Abstract
The emergence of plasmid-mediated tigecycline resistance gene tet(X4) poses a challenging threat to public health. Based on the analysis of tet(X4)-positive plasmids in the NCBI database, we found that the IncX1-type plasmid is one of the most common vectors for spreading tet(X4) gene, but the mechanisms by which these plasmids adapt to host bacteria and maintain the persistence of antibiotic resistance genes (ARGs) remain unclear. Herein, we investigated the underlying mechanisms of how host bacteria modulate the fitness cost of IncX1 plasmids carrying tet(X4) gene. Interestingly, we found that the tet(X4)-bearing IncX1 plasmids encoding H-NS protein imposed low or no fitness cost in Escherichia coli and Klebsiella pneumoniae; instead, they partially promoted the virulence and biofilm formation in host bacteria. Regression analysis revealed that the expression of hns gene in plasmids was positively linked to the relative fitness of host bacteria. Furthermore, when pCE2::hns was introduced, the fitness of tet(X4)-positive IncX1 plasmid pRF55-1 without hns gene was significantly improved, indicating that hns mediates the improvement of fitness. Finally, we showed that the expression of hns gene is negatively correlated with the expression of tet(X4) gene, suggesting that the regulatory effect of H-NS on adaptability may be attributed to its inhibitory effect on the expression of ARGs. Together, our findings suggest the important role of plasmid-encoded H-NS protein in modulating the fitness of tet(X4)-bearing IncX1 plasmids, which shed new insight into the dissemination of tet(X4) gene in a biological environment.
Keywords: H-NS protein, fitness, tet(X4), IncX1 plasmids, Gram-negative bacteria (GNB)
Introduction
Antibiotic resistance has constituted a growing threat to global public health. According to the data released by WHO, it is estimated that by 2050, the number of human deaths caused by multiple-drug resistance will increase to 10 million, surpassing the number of cancer deaths and becoming one of the leading causes of human death worldwide (Kuehn, 2011). Because of the widespread distribution of carbapenem resistance gene, blaNDM (Nordmann et al., 2011), and polymyxin resistance gene, mcr-1 (Liu et al., 2016), worldwide, the clinical efficacy of carbapenems and polymyxin, two high-priority agents for treatment of multidrug-resistant (MDR) Gram-negative bacterial infections (Kumarasamy et al., 2010; Rodríguez-Baño et al., 2018), was severely diminished. Therefore, tigecycline is recognized as the last option for treating serious infections (Tasina et al., 2011). However, with the emergence of high-level tigecycline resistance gene tet(X3/X4) (He et al., 2019; Sun et al., 2019), few choices were left for clinicians from the traditional antibiotic pipeline. Conjugable plasmid-mediated horizontal gene transfer is the dominant pathway accounting for the spread of antibiotic resistance genes (ARGs) (San Millan, 2018; Lerminiaux and Cameron, 2019). Plasmids, as mobile genetic elements, mediate the transmission of genetic information between bacteria and promote the adaptation of bacteria to various environments and the development of bacterial resistance (Lopatkin et al., 2017). Nevertheless, the high expression of plasmid-related genes would impose additional energy and metabolic burden to host bacteria, thereby resulting in fitness cost (San Millan and MacLean, 2017). Accordingly, the fitness cost elicited by resistance plasmids is closely associated with the dissemination of ARGs (Alonso-Del Valle et al., 2021). Since its first description in China in 2019, tet(X4)-bearing plasmids have been found worldwide (Ding et al., 2020; Zhang et al., 2020; Mohsin et al., 2021). For example, it is reported that tet(X4)-positive plasmids were widely distributed in the gut microbiota of Singaporeans (Ding et al., 2020). Besides, tet(X4)-bearing plasmids were identified from poultry, food, and environmental samples in South Asia (Mohsin et al., 2021). The prevalence of tet(X4) gene has posed a great challenge to human safety (Fu et al., 2021). IncX1 plasmid is one of the important vectors of tet(X4) gene (Li et al., 2020b), but the physiological mechanisms of widespread of tet(X4)-harboring IncX1 plasmids in different host strains are still unclear.
Histone-like nucleoid structuring (H-NS) protein is a kind of DNA-binding protein, which exists widely in Gram-negative bacteria (Navarre et al., 2006). H-NS protein not only participates in the regulation of cell metabolism and cellular stress response, but also responds to various environmental changes, such as pH, metal ion concentration, osmotic pressure, temperature, and stringent response (Gao et al., 2018). Notably, H-NS protein can act as a transcriptional inhibitor, silencing the expression of many genes (Dorman, 2007), including pathogenicity islands (SPIs) and fimbriae gene pef in Salmonella (Ali et al., 2014; Hurtado-Escobar et al., 2019), virulence genes virF, virB, and icsA/B/P in Shigella (Picker and Wing, 2016) and some costly conjugation genes (Doyle et al., 2007). The basis of H-NS silencing gene is its ability to target an AT-rich sequence, thus hindering its transcription (Dorman, 2014). As a global regulator, it is indicated that H-NS can regulate bacteria fitness by inhibiting the expression of certain genes. For example, the overexpression of the Salmonella pathogenicity island SPI2 generally acquired by horizontal gene transfer can lead to the growth defect of Salmonella, and H-NS inhibited the expression of SPI2 to improve the fitness of Salmonella (Lucchini et al., 2006). It was found that some plasmids also have genes encoding H-NS family proteins, and the deletion of hns decreases the fitness of host bacteria (Doyle et al., 2007), indicating that H-NS is likely to mediate the fitness cost of plasmids. A recent study showed that H-NS plays an important role in the dissemination of blaNDM–1-bearing IncX3 Plasmid in Escherichia coli by regulating the expression of plasmid-related genes (Liu et al., 2020).
In this study, we explored the role of H-NS protein in the prevalence of tet(X4)-bearing IncX1 plasmids by a series of assays, including bacterial growth curve, relative fitness evaluation, and virulence assay. Most importantly, we investigated the correlation between the expression of hns and fitness cost of tet(X4)-positive IncX1 plasmids in various host bacteria. Our data show that H-NS plays a crucial role in alleviating the fitness cost of plasmids, which may explain the successful epidemic of tet(X4)-positive IncX1 plasmids in the clinical setting and provide new insights for addressing the global bacterial resistance crisis.
Materials and Methods
Bacterial Strains and Plasmids
The accession numbers of 51 tet(X4)-positive plasmids from NCBI databases are listed in Supplementary Table 1. Three tet(X4)-positive plasmids carrying hns gene used in this experiment are shown in Supplementary Table 2 and Supplementary Figure 1, in which pRF14-1 plasmid was isolated from the slaughterhouse of Jiangsu Province, China (Li et al., 2020b), and the other two plasmids pSC4R and pHS14-2 were isolated from Chinese pork samples. Standard strains involving E. coli TOP10, Klebsiella pneumoniae ATCC700603, and Salmonella Enteritidis ATCC13076 were used in this experiment.
Plasmid Extraction, Whole Genome Sequencing, and Bioinformatics Analysis
Plasmids were extracted using the Plasmid Mini Kit I (Omega, China). Combined with the highly accurate short-read Illumina and long-read Oxford Nanopore Technologies (ONT) MinION platforms, WGS and de novo assembly using the hybrid strategy was performed. Plasmid sequences were annotated using the RAST1 automatically and modified manually. BRIG (Alikhan et al., 2011) and Easyfig (Sullivan et al., 2011) tools were used to visualize the genetic comparisons.
Preparation of Competent Cells and Electroporation Experiment
As described in a previous study (Choi et al., 2006), the E. coli TOP10, K. pneumoniae ATCC700603, and S. Enteritidis ATCC13076 were cultured in LB broth to the logarithmic growth phase at 37°C and 200 rpm. The strains were precooled for 30 min and centrifuged at 4°C and 5,000 rpm for 5 min. Then, the clumps of bacteria are washed with water once and with 10% glycerin three times. Bacterial liquid (50 ml) was condensed into three tubes of 100 μl of competent cells.
A tube of competent cells was taken and then mixed with the plasmids including pRF14-1, pSC4R, and pHS14-2 separately (the volume is not greater than 10% of the competent cells volume). Immediately after electroporation, 1 ml of LB was added to the competent cells and recovered in 37°C and 200 rpm for 1 h. Then, the positive strains were screened by agar plate with tigecycline (2 μg/ml) and polymerase chain reaction (PCR) was used to verify whether the plasmid was successfully transferred into the cell. Nine transformants were obtained.
Bacterial Growth Curve Assay
According to a previous method (Liu et al., 2020), overnight cultures of strains carrying pRF14-1, pSC4R, pHS14-2, and its recipient bacteria were diluted 1:1,000 into fresh LB broth and were incubated at 37°C with 200 rpm for 12 h. Growth curves were obtained by measuring the optical density of cultures at 620 nm every hour by Multiskan FC (Thermo Fisher Scientific). All experiments were conducted in triplicate and repeated three times independently, and the average values were used to estimate growth parameters.
Plasmid Stability Experiment
Plasmid stability testing was performed by the serial passage method for 25 consecutive days at 1:1,000 dilutions without antibiotic pressure (two generations of growth per day) (Gao et al., 2020). Strains with different plasmids were propagated in antibiotic-free LB medium at 37°C with 200 rpm and shaken for 25 days (50 generations) to determine their stability in a different strain background. PCR was conducted every 10 generations to detect the plasmid and tet(X4).
In vitro Competition Experiment
Overnight cultures of plasmid-carrying clone and corresponding recipient bacteria were diluted 1:1,000 in LB broth and mixed at 1:1 ratio. Then, this mixture was incubated for 3 days at 37°C with 200 rpm, and diluted 1:100 into fresh LB broth every 24 h. The competition mixture at 0, 24, 48, and 72 h was plated on LB agar without drug and LB agar containing tigecycline with proper dilution to count the colony numbers. The formula Wt = ln(Nf, R/Ni,R)/ln (Nf, S/Ni, S) was used to calculate the Wt, namely, relative fitness of plasmid-carrying strain (R) compared to the recipient strain (S). Ni,R and Nf,R are the numbers of cells of R at the beginning and end of the competition, and Ni, S and Nf, S are the densities of cells of S at the start and end of the competition, respectively (DelaFuente et al., 2020). Each experiment was set in three parallel and repeated three times.
Biofilm Formation Assay
Biofilm formation assay was performed using crystal violet staining (Rossi Gonçalves et al., 2017). Specifically, 200 μl of bacteria suspension containing 1 × 106 CFU/ml prepared in LB broth was added to 96-well polystyrene plates (flat bottom with cover, aseptic) and incubated at 37°C for 36 h, at which time the biofilm had been attached to the bottom and wall of the well, and the culture medium was carefully removed. The wells were washed with PBS twice and fixed 15 min with methanol of 50 μl. The methanol was removed and the 0.1% crystal purple solution of 100 μl was added to stain 30 min. After cleaning the wells with PBS, 33% glacial acetic acid solution of 100 μl was added, and 30 min was incubated at 37°C. The OD value at 570 nm of each well was determined. The same operation was carried out with the LB broth without bacteria as a negative control. Experiments were performed with three biological replicates.
Galleria mellonella Infection Model
In order to evaluate the effects of three plasmids on the virulence of different receptor bacteria, G. mellonella larvae were used as an in vivo infection model (Tsai et al., 2016). Eight healthy and uniform larvae were used for each strain. The bacterial particles were washed with aseptic saline and then they were diluted with aseptic saline to an appropriate concentration. Using the Hamilton syringe of 25 μl, the bacterial liquid of 10 μl was inoculated into the left forefoot of the last pair of larvae. There were two control groups; one was inoculated with 10 μl of sterile saline, and the other received simulated injection to ensure no physical trauma. All the larvae were incubated in a constant temperature incubator at 37°C for 5 days. During this period, the larvae were observed. If the larvae no longer move when they are touched or blacken, they are considered dead. Results were analyzed by Kaplan–Meier survival curves (GraphPad Prism version 8.3.0 statistics software).
Motility Test
The method of semisolid medium was used to determine the movement ability of bacteria (Liu et al., 2020). Several single colonies were picked and were cultured in LB broth. Then, the bacteria liquid was centrifuged and resuspended with sterile saline. Meanwhile, sterile saline was used to uniformly adjust the concentration of bacteria to 0.6 McFarland. Then, 0.3% semisolid medium (3 g/L) agar plate was prepared, and the 2 μl of bacterial solution was added to the center of the semisolid medium. After the plates were incubated at 37°C for 48 h, the size of bacterial colonies was measured.
Quantitative Real-Time PCR
The absolute expression of hns and tet(X4) was detected by RT-qPCR. Firstly, hns and tet(X4) genes were, respectively, cloned into pBAD and pCE2 to generate pBAD-hns and pCE2-tet(X4). Plasmid pBAD-hns and pCE2-tet(X4) were used as template DNA with primers hns-RNA-F, hns-RNA-R, tet(X4)-RNA-F, and tet(X4)-RNA-R, respectively, to set up the corresponding standard curve. The total RNA of bacteria was extracted by Bacteria RNA Extraction Kit (Vazyme Biotech Co., Ltd) and then reverse transcribed into cDNA by 1 μg of RNA using the HiScriptRR III RT SuperMix for qPCR (+gDNA wiper) Kits (Vazyme Biotech Co., Ltd), and the cDNA of 1 μl was used as template for RT-qPCR with primers hns-RNA-F, hns-RNA-R, tet(X4)-RNA-F, and tet(X4)-RNA-R using the ChamQTM Universal SYBRRR Color qPCR Master Mix Kits (Vazyme Biotech Co., Ltd). According to the standard curve, the absolute expression of hns and tet(X4) genes was calculated. The specific sequence of the primers is shown in Supplementary Table 3.
Determination of Uronic Acid Content
The strains in the exponential growth phase 400 μl were harvested by centrifugation at 8,000 × g for 10 min at 4°C and washed with 500 μl of 1 × PBS three times. Next, the bacteria were resuspended in 500 μl of Tris-HCl buffer (pH = 7.0) and centrifuged at 8,000 × g for 5 min at 4°C to remove the supernatant and then resuspended with 500 μl of Tris-HCl buffer again. Subsequently, 2.4 ml of 12.5 mM tetraborate solution was mixed and heated at 100°C for 5 min. All of the samples were placed in ice, and 40 μl of 15% m-hydroxybiphenyl solution was added to each sample. Thorough mixing was needed. Finally, the concentration of each sample was detected in UV-visible spectroscopy reader at 520 nm (Zheng et al., 2020).
Statistical Analysis
Data are expressed as mean ± standard deviations (SD), and analyzed by either a non-parametric one-way ANOVA, unpaired t-test, or two-way ANOVA. Statistical analysis was conducted using GraphPad Prism version 8.3.0 statistics software. Results were considered significant with p < 0.05.
Results
The Distribution of hns in IncX1-Type tet(X4)-Positive Plasmids
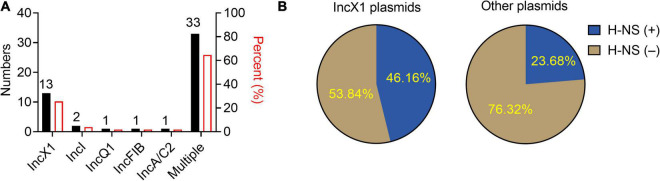
To understand the epidemic characteristics of tet(X4)-positive plasmids, the replicon types of a total of 51 tet(X4)-positive plasmids from different bacteria in NCBI database were analyzed. We found that the plasmids with multiple replicon types were the most common in tet(X4)-positive plasmids, while IncX1 plasmids were dominant in single replicon plasmid types, accounting for 72.2% (Figure 1A). Then, the genome sequences of these IncX1 plasmids were analyzed, and it was found that nearly half of the plasmids carry hns gene (46.16%) (Figure 1B). Combining these results, we hypothesized that H-NS protein may play an unappreciated role in the widespread prevalence of tet(X4)-harboring IncX1 plasmids.
FIGURE 1.
IncX1 type is dominant in tet(X4)-bearing plasmids encoding H-NS from NCBI database. (A) The histogram shows plasmid typing and proportion of tet(X4)-positive plasmids from NCBI database. The black columns represent the numbers of tet(X4)-positive plasmids in different types. The red columns represent the corresponding percentage. (B) The left side of the pie graph shows the carrying rate of hns in tet(X4)-positive IncX1 plasmids. On the right is the carrying rate of hns gene in other types of plasmids.
Analysis of Fitness of tet(X4)-Positive Plasmids Encoding Histone-Like Nucleoid Structuring Protein
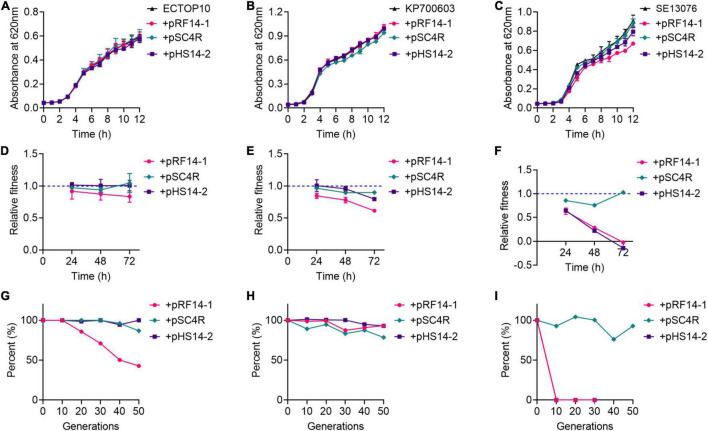
Three hns and tet(X4)-positive IncX1 plasmids, including pRF14-1, pSC4R, and pRF14-1, were applied as models to explore the role of H-NS protein in the prevalence of tet(X4) gene. Considering that fitness cost elicited by resistance plasmids directly affects the dissemination of resistance genes, we hypothesize that H-NS protein may modulate the fitness of host bacteria, which is critical for the transmission of plasmids. To test this hypothesis, these three plasmids were transferred into three recipient bacteria involving E. coli TOP10 (abbreviated as ECTOP10), K. pneumoniae ATCC700603 (abbreviated as KP700603), and S. Enteritidis ATCC13076 (abbreviated as SE13076) separately, and a total of nine transformants were obtained and designated as ECTOP10/pRF14-1, ECTOP10/pSC4R, ECTOP10/pHS14-2, KP700603/pRF14-1, KP700603/pSC4R, KP700603/pHS14-1, SE13076/pRF14-1, SE13076/pSC4R, and SE13076/pHS14-2, respectively. We analyzed the fitness of these nine transformants and their corresponding recipient bacteria, and determined their growth curve, relative fitness, and plasmid stability. Except for the slight slow growth of SE13076/pRF14-1, there was no significant difference in the growth curve in other transformants compared with recipient bacteria (Figures 2A–C). These results showed that the three tet(X4)-positive IncX1 plasmids encoding H-NS protein did not affect the growth rate of host bacteria, implying that the acquisition of these plasmids has no antagonistic effect on the growth of host cells.
FIGURE 2.
tet(X4)-positive IncX1 plasmids encoding H-NS protein displays low fitness cost in host bacteria. (A–C) Growth curves of transformants with E. coli TOP10 (A), K. pneumoniae ATCC700603 (B), or S. Enteritidis ATCC13076 (C) as recipient strains. (D–F) Relative fitness of transformants with E. coli TOP10 (D), K. pneumoniae ATCC 700603 (E), or S. Enteritidis ATCC13076 (F) as recipient strains at 24, 48, and 72 h. (G–I) Plasmid stability in three recipient strains, including E. coli TOP10 (G), K. pneumoniae ATCC700603 (H), or S. Enteritidis ATCC13076 (I). Data are representative of three biological replicates and shown as mean ± SD.
Next, the in vitro competitive test, the most direct method for fitness evaluation (DelaFuente et al., 2020), was used to assess the fitness cost of three plasmids in different hosts. In ECTOP10 and KP700603, these three plasmids did not show obvious fitness cost at both three time points (Figures 2D,E), which were in agreement with the results of growth kinetics. It is demonstrated that the fitness cost is the key factor affecting the replacement rate of drug-resistant bacteria by sensitive bacteria (De Gelder et al., 2004). These results implied that the prevalence of tet(X4)-positive IncX1 plasmids may be due to their small fitness cost on host bacteria. Moreover, we found that even without antibiotic pressure, these three plasmids still conferred a fitness advantage in ECTOP10 during 40 passages (Supplementary Figure 2). In contrast, in SE13076 host bacteria, plasmids pRF14-1 and pHS14-2 showed a high fitness cost, especially after 24 h, whereas plasmid pSC4R did not show obvious fitness cost (Figure 2F).
The high cost of resistance plasmids may also be manifested by the instability of plasmids in host bacteria (Wu et al., 2021). To fully understand the dissemination of tet(X4)-positive IncX1 plasmids, we investigated the stability of these plasmids in three host bacteria. The strains carrying plasmids were passaged for 25 days (a total of 50 generations) in the absence of drugs, and then the loss rate of plasmids at every 10 generations was detected. We found that plasmids pSC4R and pHS14-2 showed good stability in both ECTOP10 and KP7000603. The plasmid pRF14-1 had good stability in KP700603, but in ECTOP10, it began to be lost at the 10th generation, and the plasmid loss rate reached about 60% by the 50th generation (Figures 2G,H). For SE13076, the plasmid pSC4R showed good stability, while the other two plasmids were unstable and completely lost in the 10th generation (Figure 2I). The instability of these two plasmids in SE13076 may due to the low compatibility between plasmids and host bacteria. Thus, the high fitness cost of pRF14-1 and pHS14-2 in SE13076 may be attributed to their poor stability. Further conjugation experiments indicated that plasmid pRF14-1 had the highest conjugation frequency (Supplementary Figure 3), which may partly affect its stability in the recipient. Consistently, a previous study has shown that conjugation can easily offset the fitness cost of plasmids (Lopatkin et al., 2017), especially the high conjugation frequency. Combining with in vitro competition experiments and growth kinetics, these results indicate that tet(X4)-positive IncX1 plasmids carrying hns gene display great fitness advantages in various host bacteria.
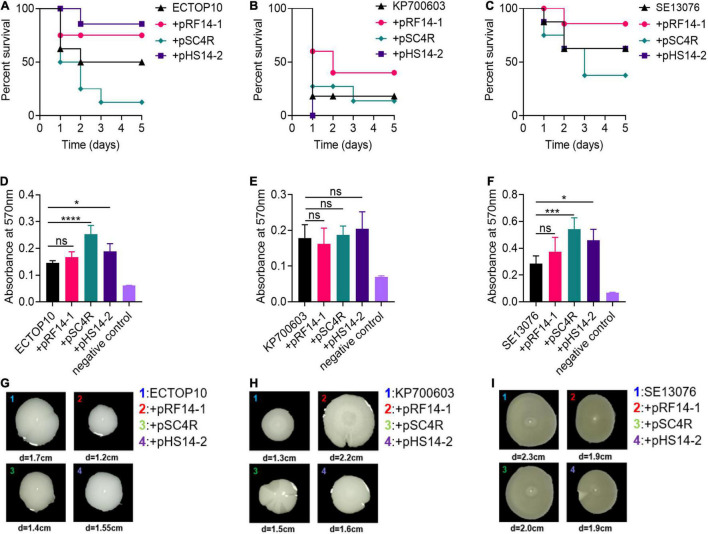
Effect of tet(X4)-Positive Plasmids on Virulence of Host Bacteria
It is suggested that the fitness cost of antibiotic resistance plasmids may result in virulence reduction of host bacteria (Yang et al., 2017). To analyze the impact of tet(X4)-positive plasmids on the virulence of host cells, we first used the Galleria mellonella infection model to evaluate the pathogenicity changes of host bacteria after obtaining resistance plasmids. Interestingly, results showed that the carriage of plasmid pSC4R remarkably enhanced the virulence of host bacteria, while plasmid pRF14-1 reduced bacterial pathogenicity to larvae. However, the adoption of plasmid pHS14-2 showed different effect in three host bacteria (Figures 3A–C).
FIGURE 3.
Effect of tet(X4)-bearing IncX1 plasmids encoding H-NS on the virulence phenotype of different recipient strains. (A–C) Survival of G. mellonella larvae infected by recipient strains and corresponding transformants, including E. coli TOP10 (A), K. pneumoniae ATCC700603 (B), or S. Enteritidis ATCC13076 (C). (D–F) Biofilm formation ability of three recipient strains and corresponding transformants, including E. coli TOP10 (D), K. pneumoniae ATCC700603 (E), or S. Enteritidis ATCC13076 (F). Biofilm mass was monitored using crystal violet staining. Data are representative of three biological replicates and shown as mean ± SD. p-values were determined using a non-parametric one-way ANOVA (*p < 0.05; ***p < 0.001; ****p < 0.0001; ns, not significant). (G–I) Swimming motility of three recipient strains and corresponding transformants. The mean diameters from three biological replicates were shown.
The formation of dormant or biofilm-grown cells has been one of the important ways for bacteria to adapt to the environmental pressure such as antibiotic killing (Flemming et al., 2016). Thus, we next investigated the effect of tet(X4)-positive IncX1 plasmids on the biofilm formation in host bacteria using crystal violet staining. These results showed that the introduction of plasmid pSC4R and pHS142 is able to significantly enhance the biofilm formation ability of ECTOP10 and SE13076, while plasmid pRF14-1 had no obvious effect on the biofilm generation in three host bacteria. In addition, the presence of three tet(X4)-positive plasmids did not influence the biofilm production in KP700603 (Figures 3D–F).
The swimming motility of bacteria is associated with the invasiveness and adhesion of pathogenic bacteria and thereby serves as an important indicator of bacterial virulence (Brunelle et al., 2017). Although the genes encoding bacterial flagella are usually on chromosomes (Samatey et al., 2001), the intake of plasmids will change the bacterial regulatory network (Li et al., 2020a), and the expression of plasmid genes will be strictly regulated accordingly. We compared the movement ability of bacteria with or without tet(X4)-positive IncX1 plasmids on 0.3% (w/v) agar media. In ECTOP10, plasmid pRF14-1 mildly reduced the movement ability of recipient bacteria, but markedly enhanced the swimming motility of KP700603 (Figures 3G,H). Considering that K. pneumoniae lack flagella, we supposed that the motility phenotype may be attributed to the capsule overproduction. Thus, we determined the capsule production of KP700603 and its transformants. Consistently, the results showed that plasmids pRF14-1 and pHS14-2 significantly increased the production of capsule in K. pneumoniae (Supplementary Figure 4). With regard to SE13076, these three plasmids had no remarkable effect on bacterial motility (Figure 3I). Collectively, our data suggest that the host bacteria commonly maintain their virulence, biofilm production, and motility after obtaining tet(X4)-positive plasmids carrying hns gene.
Histone-Like Nucleoid Structuring Protein Reduces the Fitness Cost Elicited by tet(X4)-Positive Plasmids
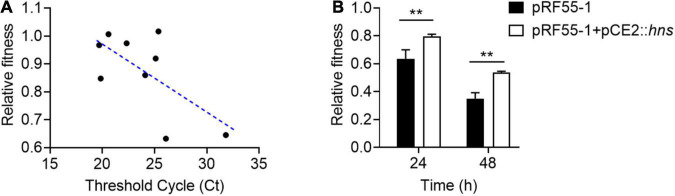
To understand whether H-NS really plays a direct role in plasmid fitness, we conducted a regression analysis of the relationship between the relative fitness of plasmid-bearing host bacteria and the expression of hns gene by RT-qPCR analysis. Ct values were used to represent the expression of hns; the smaller the Ct, the higher the expression of hns gene. Interestingly, we found that the Ct value of hns was negatively correlated with the relative fitness of tet(X4)-positive plasmids in host bacteria (Figure 4A), indicating that plasmid-encoded H-NS protein was positively linked with the relative fitness. To further verify this finding, we tried to construct the hns-deletion plasmid using both CRISPR-Cas9 or λRed homologous recombination. However, we failed to obtain the hns-deficient plasmid without changes in the plasmid backbone. We speculated that the presence of hns gene is critical for the stability of clinical tet(X4)-positive plasmids. To address this issue, we cloned hns gene into pCE2 vector and successfully constructed pCE2::hns. A tet(X4)-positive IncX1 plasmid pRF55-1 without hns gene was introduced into ECTOP10. Subsequently, we compared the fitness of ECTOP10/pRF55-1 transformant before and after the introduction of plasmid pCE2::hns. Consistent with the regression analysis, we found that the fitness of TOP10/pRF55-1 was markedly improved after the acquisition of plasmid pCE2::hns (Figure 4B). These data support our hypothesis that H-NS protein can regulate the fitness cost induced by tet(X4)-positive plasmids in bacteria.
FIGURE 4.
H-NS has a critical role in the fitness of tet(X4)-positive IncX1 plasmids. (A) Negative correlation between relative fitness and hns Ct value by quantitative real-time PCR. (B) The presence of H-NS protein improves the relative fitness of E. coli TOP10 carrying plasmid pRF55-1. Plasmid pRF55-1 is tet(X4)-bearing IncX1 plasmid without hns gene. Data are representative of three biological replicates and shown as mean ± SD. p-values were determined by unpaired t-test (**p < 0.01).
Fitness and hns Expression of Bacteria Carrying tet(X4)-Positive Plasmids Under Different Nutritional Conditions
Previous studies have reported that the expression of hns gene is negatively correlated with the concentration of intracellular (p)ppGpp (Brandi et al., 2020). Thus, it is plausible that the accumulation of (p)ppGpp under the condition of nutrient deficiency would lead to the inhibition of hns expression. To further strengthen our findings on the relationship between hns expression and fitness of tet(X4)-positive plasmids, we determined the relative fitness of host bacteria carrying tet(X4)-positive plasmids under different nutritional conditions, including brain heart infusion (BHI) high nutrition broth, LB broth, and M9CA(M9) broth low nutrition medium. Consistently, host bacteria showed the highest fitness in BHI broth, but relatively low fitness in M9 medium (Figures 5A–C). In addition, we measured the expression of hns gene in BHI high nutrition broth and LB common broth using SE13076 carrying tet(X4)-positive plasmids as a representative strain. Due to the lack of nutrition in M9 medium, the bacteria could not be expanded in enough numbers for following RNA extraction. As expected, we found that the expression of hns gene in BHI broth was higher than that in LB broth (Figure 5D). Taken together, our findings reveal that the altered hns expression in host bacteria carrying tet(X4)-positive plasmids under different nutritional conditions is related to their fitness changes.
FIGURE 5.
Fitness and hns expression of host bacteria under different nutritional conditions. (A–C) Relative fitness of transformants of E. coli TOP10 (A), K. pneumoniae ATCC700603 (B), or S. Enteritidis ATCC 13076 (C) carrying three tet(X4)-bearing IncX1 plasmids in high nutritional BHI culture, medium nutrition LB broth, and low nutrition M9 culture. p-values were determined using a non-parametric one-way ANOVA (*p < 0.05, **p < 0.01; ns, not significant). (D) Absolute expression of hns gene in transformants of S. Enteritidis ATCC13076 as recipient. Data are representative of three biological replicates and shown as mean ± SD. p-values were determined by unpaired t-test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant).
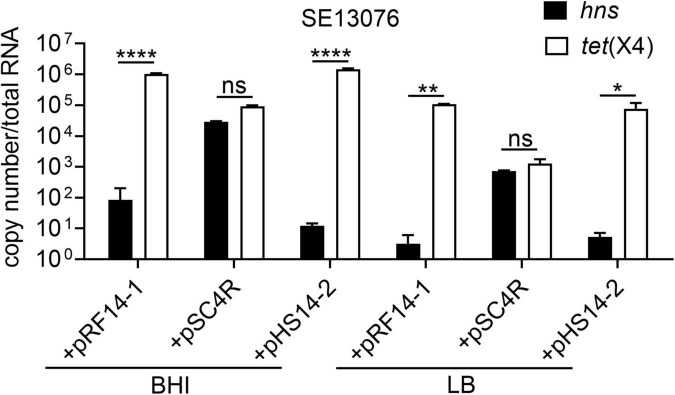
Negative Correlation Between the Expression of hns and tet(X4) Gene
Having shown that plasmid-encoded H-NS protein plays an important role in the fitness of tet(X4)-bearing IncX1 plasmids, we next explored how H-NS regulates the adaptive cost of resistance plasmids in bacteria. To this end, we determined the correlation between the expression of tet(X4) and hns genes in SE13076 transformant. Interestingly, we found that the expression of hns in SE13076 carrying plasmids pRF14-1 or pHS14-2 was negatively correlated with that of tet(X4), while there was no significant relationship between hns and tet(X4) expresssion in plasmid pSC4R (Figure 6). In line with the previous relative fitness and stability results, the low relative fitness and instability of plasmids pRF14-1 or pHS14-2 in SE13076 may be attributed by the low expression of hns and the high expression of tet(X4) gene. By contrast, the better fitness advantage and stability of plasmid pSC4R may be caused by high expression of hns gene. Together, these results suggest that H-NS modulates fitness of tet(X4)-bearing IncX1 plasmids in Gram-negative bacteria by altering the expression of tigecycline resistance gene tet(X4).
FIGURE 6.
Negative correlation between absolute expression of hns and tet(X4) gene in S. Enteritidis ATCC 13076 transformants under different nutritional cultures. The copy number of hns and tet(X4) in transformants using S. Enteritidis ATCC13076 as a recipient strain in BHI, LB, and M9 media. Data are representative of three biological replicates and shown as mean ± SD. p-values were determined by two-way ANOVA (*p < 0.05; **p < 0.01; ****p < 0.0001; ns, not significant).
Discussion
The dissemination of tet(X4)-positive IncX1 plasmids have attracted much attention because it confers high level resistance to tigecycline, a clinically relevant antibiotic for Gram-negative bacterial infections (Ding et al., 2020; Li et al., 2020c). However, it remains unclear how tet(X4)-bearing IncX1 plasmids adapt to new hosts and further spread in various Gram-negative microorganisms. In this study, we investigated the potential role of plasmid-encoded H-NS protein in the dissemination of tet(X4)-bearing IncX1 plasmid by monitoring bacterial growth, fitness, stability, and virulence. Interestingly, we found that the carriage of tet(X4)-positive plasmids encoding H-NS had low fitness cost in E. coli TOP10 and K. pneumoniae ATCC700603, rather than in S. Enteritidis ATCC13076. These findings support the fact that tet(X4) was reported in E. coli and K. pneumoniae, but not in S. Enteritidis (He et al., 2019; Yu et al., 2021), suggesting that the tet(X4)-carrying plasmids impose a high cost in Salmonella. It would be interesting to investigate the underlying mechanisms behind this phenomenon. Furthermore, we found that the expression of hns gene was positively associated with the fitness advantages of tet(X4)-bearing IncX1 plasmids, indicating that H-NS plays a key role in the prevalence of tet(X4) genes in Gram-negative pathogens.
Accordingly, high-cost resistance plasmids will produce a burden in the host such as a reduced bacterial growth (San Millan and MacLean, 2017). For example, previous studies have reported that the high cost caused by overexpression of mcr-1 can lead to bacterial growth defects (Yang et al., 2021) and the low-cost blaNDM–5-carrying IncX3 plasmid has no significant effect on the growth of bacteria (Ma et al., 2020). Consistently, our results showed that three tet(X4)-positive plasmids carrying hns gene did not bring obvious growth defects to the host bacteria, suggesting that these plasmids impose a low fitness cost. There were positive and negative effects between the acquisition of antibiotic resistance and bacterial virulence (Beceiro et al., 2013). The most worrying thing is that while antibiotic resistance is acquired, the virulence of bacteria is also increasing, which will greatly increase the difficulty of clinical treatment. In this study, plasmid pSC4R significantly enhanced the virulence of three recipient bacteria to larvae, which was consistent with the results of biofilm assay. Biofilms can adhere to cells and have an anti-phagocytic effect, thus enhancing the virulence of bacteria to the body (Koo et al., 2017). In fact, the phenotype of bacterial virulence is affected by many factors; in addition to the expression of virulence genes, it may also be affected by two-component regulatory systems, antibiotic resistance, and so on (Beceiro et al., 2013). For example, the expression of homogeneous methicillin resistance in S. aureus affects the biofilm phenotype and reduces virulence (Rudkin et al., 2012). Therefore, we supposed that plasmid pSC4R is likely to encode virulence regulators that enhance host virulence. The virulence difference of the same plasmid in various hosts implied that the virulence of bacteria may be regulated by both the host and plasmid, which requires further exploration.
It is suggested that the epidemic plasmids were stable in host bacteria, even in the absence of antibiotic pressure, and endow the host bacteria the advantage of fitness (Cottell et al., 2012; Fischer et al., 2014). Heretofore, some mechanisms have been shown to mitigate the fitness cost to host bacteria, such as compensatory mutations in host chromosomal genomes or plasmids. For example, the mutation of chromosome gene gacA/gacS, which is involved in the regulation of secondary metabolism, can alleviate the fitness cost of the pQBR103 plasmid in Pseudomonas fluorescens by reducing the expression of plasmid and chromosomal genes (Harrison et al., 2015). Another study showed that the trfA mutation in the plasmid pMS0506 leads to a significant increase in the copy number of plasmid, which greatly improved the plasmid stability and enhanced its adaptability to the host (Sota et al., 2010). In addition to compensatory mutations, there are also some regulatory factors that can directly or indirectly modulate the fitness of the plasmid-bearing host bacteria. For instance, it has been found that the plasmid pHNSHP45 carrying mcr-1 resistance gene encodes a hypothetical ProQ/FinO family protein PcnR, which can control the appropriate expression of mcr-1 by inhibiting the replication of plasmids, thereby reducing the fitness cost caused by high expression of mcr-1 (Yang et al., 2021). The toxin–antitoxin (TA) system, which was originally found on plasmids, can also maintain the stable existence of plasmids (Hayes, 2003). Besides, several studies showed that H-NS protein can inhibit the expression of high-cost genes and improve the fitness advantages of bacteria (Ali et al., 2014; Jian et al., 2016). H-NS protein can bind to the gene promoter and further inhibit the transcriptional expression of genes (Navarre et al., 2006). Reducing the cost of plasmids by inhibiting the expression of high cost genes such as ARGs on tet(X4)-bearing IncX1 plasmids may be a key factor for the persistence of tet(X4)-plasmids in bacterial populations. This view was supported by our observation that plasmids pRF14-1 and pHS14-2 with low-level of hns gene expression had a lower fitness in SE13076. In addition, our study also proved that when the plasmid pCE2:hns was introduced, the fitness of plasmid pRF55-1 without hns in ECTOP10 was significantly improved, which clearly indicated that H-NS could improve the fitness of tet(X4)-bearing IncX1 plasmids. Consistent with our findings, it is shown that H-NS family proteins can regulate the expression of genes on the plasmid and further reduce the fitness cost of plasmids (Doyle et al., 2007). Nevertheless, the detailed mechanisms underlying how H-NS protein regulate the transcription and translation of tet(X4) gene warrant further studies.
Conclusion
In conclusion, our study indicates that tet(X4)-bearing IncX1 plasmids carrying hns gene result in low fitness costs in various Gram-negative pathogens, but instead confer fitness advantages to the host such as enhanced virulence and biofilm formation. Importantly, we find that plasmid-encoded H-NS protein inhibits the expression of tet(X4) gene, thereby reducing the fitness cost of IncX1 plasmids. Together, these data highlight the important role of H-NS protein in the dissemination of tet(X4)-positive plasmids.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YL and ZW designed this study. WC and FT performed all experiments and wrote the draft manuscript. YL, WC, LJ, and RL analyzed the data. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (32172907, 32002331, and 31872526), the Agricultural Science and Technology Independent Innovation Fund of Jiangsu Province [CX(20)3091 and CX(21)2010], the China Postdoctoral Science Foundation funded project (2019M651984 and 2021T140579), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Young Elite Scientists Sponsorship Program by CAST (2020QNRC001).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.763288/full#supplementary-material
References
- Ali S. S., Soo J., Rao C., Leung A. S., Ngai D. H., Ensminger A. W., et al. (2014). Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 10:e1004500. 10.1371/journal.ppat.1004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan N.-F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Del Valle A., León-Sampedro R., Rodríguez-Beltrán J., Delafuente J., Hernández-García M., Ruiz-Garbajosa P., et al. (2021). Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nat. Commun. 12:2653. 10.1038/s41467-021-22849-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A., Tomás M., Bou G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26 185–230. 10.1128/cmr.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi A., Giangrossi M., Fabbretti A., Falconi M. (2020). The gene of is transcriptionally down-regulated by (p)ppGpp. Microorganisms 8:1558. 10.3390/microorganisms8101558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle B. W., Bearson B. L., Bearson S. M. D., Casey T. A. (2017). Multidrug-resistant Salmonella enterica serovar Typhimurium isolates are resistant to antibiotics that influence their swimming and swarming motility. mSphere 2:e00306. 10.1128/mSphere.00306-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-H., Kumar A., Schweizer H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64 391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Cottell J. L., Webber M. A., Piddock L. J. V. (2012). Persistence of transferable extended-spectrum-β-lactamase resistance in the absence of antibiotic pressure. Antimicrob. Agents Chemother. 56 4703–4706. 10.1371/journal.pone.0123039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder L., Ponciano J. M., Abdo Z., Joyce P., Forney L. J., Top E. M. (2004). Combining mathematical models and statistical methods to understand and predict the dynamics of antibiotic-sensitive mutants in a population of resistant bacteria during experimental evolution. Genetics 168 1131–1144. 10.1534/genetics.104.033431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelaFuente J., Rodriguez-Beltran J., San Millan A. (2020). Methods to study fitness and compensatory adaptation in plasmid-carrying bacteria. Methods Mol. Biol. 2075 371–382. 10.1007/978-1-4939-9877-7_26 [DOI] [PubMed] [Google Scholar]
- Ding Y., Saw W.-Y., Tan L. W. L., Moong D. K. N., Nagarajan N., Teo Y. Y., et al. (2020). Emergence of tigecycline- and eravacycline-resistant Tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J. Antimicrob. Chemother. 75 3480–3484. 10.1093/jac/dkaa372 [DOI] [PubMed] [Google Scholar]
- Dorman C. J. (2007). H-NS, the genome sentinel. Nat. Rev. Microbiol. 5 157–161. 10.1038/nrmicro1598 [DOI] [PubMed] [Google Scholar]
- Dorman C. J. (2014). H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid 75 1–11. 10.1016/j.plasmid.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Doyle M., Fookes M., Ivens A., Mangan M. W., Wain J., Dorman C. J. (2007). An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315 251–252. 10.1126/science.1137550 [DOI] [PubMed] [Google Scholar]
- Fischer E. A. J., Dierikx C. M., Van Essen-Zandbergen A., Van Roermund H. J. W., Mevius D. J., Stegeman A., et al. (2014). The IncI1 plasmid carrying the blaCTX-M-1 gene persists in in vitro culture of a Escherichia coli strain from broilers. BMC Microbiol. 14:77. 10.1186/1471-2180-14-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14 563–575. 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- Fu Y., Chen Y., Liu D., Yang D., Liu Z., Wang Y., et al. (2021). Abundance of tigecycline resistance genes and association with antibiotic residues in Chinese livestock farms. J. Hazard. Mater. 409:124921. 10.1016/j.jhazmat.2020.124921 [DOI] [PubMed] [Google Scholar]
- Gao H., Liu Y., Wang R., Wang Q., Jin L., Wang H. (2020). The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. Ebiomedicine 51:102599. 10.1016/j.ebiom.2019.102599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yang X., Li J., Zhang Y., Chen P., Lin Z. (2018). Engineered global regulator H-NS improves the acid tolerance of E. coli. Microb. Cell Fact. 17:118. 10.1186/s12934-018-0966-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E., Guymer D., Spiers A. J., Paterson S., Brockhurst M. A. (2015). Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25 2034–2039. 10.1016/j.cub.2015.06.024 [DOI] [PubMed] [Google Scholar]
- Hayes F. (2003). Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301 1496–1499. 10.1126/science.1088157 [DOI] [PubMed] [Google Scholar]
- He T., Wang R., Liu D., Walsh T. R., Zhang R., Lv Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4 1450–1456. 10.1038/s41564-019-0445-2 [DOI] [PubMed] [Google Scholar]
- Hurtado-Escobar G. A., Grépinet O., Raymond P., Abed N., Velge P., Virlogeux-Payant I. (2019). H-NS is the major repressor of Salmonella Typhimurium Pef fimbriae expression. Virulence 10 849–867. 10.1080/21505594.2019.1682752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian H., Xu G., Gai Y., Xu J., Xiao X. (2016). The histone-like nucleoid structuring protein (H-NS) is a negative regulator of the lateral flagellar system in the deep-sea bacterium Shewanella piezotolerans WP3. Appl. Environ. Microbiol. 82 2388–2398. 10.1128/AEM.00297-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Allan R. N., Howlin R. P., Stoodley P., Hall-Stoodley L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15 740–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn B. M. (2011). Proposals seek to reduce resistance, boost development of new antibiotics. JAMA 305 1845–1846. 10.1001/jama.2011.598 [DOI] [PubMed] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerminiaux N. A., Cameron A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65 34–44. 10.1139/cjm-2018-0275 [DOI] [PubMed] [Google Scholar]
- Li L., Dechesne A., Madsen J. S., Nesme J., Sørensen S. J., Smets B. F. (2020a). Plasmids persist in a microbial community by providing fitness benefit to multiple phylotypes. ISME J. 14 1170–1181. 10.1038/s41396-020-0596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Lu X., Peng K., Liu Z., Li Y., Liu Y., et al. (2020b). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems 5:e00134. 10.1128/mSystems.00134-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Peng K., Li Y., Liu Y., Wang Z. (2020c). Exploring tet(X)-bearing tigecycline-resistant bacteria of swine farming environments. Sci. Total Environ. 733:139306. 10.1016/j.scitotenv.2020.139306 [DOI] [PubMed] [Google Scholar]
- Liu B., Shui L., Zhou K., Jiang Y., Li X., Guan J., et al. (2020). Impact of plasmid-encoded H-NS-like protein on blaNDM-1-bearing IncX3 plasmid in Escherichia coli. J. Infect. Dis. 221 S229–S236. 10.1093/infdis/jiz567 [DOI] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lopatkin A. J., Meredith H. R., Srimani J. K., Pfeiffer C., Durrett R., You L. (2017). Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 8:1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S., Rowley G., Goldberg M. D., Hurd D., Harrison M., Hinton J. C. D. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. 10.1371/journal.ppat.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Fu J., Xie N., Ma S., Lei L., Zhai W., et al. (2020). Fitness Cost of blaNDM-5-carrying p3R-IncX3 plasmids in wild-type NDM-Free Enterobacteriaceae. Microorganisms 8:377. 10.3390/microorganisms8030377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin M., Hassan B., Martins W. M. B. S., Li R., Abdullah S., Sands K., et al. (2021). Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in south Asia. Sci. Total Environ. 787:147613. 10.1016/j.scitotenv.2021.147613 [DOI] [PubMed] [Google Scholar]
- Navarre W. W., Porwollik S., Wang Y., Mcclelland M., Rosen H., Libby S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313 236–238. 10.1126/science.1128794 [DOI] [PubMed] [Google Scholar]
- Nordmann P., Poirel L., Walsh T. R., Livermore D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19 588–595. 10.1016/j.tim.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Picker M. A., Wing H. J. (2016). H-NS, its family members and their regulation of virulence genes in Shigella species. Genes 7:112. 10.3390/genes7120112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Baño J., Gutiérrez-Gutiérrez B., Machuca I., Pascual A. (2018). Treatment of infections caused by extended-spectrum-beta- lactamase-, ampc-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 31:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi Gonçalves I., Dantas R. C. C., Ferreira M. L., Batistão D. W. D. F., Gontijo-Filho P. P., Ribas R. M. (2017). Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz. J. Microbiol. 48 211–217. 10.1016/j.bjm.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin J. K., Edwards A. M., Bowden M. G., Brown E. L., Pozzi C., Waters E. M., et al. (2012). Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infect. Dis. 205 798–806. 10.1093/infdis/jir845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatey F. A., Imada K., Nagashima S., Vonderviszt F., Kumasaka T., Yamamoto M., et al. (2001). Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410 331–337. 10.1038/35066504 [DOI] [PubMed] [Google Scholar]
- San Millan A. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26 978–985. [DOI] [PubMed] [Google Scholar]
- San Millan A., MacLean R. C. (2017). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5:16. [DOI] [PubMed] [Google Scholar]
- Sota M., Yano H., Hughes J. M., Daughdrill G. W., Abdo Z., Forney L. J., et al. (2010). Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 4 1568–1580. 10.1038/ismej.2010.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Chen C., Cui C.-Y., Zhang Y., Liu X., Cui Z.-H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4 1457–1464. 10.1038/s41564-019-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasina E., Haidich A.-B., Kokkali S., Arvanitidou M. (2011). Efficacy and safety of tigecycline for the treatment of infectious diseases: a meta-analysis. Lancet Infect. Dis. 11 834–844. 10.1016/s1473-3099(11)70177-3 [DOI] [PubMed] [Google Scholar]
- Tsai C. J.-Y., Loh J. M. S., Proft T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7 214–229. 10.1080/21505594.2015.1135289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Dong X., Zhang L., Lin Y., Yang K. (2021). Reversing antibiotic resistance caused by mobile resistance genes of high fitness cost. mSphere 6:e0035621. 10.1128/mSphere.00356-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang H.-H., Lu Y., Yi L.-X., Deng Y., Lv L., et al. (2021). A ProQ/FinO family protein involved in plasmid copy number control favours fitness of bacteria carrying mcr-1-bearing IncI2 plasmids. Nucleic Acids Res. 49 3981–3996. 10.1093/nar/gkab149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li M., Spiller O. B., Andrey D. O., Hinchliffe P., Li H., et al. (2017). Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 8:2054. 10.1038/s41467-017-02149-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Cui C. Y., Kuang X., Chen C., Wang M. G., Liao X. P., et al. (2021). Prevalence of tet(X4) in Escherichia coli from duck farms in southeast China. Front. Microbiol. 12:716393. 10.3389/fmicb.2021.716393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Dong N., Zeng Y., Shen Z., Lu J., Liu C., et al. (2020). Chromosomal and plasmid-borne tigecycline resistance genes tet(X3) and tet(X4) in dairy cows on a Chinese farm. Antimicrob. Agents Chemother. 64:e674–e620. 10.1128/AAC.00674-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.-D., Pan Y., He K., Li N., Yang D., Du G.-F., et al. (2020). SPD_1495 contributes to capsular polysaccharide synthesis and virulence in Streptococcus pneumoniae. mSystems 5:e25–e20. 10.1128/mSystems.00025-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.