrifuged for 2 h 15 min at 39,000 rpm and 4°C in an SW41 Ti rotor (Beckman Coulter). Gradients were pumped out with 50% sucrose and polysome profiles were monitored at 254 nm using the Brandel density gradient fractionation system. Total and polysome-associated RNA was isolated with the hot-phenol method [30].
Analysis of RNAseq data
The used reference for the high throughput sequencing dataset was the strain: BY4742, downloaded from the Saccharomyces Genome Database [31]. The sequenced raw reads were aligned to the reference using Hisat2 software with the default settings [32]. The reads aligned with high rates to the reference genome ~ 95% across all samples.
The reads aligned over genomic features were counted using featureCounts software [33]. Downstream analysis was performed with R scripts. DESeq2 [34] was used for reads normalization across libraries and differential abundance by contrasting polysomal mRNA (P) to total mRNA (T) (for determining translation efficiency TE). For each mRNA in a particular condition (either mock, rancRNA_18 or rancRNA_18-M2), a P/T ratio of reads was calculated, followed by a log2 transformation. A negative P/T ratio indicated that the corresponding mRNA was poorly translated while a positive ratio indicated efficient translation. r2 coefficients for fold-changes between the mock and the rancRNA_18 (or rancRNA_18-M2) data were determined by the double-sided t test for association between paired samples, using Pearson’s product moment correlation coefficient [35].
Cryo-EM
For Cryo-EM studies, we used gradient purified 80S ribosomes from trm10Δ cells in a final concentration of 0.1 µg/µl together with 800 times molar excess of rancRNA_18 in binding buffer. Binding was done at room temperature (RT) for 30 min. Vitrification of the sample for cryo-EM was performed using an FEI Vitrobot Mach 4 on Quantifoil 2/1 grids with or without 2 nm carbon film at 4°C and 100% chamber humidity. The grids were briefly glow discharged in a Balzers CTA 010 prior to plunging (15 seconds, 10–15 mA). 3 µl of the ribosome sample at 0.1 mg/ml was added to the grids and then the Vitrobot procedure was performed with 10 seconds waiting time, blot force −1 and 2–4 seconds blot times. Grids were stored in liquid nitrogen until acquisition.
The acquisition was performed on an FEI Tecnai F20 microscope equipped with a Falcon III direct detector using a Gatan 626 cryo-holder. All acquisitions were performed in nanoprobe mode for both overview and exposure. Microscope alignment was carefully adjusted for maximum resolution. Parallel illumination was set for the exposure, focus and drift modes. Refilling of the cryo-holder was performed using an in-house built robot every 3 h. In order to obtain the best possible results, beam alignments (including astigmatism and coma-free) were performed every 3–4 h during daily acquisition and after 7–8 h overnight acquisition. As rough, on the fly control for the alignment images on the carbon area were acquired. An acceptable alignment was considered when we could observe thon rings up to 3 Å in the Fourier Transform. Images were acquired in movie mode using FEI EPU and a total dose of 65–75 e-/Å2 with a pixel size of 1.052 (100,000x magnification).
Image processing was performed using Relion [36], Motioncor [37] and Ctffind [38]. Following motion correction and CTF estimation, the images were selected based on the resolution estimated by ctffind followed by visual inspection. Relion automatic picking was performed using classes obtained from 2000 manually picked particles. Following manual and automatic picking, particles were extracted with a box size of 450 pixels and reference-free 2D classification was performed (100 classes, 25 iterations, 360 Å mask, default Relion parameters). Several approaches were tested to separate different conformations present in the sample. Best results were achieved by performing an initial refinement run with the particles belonging to the good classes from the initial 2D classification run (427,367 particles for the Control sample and 507,761 particles for the rancRNA_18 sample). The initial round of refinement was performed with a reference map low pass filtered at 50 Å and a soft circular mask with a diameter of 360 Å. After the refinement of all the particles, maps with a final resolution of 6.5 Å and 5.2 Å were obtained for control and rancRNA_18 samples, respectively. Following refinement, all the particles were subjected to a round of 3D Classification with their respective reference. The particles were classified in 10 classes without image alignment, with a soft circular mask of 360 Å. All the classes were compared and inspected. The particles from the four best classes from each sample were then selected and separately refined as 80S or with the density corresponding to the 40S subtracted from each image using the implemented particle subtraction procedure from Relion [39] and a mask for the 40S subunit. All segmentations of the maps (classes and refinements) were performed in Chimera [40] and masks were created using Relion mask creation procedure. In addition, for refinements of the 60S particles, soft masks made from the maps of the 3D classification were used instead of only a soft circular mask. A separate round of refinement of all subtracted particles excluding the particles from tRNA containing classes yielded maps with final resolutions of 4.4 Å and 5.1 Å for rancRNA_18 and control samples, respectively.
Multibody refinement was performed in Relion as four rigid bodies after binning by 2. In brief, all particles were processed into a consensus refinement and four masks were created in Chimera to cover the large subunit (LSU – body 1), the small subunit (SSU – body 2) the head domain (Head – body 3) and the L1 stalk (body 4). The multibody refinement was performed with respect to the LSU on the main classes identified by 3D classification. Principal component analysis performed by Relion on the orientation of the bodies in respect to the LSU resulted in a series of ten maps for the first principal components. Movies representing motions described by the principal components were created using Chimera. In preparing the figures emphasizing the observed motions, only the extreme conformations (first and last) are represented. The movement of the three main components of selected classes was quantified as follows. First, the distance of selected components between the two extreme conformations (‘displacement range’ in Supplementary Table S2) was measured using Chimera ‘Distance’ feature. Subsequently, the weighted average displacement was calculated by integrating the element-wise product of the conformation frequency distribution and the absolute distance of the conformation to the mean conformation.
Results
rancRNA_18 has one ribosomal binding site on the large subunit
In our previous study, we showed that rancRNA_18 derives from the TRM10 mRNA transcript, is primarily bound to 80S and 60S ribosomal particles in vivo and is capable of inhibiting protein synthesis both in vivo and in vitro [11]. Yeast cells lacking rancRNA_18 expression showed an extended lag phase under hyperosmotic conditions and genetic analyses demonstrated that Trm10, a tRNA methyltransferase, does not contribute to rancRNA_18 functionality during high salt stress. During hyperosmotic conditions, rancRNA_18 shifts from the 60S subunits and 80S monosomes into the polysomal fractions. Expression of a mutant version of rancRNA_18 harbouring three nucleotide substitutions (referred to as rancRNA_18-M2) failed to affect protein biosynthesis [11]. To better characterize the rancRNA_18 interaction with the ribosome, binding saturation and microscale thermophoresis experiments with Cy5-labelled rancRNAs were performed employing ribosomes isolated from the trm10Δ strain, thus guaranteeing a vacant binding site (Supplementary Figure S1A and B). The inhibitory activity of Cy5-labelled rancRNA_18 on translation was verified by metabolic labelling and turned out to be similar to the unmodified rancRNA_18 control (Supplementary Figure S1C). Microscale thermophoresis allows to accurately measure dissociation constants based on the phenomenon that unbound and ribosome-bound Cy5-labled rancRNA_18 moves differently in the established microscale temperature gradient [41]. The in vitro binding assays revealed a single rancRNA_18 binding site per ribosome and an apparent Kd of 0.6 µM (Supplementary Figure S1B). This binding affinity is in a physiologically reasonable range, considering the previously determined in vivo concentrations of rancRNA_18 of 1.1 µM [11]. The mutated rancRNA_18-M2, which failed to inhibit protein synthesis and metabolic labelling, had an almost 30-fold reduced binding affinity to yeast ribosomes (Supplementary Figure S1B).
rancRNA_18 is a bona fide ncRNA
rancRNA_18 is located within the TRM10 gene, which encodes a tRNA methyltransferase [42]. Deletion of the entire TRM10 locus resulted in the loss of rancRNA_18 expression as well as of the tRNA methyltransferase activity and resulted in a yeast strain with retarded growth rate under hyperosmotic conditions [11]. Subsequent genetic investigations demonstrated that the growth phenotype of the trm10Δ strain originated from the lack of the rancRNA_18 rather than from the missing methyltransferase activity [11]. To unravel the minimal genomic region required to express functionally active rancRNA_18, nine different 3ʹ truncated versions of the TRM10 locus were expressed from plasmids in the trm10Δ strain. In all truncated versions the endogenous TRM10 promoter was used and the ATG start codon of the TRM10 gene was mutated to CTG in order to render the transcripts non-translatable [11]. Northern blot analysis demonstrated that the shortest construct that gave rise to rancRNA_18 is 105 nucleotides long (when position 1 is defined as the first nucleotide of the TRM10 start codon) (Figure 1A). Concomitantly to the rancRNA_18 appearance a larger band became evident in northern blot analyses (ref. 11 and Figure 1A), which possibly represents a 49 nucleotide long precursor as determined by 3ʹ RACE (rapid amplification of cDNA ends) experiments (Supplementary Figure S2A). This 49-mer, however, does not affect translation as assessed by metabolic labelling (Supplementary Figure S2B). Thus, a transcript of about 100 residues is necessary and sufficient for proper processing of rancRNA_18.
Figure 1.
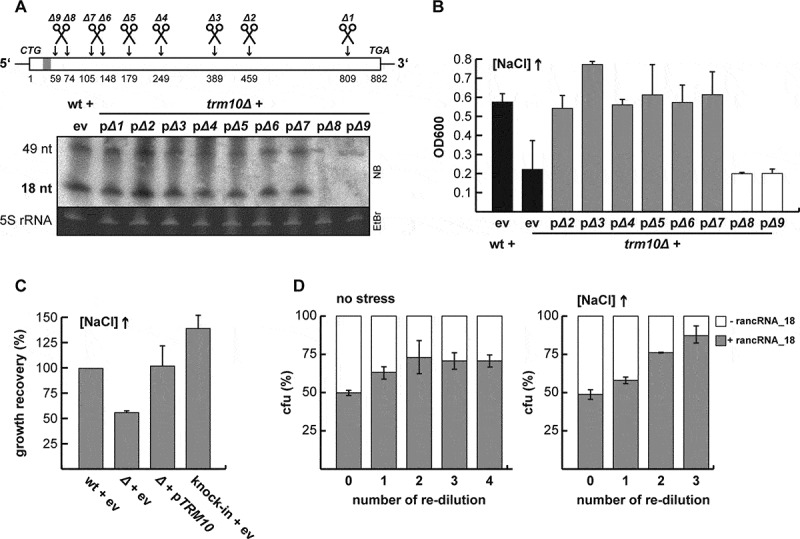
rancRNA_18 is a bona fide ncRNA in S. cerevisiae. (A) schematic overview of the truncated versions (Δ1 – Δ9) of the TRM10 locus. the grey bar close to the 5ʹ end depicts the location of rancRNA_18. the presence of rancRNA_18, and the putative 49-mer processing intermediate, was assessed via northern blot analysis (5S rRNA served as loading control). all constructs were expressed from a plasmid in the trm10Δ strain, whereas the original start codon was replaced by a CTG codon to prevent translation of the transcripts (11). ev, empty vector control. (B) growth recovery after 1,050 minutes under hyper-osmotic conditions of wildtype (wt) and trm10Δ cells (black bars) was compared to trm10Δ cells expressing truncated versions of the TRM10 locus. n = 2, mean ± SD. (C) growth of the knock-in strain that expresses the truncated TRM10 construct Δ6 from a novel genomic location (knock-in + ev) in high salt media was compared to the wt cells or to the strain carrying either no additional plasmid (ev) or the plasmid encoding the full-length TRM10 locus (pTRM10). n = 6, mean ± SD. (D), growth competition between a mixture of equal starting amounts of two strains differing only in the absence (white) or presence (grey) of rancRNA_18 expression in normal (no stress) or high salt containing media. cultures were grown to stationary phase and colony forming units (cfu) determined. cultures were then re-diluted and cfu determined at each stationary phase. n = 3, mean ± SD
To test if the rancRNA_18 molecules expressed from truncated TRM10 constructs are biologically active during hyperosmotic stress conditions, growth recovery at high NaCl concentrations was performed as described previously [11]. All cells expressing the rancRNA_18 from truncated TRM10 constructs were able to recover under high salt conditions like the full-length wild type control (Figure 1B). In contrast, cells lacking rancRNA_18 expression (truncated construct Δ9) could not recover from high salt stress in the same time frame and thus behaved like the trm10Δ strain (Figure 1B).
In order to investigate if the genomic localization of rancRNA_18 expression matters and to also physically separate the rancRNA_18 gene construct from the TRM10 locus, we knocked-in the truncated construct Δ6 (Figure 1A) into the trm10Δ strain. According to a previous study that characterized chromosomal integration sites in S. cerevisiae [27], chromosome V was chosen as a suitable insertion of the Δ6 construct. Expression of the chromosomally integrated Δ6 construct was verified using RT-PCR with construct-specific primers (Supplementary Figure S3). To ascertain the functionality of rancRNA_18 in the Δ6 knock-in strain, growth was monitored at elevated salt concentration. While wild type cells, as well as trm10Δ cells complemented with the wt TRM10 gene expressed from a plasmid (pTRM10) were able to resume growth in hyperosmotic medium, cells lacking rancRNA_18 were not (Figure 1C). Importantly, Δ6 knock-in cells expressing rancRNA_18 from the new host chromosome V (but lack any Trm10 tRNA methyltransferase expression), were also tolerant to high salt stress (Figure 1C). Thus, rancRNA_18 expression confers efficient growth under hyperosmotic stress regardless of its genomic location and is independent of the TRM10 methyltransferase locus. Therefore, the Δ6 sequence encoding rancRNA_18 behaves like a bona fide ncRNA gene.
To examine if the presence of rancRNA_18 has an influence on the biological fitness of yeast cells, we performed growth competition experiments between two strains, which only differ by the presence or absence of rancRNA_18. Therefore, we made use of the Δ6 knock-in strain, which expresses a functionally active rancRNA_18 (Figure 1C), and transformed it with a plasmid encoding the M2_TRM10 gene locus. This construct contains three synonymous mutations in the TRM10 open reading frame shown not to alter tRNA methyltransferase activity of the encoded enzyme, but destroying rancRNA_18 activity in vitro and in vivo (Supplementary Figure S1 and ref. 11). The other strain used in the growth competition set up was the trm10Δ strain transformed with the same plasmid harbouring the M2_TRM10 construct. In contrast to the first strain, this strain only produces a fully functional Trm10 enzyme but lacks any functional rancRNA_18 transcripts. For the growth competition experiment, both strains were mixed in a 1:1 ratio normalized for OD600 and growth was monitored in normal and high salt media. At stationary phase, cells were plated and colony forming units were determined. Stationary phase cultures were subsequently re-diluted into fresh media and allowed to grow to stationary phase again. This procedure was repeated in total three times. The obtained data suggest that after several re-dilutions, the strain expressing functional rancRNA_18 out-competed the strain lacking rancRNA_18 indicating that the presence of this ncRNA increases biological fitness (Figure 1D). This growth advantage was apparent even under unstressed conditions but became much more pronounced in high salt media (Figure 1D). In summary, the data obtained demonstrate that rancRNA_18 is a functional ncRNA that provides a physiological advantage to S. cerevisiae and reveals its chromosomal locus as bona fide ncRNA gene.
The RNase III homolog Rnt1 is involved in rancRNA_18 biogenesis
In yeast, Rnt1 is the only known RNase III [43] and is known to be involved in the processing of small nucleolar RNAs (snoRNAs), ribosomal RNAs (rRNAs) and has been shown also to cleave RNA in trans analogous to Argonaut proteins [44]. Argonaut endonucleases, however, are absent in S. cerevisiae. Rnt1 is an essential gene in this strain background and can therefore not be deleted from the chromosome. Thus, in order to investigate if Rnt1 is involved in the processing of rancRNA_18 from the TRM10 mRNA transcript, we made use of a tetracycline-inducible depletion strain of Rnt1. We first checked the growth rate of this strain after tetracycline-mediated Rnt1 depletion and compared it to the un-induced strain as well as to the wt strain. Cells depleted of Rnt1 show a severely compromised growth phenotype with a 7-fold slower rate compared to both control strains (Figure 2A,B). To explore if Rnt1 is involved in rancRNA processing, total RNA was isolated and tested for rancRNA_18 expression by northern blot analysis. Upon Rnt1 depletion loss of the rancRNA_18 signal as well as of the 49-mer precursor signal became evident (Figure 2C). To investigate whether this rancRNA_18 loss is specific to Rnt1 depletion or the consequence of general cell stress upon depletion of an important RNase, we included the two RNase deletion strains lacking either XRN1 or RRP6 in the northern blot analyses. In both of these strains, the expression of the rancRNA_18 was not influenced, indicating that Rnt1 is specifically involved in the biogenesis of the rancRNA_18 (Figure 2C).
Figure 2.
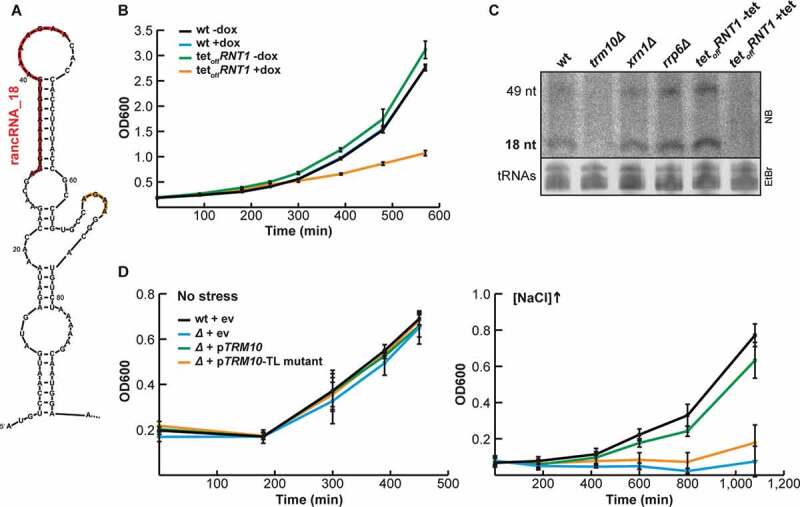
Rnt1 is involved in rancRNA_18 biogenesis. (A) secondary structure prediction of the 5 ’end of the TRM10 mRNA with the rancRNA_18 sequence indicated in red and a downstream AGNN tetraloop in orange. (B) growth analysis of a tetracycline (tet)-inducible rnt1 depletion strain (tetoff) in the presence (orange) or absence (green) of doxycycline compared to wildtype (wt, black and blue). n = 6, mean ± SD (C) northern blot analysis on total RNA isolated from wt, trm10Δ, xrn1Δ, rrp6Δ and Rnt1::tet cells either in the absence (-tet) or presence (+tet) of tetracycline. the locations of rancRNA_18 and the putative 49 nucleotides long precursor are indicated. the ethidium-bromide stained tRNAs serve as a loading control. (D) growth of the tetraloop mutant strain (orange) compared to wildtype (black) and trm10Δ (blue) strains was monitored either in normal (left) or high salt media. as a complementation control the trm10Δ strain transformed with a plasmid containing the wt TRM10 locus (green). n = 3, mean ± SD
The experiment with truncated versions of the TRM10 locus revealed that a minimal length of 105 nucleotides is needed for proper rancRNA_18 expression in vivo (Figure 1A,B). Secondary structure prediction of this 105 residues long TRM10-mRNA fragment suggested rancRNA_18 of being located in a hairpin structure followed by a short hairpin structure closed by a 5ʹ-AGAA-3ʹ tetraloop (Figure 2A). A tetraloop of the 5ʹ-AGNN-3ʹ family has been described as one of the primary RNA binding motifs of the Rnt1 enzyme [45]. To corroborate the finding about a possible involvement of Rnt1 in rancRNA_18 biogenesis, the tetraloop was mutated from 5ʹ-AGAA-3ʹ to 5ʹ-GAAA-3ʹ in the context of the Δ6 construct (Figure 1A). Both the wt and the tetraloop mutant (TL) were expressed from a plasmid and growth was monitored at high salt concentrations. Under these conditions, rancRNA_18 expression is crucial for optimal growth (ref. 11 and Figure 1B,D). Under these hyperosmotic conditions, the 5ʹ-GAAA-3ʹ tetraloop mutant strain loses its ability to grow efficiently and behaves essentially like the trm10Δ strain (Figure 2D). Importantly, performing the same experiment in normal media, conditions under which rancRNA_18 is not pivotal for effective growth [11], all strains, including the 5ʹ-GAAA-3ʹ tetraloop mutant and the trm10Δ strain, showed wt-like doubling times (Figure 2D). These data are compatible with the view of Rnt1 being involved in processing rancRNA_18 from the TRM10 mRNA transcript.
mRNA-Seq reveals rancRNA_18 as global translation inhibitor
Our recent study suggests that rancRNA_18 is needed for a fast reduction of translational activity in response to hyperosmotic stress, which allows adaptation processes to be established [11]. Upon high salt stress rancRNA_18 has been shown to shift from the 60S ribosomal subunits and 80S monosomes into the polysomal fractions accompanied by reduced translational activities. Here we wanted to clarify if rancRNA_18-mediated translation inhibition happens on a global scale or whether certain mRNAs are preferentially translated by rancRNA_18-containing ribosomes under high salt conditions. If rancRNA_18 is a global inhibitor of protein production affecting all mRNAs to the same extent, the same set of mRNAs are expected to be found in polysomes either in the presence or absence of rancRNA_18. On the other hand, if the association of rancRNA_18 to ribosomes and polysomes leads to the production of salt stress-relevant proteins, the mRNA pool in polysomes will differ in the presence of rancRNA_18.
In order to investigate which of the two possible scenarios is true, spheroplasts derived from trm10Δ cells were electroporated with synthetic rancRNA_18 or the inactive rancRNA_18-M2 variant. Northern blot analysis and metabolic labelling confirmed that under these conditions the synthetic rancRNA_18 associates with 80S and polysomes and inhibits protein synthesis (Figure 3A,B), thus mimicking high salt stress conditions [11]. Subsequently, the mRNA pools from the polysomal fractions as well as from total mRNA were isolated and analysed by RNA-Seq. mRNA levels were quantified from two biological replicates and the translation efficiency (log2 fold-changes of polysomal mRNA/total mRNA) was determined. Comparison of the translation efficiencies between the sample electroporated in the absence of any synthetic RNA (mock) and the rancRNA_18 sample showed a high correlation (r2 = 0.699) (Figure 3C). Similar unchanged translation efficiencies were obtained when mRNA-Seq data were compared between the mock and the rancRNA_18-M2 samples (Figure 3D). These mRNA-Seq analyses do not support the scenario where rancRNA_18 induces specialized translation. The data are therefore compatible with the view that rancRNA_18 inhibits protein biosynthesis on a global scale.
Figure 3.
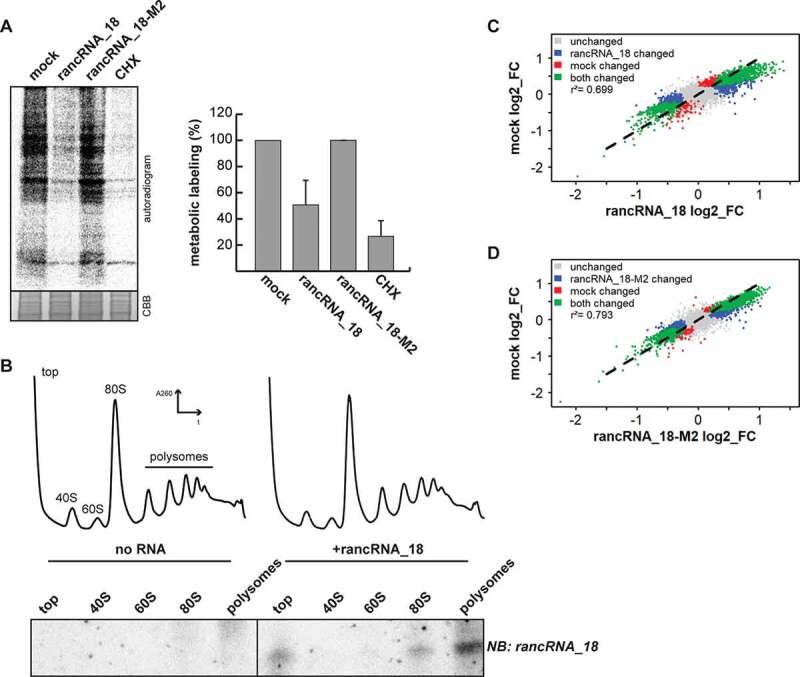
rancRNA_18 is a global translational inhibitor. (A) spheroplasts from trm10Δ cells were electroporated with synthetic rancRNA_18 or the mutant version rancRNA-18-M2 and used for metabolic labelling. electroporation in the absence of synthetic RNA (mock) and samples containing the translational inhibitor cycloheximide (CHX) served as controls. radioactively labelled proteins were analysed with SDS-PAGE and autoradiography. coomassie brilliant blue (CBB) stained proteins serve as loading control. signals were quantified and normalized to the no RNA (mock) control. n = 2, mean ± SD. (B) polysome profiling was performed with recovered spheroplasts, RNA was isolated from collected fractions and separated on a denaturing polyacrylamide gel. the distribution of the synthetic rancRNA_18 in the retrieved fractions was visualized by northern blot analysis. (C and D) scatter plots showing translation efficiencies (log2 fold-changes; FC) of sequencing data from total (T) and polysome-associated (P) mRNA from spheroplasts electroporated with no RNA (mock), synthetic rancRNA_18 or rancRNA_18-M2, respectively. different colours indicate mRNAs that show a different association to polysomes in both conditions (green), in samples electroporated with rancRNA_18 only or rancRNA_18-M2 only (blue), in mock samples only (red), or in none of the conditions (grey). p-values for control vs rancRNA_18 data and control vs rancRNA_18-M2 data were <2.2 •10−16. correlation coefficients are indicated as r2
RancRNA_18 affects tRNA binding to the ribosome
Since rancRNA_18 globally inhibits protein biosynthesis in an efficient manner (Figure 3A), we assumed that it binds to a functionally important region on the ribosome. To gain insight into ribosome association behaviour, in vitro binding competition experiments were performed with radiolabeled rancRNA_18 and unlabelled mRNA or tRNAs. Constant levels of gradient-purified yeast 80S ribosomes were incubated with 32P-labelled rancRNA_18 and increasing amounts of in vitro transcribed LEU2 mRNA or yeast bulk tRNA and analysed by filter binding. While the binding affinity of the rancRNA_18 was not affected by mRNA (Figure 4A), deacylated yeast tRNAs markedly reduced ribosome binding with an IC50 of 1.9 µM (Figure 4B). The reciprocal experiment, when 32P-labelled tRNAs were chased by increasing amounts of unlabelled rancRNA_18 showed the same competition effect (Supplementary Figure S4A). On the other hand, rancRNA_18-M2 failed to compete with tRNA for ribosome binding (Supplementary Figure S4B), which highlights the sequence specificity of this effect. To explore the competition in more detail, we designed experiments to investigate specific tRNA binding sites on the ribosome. For the P-site competition experiment, increasing amounts of deacylated tRNAPhe were bound to ribosomes programmed with a heteropolymeric mRNA analogue harbouring one unique Phe codon in the presence of radiolabeled rancRNA_18 (Figure 4C). Even at the highest tRNAPhe concentration, rancRNA_18 binding was unaffected, suggesting no binding overlap at the P-site. Unaffected peptide bond formation activities in the presence of rancRNA_18 between the peptidyl-tRNA analogue Ac-[3H]Phe-tRNAPhe and puromycin in P- and A-site, respectively, support this interpretation (Figure 4D). To assess possible A-site binding effects we constructed a pre-translocation ribosomal complex (Figure 4E). To this end, the P-site of poly(U)-programmed ribosomes was first blocked by saturating amounts of deacylated tRNAPhe followed by binding of Ac-[3H]Phe-tRNAPhe to the A-site either in the absence or presence of increasing amounts of unlabelled rancRNA_18 (Figure 4E). Additionally, an analogous pre-translocation complex was constructed employing a physiologically more relevant heteropolymeric mRNA possessing unique Met and Phe codons with deacylated tRNAfMet and Ac-[3H]Phe-tRNAPhe in the P- and A-sites, respectively (Supplementary Figure S5). The authenticities of the pre-translocation complexes using two different mRNAs were verified by the puromycin reaction (Supplementary Figure S5A and B). With the addition of increasing amounts of rancRNA_18 binding of Ac-[3H]Phe-tRNAPhe to the A-site was markedly reduced in both cases in a dose-dependent manner with apparent IC50 values between 0.1 and ~1 µM (Figure 4E and Supplementary Figure S5C). Also in this setting, the mutant version rancRNA_18-M2 did no effect on tRNA/ribosome complex formation (Fig. 4F). Binding competition experiments with cycloheximide, a small molecule translation inhibitor, which binds to the ribosomal E-site and is a known translation elongation blocker [46] did not show any effect either (Supplementary Figure S4C). Collectively the data suggest that binding of rancRNA_18 to the yeast ribosome has an effect on A-site tRNA binding.
Figure 4.
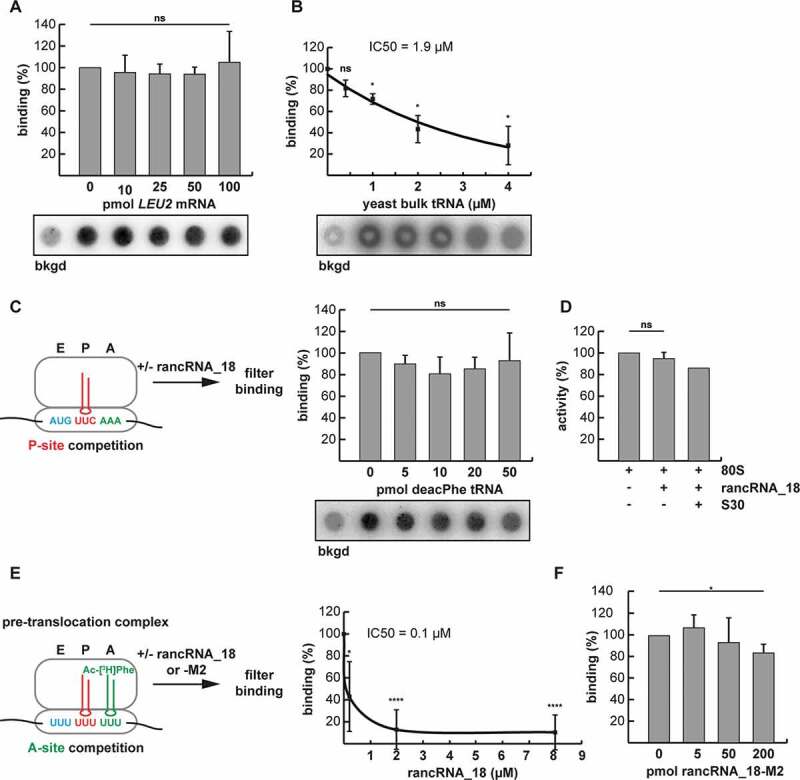
RancRNA_18 has an influence on A-site tRNA binding. (A) dot blot filter binding analysis was used to analyse binding competition of radiolabeled rancRNA_18 with increasing amounts of LEU2 mRNA to 5 pmol purified yeast ribosomes. the signal in the absence of 80S particles (bkgd) was subtracted. ns, not significant. n = 2; mean ± SD. (B) as in (A) but with increasing amounts of yeast bulk tRNA. The rancRNA_18 binding data (n = 3; mean ± SD; * p < 0.05) are blotted as function of increasing tRNA competitor concentration and the trend-line is depicted. (C) dot blot approach to investigate ribosome binding competition between radiolabeled rancRNA_18 and P-site bound deacylated tRNAPhe (red). n = 3; mean ± SD. (D) peptidyl transfer between P-site bound N-acetyl-[3H]Phe-tRNAPhe and A-site located puromycin was assessed in the absence or presence of rancRNA_18. S30 cell extract was used as a control if additional factors are needed for rancRNA_18 functionality. n = 2; mean ± SD. (E) A pre-translocation complex formed on poly(U) mRNA analogues carrying deacylated tRNAPhe in the P-site (red) and N-acetyl-[3H]Phe-tRNAPhe in the A-site (green) was used to investigate effects of unlabelled rancRNA_18 addition on tRNA occupancy. extend bound A-site tRNA was assessed via filter binding. n = 6; mean ± SD; * p < 0.05, **** p < 0.0001. (F) as in (E) but in the presence of increasing amounts of rancRNA_18-M2
Cryo-EM structure of 80S ribosomes with bound rancRNA_18
In order to better understand the changes to the ribosome structure upon binding of rancRNA_18 and to characterize the rancRNA_18 binding site in more detail, we decided to directly visualize 80S particles using cryo-EM [47]. To this purpose we used gradient-purified 80S ribosomes from the trm10Δ strain, to which we added rancRNA_18 carrying the 4-thio-U modifications and crosslinked the complex at 366 nm. As negative control, ribosomes without the addition of rancRNA_18 were used. An initial refinement of all particles showed no large-scale differences between the control and the rancRNA_18 samples. At the obtained resolution of 4.4–5.1 Å the map comparison revealed small changes mostly in the L1 stalk region suggesting the presence of different ribosome conformations (Supplementary Figure S6). Following an initial reference-free 2D classification and 3D refinement, the aligned particles were subjected to a round of 3D classification into eight sub-classes (Supplementary Figure S7). Interestingly, already from this classification, the two datasets showed several differences. On the one hand, in the control sample, three out of the four best classes (~58% of all particles; classes Ia-Ic) had empty A-, P-, and E-sites, and the fourth class (9.8% of all particles; class II) containing density in the P- and E-sites. On the other hand, in the sample containing rancRNA_18, only 23.4% of all particles were empty (class Ia; Supplementary Figure S7). In the rancRNA_18 containing sample, a new class of particles became evident (18.5% of all particles) that showed a faint extra density between the P- and E-tRNA binding site regions (class III). Two minor classes populated by 13.7% and 12%, respectively, showed density in the P- and E-sites (class II) or only in the P-site (class IV) (Supplementary Figure S7). Comparison of completely empty ribosomes (class I) with particles possessing this rancRNA_18-dependent extra density (class III) showed a slight inward shift of the L1 stalk towards the E-site in particles carrying rancRNA_18 (Supplementary Figure S8). Refinement of class III did not significantly improve the extra density resolution, suggesting that rancRNA_18 is somewhat flexible. The obtained structural data can be interpreted in several ways. Either rancRNA_18 interacts with the 60S particles in a site located in between the P- and E-tRNA binding sites (Figure 5A and class III of Supplementary Figure S7), or the interaction of rancRNA_18 with the large ribosomal subunit leads to a higher percentage of ribosomes locked in conformations with tRNA either in both E- and P-sites or in an intermediate P/E state (classes II and IV of Supplementary Figure S7). While the cryo-EM data alone do not allow discriminating between these scenarios, our biochemical evidence (Figure 4B,C) supports rather the first scenario where rancRNA_18 interacts with the ribosome at a site located in between the empty P- and E-tRNA binding sites (class III in Supplementary Figure S7 and Figure 5B).
Figure 5.
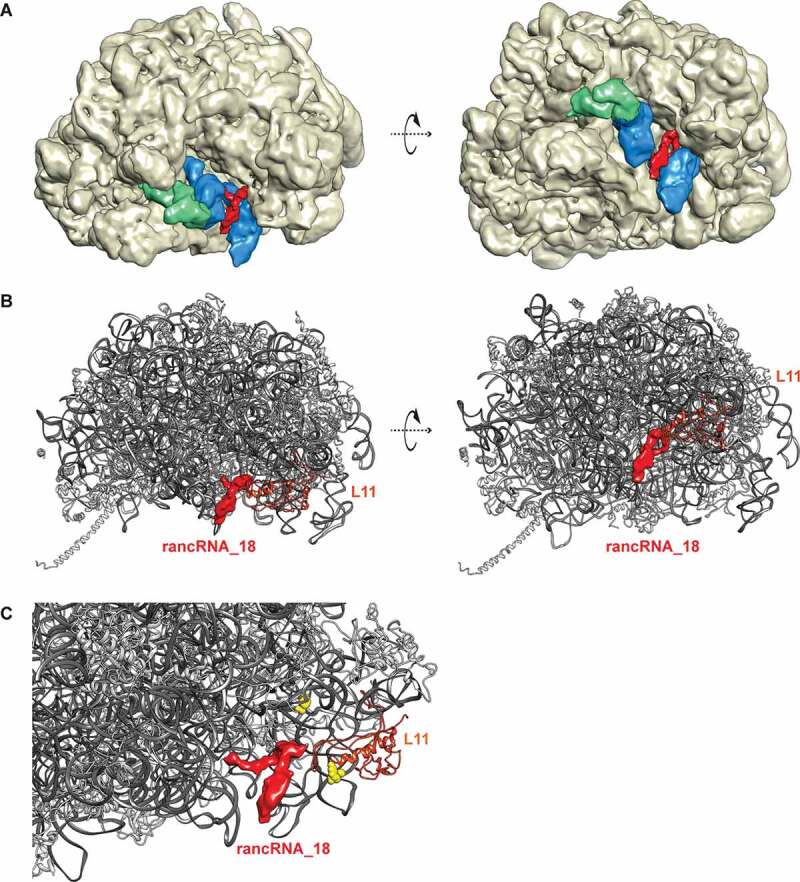
Cryo-EM structure of yeast ribosomes with bound rancRNA_18. (A) structure of the 60S ribosomal subunit (wheat) with highlighted rancRNA_18-dependent extra density (red), overlaid with the observed densities for P- and E-site tRNAs (blue) and the L1 stalk (green) in its closed conformation at 4.4 to 5.1 Å resolution. (B) representation of the 60S ribosomal subunit with rRNA in dark grey and ribosomal proteins in light grey. the observed rancRNA_18-dependent extra density (red), the two observed crosslinking site (C2675 and C2693, yellow),) and the L1 stalk (green) in its slight inward position are highlighted. (C) zoom into the rancRNA_18-dependent extra density (red) in close proximity to the observed crosslinking sites (C2675 and C2693, yellow) and r-protein L11 (uL5) (orange)
To substantiate this interpretation, we decided to employ the newly implemented multibody refinement and principal component analysis of Relion [48] for the class of particles containing the extra density near the 60S E-site tRNA which provided a characterization of the motions in the complex. As control we used the empty class obtained from the dataset of ribosomes incubated with rancRNA_18 (Supplementary Figure S7; class Ia) as well as the empty class from the control dataset obtained in the absence of rancRNA_18. Analysing the behaviour of the particles described by the first three principal components (Supplementary Figure S9) revealed a marked reduction in the dynamics of ribosomes carrying the rancRNA_18-dependent extra density in particular at the L1 stalk (Supplementary Figure S10, Supplementary Movie S1, and Supplementary Table S2). As expected, in both controls the first three principal components describe motions related with the head domain of the small subunit and the L1 stalk region of the large ribosomal subunit. In contrast, the analysis of the rancRNA_18 treated sample showed no significant motion of the L1 stalk density suggesting that the ribosome-bound rancRNA_18 significantly restricts this otherwise highly mobile region. The small subunit head domain of the particles carrying rancRNA_18 behaved similarly to the control.
RancRNA_18 cross-links to the 60S E-site region
In order to confirm that the extra density obtained in the class III particles (Supplementary Figure S7) corresponds to rancRNA_18, cross-linking experiments with rancRNA_18 carrying two 4-thio-U modifications to its ribosomal-binding site were performed. Biological activity of the 4-thio-U modified rancRNA_18 was assessed by a metabolic labelling assay (Supplementary Figure S11). Guided by the tRNA binding competition (Figure 4B,E) and cryo-EM data (Figure 5A,B) we focused the detection of putative rancRNA_18 crosslinking sites via primer extension analysis to the tRNA binding sites on the large subunit rRNA. While no primer extension stops were detected in the 60S A-site region, two rancRNA_18 crosslinking sites were detected at positions C2675 and C2693 located in domain V of 25S rRNA (Fig. 6). Since we cannot exclude the possibility that also ribosomal proteins contribute to the rancRNA_18 binding site, the crosslinking experiment was repeated with 5ʹ-[32P] labelled 4-thio-U rancRNA_18. Subsequent to irradiation of the 80S yeast ribosomes, the r-proteins were separated on an SDS gel and exposed to a phosphorimager screen. A single prominent radiolabeled protein band in the size range of 22–26 kDa became apparent (Supplementary Figure S12). In this size range, r-proteins L1 and L11 (uL1 and uL5, respectively, according to the novel nomenclature of r-proteins [49]) are expected. Displaying the observed rRNA crosslinking sites at 25S rRNA residues C2675 and C2693 on the 3D structure of the yeast ribosome reveals a single putative rancRNA_18 binding site at the E-site region of the large ribosomal subunit close to the base of the L1 stalk. The ribosomal protein that approaches these crosslinking sites most closely is r-protein L11 (uL5) (Figure 5C). Combining the cryo-EM data showing an extra density in the E-site region (class III particles) with the 4-thio-U rancRNA_18 crosslinking results, revealed an immediate proximity of the crosslinking sites at 25S rRNA positions C2675 and C2693 and the rancRNA_18-dependent extra density (Figure 5B,C). The nearest r-protein located to this extra density is L11 (uL5) (Figure 5C).
Figure 6.
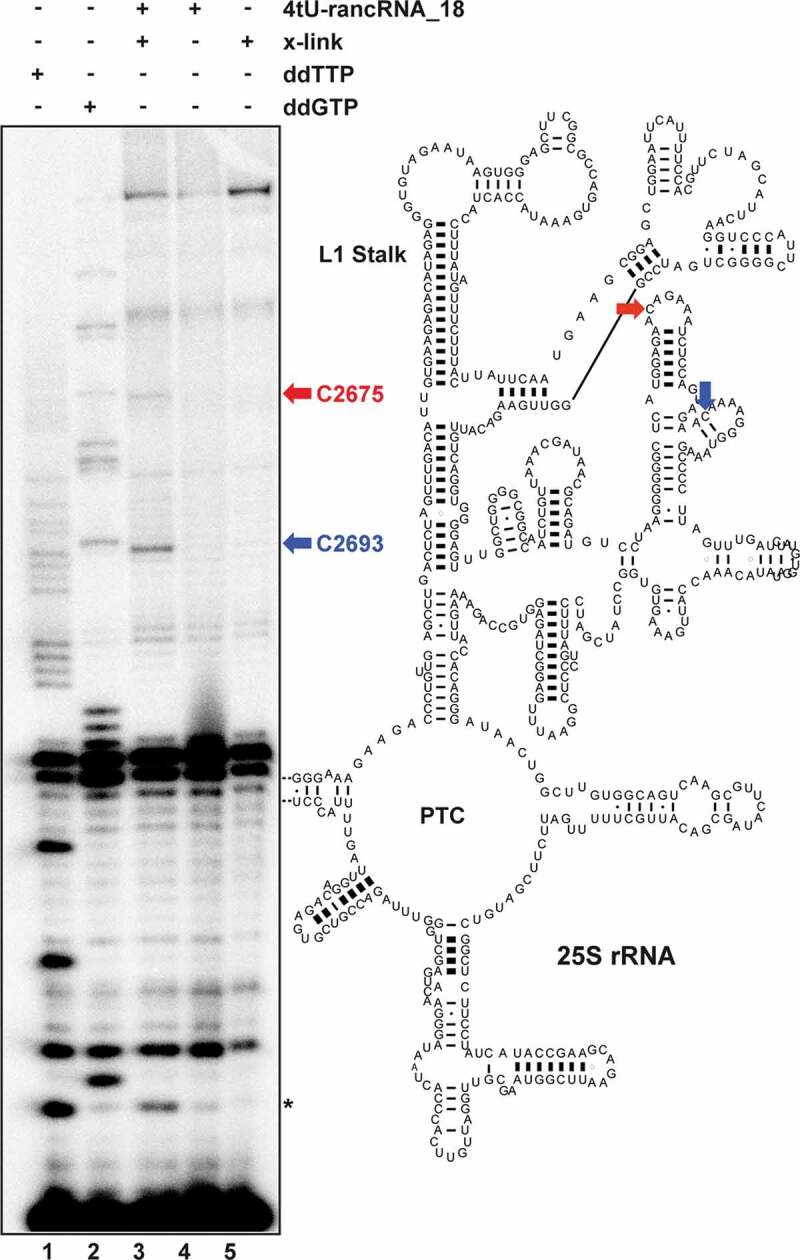
RancRNA_18 cross-links to the 60S E-site region. the sites of cross-linking (x-link) of rancRNA_18 carrying two 4-thio-U modifications (4tU-rancRNA_18) to 80S ribosomes were monitored by primer extension analysis and revealed cross-linking sites to C2675 (red) and C2693 (blue) of helix 84 of the 25S rRNA. experiments in the presence of 4tU-rancRNA_18 without irradiation (lane 4) or in the absence of 4tU-rancRNA_18 with irradiation (lane 5) served as controls. cross-linking experiments and primer extension analyses were performed on two biological replicates. the primer extension stop marked with an asterisk could not always be detected using different primers and thus was excluded from further discussions. Lane 1 (ddTTP) and 2 (ddGTP) indicate dideoxy sequencing lanes. location of cross-linking sites (arrows) are shown in the secondary structure model of yeast 25S rRNA (PTC, peptidyl transferase centre)
Thus, within the limitations of the obtained cryo-EM resolution, the combination of the structural data with the cross-linking results supports our interpretation that this additional electron density corresponds to ribosome-bound rancRNA_18.
Discussion
Here we present biochemical, genetic and structural evidence for the biogenesis, mode of action and target site of a small ribosome-associated ncRNA, called rancRNA_18, in S. cerevisiae. rancRNA_18 is an mRNA-derived regulatory ncRNA whose biogenesis involves the RNase III homolog Rnt1 (Fig. 2). rancRNA_18 binds to the large ribosomal subunit and 80S monosomes during optimal growth but shifts into polysomes upon hyperosmotic stress [11]. Ribosome-bound rancRNA_18 rapidly shuts down protein biosynthesis in vivo and in vitro. Yeast cells devoid of rancRNA_18 are hampered in their biological fitness during hyperosmotic conditions as documented in growth competition experiments with an otherwise isogenic yeast strain (Figure 1D). Even under optimal growth conditions, the presence of rancRNA_18 provides an advantage to some extent, thus indicating that this rancRNA-mediated translation regulation system is not solely beneficial during challenging environmental conditions for yeast. During hyperosmotic stress, how can such a small RNA molecule affect the much larger ribosome during protein synthesis in such an efficient and rapid manner? Is inhibition of translation a global phenomenon or are some mRNAs immune to rancRNA_18 inhibition that might be crucial for salt stress adaptation?
In order to address these two fundamental questions, we have analysed the translatome of yeast spheroplasts in the absence or presence of rancRNA_18 by deep sequencing polysome-associated mRNAs. The results of this transcriptome-wide analysis is clear and identified rancRNA_18 as a global inhibitor of translation, since the mRNA pool in polysomes does not change upon rancRNA_18 binding (Fig. 3). To uncover the molecular mechanism, we elucidated the rancRNA_18 binding site on the large ribosomal subunit. Based on the fact that yeast rancRNA_18 can also inhibit plant, mammalian and bacterial ribosomes [11], a highly conserved binding pocket was expected. Indeed, the cryo-EM structure of rancRNA/80S complexes revealed extra density near the tRNA E-site region of the large subunit (Fig. 5, Supplementary Figure S9B). Crosslinking experiments with rancRNA_18 carrying the zero-length photo-reactive group 4-thio-U provided further evidence for a binding site in the evolutionarily highly constrained domain V of 25S rRNA near the tRNA E-site region (Fig. 6). The structure of the rancRNA/80S complexes gave more detailed insight into the molecular interactions. Ribosome-bound rancRNA_18 localizes close to, but not entirely overlapping, the P- and E-site tRNAs (Figure 5A). In particles with bound rancRNA_18, the highly dynamic L1 stalk region of the large ribosomal subunit adopts a slightly inward conformation and the density also becomes to some extent more ordered (Supplementary Figure S8). However, our structural analysis reveals that the density we assigned to ribosome-bound rancRNA_18 is only detectable in particles lacking P- and E-site tRNA densities (Figure 5B and Supplementary Figure S7; class III). The L1 stalk region is known for its flexibility, and it has been shown to be involved in E-tRNA ejection from the ribosome [50]. Thus it is possible that rancRNA_18 restricts the motion of the L1 stalk and thereby freezes this dynamic part of the ribosome in a particular state, which is incompatible with efficient tRNA release. This interpretation is supported by our cryo-EM multibody refinement analysis, which shows drastically reduced L1 motion in ribosomes with bound rancRNA_18 (class III, Supplementary Figure S10, Supplementary Movie S1, and Supplementary Table S2). The small residual L1 motion is a swing up-down, and not the functionally relevant swing into and out of the inter-subunit cavity, which furthers support our model.
Since rancRNA_18 and E-site tRNA cannot co-exist on the same ribosome, it is possible that rancRNA_18 binds the large subunit every time the E-site is vacant, such as during translation initiation. This interpretation is compatible with our previous findings showing that rancRNA_18 binding leads to a concomitant increase of monosomes and decrease of polysomes, a pattern indicative of an initiation block [11]. Furthermore, we previously demonstrated that rancRNA_18 does not inhibit ribosomes during the elongation phase [11], again hinting at initiation as the primarily affected step of protein biosynthesis. P-site binding competition and transpeptidation data (Figure 4C,D), showing no inhibition of peptide bond formation between P-site located peptidyl-tRNA and A-site bound puromycin, are in line with the cryo-EM structure revealing rancRNA_18 binding near the E-site region of the 60S subunit (Fig. 5). However, the binding competition between rancRNA_18 and A-site bound tRNA (Figure 4E and Supplementary Figure S5C) appears in conflict with a rancRNA_18 binding site close to the E-site. Yet, biochemical evidence exists that argues for a functional link between the ribosomal A- and the E-sites. Predominately Nierhaus and co-workers entertained the idea that an occupied E-site results in an A-site with decreased tRNA affinity (and vice versa) [51]. RancRNA_18 photo-crosslinks to two residues in helices 84 (C2675) and 85 (C2693) of 25S rRNA (Fig. 6). Ribosomal protein L11 (uL5) in S. cerevisiae has previously been shown to possess a dynamic loop region which can modulate the solvent accessibility of helix 84 [52] a helix we show here to be cross-linked to rancRNA_18. Interestingly, an L11 (uL5) mutant that restricts L11-loop dynamics by freezing its interaction with helix 84 negatively affected A-site tRNA occupancy. It is therefore possible that rancRNA_18, when bound to its binding site near helix 84/85, similarly restricts L11 (uL5) dynamics and thereby reduces the affinity of the A-site for incoming tRNAs. More recent structural and single molecule FRET evidence showed that E-site occupancy on the large ribosomal subunit influences the dynamic rotational movement between the two ribosomal subunits which shapes the A-site conformation [53–55]. In 2014 another relevant ribosomal dynamic in eukarya was uncovered, namely subunit rolling [56]. During this conformational change, the small ribosomal subunit rolls in the direction of the L1 stalk (in the post-translocational state) thus opening the A-site region. Therefore, subunit rolling reciprocally opens and closes the A- and E-site regions of the ribosome during its oscillation between the pre- and post-translocational states. It is possible that bound ligands (such as rancRNA_18 close to the E-site) might influence these conformational changes. Our analyses on ribosomal structural dynamics are in line with this interpretation since we observe a severe inhibition of L1 stalk movement in the ribosomal complexes carrying rancRNA_18 (Supplementary Movie S1, Supplementary Figure S10 and Supplementary Table S2). In the light of these data, it is conceivable that rancRNA_18 binding close to the E-site affects A-site affinity of tRNAs, thus explaining our filter binding observation (Figure 4E and Supplementary Figure S5C). Cumulatively, the acquired structural and biochemical data support a model, where rancRNA_18 binding close to the ribosomal E-site region hampers 60S subunit dynamics, decreases A-site tRNA binding affinity and interferes with the structural mobility of the L1 stalk. This whole network of rancRNA_18-dependent alterations on the ribosome prevents the efficient transition from the initiation to the elongation phase of protein synthesis thus resulting in a global down-regulation of translation and consequently of all metabolic activities. To the best of our knowledge the rancRNA_18 binding site on the large ribosomal subunit (involving 25S rRNA helices 84 and 85 close to r-protein L11 (uL5)) does not overlap with interaction pockets of known small molecule translation inhibitors (e.g. cycloheximide; Supplementary Figure S4C) thus representing an unexplored target site for potential future design of small molecule inhibitors.
rancRNA_18 is the first of its class whose ribosome-binding site has been identified in molecular detail. In combination with the available biochemical and genetic evidence, its mode of action as a potent and swift inhibitor of global protein biosynthesis can be better understood. The class of rancRNAs includes small as well as long ncRNAs [7,17] and has been shown to be functionally very heterogeneous. Beside global inhibitors of translation [9,10,13,14,16] and this study), recent studies identified mRNA-specific rancRNA in the halophilic archaeon H. volcanii [12] and in the human parasite T. brucei [16]. Inhibiting protein biosynthesis is, however, not the only function a rancRNA can possess. Very recently, a ribosome-associated tRNA 3ʹ half has been uncovered in the human parasite T. brucei that stimulates global protein synthesis during starvation stress recovery [8]. rancRNAs have been found in all three domains of life and all so far functionally characterized members perform their particular roles during specific stress situations. rancRNAs affect protein production within minutes upon environmental challenges thus arguing for a regulatory role during the first wave of cellular response. It therefore seems that rancRNA-mediated translation control is multifaceted, stress-specific and swift, thus adding a so far largely unknown layer of gene expression regulation acting directly at the ribosome.
Supplementary Material
Acknowledgements
We would like to thank Denis Lafontaine for valuable suggestions and for providing the Rnt1 depletion construct. Jon Hall is acknowledged for his continuous support of chemically synthesized modified RNA strands and Robert Rauscher for help with RNA-seq figures. We are grateful to Alexander Leitner from the Structural Mass Spectrometry Platform of the NCCR “RNA & Disease”. The imaging was performed on equipment supported by the Microscopy Imaging Center (MIC), University of Bern, Switzerland.
Author contributions: N.P. conceived the project and designed the research (with help from B.Z). L.S. and J.R. collected most of the biochemical, genetic and transcriptomic data, I.I. performed the cryo-EM experiments and data refinements (under the supervision of B.Z.). A.P. was involved in the initial phase of this project and contributed to the biochemical characterization. W.G. performed the bioinformatics analyses. All authors contributed to data interpretation, analysis and commented on the manuscript. N.P. supervised the overall work and wrote the manuscript (based on a first draft written by L.S. and J.R.).
Funding Statement
This work was supported by the Swiss National Science Foundation [31003A_166527 and 310030_188969 to N.P. and 163761 and 179520 to B.Z.] and in part by the NCCR ‘RNA & Disease’ funded by the Swiss National Science Foundation. J.R. is a recipient of a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft DFG [RE 4041/1–1] and I.I. is a recipient of a grant of the Berne University Research Foundation.Conflict of interest statement. None declaredSchweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung [166527];Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung [163761, 179520];Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung [188969].
Disclosure of statement
No potential conflict of interest was reported by the author(s).
Data availability
mRNA-Seq sequencing data have been deposited at GEO database under the accession number GSE135475. Cryo-EM maps and data have been deposited with accession codes EMD-12,986 (-rancRNA_18) and EMD-12,988 (+rancRNA_18).
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Lane N, Martin W.. The energetics of genome complexity. Nature. 2010;467(7318):929. [DOI] [PubMed] [Google Scholar]
- [2].Gonskikh Y, Polacek N.. Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev. 2017;168:30–36. [DOI] [PubMed] [Google Scholar]
- [3].Sherman MY, Qian SB. Less is more: improving proteostasis by translation slow down. Trends Biochem Sci. 2013;38(12):585–591. [DOI] [PubMed] [Google Scholar]
- [4].Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev. 2010;11(9):597–610. [DOI] [PubMed] [Google Scholar]
- [5].Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9(9):673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014;49(1):16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pircher A, Gebetsberger J, Polacek N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol. 2014;11(11):1335–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fricker R, Brogli R, Luidalepp H, et al. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun. 2019;10(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gebetsberger J, Wyss L, Mleczko AM, et al. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017;14(10):1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gebetsberger J, Zywicki M, Kunzi A, et al. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pircher A, Bakowska-Zywicka K, Schneider L, et al. An mRNA-Derived Noncoding RNA Targets and Regulates the Ribosome. Mol Cell. 2014;54(1):147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wyss L, Waser M, Gebetsberger J, et al. mRNA-specific translation regulation by a ribosome-associated ncRNA in Haloferax volcanii. Sci Rep. 2018;8(1):12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bakowska-Zywicka K, Kasprzyk M, Twardowski T. tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res. 2016;16(6):fow077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gonskikh Y, Gerstl M, Kos M, et al. Modulation of mammalian translation by a ribosome-associated tRNA half. RNA Biol. 2020;17(8):1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rajan KS, Doniger T, Cohen-Chalamish S, et al. Developmentally regulated novel non-coding anti-sense regulators of mRNA translation in Trypanosoma brucei. iScience. 2020;23(12):101780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pinel-Marie ML, Brielle R, Riffaud C, et al. RNA antitoxin SprF1 binds ribosomes to attenuate translation and promote persister cell formation in Staphylococcus aureus. Nat Microbiol. 2021;6(2):209–220. [DOI] [PubMed] [Google Scholar]
- [17].Carvalho Barbosa C, Calhoun SH, Wieden H-J. Non-coding RNAs: what are we missing? Biochem. Cell Biol. 2020;98:23–30. [DOI] [PubMed] [Google Scholar]
- [18].Lintner NG, Cate JHD. Regulating the ribosome: a spotlight on RNA dark matter. Mol Cell. 2014;54(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Uesono Y, Toh EA. Transient inhibition of translation initiation by osmotic stress . J Biol Chem. 2002;277(16):13848–13855. [DOI] [PubMed] [Google Scholar]
- [21].Pascual-Ahuir A, Manzanares-Estreder S, Timon-Gomez A, et al. Ask yeast how to burn your fats: lessons learned from the metabolic adaptation to salt stress. Current Gen. 2018;64(1):63–69. [DOI] [PubMed] [Google Scholar]
- [22].O’Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15(2):532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vanacloig-Pedros E, Bets-Plasencia C, Pascual-Ahuir A, et al. Coordinated gene regulation in the initial phase of salt stress adaptation. J Biol Chem. 2015;290(16):10163–10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jin N, Jin Y, Weisman LS. Early protection to stress mediated by CDK-dependent PI3,5P2 signaling from the vacuole/lysosome. J Cell Biol. 2017;216(7):2075–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Qi D, Scholthof KB. A one-step PCR-based method for rapid and efficient site-directed fragment deletion, insertion, and substitution mutagenesis. J. Virol.l Meth. 2008;149(1):85–90. [DOI] [PubMed] [Google Scholar]
- [26].Gueldener U, Heinisch J, Koehler GJ, et al. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30(6):e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Flagfeldt DB, Siewers V, Huang L, et al. Characterization of chromosomal integration sites for heterologous gene expression inSaccharomyces cerevisiae. Yeast. 2009;26(10):545–551. [DOI] [PubMed] [Google Scholar]
- [28].Russell PJ, Hambidge SJ, Kirkegaard K. Direct introduction and transient expression of capped and non-capped RNA in Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19(18):4949–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Willi J, Kupfer P, Evequoz D, et al. Oxidative stress damages rRNA inside the ribosome and differentially affects the catalytic center. Nucleic Acids Res. 2018;46(4):1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18(10):3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cherry JM, Hong EL, Amundsen C, et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40(D1):D700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- [34].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hollander M, Wolfe DA. Nonparametric Statistical Methods. New York: John Wiley & sons, inc.; 1973. 185–194. [Google Scholar]
- [36].Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zheng SQ, Palovcak E, Armache JP, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Meth. 2017;14(4):331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bai XC, Rajendra E, Yang G, et al. Sampling the conformational space of the catalytic subunit of human gamma-secretase. eLife. 2015;4:e11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- [41].Jerabek-Willemsen M, Wienken CJ, Braun D, et al. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Techn. 2011;9(4):342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jackman JE, Montange RK, Malik HS, et al. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9(5):574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bernstein DA, Vyas VK, Fink GR. Genes come and go: the evolutionarily plastic path of budding yeast RNase III enzymes. RNA Biol. 2012;9(9):1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lamontagne B, Abou Elela S. Short RNA guides cleavage by eukaryotic RNase III. PloS One. 2007;2(5):e472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lamontagne B, Elela SA. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J Biol Chem. 2004;279(3):2231–2241. [DOI] [PubMed] [Google Scholar]
- [46].Schneider-Poetsch T, Ju J, Eyler DE, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6(3):209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dubochet J, Adrian M, Chang JJ, et al. Cryo-electron microscopy of vitrified specimens. Quart Rev Biophys. 1988;21(2):129–228. [DOI] [PubMed] [Google Scholar]
- [48].Nakane T, Kimanius D, Lindahl E, et al. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife. 2018;7:e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ban N, Beckmann R, Cate JH, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mohan S, Noller HF. Recurring RNA structural motifs underlie the mechanics of L1 stalk movement. Nat Comm. 2017;8(1):14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29(21):4997–5008. [DOI] [PubMed] [Google Scholar]
- [52].Rhodin MH, Dinman JD. A flexible loop in yeast ribosomal protein L11 coordinates P-site tRNA binding. Nucleic Acids Res. 2010;38(22):8377–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Frank J. Intermediate states during mRNA-tRNA translocation. Curr Opin Struct Biol. 2012;22(6):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cornish PV, Ermolenko DN, Noller HF, et al. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ferguson A, Wang L, Altman RB, et al. Functional dynamics within the human ribosome regulate the rate of active protein synthesis. Mol Cell. 2015;60(3):475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Budkevich TV, Giesebrecht J, Behrmann E, et al. Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell. 2014;158(1):121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
mRNA-Seq sequencing data have been deposited at GEO database under the accession number GSE135475. Cryo-EM maps and data have been deposited with accession codes EMD-12,986 (-rancRNA_18) and EMD-12,988 (+rancRNA_18).


