ABSTRACT
The subcellular localization of RNAs correlates with their function and how they are regulated. Most protein-coding mRNAs are exported into the cytoplasm for protein synthesis, while some mRNA species, long noncoding RNAs, and some regulatory element-associated unstable transcripts tend to be retained in the nucleus, where they function as a regulatory unit and/or are regulated by nuclear surveillance pathways. While the mechanisms regulating mRNA export and localization have been well summarized, the mechanisms governing nuclear retention of RNAs, especially of noncoding RNAs, are seldomly reviewed. In this review, we summarize recent advances in the mechanistic study of RNA nuclear retention, especially for noncoding RNAs, from the angle of cis-acting elements embedded in RNA transcripts and their interaction with trans-acting factors. We also try to illustrate the general principles of RNA nuclear retention and we discuss potential areas for future investigation.
KEYWORDS: Noncoding RNA, nuclear retention, chromatin association, u1 snRNP, splicing, hnrnpk, repeat, xist, RNA decay
Introduction
Pervasive transcription of mammalian genomes produces hundreds of thousands of protein-coding mRNAs and long noncoding RNA (lncRNA) transcripts [1,2]. Also, divergent transcription of active regulatory elements like promoters and enhancers often generates sense and antisense unstable transcripts like upstream antisense RNAs (uaRNAs), and enhancer RNAs (eRNAs) [3–5]. Besides functional differences, the overall subcellular localization of protein-coding mRNAs and these noncoding RNAs (ncRNAs) also differs greatly. Most mRNAs are located in the cytoplasm, while the majority of the ncRNAs tend to be retained in the nucleus [1,5,6]. The mechanisms governing the localization of mRNAs, especially for their processing and export, have been well studied and summarized (see reviews in [7,8]). However, studies of the mechanisms underlying nuclear retention of RNAs, especially of ncRNAs, were relatively scarce until recent years [9–13].
There are several excellent reviews that summarize the mechanism of nuclear retention of lncRNAs and mRNAs [14–16]. In this review, we focus on RNA nuclear retention mainly from the angle of cis-acting elements within the RNA and the trans-acting factors that interact with these elements. We also try to connect nuclear localization of RNA with its function and other processes related to RNA synthesis and processing. In particular, we emphasize one type of RNA nuclear retention – RNA chromatin association – over other types of nuclear retention in a handful of papers that the authors have probed RNA–chromatin interaction. It was believed that chromatin association of RNAs, especially of lncRNAs, might correlate with their function, in regulating gene expression and chromatin structure [17,18]. Non-chromatin-associated RNAs are mainly localized in the nucleoplasm and nuclear bodies such as nuclear speckles [19], which appear to be associated with the processing or degradation of the RNA. To be noted, recent advances also suggest that nuclear speckles can dynamically associate with chromatin, in a manner dependent on and correlated with transcription [20,21], and it was reported that the marker RNAs for nuclear bodies, like MALAT1 or NEAT1 (mainly located in nuclear speckles or paraspeckles, respectively, see below), bind to the chromatin of thousands of active genes [22,23]. These studies indicate that RNA-chromatin association and RNA-nuclear body association might be two highly correlated processes, and the boundary between these two forms of RNA nuclear retention is relatively fuzzy.
We begin with an introduction to how nuclear localization of RNA correlates with its function and regulation, and we illustrate the importance of proper RNA localization for the function of both protein-coding mRNAs and noncoding RNAs. Subsequently, we review mechanisms by which RNAs are retained in the nucleus or on chromatin, from the angles of both cis-acting elements and related trans-acting factors. At last, we discuss the importance of integrating RNA localization with other cellular processes, such as transcription and RNA processing, to better understand the pattern and physiological significance of nuclear retention of noncoding RNA. We also discuss future challenges for studying RNA localization and the function of noncoding RNAs.
Nuclear localization of RNA correlates with its function and regulation
The subcellular localization of RNAs often correlates with their function and regulation. In general, for most protein-coding mRNAs, the mature transcripts are mainly located in the cytoplasm, where they can be occupied by ribosomes for protein synthesis [8]. However, for some mRNAs, their localization is more elaborately regulated in order to modulate the abundance and localization of protein products. For example, it estimated that as many as three-quarters of mammalian multi-exonic genes are transcribed to generate intron-retaining isoforms, and these intron-retaining transcripts are usually retained in the nucleus where they act widely to reduce the expression of host genes [24,25]. Notably, this intron-retention-mediated form of RNA nuclear retention also acts as a means for cells to quickly respond to stimuli such as heat shock, DNA damage, neuronal activation, etc [26–28]. In the mouse neocortex, a subset of well-polyadenylated transcripts retain certain introns and stably accumulate in the nucleus. In response to stimulation, these retained introns undergo splicing and the mRNAs are exported to the cytoplasm for immediate protein synthesis [28] (Fig. 1A). When cells encounter stress conditions such as heat shock, widespread intron retention occurs, and these intron-retaining transcripts are kept in the nucleus to prevent them from engaging with ribosomes. Interestingly, the transcripts from a subset of functionally defined genes can escape this nuclear retention mechanism for normal protein synthesis [27]. Thus, nuclear retention can function as a fast-acting post-transcriptional regulatory mechanism for cells that encounter stress or stimuli.
Figure 1.
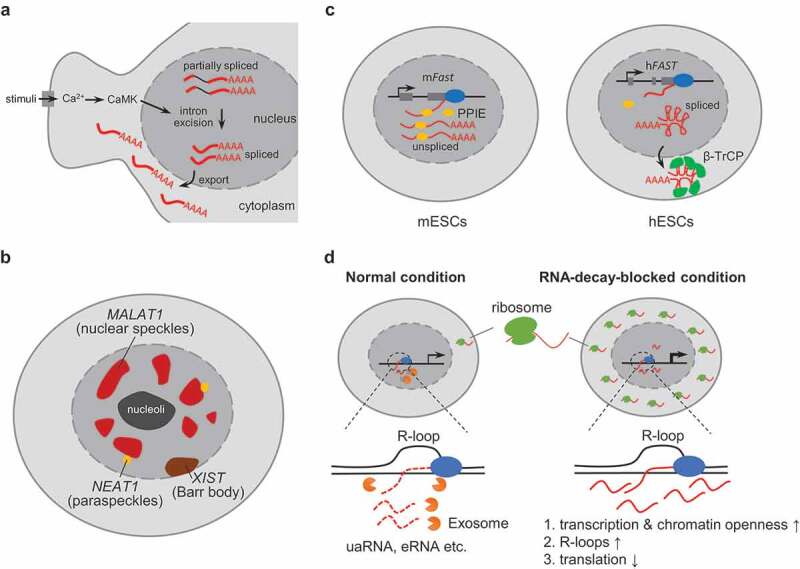
Nuclear localization of an RNA correlates with its function and regulation. (A) in the mouse neocortex, a subset of well-polyadenylated transcripts retain selected introns and are retained in the nucleus. These transcripts undergo further posttranscriptional splicing upon neuronal stimulation, and the fully spliced transcript is then exported into the cytoplasm for further protein synthesis. (B) the diagram shows the nuclear location of XIST (on the inactive X chromosome, which forms a condensed structure called the barr body, close to the nuclear membrane), MALAT1 (in nuclear speckles, located in interchromatin regions of the nucleoplasm), and NEAT1 (in paraspeckles, which frequently associate with nuclear speckles). (C) RNA nuclear retention correlates with the distinct functions of the lncRNA FAST in mESCs and hESCs. left: in mESCs, the high abundance of PPIE suppresses the splicing of lncRNA mFast, and these unspliced transcripts are retained in the nucleus; right: the abundance of PPIE in hESCs is low, so hFAST can be fully processed and exported into the cytoplasm, where it interacts with β-TrCP to promote pluripotency maintenance. (D) RNA decay modulates the localization and function of uaRNAs and eRNAs. left: under normal conditions, uaRNAs and eRNAs are removed by the RNA-decay pathway (e.g. by exosomes) during or immediately after their synthesis, before they have a chance to export to move to the cytoplasm. right: after the RNA-decay pathway is blocked, these transcripts accumulate and more RNAs are transferred into the cytoplasm. In the nucleus, the RNA form more R-loop structures and increase genome instability, transcription rate, and chromatin openness; in the cytoplasm, they will occupy more ribosomes and impair the overall translation rate
Unlike protein-coding mRNAs, most lncRNAs are located in the nucleus, and many of them are associated with chromatin [1,29]. The nuclear retention and chromatin association of lncRNAs are often correlated with their function in regulating gene transcription and RNA processing, and also with how they are regulated and processed [17,30–33]. One of the most well-studied lncRNAs is XIST, which mediates the inactivation of one of the X chromosomes in mammalian females and is transcribed from the inactive X chromosome (Xi) [34]. Accordingly, the majority of the XIST RNA ‘coat’ is on the inactivated X chromosome (Fig. 1B) and forms a ‘cloud’-like pattern as viewed by RNA fluorescent in situ hybridization (FISH) [35]. Impairing the chromatin association of XIST abolishes its function in regulating X chromosome inactivation [36,37]. Another example is provided by the lncRNA MALAT1, which was reported to function in regulating gene expression and RNA processing [38,39]. Consistent with this, MALAT1 RNAs mainly locate in nuclear speckles, which are enriched with RNA processing factors (Fig. 1B).
Compared to protein-coding mRNAs, the expression of lncRNAs is more likely to be cell type and tissue-specific [40], implying a functional relevance of lncRNAs in development and cell fate determination [17]. Indeed, an increasing number of studies have revealed that many lncRNAs play roles in development and pathogenesis [41]. Intriguingly, some lncRNAs fulfil their function in these processes by altering their nuclear retention status. For example, TINCR is a lncRNA required for proper epidermal differentiation. The function of TINCR is correlated with changes of its subcellular localization. In undifferentiated epidermal cells, TINCR is mainly located in the nucleus. During differentiation, TINCR levels are elevated and TINCR RNAs are translocated into the cytoplasm, where they interact with and stabilize a range of mRNAs of differentiation-related genes, thus contributing to proper epidermal differentiation [42]. A recent study in human and mouse embryonic stem cells (hESCs) suggests that the distinct subcellular localization pattern of the lncRNA FAST in the two species contributes to the non-conserved function of FAST in ESC pluripotency maintenance [13]. The FAST gene is positionally conserved and specifically expressed in ESCs. Intriguingly, human FAST (hFAST) RNA is mainly localized in the cytoplasm, where it binds to β-TrCP and is required for the pluripotency maintenance of hESCs. Mouse Fast (mFast) RNA, on the other hand, is mainly located in the nucleus, and is not required for the pluripotency maintenance of mESCs. The author found that the nuclear retention of mFast is regulated by the splicing factor PPIE, which is highly expressed in mESCs but not hESCs. Highly expressed PPIE binds to the nascent mFast transcripts and suppresses their splicing, thus promoting their nuclear retention [13] (Fig. 1C).
For some short-lived lncRNAs and regulatory element-associated unstable transcripts like uaRNAs and eRNAs, their subcellular localization correlates with how they are regulated. Experimentally, these noncoding RNAs are usually located in the nucleus and enriched in the chromatin fraction [5,31]. Their nuclear retention at least partially correlates with their short half-lives – they are degraded co-transcriptionally, or immediately after synthesis, mainly through nuclear surveillance pathways [31,43,44], before they have a chance to be exported to the cytoplasm. Although they are short-lived, some of them were proved to play roles in regulating the expression of nearby genes through their transcripts or transcription [5,45,46]. This RNA degradation-mediated nuclear retention may be important for two reasons: The first is that the degradation of these RNAs keeps them at a suitable level for genomic stability and to modulate the proper activity of the regulatory elements which they are associated with. Impeding the degradation of these RNAs by knocking out RNA exosome components alters the activity of regulatory elements like enhancers and promoters, and increases R-loop structures and genomic instability [47,48]. The second reason is that nuclear decay of these ncRNAs prevents their export to the cytoplasm, where they may swap the protein synthesis machinery. Artificial stabilization of these RNAs by blocking the RNA degradation pathway promotes their cytoplasmic localization, which will overwhelm ribosomes and later result in global translational repression [49] (Fig. 1D). Thus, the proper localization of these short-lived ncRNAs correlates with their regulation and prevents them from interfering with other cellular processes.
Mechanisms for nuclear localization and chromatin association of RNA
Both cytoplasmic protein-coding mRNAs and nuclear ncRNAs utilize RNA polymerase II (RNA Pol II) and a similar set of RNA processing factors for their synthesis, packaging, processing, and turnover [1,29]. They may diverge at some point along their maturation pathway before reaching their final destination. Intriguingly, when a nuclear-retained transcript is fused with a cytoplasm located mRNA fragment, the chimeric transcript is mainly retained in the nucleus [50], which suggest that the ‘code’ responsible for RNA localization might be embedded in the RNA sequence. Indeed, studies show that some intrinsic mRNA elements, like the constitutive transport element (CTE) and the RNA transport element (RTE), serve as the ‘postage’ for RNA export and cytoplasmic localization of some viral RNAs and a small subset of mRNAs [51,52]. In addition, it has been reported that an expression and nuclear retention element (ENE) promotes the nuclear retention and stability of PAN transcripts derived from Kaposi’s sarcoma-associated herpesvirus [53]. Thus, identifying key sequences that participate in regulating RNA subcellular localization may serve as an entry point for investigating the mechanism of RNA nuclear retention.
Traditional strategies to identify cis-elements responsible for RNA subcellular localization are mainly based on two principles: 1) truncate the candidate RNA into small fragments, fuse them with a cytoplasm located reporter RNA, and analyse the localization of each fragment to identify the ones that exhibit nuclear localization patterns or promote the nuclear retention of the fused reporter RNA; and 2) serially delete different parts of a candidate RNA to identify key regions that impair the proper localization of the candidate RNA. By utilizing these methods, it has been possible to identify several fragments and cis-elements that are required for nuclear retention of some lncRNAs [38,54–57]. For instance, in the lncRNA MALAT1, two regions called E and M, around 600–1000 nt in length, were identified as being responsible for locating the lncRNA to nuclear speckles [57]. However, these are low-throughput methods, and the identified RNA fragments are usually are long. The low-resolution of these techniques makes it hard to identify the key sequences responsible for the nuclear retention of host RNAs.
New approaches were needed to solve these problems. High-throughput techniques, such as massively parallel reporter assays (MPRAs) or self-transcribing active regulatory region sequencing (STARR-seq), were developed to identify cis-elements that contribute to transcriptional regulation [58,59]. Recently, three groups combined these strategies with subcellular fractionation of RNA, to identify potential cis-elements regulating the nuclear retention of some nucleus-localized RNAs [9–11]. The basic ideas are similar: The first step is to construct MPRA libraries by fusing a cytoplasm-localized reporter gene with tens of thousands of insert sequences derived from synthetic DNAs or randomly fragmented endogenous sequences. These inserts cover all the mature transcript sequences of candidate nuclear-localized RNAs. Next, the reporter library is transfected into cells, followed by subcellular fractionation and isolation of RNAs from the different fractions. Finally, the relative enrichment of reporter-insert-fusion RNAs in each fraction is measured by deep sequencing. It is then possible to identify inserts containing potential cis-elements that promote nuclear retention or chromatin association of RNA. Using this approach, the three groups identified several cis-elements [9–11]. Here we summarize these studies and integrate their discoveries into previously known mechanisms. We will not specifically discuss the mechanism of nuclear surveillance pathway-mediated nuclear retention of unstable nuclear transcripts mentioned above, as it has been well summarized and discussed by previous reviews [44,60,61]. The cis-element-based mechanisms that regulate nuclear retention of RNA can be roughly divided into seven groups (Table 1). These will be described in the following sections.
Table 1.
A list of cis-elements and their interacting trans-factors that participate in regulating RNA nuclear retention
| Categories | Cis-elements | Trans-factors | Function | Representative RNAs | References |
|---|---|---|---|---|---|
| U1 recognition and splicing | U1-recognition motif (potential 5’ss) | U1 snRNP | nuclear retention, chromatin association, RNA decay | MALAT1, MEG3, Tsix, uaRNAs, eRNA, etc. | [11] |
| 5’ss | U1 snRNP | nuclear retention | ftz, mvp2, sd4, gag-pol | [67–70] | |
| NRE | U1 snRNP | nuclear retention | MEG3 | [66] | |
| MALAT1 regions E and M | SON, RNPS1, U1 snRNP | nuclear retention, chromatin association, localization in nuclear speckles | MALAT1 | [11,38,57] | |
| SINE-derived elements and C-rich motifs | SIRLON | HNRNPK | nuclear retention | JPX, PVT, etc. | [9] |
| HNRNPK binding site (C-rich) | HNRNPK | nuclear retention | MLXIPL, etc. | [9] | |
| AGCCC | HNRNPK? | nuclear retention | BORG | [54] | |
| 15nt C-rich motif | HNRNPK? | nuclear retention | MALAT1, lincSFPQ, etc. | [10] | |
| IRAlu and paraspeckles | IRAlu | paraspeckles (NONO) | nuclear retention, localization in paraspeckles | CTN-RNA, Linc-p21, etc. | [88,98,99] |
| Other genomic repeat elements | L1PA16, L2b, MIRb, and MIRc | unknown | nuclear retention | RP11-5407, LINC00173, RP4-806M20.4, etc. | [12] |
| CTG expansion | CELF1, MBNL1 | nuclear retention, localization in nuclear speckles | DMPK, MBNL1 | [113,116,120] | |
| CAG expansion | MBNL1, U2AF65 | nuclear retention, localization in nuclear speckles | HTT | [119,122] | |
| GGGGCC expansion | TDP43, HNRNPH, MBNL1 | nuclear retention, localization in nuclear speckles | noncoding region of C9orf72 | [117,118,122] | |
| Tandem repetitive sequences | XIST A-repeat | SPEN | nuclear retention, chromatin association | XIST | [11,126,127] |
| XIST LBR binding site | LBR | Lamin localization | XIST | [124] | |
| XIST B-repeat | HNRNPK, polycomb proteins | nuclear retention, chromatin association, X-chromosome painting | XIST | [90,91,134] | |
| XIST E-repeat | CIZ1, PTBP1, MATR3, TDP43, CELF | nuclear retention, chromatin association | XIST | [36,37] | |
| Multiple broad regions in XIST | HNRNPU | nuclear retention, chromatin association | XIST | [37,140] | |
| RRD | HNRNPU | nuclear retention, chromatin association | FIRRE | [141,142] | |
| R-loop and RNA-DNA Triplex | R-loop | RAD51, Cas9 | nuclear retention, chromatin association, telomere targeting | TERRA | [151,152,155] |
| RNA/DNA triplex | unknown | nuclear retention, chromatin association | MEG3, Khps1, etc. | [156–160] | |
| SnoRNA-end | snoRNA-end | snoRNA-binding proteins? | nuclear retention, localization in nucleoli and Cajal bodies | sno-lncRNAs | [163,165,166] |
3.1. U1 recognition and splicing
The U1-recognition site, which refers to the binding site of U1 snRNP, is also known as the potential 5ʹ splice site (5’ss), and is well known for its function in pre-mRNA splicing [62]. RNA splicing is performed by the spliceosome that assembles on each intron and predominantly comprises small nuclear ribonucleoproteins (snRNPs), including U1, U2, U4, U5, and U6 snRNPs in equal stoichiometry [62,63]. The binding of U1 snRNP to 5’ss is the first step of spliceosome assembly, and it further promotes the assembly of the pre-catalytic spliceosome together with U2 snRNP [64]. Notably, the abundance of U1 snRNP far exceeds that of other splicing-related snRNPs [65], and a transcriptome-wide survey of its RNA binding targets indicates that it associates with a large number of sites located in noncoding regions beyond 5’ss [22]. The high abundance of U1 snRNP and its prevalent binding on RNA transcripts imply non-canonical functions of U1 snRNP beyond splicing.
Recently, it has been reported that U1 snRNP plays a role in promoting the chromatin retention of lncRNAs as well as short-lived noncoding transcripts such as uaRNAs and eRNAs [11]. In this study, Yin et al developed a method named RNA elements for subcellular localization by sequencing (REL-seq), which is strategically similar to the MPRA and STARR-seq-based approaches mentioned above. Different types of reporter were integrated into the genome through the piggyBac system, then a screen was carried out to find cis-elements that promoted the chromatin retention of reporter RNAs. By utilizing this method, they identified 26 chromatin-enriched fragments from 9 representative nucleus-retained transcripts in mouse. For one chromatin enriched 162-nt RNA fragment, which was identified from a retained intron of the gene NXF1, the authors performed random mutagenesis and used the mutated fragments as a new library for REL-seq. In this way, they identified that the U1-recognition motif embedded in the fragment strongly promotes the chromatin retention of reporter RNAs [11].
Notably, they also found that the occurrence of the U1-recognition site and the binding of U1 snRNP is higher in lncRNA transcripts than in protein-coding mRNAs. Impairing the function of U1 snRNP reduces the chromatin association of nearly half of the lncRNAs expressed in mESCs [11]. Consistent with this study, another group also found that knockdown of the U1 snRNP-related protein components impairs the nuclear retention of several lncRNAs, for example MEG3 [66]. Besides, several other studies have revealed that the 5’ss is involved in the nuclear retention of a handful of RNA transcripts encoded by the gene ftz and the viral genes mvp2, sd4, and gag-pol [67–70]. This suggests that U1 recognition is universally employed by many noncoding RNA transcripts, and intron-retaining mRNA transcripts to regulate their chromatin retention.
A systematic comparison of RNA processing between mRNAs and lncRNAs suggests that the overall splicing efficiency of lncRNAs is lower than for mRNAs [6,71] even though lncRNA transcripts harbour a higher density of predicted U1-recognition sites and U1 binding than mRNAs [11,22]. This raises an intriguing question about U1-mediated chromatin retention of lncRNAs: why do these U1-tethered nuclear RNAs not undergo splicing? At least two factors have been reported that may contribute to the inefficient splicing of lncRNAs. The first is a lack of efficient 3ʹ splice sites (3’ss). A genome-wide survey of the distribution of predicted potential 3ʹ ss indicates that compared to mRNA genes, lncRNA genes harbour a substantially lower density of putative 3’ss [11], and the overall strength of 3’ss in lncRNAs seems lower than that in mRNAs [71,72]. The uneven distribution of 5’ss and 3’ss, and the weak internal 3’ss signal may partially account for the inefficient splicing of lncRNA transcripts. The second factor which may affect the splicing efficiency of lncRNA transcript is the binding and abundance of splicing regulators. Consistent with the observation of weak internal 3’ss in lncRNAs, the binding density of the splicing activator U2AF65 on lncRNAs is less than that on mRNAs [71]. In addition, lncRNA exons harbour fewer exonic splicing enhancers (ESEs), which lead to the binding of SR proteins at a lower level in lncRNAs than that in protein-coding mRNAs [72]. Thus, both cis- and trans- elements work in concert to contribute to the inefficient splicing of lncRNAs, which may partially contribute to the constant binding of U1 snRNP. Another intriguing question is how U1 snRNP promote the chromatin retention of lncRNA? U1 snRNP can physically interact with transcriptionally engaged RNA polymerase II (RNA Pol II) [11,73], and the chromatin association of both U1 snRNP and its interacting lncRNAs depends on an active transcription status [11]. It is possible that U1 snRNP tethers and mobilizes its associated lncRNA on chromatin through its interaction with transcriptionally engaged Pol II [11] (Fig. 2A).
Figure 2.
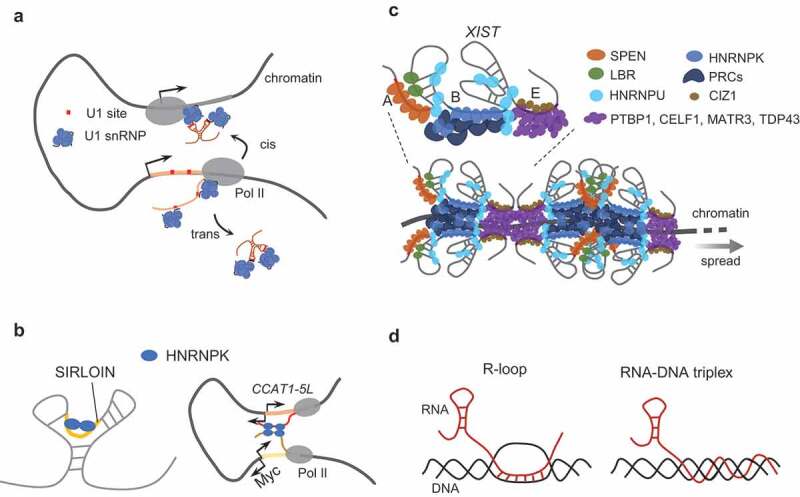
Representative mechanisms regulating RNA nuclear retention and chromatin association. (A) U1 snRNP regulates the chromatin retention of lncRNAs. U1 snRNP binds to U1-recognition motifs embedded in lncRNA transcripts, and further tethers the lncRNA to cis and/or trans chromatin through interaction with transcriptionally engaged Pol II. (B) C-rich motifs interact with HNRNPK to promote RNA nuclear retention. left: HNRNPK interacts with long RNAs harbouring C-rich motifs or SIRLOIN (SINE-derived nuclear RNA LOcalizatIoN, which harbours two C-rich motifs) and promotes their nuclear retention; right: HNRNPK interacts with the super-enhancer (SE)-associated lncRNA CCAT-5 L and with MYC promoter RNA, and promotes chromatin looping between the SE and the MYC promoter through its oligomerization. (C) tandem repetitive sequences interact with multiple regulators to promote the chromatin tethering and spreading of XIST. A-, B-, and E-repeats provide a multivalent platform for recruiting multiple RNA binding proteins and chromatin regulators to promote the chromatin retention of XIST RNA. The oligomerization and phase-separation-potential properties of B-repeat interactors (HNRNPK, polycomb proteins, etc.) and E-repeat interactors (PTBP1, CELF1, MATR3, TDP43, etc.) may promote the spreading of XIST RNA along the X chromosome through a feed-forward mechanism. (D) R-loops (left) and RNA-DNA triplexes (right) regulate the chromatin tethering of RNAs
Another intriguing question related to this mechanism is what happens to Pol II when U1 snRNP cannot be efficiently released? Studies of intron-retaining transcripts in yeast suggest that these unspliced transcripts will inhibit polyadenylation and stall Pol II on chromatin, and promote the degradation of the stalled pre-mRNAs [74]. It is proposed that mammalian cells may use different mechanisms as most of their genes harbour multiple introns [16]. However, as most lncRNAs contain fewer introns than protein-coding mRNAs, and it was reported that cotranscriptional RNA decay is frequently observed for lncRNA [31], it is possible that the constantly binding of U1 snRNP may affect the RNA termination process and promote the decay of some lncRNAs. Consistent with this, when the U1-recognition site is inserted just upstream of an authentic polyadenylation site (PAS), it will inhibit the PAS and trigger RNA decay, possibly through nuclear RNA surveillance-related pathways [11,44,75]. Impairing the function of U1 snRNP indeed increases the RNA abundance and cytoplasmic localization of some lncRNAs, uaRNAs, and eRNAs [11,76]. It has also been reported that U1 snRNP represses cryptic PASs in introns and protects pre-mRNAs from premature cleavage and polyadenylation; furthermore, it was reported that the U1-PAS axis enforces transcriptional directionality [63,77,78]. It is not fully clear how U1 snRNP coordinates the fate of its RNA targets amid all these regulatory events, which occur on chromatin and in a transcription-dependent manner. As U1 snRNP physically interacts with transcriptionally engaged RNA polymerase II [11], U1 snRNP may work in concert with transcriptional machinery and transcription status to ensure proper expression, localization, and turnover of its RNA targets.
3.2. SINE-derived elements and C-rich motifs
Short interspersed elements (SINEs) are a group of retrotransposon elements that widely exist in the mammalian genome. In humans, the most abundant SINEs are Alu elements, which have more than 1 million copies and contribute more than 10% of the genome [79]. Alu elements are unevenly distributed in the genome with a strong preference towards gene-rich regions [80,81]. Most Alu elements are located in the intronic regions of the human genome, where some of them play roles in modulating RNA processing events such as splicing, biogenesis of circular RNA, etc [82–85]. A subset of Alu elements can be exonized by splicing-mediated insertion [85,86]. For most protein-coding genes, this exonization is suppressed [87] and only hundreds of mRNAs harbour Alu elements in their 3ʹUTR [88]. In contrast, Alu elements are commonly present in mature lncRNA transcripts, and these Alu-containing lncRNAs usually exhibit a higher expression level but less tissue specificity [89].
A recent study by Lubelsky and Ulitsky indicated that Alu elements can contribute to the nuclear localization of their long host RNAs in human cells [9]. In their study, the authors constructed an MPRA library containing 5511 sequences derived from 50 nucleus-localized transcripts, including 37 human lncRNAs and 13 3ʹUTRs of mouse mRNAs. Through the screen, they identified three nucleus-enriched regions that overlapped with Alu repeat sequences. They further analysed the sequences and determined that a 42 nt sequence which they named as SIRLOIN (SINE-derived nuclear RNA LOcalizatIoN) can both promote the nuclear retention of reporter RNAs and attenuate their expression [9]. The SIRLOIN elements contain two C-rich consensus sequence motifs (RCCTCCC, R = A/G), and mutation of these motifs abolished the effect of SIRLOIN on both localization and expression level [9] (Fig. 2B, left panel). In another study, Shukla et al. used a similar strategy to identify a C-rich motif, and a similar C-rich pentamer motif (AGCCC) was previously reported as necessary and sufficient for nuclear localization of cytoplasm-localized RNA [10,54]. Moreover, it was reported that XIST harbours multiple C-rich motifs which promote the chromatin tethering of XIST RNA [90,91] (see below). However, the C-rich motif of SIRLON alone failed to promote RNA nuclear retention [9]. Furthermore, the three groups who performed cis-acting element screens did not identify the C-rich motif of XIST, despite the fact that their libraries all contained XIST sequences [9–11]. These observations suggest that C-rich motifs promote nuclear retention of RNA in a context-dependent manner. It is possible that other sequences, possibly related to RNA structure or the binding of other factors, and chromatin environment, may also play a role in this process.
Mechanistic study of SIRLOIN and XIST suggests that these C-rich motifs promote RNA nuclear retention through interaction with an RNA binding protein (RBP), HNRNPK, which is mainly localized in the nucleus. HNRNPK also mediates the nuclear accumulation of its bound RNA outside Alu elements [9]. Moreover, HNRNPK was also reported to interact with the super-enhancer (SE)-associated lncRNA CCAT1-5 L and with MYC promoter RNAs. This interaction further promotes chromatin looping between the SE and the MYC promoter, possibly together with the chromatin organizer CTCF [92], to enhance the Pol II occupancy and transcription of MYC [93] (Fig. 2B, right panel). These studies imply that HNRNPK might serve as a general trans-factor in promoting the nuclear retention of its associated RNAs. An intriguing question is how does HNRNPK promote the nuclear retention of its interacting RNA? A genome-wide survey of HNRNPK binding regions revealed that it mainly binds to gene bodies and open chromatin regions, which indicates that it associates with chromatin, directly or indirectly [94,95]. It has also been shown that the nuclear localization of HNRNPK is dynamically regulated: virus infection or ERK kinase (MEK1) mediated phosphorylation can promote its cytoplasmic translocation [96,97]. However, it is still unknown whether HNRNPK is accompanied by its interacting RNAs when it moves between compartments, and it is unclear how RNA binding affects the function and regulation of HNRNPK. These are some interesting questions that await to be answered in the future.
3.3. Inverted repeated Alu and paraspeckle-related components
Alu elements also promote the nuclear retention of RNAs in another scenario. Specific pairs of Alu elements – inverted repeated Alu (IRAlu) elements, were reported to promote the nuclear retention of their host mRNAs and the lncRNA hLincRNA-p21 [88,98,99]. This regulation is mainly mediated by the interaction between IRAlu and the RNA binding protein p54nrb/NONO and depends on the secondary structure formed by the IRAlu elements [88,98,99]. The IRAlu RNAs are mainly located in the nucleus and some of them associate with speckle-like, membraneless organelles called ‘paraspeckles’. The paraspeckles frequently associate with nuclear speckles and consist of more than 40 different RBPs, including NONO mentioned above, and an architectural RNA NEAT1 [100–103] (Fig. 1B). NEAT1 and some of the paraspeckle component proteins, such as NONO, PSP, PSF, etc., are essential for the formation of the paraspeckle structure. The morphology and numbers of paraspeckles are regulated by the abundance of NEAT1 RNA. Serial deletion analysis of NEAT1 suggests that its middle domain is required for proper paraspeckle assembly, mainly by recruiting NONO dimers and initiating the assembly of paraspeckles through a phase separation mechanism [55]. The phase-separation process involves interactions between multivalent macromolecules, including RNAs and proteins, which generate micron-sized liquid droplets in aqueous solution [104]. However, studies of CTN-RNA, which is an mRNA isoform of mouse cationic amino acid transporter 2 (mCAT2) gene with a longer 3ʹUTR harbouring an IRAlu element [98], suggest that neither disrupting paraspeckle structure nor deleting NEAT1 RNA obviously impairs the nuclear retention of IRAlu-containing RNAs, but mainly alters their intranuclear distribution [105]. As CTN-RNA interacts with some of the paraspeckle-related protein components independent of NEAT1 RNA or paraspeckle structure [105], it is possible that paraspeckle-related proteins, possibly together with some other proteins, play major roles in promoting the nuclear retention of IRAlu-containing RNAs. NEAT1 RNA, which mainly provides a multivalent platform to facilitate the formation of paraspeckle structure, modulates the interaction between paraspeckle-related proteins and IRAlu-containing RNAs, and influences the intranuclear compartmentalization of IRAlu-containing RNAs.
The nuclear retention of IRAlu elements is dynamically regulated, and this regulation is mainly achieved through three counteracting mechanisms: 1) The IRAlu element can be removed through alternative polyadenylation. Under normal conditions, CTN-RNA is the major isoform and is mainly located in the nucleus, while under stress conditions, CTN-RNA is post-transcriptionally cleaved through alternative polyadenylation to generate an mCAT2 isoform with a shorter 3ʹUTR lacking the IRAlu element. Thus, the mCAT2 RNA will be exported into the cytoplasm for protein synthesis [98]. 2) For a subset of IRAlu-containing mRNAs, the dsRNA-binding protein STAU1 counteracts the binding of NONO to IRAlu, and promotes the export and translation of IRAlu-containing RNAs [106]. 3) The arginine methyltransferase CARM1 dynamically regulates NONO and IRAlu interaction and the abundance of paraspeckles in response to cellular stresses. Under normal physiological conditions, CARM1 on the one hand methylates NONO and reduces its binding to IRAlu-containing transcripts; on the other hand, CARM1 inhibits the formation of paraspeckles by suppressing the expression of NEAT1. These two effects synergistically reduce the nuclear retention effect mediated by IRAlus [107]. In contrast, under certain cellular stresses, the action of CARM1 is attenuated, which increases the level of unmethylated NONO and NEAT1 expression, thus resulting in enhanced nuclear retention of mRNAs containing IRAlus. Besides, under mitochondrial stress conditions, paraspeckles also participate in regulating the nuclear retention of some mitochondrial mRNAs, a process that depends on the presence of IRAlus or AG- or U-rich motifs in these mRNAs. This regulation mainly occurs via increasing NEAT1 transcription and altering the morphology and numbers of paraspeckles [108]. Thus, NEAT1 and paraspeckle-mediated RNA nuclear retention are dynamically regulated as a part of the cell’s responses when stress conditions are encountered.
3.4. Other genomic repeat elements
Besides SINE repeats, many other transposable elements (TEs) were also reported to exist in mature transcripts, especially in the transcripts of lncRNA genes. A survey of 9241 human lncRNAs suggests that about 83% of lncRNAs contain a TE, and TEs comprise 42% of lncRNA sequences [89]. It is hypothesized that some of these inserted TEs might form functional domains that mediate interaction with proteins or nucleic acids, thus contributing to the function and regulation of their host lncRNAs [109–111]. Based on this hypothesis, Carlevaro-Fita et al thoroughly studied the relationship between TEs and lncRNA subcellular localization in an in-silico approach [12]. They uncovered a significant correlation between four repeat elements (L1PA16, L2b, MIRb, and MIRc) and nuclear retention of RNA. The authors validated three lncRNAs harbouring repeat elements L2b, MIRb, and MIRc and confirmed that the nuclear localization of these lncRNAs depends on the presence of wild-type TE [12]. Intriguingly, the authors also uncovered a dose-dependent effect of TEs on nuclear retention of RNAs. These TEs may cooperate with other nuclear retention elements, synergistically promoting the nuclear retention of their host RNAs.
Microsatellite, also called simple sequence repeats (SSRs) or short tandem repeats (STRs), are another type of genomic repeat that comprises about 3% of the human genome [79]. In microsatellites, a short DNA sequence motif, with lengths ranging from one to six nucleotides, is repeated from 5 to more than 50 times [112]. Abnormal expansion of these repeats is responsible for more than 20 neurological and neuromuscular disorders like myotonic dystrophy (DM), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD) etc [113–115]. For at least some of these diseases, their pathogenesis is partially correlated with nuclear retention of disease gene transcripts, mediated by the presence of the expanded repeat in the RNA [113,116–119] (Table 1). For example, myotonic dystrophy type 1 (DM1) is caused by a CTG trinucleotide expansion in the 3ʹUTR of the Myotonin-protein kinase (DMPK) gene. The RNA transcripts harbouring the expanded CTG repeat are mainly retained in the nucleus and aggregate into nuclear foci. The mutant RNAs contribute to disease pathogenesis by reducing the DMPK protein level and by associating with multiple RBPs involved in RNA processing, thus reducing the availability of these RBPs [113,116]. Interestingly, some RNAs with expanded repeats can form a gel-like structure in vitro and in vivo. The expanded RNAs mainly associate with nuclear speckles, which may contribute to the retention of RNA in the nucleus via a phase-separation mechanism [120].
3.5. Tandem repetitive sequences
The XIST RNA transcript comprises six interspersed repeats: A, B, C, D, E, and F [121]. These repeats are required for the proper function of XIST and X-chromosome inactivation (XCI) [36,37,56,90,122–125]. Cis-fragment deletions and reporter assay analyses suggest that some of these repeats contribute to the nuclear localization and proper chromatin association of XIST [10,11,36,37,90,91,122,126,127]. For instance, deletion of the conserved A-repeat, which consists of 7–8 copies of GC-rich sequences, severely impairs XCI [56]. This deletion does not severely affect the overall X chromosome painting pattern of XIST RNAs detected by RNA FISH [56,128]. However, detailed analysis of RNA–chromatin interactions suggests that loss of the A-repeat impairs the XIST chromatin association, especially at some active gene-dense regions on the X chromosome [126]. Moreover, expressing a 950-nt 5ʹ fragment of XIST containing the A-repeat alone, or fusing the A-repeat with a cytoplasm-localized reporter RNA, causes the majority of RNA transcripts to localize in the nucleus. This indicates that the A-repeat also plays a role in promoting the nuclear retention of XIST RNA [11,127]. The A-repeat mainly interacts with a transcriptional co-repressor protein, SPEN (also known as SHARP), which further recruits the HDAC3 complex to mediate gene silencing and RNA Pol II exclusion [129,130]. Meanwhile, SPEN may also promote the chromatin association of XIST, as depletion of SPEN impairs the chromatin retention of A-repeat-fused GFP reporter RNAs [11]. Also, an LBR (Lamin B receptor) binding region, which located at the downstream of A-repeat and overlap with F-repeat, is required for XCI and targeting of XIST to active genes on X chromosome, mainly through tethering the XIST-coated inactive X chromosome to nuclear lamins [124].
Unlike some other abundant nuclear lncRNAs, which mainly associate with the peripheral chromatin where they are generated (e.g. Haunt [33], Evx1as [32]), or trans-associate with many genomic loci along with nuclear speckle-like structures (e.g. MALAT1 [22,23], NEAT1 [23]), most XIST RNA transcripts ‘spread’ along and ‘coat’ the X chromosome to silence more than 80% of X-linked genes [131]. Intriguingly, RNAs expressed from an ectopic XIST locus on an autosome constantly associate with and mediate the inactivation of the surrounding chromosomal region [132]. This implies that the XIST transcript itself, rather than the X chromosome, or the chromatin environment, plays a major role in determining the specificity of its chromatin association. An interesting question is how is this unique chromatin association pattern achieved? Several studies have systematically investigated the protein interactome of XIST RNA, and revealed that multiple RBPs, transcription regulators, chromatin organizers, etc. interact with XIST RNA directly or indirectly [129,130,133] (Fig. 2C, upper panel). As none of these trans-interacting proteins exclusively interact with XIST, the unique binding pattern of XIST must be regulated by a cooperative regulatory mechanism between these trans-factors and the cis-elements of XIST RNA. Indeed, recently, several studies have indicated that the XIST B and E repeats and their interacting proteins are required for the proper association and spreading of XIST RNA on the X chromosome [36,37,90,91].
Upon depletion of the B-repeat, the ‘cloud-like’ morphology of XIST RNA around the inactivated X chromosome becomes more diffuse, and the spreading of XIST RNA on the X chromosome is severely impaired [90]. The B-repeat consists of multiple C-rich motifs, which are directly bound by HNRNPK, and further recruit the chromatin regulator complexes PRC2 and PRC1 [90,91,134]. Both of these repressive polycomb complexes and the interaction between HNRNPK and the B-repeat are required for the spreading of XIST [90,91]. Intriguingly, it was reported that the polycomb complexes and their marked repressive chromatin domains can be propagated to nearby regions from the nucleation centre where they were initially targeted, partially through modulating nucleosome compaction via a phase separation mechanism [104,135–137]. It has also been reported that HNRNPK can be oligomerized [93,138], and can participate in the formation of paraspeckles [101]. It is possible that the multivalent-interactions of B-repeat-containing RNA, HNRNPK, and the polycomb complexes, create a feed-forward regulatory loop to promote the spreading of XIST RNAs on the X chromosome (Fig. 2C).
Besides the B-repeat, the E-repeat is also required for proper X chromosome association of XIST RNA. Deletion of the E-repeat causes XIST to delocalize from the X chromosome and disperse into the nucleoplasm [36,37]. Mechanistically, the E-repeat acts in two ways. On the one hand, it directly interacts with the nuclear matrix protein CIZ1 (CDKN1-interacting zinc finger protein) to ensure a stable association of XIST RNA with the X chromosome [36,37]. On the other hand, the E-repeat provides a multivalent platform for assembly of the RBPs PTBP1, MATR3, TDP-43, and CELF. These proteins further promote the anchoring of XIST to Xi territory through a phase-separation mechanism [139]. As both the B-repeat and E-repeat are required for proper spreading of XIST RNA along the X chromosome, these two phase-separation-related mechanisms may work in concert with each other to ensure that Xi is coated with XIST RNAs (Fig. 2C, bottom panel).
Depleting another nuclear matrix protein HNRNPU also impaired proper XIST chromatin tethering and abolished XCI, in a mechanism independent of CIZ1 [37,140]. It has been reported that HNRNPU participates in regulating the nuclear retention or chromatin association of other RNAs. For instance, another X chromosome-linked lncRNA, FIRRE, which interacts with HNRNPU through multiple intrinsically repeating RNA domains (RRD) embedded in its transcripts. The interaction between HNRNPU and RRDs promotes nuclear retention and trans-targeting of FIRRE to other chromosomes [141,142]. Moreover, HNRNPU can pervasively interact with chromatin-associated RNAs, and further modulates chromatin structure through its oligomerization in a transcription-dependent manner [143]. The mechanism of how HNRNPU promotes the chromatin tethering of these RNAs is still elusive. However, it prevalently binds to thousands of RNAs derived from both autosomes and inactivated X chromosomes [143,144], and, in contrast to other XCI regulators, it does not exhibit an obviously enriched binding signal on the inactivated X chromosome [37]. These observations imply that HNRNPU may be a general regulator of RNA chromatin tethering, or may act indirectly through modulating the overall nuclear organization. This is consistent with the fact that HNRNPU function was initially reported to function as a nuclear scaffold attachment factor [145].
3.6. R-loop and RNA-DNA triplex
The above-mentioned mechanisms for retaining RNA in nuclei and on chromatin mainly occur indirectly, through interactions between the RNAs and other factors. It has been reported that some RNAs can associate with their chromatin target directly, mainly through forming R-loop or RNA-DNA triplex structures. Both of these structures are formed by three strands of nucleic acid, including two complementary DNA strands and one single-stranded RNA. Within an R-loop the RNA hybridizes with one of the DNA strands, leaving the other one single-stranded [146]. In the RNA-DNA triplex, on the other hand, the complementary DNA strands hybridize, and the RNA inserts into the major groove of the duplex structure with sequence specificity [147] (Fig. 2D).
The most well-known example of R-loop-mediated RNA-chromatin association and targeting is the CRISPR/Cas system, which is a natural part of the bacterial adaptive immune response against virus infection [148]. The system is now widely used for genome editing and gene expression regulation (reviewed in [149,150]). The most attractive part of this system is that the specificity of the targeting site is controlled by a short RNA sequence, the guide RNA (gRNA), which mediates targeting of the CRISPR/Cas complex through RNA-DNA base-pairing, and finally forms an R-loop structure with the target DNA [151,152]. Intriguingly, this system was engineered to fuse lncRNA transcripts with the gRNA, in order to target lncRNAs to specific chromatin sites [153]. In eukaryotic cells, the R-loop mechanism has been adopted by a few genes for chromatin association of their transcripts. For example, a recent study suggests that TERRA, a lncRNA transcribed from chromosome ends [154], is recruited to chromosome ends through an R-loop-dependent mechanism [155]. However, as the formation of R-loops may trigger genome instability [47,146], cases of RNA targeting via this mechanism are relatively scarce.
Some lncRNAs can also directly associate with their chromatin targets through forming RNA-DNA triplex structures [147,156–160]. This structure usually depends on specific sequence features such as GA-rich homopurine sequences [147]. The RNA-DNA triplex mechanism can mediate the chromatin targeting of RNAs both in cis and in trans. For instance, the human gene encoding dihydrofolate reductase (DHFR) contains two promoters. In quiescent cells, the major promoter of DHFR is repressed, in a manner dependent on a non-coding transcript transcribed from the upstream minor promoter. The noncoding transcripts specifically target the major promoter, which is GC-rich and contains several G-track sequences, by forming a stable triplex structure with it [160]. The targeted non-coding transcripts further repress transcription initiation from the major promoter through interaction with transcription factor TFIIB to dissociate the preinitiation complex from the major promoter [160]. In another example, lncRNA Khps1, which is transcribed in an antisense orientation from the proto-oncogene SPHK1, anchors the promoter region of SPHK1 by forming an RNA-DNA triplex structure with a homopurine stretch upstream of the transcription start site of SPHK1 [157]. For the lncRNA MEG3, on the other hand, the RNA-DNA triplex guides its transcripts to bind to its chromatin targets in trans. Analysis of the targeted chromatin binding sites of MEG3 suggests that they are GA-rich sequences, and the triplex structures are mainly formed by the target sequences and the triplex-forming oligonucleotides (TFOs) located at the 5ʹ of MEG3 RNA [156]. In addition, motif analysis of the lncRNAs HOTAIR and roX2 also found that their targeting sites contain GA-enriched motifs [158], which implies that RNA-DNA triplex structures might also participate in their chromatin association. Nevertheless, GA-rich homopurine sequences are prevalently distributed in the genome, which suggests that other mechanisms are also required to cooperatively determine the specificity of the targeting, especially for the trans-targeting of lncRNAs.
3.7. SnoRNA end
Besides the principal mechanisms that may play general roles in regulating RNA nuclear retention mentioned above, additional mechanisms have also been reported that are related to the nuclear retention of specific sets of RNAs or a specific RNA. For instance, small nucleolar RNAs (snoRNAs) are a family of conserved, nucleus-localized noncoding RNAs that function in RNA modification and rRNA processing [161]. The majority of snoRNA are generated from the introns of snoRNA host genes and protein-coding genes [162]. Intriguingly, the processing of some intronic snoRNAs will generate a class of sno-lncRNAs, whose ends correspond to the positions of intronic snoRNAs. These sno-lncRNAs mainly localize in the nucleus and may associate with specific nuclear bodies such as Cajal bodies and the nucleolus [163]. Notably, fusing snoRNAs to the ends of cytoplasm-localized transcripts will promote the nuclear retention of the fusion-RNAs [164]. Mechanistically, the nuclear retention of sno-lncRNAs depends on their snoRNA ends, which likely recruit interacting partners involved in the function and processing of snoRNAs [163,165,166].
Conclusion and perspectives
In summary, the mechanisms regulating the nuclear retention of noncoding RNAs can be mainly divided into three main categories: 1) association of RNA transcript, via specific nucleotide sequence motifs, with nucleus-localized factors like U1 snRNP, HNRNPK etc., to promote nuclear retention or chromatin association of the RNA (Fig.s 2A-Fig.s 2C); 2) RNA–DNA interactions, which mainly occur through the formation of R-loop or RNA-DNA triplex structures (Fig. 2D); and 3) RNA decay mediated by nuclear surveillance pathways, which degrade some noncoding transcripts before they have a chance to be exported to the cytoplasm (Fig. 1D). Notably, these mechanisms often work together to synergistically promote nuclear localization of RNA. For instance, U1 snRNP can function to promote both chromatin tethering and decay for some of its RNA targets [11]. SPEN-mediated nuclear retention of A-repeat-fused reporter RNA also decreases the expression level of the reporter [11]. It has been proposed that decay is a default fate for nuclear RNA [44]. However, for some stable lncRNAs like MALAT1, NEAT1, etc., other elements, such as those mediating triple-helix structures, may stabilize their transcripts [167,168].
The nuclear retention of an RNA is often regulated by multiple regulators. The best example is XIST. XIST harbours multiple exonic tandem repeat sequences, including the A-, B-, and E- repeats as well as some sequences located downstream of the E-repeat in the last exon, which promote XIST-chromatin association through interaction with different factors [11,36,37,90,91]. These regulators function synergistically to promote proper chromatin tethering and spreading of XIST RNA. Another important observation is that most of the regulators participating in RNA nuclear retention harbour low-complexity sequences (LCS), and some of them can form droplet- or puncta-like structures in vitro and in vivo [55,135,169–171]. Furthermore, many nucleus-retained RNAs are associated with membraneless nuclear organelles like nuclear speckles and paraspeckles [38,55,102]. These findings suggest that a phase-separation mechanism may participate in the regulation of RNA nuclear retention. The detailed mechanism of how phase separation participates in regulating nuclear retention, especially in chromatin tethering, needs to be investigated in the future.
It should be noted that this review focuses on known cis-element and trans-factors, and therefore summarizes only a small portion of the mechanisms, that regulate of RNA nuclear retention. Other factors, such as transcriptional regulation, chromatin environment, RNA modifications, and cellular conditions etc., are also reported or presumed to be involved in this process [172]. Future studies are required to integrate the information derived from all these dimensions, in order to systematically investigate the mechanism and regulation of RNA nuclear retention.
Acknowledgments
We thank Xiaoyu Li, Yantao Hong, Tong Li, and Lingqing Xing for critical reading and helpful discussions. We regret that it is not possible to cite all the publications related to this review due to space limitations. This work was supported by grants 31900439 from the National Natural Science Foundation of China, and 419000-11111/043 from Zhejiang University Education Foundation.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31900439]; Zhejiang University Education Foundation [419000-11111/043].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hon CC, Ramilowski JA, Harshbarger J, et al. An atlas of human long non-coding RNAs with accurate 5ʹ ends. Nature. 2017;543(7644):199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Flynn RA, Almada AE, Zamudio JR, et al. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A. 2011;108(26):10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Seila AC, Calabrese JM, Levine SS, et al. Divergent transcription from active promoters. Science. 2008;322(5909):1849–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li W, Notani D, Rosenfeld MG.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17(4):207–223. [DOI] [PubMed] [Google Scholar]
- [6].Tilgner H, Knowles DG, Johnson R, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22(9):1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kohler A, Hurt E.. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8(10):761–773. [DOI] [PubMed] [Google Scholar]
- [8].Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16(7):431–442. [DOI] [PubMed] [Google Scholar]
- [9].Lubelsky Y, Ulitsky I. Sequences enriched in alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555(7694):107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shukla CJ, McCorkindale AL, Gerhardinger C, et al. High-throughput identification of RNA nuclear enrichment sequences. Embo J. 2018;37(6):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yin Y, Lu JY, Zhang X, et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature. 2020;580(7801):147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carlevaro-Fita J, Polidori T, Das M, et al. Ancient exapted transposable elements promote nuclear enrichment of human long noncoding RNAs. Genome Res. 2019;29(2):208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo CJ, Ma XK, Xing YH, et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell. 2020;181(3):621–636 e22. [DOI] [PubMed] [Google Scholar]
- [14].Guo CJ, Xu G, Chen LL. Mechanisms of long noncoding RNA nuclear retention. Trends Biochem Sci. 2020;45(11):947–960. [DOI] [PubMed] [Google Scholar]
- [15].Palazzo AF, Lee ES. sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet. 2018;9:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wegener M, Muller-McNicoll M. Nuclear retention of mRNAs - quality control, gene regulation and human disease. Semin Cell Dev Biol. 2018;79:131–142. [DOI] [PubMed] [Google Scholar]
- [17].Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kopp F, Mendell JT, Classification F. experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3(2):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Quinodoz SA, Ollikainen N, Tabak B, et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174(3):744–757 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo YE, Manteiga JC, Henninger JE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572(7770):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Engreitz JM, Sirokman K, McDonel P, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].West JA, Davis CP, Sunwoo H, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55(5):791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Braunschweig U, Barbosa-Morais NL, Pan Q, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24(11):1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yap K, Lim ZQ, Khandelia P, et al. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26(11):1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29(1):63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shalgi R, Hurt JA, Lindquist S, et al. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7(5):1362–1370. [DOI] [PubMed] [Google Scholar]
- [28].Mauger O, Lemoine F, Scheiffele P. targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016;92(6):1266–1278. [DOI] [PubMed] [Google Scholar]
- [29].Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41(9):761–772. [DOI] [PubMed] [Google Scholar]
- [31].Schlackow M, Nojima T, Gomes T, et al. distinctive patterns of transcription and rna processing for human lincRNAs. Mol Cell. 2017;65(1):25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Luo S, Lu Y, Liu L, et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18(5):637–652. [DOI] [PubMed] [Google Scholar]
- [33].Yin Y, Yan P, Lu J, et al. Opposing roles for the lncRNA haunt and its genomic locus in regulating hoxa gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015;16(5):504–516. [DOI] [PubMed] [Google Scholar]
- [34].Penny GD, Kay GF, Sheardown SA, et al. Requirement for XIST in X chromosome inactivation. Nature. 1996;379(6561):131–137. [DOI] [PubMed] [Google Scholar]
- [35].Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. [DOI] [PubMed] [Google Scholar]
- [36].Ridings-Figueroa R, Stewart ER, Nesterova TB, et al. The nuclear matrix protein CIZ1 facilitates localization of XIST RNA to the inactive X-chromosome territory. Genes Dev. 2017;31(9):876–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sunwoo H, Colognori D, Froberg JE, et al. Repeat E anchors XIST RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc Natl Acad Sci U S A. 2017;114(40):10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154(5):996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bresson S, Tollervey D. Surveillance-ready transcription: nuclear RNA decay as a default fate. Open Biol. 2018;8(3):170270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking XIST RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci U S A. 2001;98(18):10232–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pefanis E, Wang J, Rothschild G, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161(4):774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu J, Dou X, Chen C, et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367(6477):580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ogami K, Richard P, Chen Y, et al. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017;31(12):1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cohen HR, Panning B. XIST RNA exhibits nuclear retention and exhibits reduced association with the export factor TAP/NXF1. Chromosoma. 2007;116(4):373–383. [DOI] [PubMed] [Google Scholar]
- [51].Nappi F, Schneider R, Zolotukhin A, et al. Identification of a novel posttranscriptional regulatory element by using a rev- and RRE-mutated human immunodeficiency virus type 1 DNA proviral clone as a molecular trap. J Virol. 2001;75(10):4558–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hammarskjold ML. Constitutive transport element-mediated nuclear export. Curr Top Microbiol Immunol. 2001;259:77–93. [DOI] [PubMed] [Google Scholar]
- [53].Conrad NK, Steitz JA, Kaposi’s A. sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. Embo J. 2005;24(10):1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang B, Gunawardane L, Niazi F, et al. A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA. Mol Cell Biol. 2014;34(12):2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yamazaki T, Souquere S, Chujo T, et al. Functional domains of NEAT1 architectural lncrna induce paraspeckle assembly through phase separation. Mol Cell. 2018;70(6):1038–1053 e7. [DOI] [PubMed] [Google Scholar]
- [56].Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of XIST RNA. Nat Genet. 2002;30(2):167–174. [DOI] [PubMed] [Google Scholar]
- [57].Miyagawa R, Tano K, Mizuno R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18(4):738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Melnikov A, Murugan A, Zhang X, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30(3):271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Arnold CD, Gerlach D, Stelzer C, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339(6123):1074–1077. [DOI] [PubMed] [Google Scholar]
- [60].Ogami K, Chen Y, Manley JL. RNA surveillance by the nuclear RNA exosome: mechanisms and significance. Noncoding RNA. 2018;4(1). DOI: 10.3390/ncrna4010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Garland W, Jensen TH. Nuclear sorting of RNA. Wiley Interdiscip Rev RNA. 2020;11(2):e1572. [DOI] [PubMed] [Google Scholar]
- [62].Lerner MR, Boyle JA, Mount SM, et al. Are snRNPs involved in splicing? Nature. 1980;283(5743):220–224. [DOI] [PubMed] [Google Scholar]
- [63].Kaida D, Berg MG, Younis I, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468(7324):664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7):a003707-a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Baserga SJ, Steitz JA. The diverse world of small ribonucleoproteins. In: The RNA World. Gesteland RF and Atkins JF, eds. (Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press). 1993. p. 359–381. [Google Scholar]
- [66].Azam S, Hou S, Zhu B, et al. Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus. RNA Biol. 2019;16(8):1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee ES, Akef A, Mahadevan K, et al. The consensus 5ʹ splice site motif inhibits mRNA nuclear export. PLoS One. 2015;10(3):e0122743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kammler S, Leurs C, Freund M, et al. The sequence complementarity between HIV-1 5ʹ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA. 2001;7(3):421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lutzelberger M, Reinert LS, Das AT, et al. A novel splice donor site in the gag-pol gene is required for HIV-1 RNA stability. J Biol Chem. 2006;281(27):18644–18651. [DOI] [PubMed] [Google Scholar]
- [70].Huang Y, Carmichael GG. A suboptimal 5ʹ splice site is a cis-acting determinant of nuclear export of polyomavirus late mRNAs. Mol Cell Biol. 1996;16(11):6046–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mele M, Mattioli K, Mallard W, et al. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27(1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Krchnakova Z, Thakur PK, Krausova M, et al. Splicing of long non-coding RNAs primarily depends on polypyrimidine tract and 5ʹ splice-site sequences due to weak interactions with SR proteins. Nucleic Acids Res. 2019;47(2):911–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nojima T, Rebelo K, Gomes T, et al. RNA polymerase II phosphorylated on CTD serine 5 interacts with the spliceosome during co-transcriptional splicing. Mol Cell. 2018;72(2):369–379 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol. 2011;12(6):377–384. [DOI] [PubMed] [Google Scholar]
- [75].Boelens WC, Jansen EJ, Van Venrooij WJ, et al. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72(6):881–892. [DOI] [PubMed] [Google Scholar]
- [76].Chiu AC, Suzuki HI, Wu X, et al. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol Cell. 2018;69(4):648–663 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Berg MG, Singh LN, Younis I, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150(1):53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Almada AE, Wu X, Kriz AJ, et al. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499(7458):360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- [80].Sela N, Mersch B, Gal-Mark N, et al. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8(6):R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Versteeg R, Van Schaik BD, Van Batenburg MF, et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13(9):1998–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. [DOI] [PubMed] [Google Scholar]
- [83].Daniel C, Behm M, Ohman M. The role of Alu elements in the cis-regulation of RNA processing. Cell Mol Life Sci. 2015;72(21):4063–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12(7):1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Makalowski W, Mitchell GA, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10(6):188–193. [DOI] [PubMed] [Google Scholar]
- [86].Lev-Maor G, Sorek R, Shomron N, et al. The birth of an alternatively spliced exon: 3ʹ splice-site selection in Alu exons. Science. 2003;300(5623):1288–1291. [DOI] [PubMed] [Google Scholar]
- [87].Zarnack K, Konig J, Tajnik M, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. Embo J. 2008;27(12):1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13(11):R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Colognori D, Sunwoo H, Kriz AJ, et al. XIST deletional analysis reveals an interdependency between XIST RNA and polycomb complexes for spreading along the inactive X. Mol Cell. 2019;74(1):101–117 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Pintacuda G, Wei G, Roustan C, et al. hnRNPK Recruits PCGF3/5-PRC1 to the XIST RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell. 2017;68(5):955–969 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cai Z, Cao C, Ji L, et al. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature. 2020;582(7812):432–437. [DOI] [PubMed] [Google Scholar]
- [94].Mikula M, Bomsztyk K, Goryca K, et al. Heterogeneous nuclear ribonucleoprotein (HnRNP) K genome-wide binding survey reveals its role in regulating 3ʹ-end RNA processing and transcription termination at the early growth response 1 (EGR1) gene through XRN2 exonuclease. J Biol Chem. 2013;288(34):24788–24798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li J, Chen Y, Xu X, et al. HNRNPK maintains epidermal progenitor function through transcription of proliferation genes and degrading differentiation promoting mRNAs. Nat Commun. 2019;10(1):4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Habelhah H, Shah K, Huang L, et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3(3):325–330. [DOI] [PubMed] [Google Scholar]
- [97].Pettit Kneller EL, Connor JH, D.S. Lyles, hnRNPs Relocalize to the cytoplasm following infection with vesicular stomatitis virus. J Virol. 2009;83(2):770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Prasanth KV, Prasanth SG, Xuan Z, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. [DOI] [PubMed] [Google Scholar]
- [99].Chillon I, Pyle AM. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 2016;44(19):9462–9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16(11):5304–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Naganuma T, Nakagawa S, Tanigawa A, et al. Alternative 3ʹ-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. Embo J. 2012;31(20):4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li P, Banjade S, Cheng HC, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Anantharaman A, Jadaliha M, Tripathi V, et al. Paraspeckles modulate the intranuclear distribution of paraspeckle-associated Ctn RNA. Sci Rep. 2016;6(1):34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Elbarbary RA, Li W, Tian B, et al. STAU1 binding 3ʹ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27(13):1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hu SB, Xiang JF, Li X, et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 2015;29(6):630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang Y, Hu SB, Wang MR, et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol. 2018;20(10):1145–1158. [DOI] [PubMed] [Google Scholar]
- [109].Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Johnson R, Guigo R. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA. 2014;20(7):959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. [DOI] [PubMed] [Google Scholar]
- [112].Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. [DOI] [PubMed] [Google Scholar]
- [113].La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11(4):247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Davis BM, McCurrach ME, Taneja KL, et al. Expansion of a CUG trinucleotide repeat in the 3ʹ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A. 1997;94(14):7388–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Chew J, Gendron TF, Prudencio M, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348(6239):1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Conlon EG, Lu L, Sharma A, et al. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife. 2016;5. DOI: 10.7554/eLife.17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sun X, Li PP, Zhu S, et al. Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci Rep. 2015;5(1):12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546(7657):243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Brockdorff N, Ashworth A, Kay GF, et al. tThe product of the mouse XIST gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. [DOI] [PubMed] [Google Scholar]
- [122].Yamada N, Hasegawa Y, Yue M, et al. XIST Exon 7 Contributes to the Stable Localization of XIST RNA on the Inactive X-Chromosome. PLoS Genet. 2015;11(8):e1005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lv Q, Yuan L, Song Y, et al. D-repeat in the XIST gene is required for X chromosome inactivation. RNA Biol. 2016;13(2):172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Chen CK, Blanco M, Jackson C, et al. XIST recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354(6311):468–472. [DOI] [PubMed] [Google Scholar]
- [125].Jeon Y, Lee JT. YY1 tethers XIST RNA to the inactive X nucleation center. Cell. 2011;146(1):119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Engreitz JM, Pandya-Jones A, McDonel P, et al. The XIST lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Chigi Y, Sasaki H, Sado T. The 5ʹ region of XIST RNA has the potential to associate with chromatin through the A-repeat. RNA. 2017;23(12):1894–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Chaumeil J, Le Baccon P, Wutz A, et al. A novel role for XIST RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].McHugh CA, Chen CK, Chow A, et al. The XIST lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chu C, Zhang QC, Da Rocha ST, et al. Systematic discovery of XIST RNA binding proteins. Cell. 2015;161(2):404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. [DOI] [PubMed] [Google Scholar]
- [132].Hall LL, Byron M, Sakai K, et al. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U S A. 2002;99(13):8677–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Minajigi A, Froberg J, Wei C, et al. Chromosomes. A comprehensive XIST interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245):6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Almeida M, Pintacuda G, Masui O, et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356(6342):1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Plys AJ, Davis CP, Kim J, et al. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33(13–14):799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Oksuz O, Narendra V, Lee CH, et al. Capturing the Onset of PRC2-mediated repressive domain formation. Mol Cell. 2018;70(6):1149–1162 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Yuan W, Wu T, Fu H, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337(6097):971–975. [DOI] [PubMed] [Google Scholar]
- [138].Backe PH, Messias AC, Ravelli RB, et al. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13(7):1055–1067. [DOI] [PubMed] [Google Scholar]
- [139].Pandya-Jones A, Markaki Y, Serizay J, et al. A protein assembly mediates XIST localization and gene silencing. Nature. 2020;587(7832):145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hasegawa Y, Brockdorff N, Kawano S, et al. The matrix protein hnRNP U is required for chromosomal localization of XIST RNA. Dev Cell. 2010;19(3):469–476. [DOI] [PubMed] [Google Scholar]
- [141].Hacisuleyman E, Shukla CJ, Weiner CL, et al. Function and evolution of local repeats in the FIRRE locus. Nat Commun. 2016;7(1):11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Hacisuleyman E, Goff LA, Trapnell C, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA FIRRE. Nat Struct Mol Biol. 2014;21(2):198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Nozawa RS, Boteva L, Soares DC, et al. SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell. 2017;169(7):1214–1227 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Van Nostrand EL, Pratt GA, Yee BA, et al. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol. 2020;21(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Romig H, Fackelmayer FO, Renz A, et al. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. Embo J. 1992;11(9):3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: r loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28(13):1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Li Y, Syed J, Sugiyama H, et al. Formation by long noncoding RNAs. Cell Chem Biol. 2016;23(11):1325–1333. [DOI] [PubMed] [Google Scholar]
- [148].Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [DOI] [PubMed] [Google Scholar]
- [151].Jiang F, Taylor DW, Chen JS, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351(6275):867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Szczelkun MD, Tikhomirova MS, Sinkunas T, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111(27):9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Shechner DM, Hacisuleyman E, Younger ST, et al. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12(7):664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Azzalin CM, Reichenbach P, Khoriauli L, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. [DOI] [PubMed] [Google Scholar]
- [155].Feretzaki M, Pospisilova M, Valador Fernandes R, et al. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. 2020;587(7833):303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Mondal T, Subhash S, Vaid R, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6(1):7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell. 2015;60(4):626–636. [DOI] [PubMed] [Google Scholar]
- [158].Chu C, Qu K, Zhong FL, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Schmitz KM, Mayer C, Postepska A, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24(20):2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. [DOI] [PubMed] [Google Scholar]
- [161].Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8(3):209–220. [DOI] [PubMed] [Google Scholar]
- [162].Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14(3):319–327. [DOI] [PubMed] [Google Scholar]
- [163].Yin QF, Yang L, Zhang Y, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48(2):219–230. [DOI] [PubMed] [Google Scholar]
- [164].Yin Q-F, Hu S-B, Xu Y-F, et al. SnoVectors for nuclear expression of RNA. Nucleic Acids Res. 2015;43(1):e5–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Wu H, Yin QF, Luo Z, et al. Unusual Processing Generates SPA LncRNAs that Sequester Multiple RNA Binding Proteins. Mol Cell. 2016;64(3):534–548. [DOI] [PubMed] [Google Scholar]
- [166].Xing YH, Yao RW, Zhang Y, et al. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell. 2017;169(4):664–678 e16. [DOI] [PubMed] [Google Scholar]
- [167].Brown JA, Valenstein ML, Yario TA, et al. Formation of triple-helical structures by the 3ʹ-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109(47):19202–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Wilusz JE, JnBaptiste CK, Lu LY, et al. A triple helix stabilizes the 3ʹ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26(21):2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Cerase A, Armaos A, Neumayer C, et al. Phase separation drives X-chromosome inactivation: a hypothesis. Nat Struct Mol Biol. 2019;26(5):331–334. [DOI] [PubMed] [Google Scholar]
- [170].Greig JA, Nguyen TA, Lee M, et al. Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol Cell. 2020;77(6):1237–1250 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Xue S, Gong R, He F, et al. Low-complexity domain of U1-70K modulates phase separation and aggregation through distinctive basic-acidic motifs. Sci Adv. 2019;5(11):eaax5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Gandhi M, Caudron-Herger M, Diederichs S. RNA motifs and combinatorial prediction of interactions, stability and localization of noncoding RNAs. Nat Struct Mol Biol. 2018;12:1070–1076. DOI: 10.1038/s41594-018-0155-0 [DOI] [PubMed] [Google Scholar]


