Graphical abstract
Keywords: Aging, Lipoproteins, NMR, Metabolomics, Glutamate
Abstract
Aging is a major risk factor for metabolic impairment that may lead to age-related diseases such as cardiovascular disease. Different mechanisms that may explain the interplay between aging and lipoproteins, and between aging and low-molecular-weight metabolites (LMWMs), in the metabolic dysregulation associated with age-related diseases have been described separately. Here, we statistically evaluated the possible mediation effects of LMWMs on the relationships between chronological age and lipoprotein concentrations in healthy men ranging from 19 to 75 years of age. Relative and absolute concentrations of LMWMs and lipoproteins, respectively, were assessed by nuclear magnetic resonance (NMR) spectroscopy. Multivariate linear regression and mediation analysis were conducted to explore the associations between age, lipoproteins and LMWMs. The statistical significance of the identified mediation effects was evaluated using the bootstrapping technique, and the identified mediation effects were validated on a publicly available dataset. Chronological age was statistically associated with five lipoprotein classes and subclasses. The mediation analysis showed that serine mediated 24.1% (95% CI: 22.9 – 24.7) of the effect of age on LDL-P, and glutamate mediated 17.9% (95% CI: 17.6 – 18.5) of the effect of age on large LDL-P. In the publicly available data, glutamate mediated the relationship between age and an NMR-derived surrogate of cholesterol. Our results suggest that the age-related increase in LDL particles may be mediated by a decrease in the nonessential amino acid glutamate. Future studies may contribute to a better understanding of the potential biological role of glutamate and LDL particles in aging mechanisms and age-related diseases.
1. Introduction
Aging is a major nonmodifiable risk factor for chronic diseases such as type 2 diabetes mellitus and cardiovascular disease [1]. Indeed, an array of physiological processes and signaling pathways have been associated with aging, with metabolic dysregulation being one of its seven key hallmarks [2], [3]. From our point of view, the integration of data containing the concentration of different molecular species (e.g., amino acids, lipids or lipoproteins) from a single sample and experiment, will result in a greater coverage of the metabolome, thus facilitating the study of their interconnection in aging and disease progression as it has been evidenced previously. For example, different mechanisms that may explain the interplay between amino acids and lipids in the onset of insulin resistance and metabolic diseases have been described. One example is a model described by Newgard et al. showing that an accumulation of branched-chain amino acids may lead to an increase in the circulating levels of certain lipid species in humans with insulin resistance [4]. Therefore, understanding the metabolic alterations (and their interrelationships) that occur in living organisms and that have a central role in the mechanistic contribution to both lifespan and disease course in humans is of paramount importance.
Current approaches to better understand the metabolic alterations linked to aging include the search for statistical associations between chronological age and metabolome profiles provided by high-throughput metabolomic technologies [3], [5]. Along this line, mass spectrometry (MS)-based platforms allow for the profiling of a large number of metabolites including amino acids and lipid species, the so-called low-molecular-weight metabolite (LMWM) window [6]. For example, Wang et al. investigated age-specific statistical associations in a set of blood plasma metabolites obtained by electrospray ionization tandem MS [7]. On the other hand, nuclear magnetic resonance (NMR) spectroscopy allows for the simultaneous profiling of lipoprotein classes and subclasses, the so-called LIPO window, in addition to the profiling of a smaller set of LMWMs (smaller compared to MS-based platforms) [8]. In this case, the seminal work by Freedman et al. is a comprehensive study on the effects of age and sex on NMR-based advanced lipoprotein profiles. In their study, the authors investigated the statistical relationship between chronological age and lipoprotein concentrations as determined by NMR spectroscopy [9]. Of note, most of the studies reported to date that have investigated the effect of chronological age on metabolome profiles have used MS-based platforms for the metabolic profiling of blood samples (i.e., they have used the LMWM window as the phenotype of choice) [7], [10], [11], [12], [13], [14] whereas NMR-based studies have only evaluated the effect of age on lipoprotein profiles (i.e., they have used the LIPO window as the phenotype of choice) [9]. Indeed, we have found few studies investigating the metabolic correlates of aging that included both MS- and NMR-based profiles [13], [15].
Overall, these and other studies highlight the potential of metabolic profiling for the study of the associations between aging, metabolism, and disease. However, studies evaluating the effect of age on MS-based metabolic profiles have not explored more complex statistical relationships such as those involving mediation effects (e.g., they have not explored whether the statistical relationships between age and lipid species are statistically mediated by amino acids). In our opinion, the examination of the statistical mediation between age and metabolome profiles could contribute to furthering our biological understanding of aging, metabolism, and disease. For this purpose, we aimed to expand the existing literature by examining the mediation effects in the statistical analyses evaluating the effect of age on metabolome profiles. Specifically, we investigated whether statistical mediation of the relationship between lipoproteins and age by LMWMs in the absence of any pathological condition existed. To this end, both lipoprotein and metabolic profiles were assessed by NMR spectroscopy in healthy men ranging from 19 to 75 years, and a publicly available dataset was used for validation purposes. The identified mediation effects may be further investigated in future studies to establish whether they are associated with aging mechanisms and disease conditions.
2. Materials and methods
2.1. Subjects
Subjects were recruited in Clermont-Ferrand, France (n = 77) and Reus, Spain (n = 96) as part of a European Commission-funded research and technology development project [16]. The methodology was standardized between recruitment centers. A physician conducted personal interviews with the potential participants to gather information on anthropometric data, personal history, lifestyle, medication use, physical activity, smoking habits, and use of dietary supplements containing vitamins or trace elements. The exclusion criteria were strict and included familial hypercholesterolemia, chronic diseases (including diabetes; cancer; cardiac insufficiency; neurological diseases; inflammatory diseases and chronic diseases of the liver, lung, or thyroid; unstable hypertension; dementia; and infectious diseases known to affect the immune system such as human immunodeficiency virus and hepatitis C), vaccination during the previous 2 months, alcohol abuse or drug addiction, competitive sports activities, and the consumption of special diets or dietary supplements in the previous 3 months. The ethics committee of the two recruiting centers approved the study protocol, and written informed consent was obtained from all volunteers.
2.2. Blood sample collection and biochemical analyses
Fasting venous blood samples were collected in EDTA tubes and centrifuged immediately for 15 min at 4 °C at 1,500 g. The plasma samples were then kept at −80 °C until further analysis. Biochemical parameters such as glucose, triglycerides, total cholesterol, LDL cholesterol (calculated by the Friedewald formula), and HDL cholesterol were measured using colorimetric and enzymatic assays (Spinreact, SA, Spain; Wako Chemicals GmbH, Germany; Polymedco, NY, US; CV < 4%) that were adapted to a Cobas Mira Plus Autoanalyzer (Roche Diagnostics, Spain). The standard lipid profile was analyzed according to Spintrol “H” CAL (Spinreact, SA, Spain) GC–MS reference methods. Spintrol “H” Normal (Spinreact, SA, Spain) was used as a quality control.
2.3. NMR-based metabolomics
1H NMR spectroscopy. A total of 430 µl of plasma samples was transferred to a 5-mm NMR tube. A double tube system was used. The external reference tube (O.D. 2 mm, supported by a Teflon adapter) containing the reference substance (9.9 mmol/l sodium 3-trimethylsilyl[2,2,3,3-d4]propionate (TSP) and 0.47 mmol/l MnSO4 in 99.9\% D2O) was placed coaxially into the NMR sample tube (O.D. 5 mm). This double tube system was kept at 4 °C in the sample changer until the moment of analysis. 1H NMR spectra were recorded at 37 °C on a Bruker Avance III 600 spectrometer operating at a proton frequency of 600.20 MHz (14.1 T). The Carr-Purcell-Meiboom-Gill (CPMG) sequence was used to attenuate the signals from the macromolecules. A spin-echo of 100 ms was used. The acquisition time was approximately 8 min per sample. The NMR spectra were phase- and baseline-corrected using TOPSPIN and then imported into MATLAB using in-house scripts. The imported NMR spectra were referenced to the alanine doublet at 1.465 ppm.
NMR feature extraction. To reduce the dimensionality of (and the redundancy in) the data, we applied a feature extraction approach (peak picking) implemented in the Focus software [17]. To run Focus, the entire NMR spectrum was divided into two main windows: 8.5–5.15 and 4.671–0.8 ppm. We defined a peak intensity threshold to be 6 times the standard deviation of the noise, a frequency subsample of 2 data points, and a window length of 0.1 ppm. Finally, we defined the noise level threshold as 4 times the average standard deviation (SD) from the noisy region between 11 and 10 ppm and kept the selected peaks with a peak intensity above this threshold as metabolomic features for every selected peak. The resulting metabolomic features were represented by a 1H chemical shift in ppm units, which were used in the subsequent regression analyses. Statistically significant features (see the statistical analyses section) were manually annotated using existing literature and databases [18], [19], [20] as well as correlation analysis (Supplementary Figs. S4-S5).
2.4. NMR-based lipoprotein profiling
Lipoprotein particle sizes and concentrations were obtained by NMR spectroscopy by means of the LP2 software (Liposcience, USA). This software simultaneously quantifies subclasses of lipoproteins, lipid content, and average particle size [21]. This technique allows for the determination of the average sizes and particle concentrations of VLDL, LDL, and HDL and the particle concentrations for ten subclasses: three subclasses for VLDL, four subclasses for LDL, and three subclasses for HDL.
2.5. Validation dataset
To validate possible mediation effects from the previously described dataset, we made use of a publicly available dataset from a multi-site, cross-sectional study, including subjects with age-related macular degeneration (early, intermediate and late disease) and a control group of subjects without any macular diseases [22]. Briefly, this dataset consisted of 396 1H NMR CPMG and diffusion-based spectra of plasma samples obtained from 253 female and 143 male subjects. Only data from the male group were included in our validation step. For each individual, age, smoking status, body mass index, disease status and city of origin (Boston or Coimbra) were also available. This data is available at the NIH Common Fund's National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned Project ID ST000590. The data can be accessed directly via it's Project https://doi.org/10.21228/M8BK5R. This work is supported by NIH grant U2C-DK119886.
2.6. Statistical analyses
General data processing procedures. Except when otherwise stated, all data are presented as the mean ± SD for continuous variables. Samples with missing values for clinical traits were discarded. The normal distribution of clinical traits was assessed using the Lilliefors test. All continuous traits passed the test, except for triglyceride levels, which only passed after its log transformation.
Characteristics of the sample set and covariate analysis. Differences in the mean values of clinical and biochemical variables between the two countries were assessed using Student’s t-test. Covariate analysis was performed using univariate and forward stepwise multiple linear regression analyses to evaluate the association between potential confounders (BMI and SBP) and age, metabolomic features and lipoprotein parameters.
NMR-specific data scaling and transformation. For the metabolomic data, values <= 0 were set to NaN (not-a-number), which were ignored. Additionally, metabolomic features were log transformed, normalized (Z-score row-wise) and standardized (Z-score column-wise). The Z-score transformation of all metabolomic features per individual is comparable to common biological normalizations (e.g., normalization by total metabolite content). Additionally, we applied the Z-score transformation for all metabolomic features individually to yield comparable effect sizes. For lipoproteins, outliers (>= 4 SD from the mean) were removed.
Correction for multiple testing. To note, because a metabolite can give rise to multiple peaks in an NMR spectrum, we computed the effective number of independent components, that is, the number of principal components that explain 95% of the data, as a surrogate of the number of LMWMs included in this dataset. Thus, we corrected the significance level α for multiple testing using the Bonferroni correction when associating age with metabolomic features, i.e., we divided the nominal significance level (α = 0.05) by the number of independent components in the NMR-based metabolomic dataset and followed this same approach when associating age with lipoprotein concentrations (see the Results section “3.2. Effect of age on lipoprotein profiles and metabolomic features”).
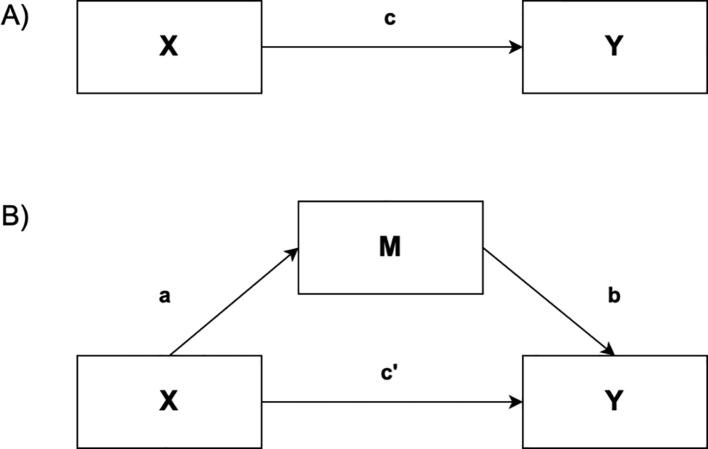
Statistical mediation analysis. Here, we aimed to identify whether statistically significant associations between age and lipoprotein concentrations (Fig. 1A) could be statistically mediated by metabolomic features (Fig. 1B), i.e., that metabolomic features could be in the causal path between aging and lipoprotein levels. Notably, in order for statistical mediation to occur, the predictor of interest (age) has to predict the mediator (metabolomic features). Thus, we first determined the effect of age on metabolomic features and lipoprotein profiles. Forward stepwise multiple linear regression analyses were performed to determine the contributions of age, BMI, and SBP (explanatory variables) to the variability in metabolomic features and lipoprotein parameters (outcome variables) separately. Next, we also explored the correlations between metabolomic and lipoprotein profiles using Pearson correlation analysis. Finally, we investigated the statistical mediation of the relationship between age and lipoprotein concentrations by metabolomic features. To this end, we built statistical models that included age and metabolomic features as predictors and lipoprotein concentrations as outcomes (Fig. 1B). We hypothesized that the addition of age- and lipoprotein-related metabolomic features to the multipredictor models showing statistically significant associations between age and lipoprotein concentrations would attenuate the estimated coefficient for age, i.e., a mediation effect was present. We used the bootstrapping technique to test the statistical significance of the identified mediation effects. Specifically, using the original dataset of 173 individuals, we created a bootstrap sample of 173 individuals by random sampling with replacement 1000 times. Then, we calculated the sample standard error of the indirect or mediated effect a*b and the corresponding 95% confidence intervals (CIs).
Fig. 1.
A) Direct effect model between predictor X and outcomes Y and B) mediation model of the relationship between predictor X and outcome Y by mediator M. a, b, c, and c’ represent the different effects (regression coefficients).
MATLAB (MathWorks) R2018b Update 7 (9.5.0.1298439) was used for all statistical analyses.
3. Results
3.1. Study subjects and covariate analysis
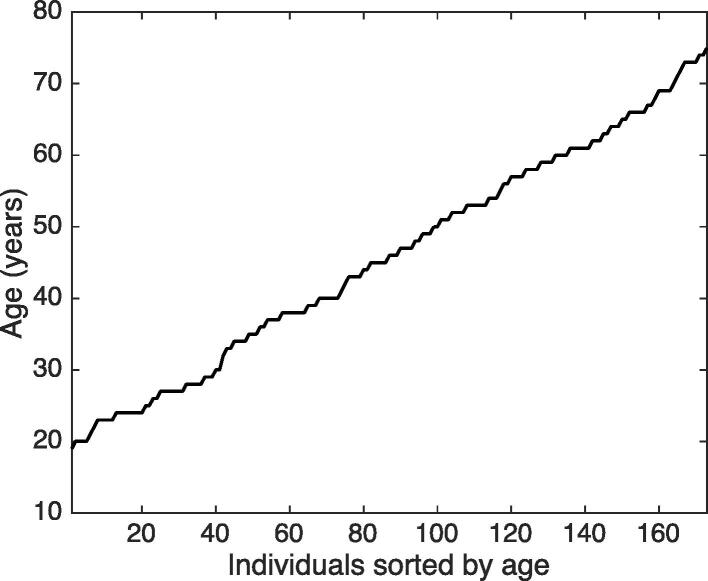
Characteristics of the sample set. The sample set comprised 173 healthy, nonsmoking males (0 cigarettes/day for > 6 months) that were aged 19–75 years with all ages being proportionally represented (Fig. 2) and free of any chronic disease or condition known to alter the concentrations of circulating LMWMs and lipoproteins. Although there were statistically significant differences in systolic blood pressure (SBP) and glucose levels between the two countries, there were no significant differences in mean age or plasma cholesterol or triglyceride levels (Table 1). Hence, the data from all subjects were pooled for the statistical analyses.
Fig. 2.
Study subjects were recruited to represent all ages proportional to the general population.
Table 1.
Characteristics of the study sample set.
| All (n = 173) | France (n = 77) | Spain (n = 96) | p-value | |
|---|---|---|---|---|
| Age, years | ||||
| Mean (SD) | 45.82 ± 15.61 | 45.19 ± 15.23 | 46.31 ± 15.97 | 0.641 |
| Range | 19–75 | 20–74 | 19–75 | |
| Body mass index, kg/m2 | 24.78 ± 2.64 | 24.33 ± 2.63 | 25.14 ± 2.62 | 0.044 |
| Systolic blood pressure, mm Hg | 129.19 ± 12.66 | 135.34 ± 13.73 | 124.26 ± 9.18 | <0.001 |
| Diastolic blood pressure, mm Hg | 78.68 ± 8.53 | 79.62 ± 9.25 | 77.92 ± 7.88 | 0.192 |
| Glucose, mmol/L | 5.02 ± 0.66 | 4.66 ± 0.57 | 5.30 ± 0.58 | <0.001 |
| Triglycerides, mmol/L | 1.06 ± 0.52 | 0.98 ± 0.54 | 1.13 ± 0.48 | 0.052 |
| Total cholesterol, mmol/L | 4.82 ± 0.88 | 4.92 ± 0.97 | 4.75 ± 0.79 | 0.194 |
| LDL cholesterol, mmol/L | 2.69 ± 0.72 | 2.75 ± 0.82 | 2.64 ± 0.62 | 0.329 |
| HDL cholesterol, mmol/L | 1.29 ± 0.3 | 1.32 ± 0.3 | 1.27 ± 0.31 | 0.222 |
Covariate analysis. First, we identified the potential confounders of the relationship between age and metabolome profiles (metabolomic features and lipoprotein concentrations) using univariate regression analysis (Table 2). In the univariate analyses, body mass index (BMI) was statistically associated with 39.3% of the metabolomic features, with 76.5% of the lipoprotein parameters, and with age (p-value = 0.024 for the latter). SBP was statistically associated with 18.5% of the metabolomic features and with age (p-value < 0.001 for the latter). In the stepwise regression analyses, the percentage of metabolomic features associated with BMI and SBP dropped to<10%, and none of the lipoprotein parameters remained associated with SBP or glucose. Notably, glucose levels were statistically associated with 25.4% of the metabolomic features in the stepwise regression analysis. However, the NMR spectrum of serum or plasma samples contains several NMR signals arising from the glucose molecules present in the sample, making it difficult to evaluate the impact of glucose signals on these associations. Therefore, we decided to not use glucose as a covariate in subsequent analyses and checked whether glucose levels were associated with our LMWMs of interest a posteriori (see the paragraph “Relationship between age and metabolomic features” in the following section). Additionally, we only considered BMI and SBP as potential confounders in subsequent analyses, and they were only included in those models involving the metabolomic features or lipoprotein parameters for which they showed a statistical association in the aforementioned stepwise regression analysis.
Table 2.
Percentage (%) of metabolomic features and lipoprotein parameters statistically associated (p-value < 0.05) with different potential confounders using univariate and multivariate (stepwise) approaches.
| Molecular window | Model | BMI | SBP | Glucose |
|---|---|---|---|---|
| LMWM | Univariate | 39.3 | 18.5 | 38.2 |
| Stepwise | 5.8 | 6.9 | 25.4 | |
| LIPO | Univariate | 76.5 | 0 | 47.1 |
| Stepwise | 41.2 | 0 | 0 |
BMI, body mass index; SBP, systolic blood pressure.
3.2. Effect of age on lipoprotein profiles and metabolomic features
Relationship between age lipoprotein concentrations. For lipoproteins, age was nominally associated (p-value < 0.05) with 7 lipoprotein parameters, including BMI and SBP as potential covariates, in the stepwise regression models (Table 3). In this case, because the different lipoprotein parameters can also be correlated, we also computed the number of independent lipoprotein parameters. We found that this number was 7. After correcting the significance level for multiple testing (α = 0.05/7, see the statistical analyses paragraph “Correction for multiple testing”), age remained statistically associated with 5 lipoprotein parameters, namely, small VLDL-P (p-value = 0.0004), LDL-P (p-value = 0.0062), large LDL-P (p-value = 0.0022), total HDL-P (p-value = 0.0006), and small HDL-P (p-value = 0.0007). Notably, age was positively associated with all lipoprotein parameters. Additionally, not all the models finally incorporated BMI as a covariate when associating age and the different lipoprotein parameters. Finally, SBP was not included in any of the stepwise regression models, as expected from the previous covariate analyses (Table 2).
Table 3.
Association statistics between age and all lipoprotein parameters. Significant associations (p-value < 0.05/7) are highlighted in bold.
| Lipoprotein parameter | Covariates | β | SE | p-value |
|---|---|---|---|---|
| VLDL-P | BMI | 0.203 | 0.127 | 0.1123 |
| Large VLDL-P | BMI | −0.004 | 0.003 | 0.1887 |
| Medium VLDL-P | BMI | −0.001 | 0.002 | 0.5394 |
| Small VLDL-P | – | 0.273 | 0.075 | 0.0004 |
| LDL-P | BMI | 0.002 | 0.001 | 0.0062 |
| IDL-P | BMI | 0.002 | 0.002 | 0.4028 |
| Large LDL-P | BMI | 0.003 | 0.001 | 0.0022 |
| Small LDL-P | BMI | 0 | 0.001 | 0.9436 |
| Medium-small LDL-P | BMI | 0 | 0.001 | 0.8042 |
| Very-small LDL-P | BMI | 0 | 0.002 | 0.9352 |
| HDL-P | – | 0.071 | 0.02 | 0.0006 |
| Large HDL-P | BMI | 0.02 | 0.014 | 0.1712 |
| Medium HDL-P | BMI | −0.002 | 0.003 | 0.3903 |
| Small HDL-P | – | 0.074 | 0.021 | 0.0007 |
| VLDL Size | BMI | 0 | 0 | 0.1976 |
| LDL Size | BMI | 0.006 | 0.003 | 0.0867 |
| HDL Size | BMI | 0 | 0 | 0.0327 |
β: regression coefficient; SE: standard error; BMI: body mass index; VLDL: very low-density lipoproteins; LDL: low-density lipoproteins; HDL: high-density lipoproteins.
Relationships between age and metabolomic features. The raw NMR spectral data comprised 64 K spectral data points within a spectral window from 15 to −5 ppm. Once the original dataset was reduced by means of Focus, the resulting dataset included 173 metabolomic features (i.e., variables) representing the signal intensities of NMR visible metabolites. We ran stepwise regression analyses to consider BMI and SBP as potential confounders when associating age with these metabolomic features. In total, age was nominally associated (p-value < 0.05) with 27 metabolomic features (Supplementary Table S1). After correcting the significance level for multiple testing (α = 0.05/31, see the statistical analyses paragraph “Correction for multiple testing”), age remained statistically associated with 9 metabolomic features (Table 4 and Supplementary Figs. S1-S3), which were manually annotated to known LMWMs using existing literature and databases and cross-correlation analysis (Supplementary Figs. S4-S5). Among these features, the metabolomic features at 2.04, 1.05, and 0.993 ppm were considered to be independent and were annotated as glutamate, isobutyrate, and isoleucine, respectively. To note, the signal at 2.324 ppm was nominally associated with age, and it also showed a statistically significant correlation with the signal at 2.04 ppm (Supplementary Figs. S4-S5), suggesting that they arise from the same molecule (i.e., glutamate). Additionally, the signals at 3.977 and 3.949 ppm were assigned to serine, and the signals at 1.026, 1.015, 0.975, and 0.963 ppm were assigned to valine. Thus, after averaging the two latter sets of signals to yield average measures for serine and valine, respectively, the final set of age-related LMWMs that passed the Bonferroni threshold was reduced to 5 variables (Table 4). All LMWMs, but isobutyrate, were negatively associated with age. Importantly, glucose levels were not statistically associated with serine or glutamate levels and were only nominally (0.05/31 < p-value < 0.05) associated with valine and isoleucine levels in the initial univariate analyses.
Table 4.
List of significant associations (p-value < 0.05/31) between age and metabolomic features, including the annotated LMWMs and the covariates that were kept in the final regression model.
| Metabolomic feature (ppm) | Annotated LMWM | Covariates | β | SE | p-value |
|---|---|---|---|---|---|
| 3.977 | Serine | BMI | −0.018 | 0.005 | 0.0002 |
| 3.949 | Serine | BMI, SBP | −0.018 | 0.005 | 0.0003 |
| 2.047 | Glutamate | SBP | −0.017 | 0.005 | 0.0011 |
| 1.057 | Isobutyrate | SBP | 0.016 | 0.005 | 0.0012 |
| 1.026 | Valine | BMI | −0.018 | 0.005 | 0.0001 |
| 1.015 | Valine | BMI | −0.019 | 0.005 | 0.0001 |
| 0.993 | Isoleucine | BMI | −0.018 | 0.005 | 0.0002 |
| 0.975 | Valine | BMI | −0.017 | 0.005 | 0.0003 |
| 0.963 | Valine | BMI | −0.018 | 0.005 | 0.0001 |
LMWM: low molecular weight metabolite; β: regression coefficient; SE: standard error; BMI: body mass index; SBP: systolic blood pressure.
3.3. Statistical mediation analysis
Correlation analysis between LMWMs and lipoprotein profiles. Before evaluating the possible statistical mediation effects, we evaluated the existence of correlations between the identified age-related metabolome profiles. We found eight pairwise correlations among LMWM levels and lipoprotein concentrations that were associated with age in the previous step (Table 5). Serine was correlated with LDL-P (r = -0.30, p-value < 0.001), glutamate was associated with large LDL-P (r = -0.19, p-value = 0.014), valine was associated with large LDL-P (r = -0.27, p-value < 0.001) and with HDL-P (r = -0.20, p-value = 0.008), isoleucine was associated with HDL-P (r = -0.16, p-value = 0.032) and with large LDL-P (r = -0.29, p-value < 0.001), and isobutyrate was associated with LDL-P (r = 0.17, p-value = 0.022). All the associations maintained the same association trends after adjustment for BMI (data not shown).
Table 5.
Correlation analysis between age-related LMWMs and lipoprotein profiles. p-values are shown in parentheses. Nominally significant correlations (p-value < 0.05) are highlighted.
| Small VLDL-P | LDL-P | Large LDL-P | HDL-P | Small HDL-P | |
|---|---|---|---|---|---|
| Serine | −0.12 (0.125) | −0.30 (<0.001) | 0.12 (0.111) | −0.14 (0.068) | −0.15 (0.046) |
| Glutamate | 0.05 (0.497) | 0.12 (0.110) | −0.19 (0.014) | −0.02 (0.828) | −0.06 (0.414) |
| Valine | −0.07 (0.392) | 0.01 (0.851) | −0.27 (<0.001) | −0.2 (0.008) | 0.03 (0.734) |
| Isoleucine | −0.12 (0.105) | 0.0 (0.959) | −0.29 (<0.001) | −0.16 (0.032) | 0.01 (0.904) |
| Isobutyrate | 0.05 (0.552) | 0.17 (0.022) | −0.06 (0.443) | 0.0 (0.950) | 0.03 (0.664) |
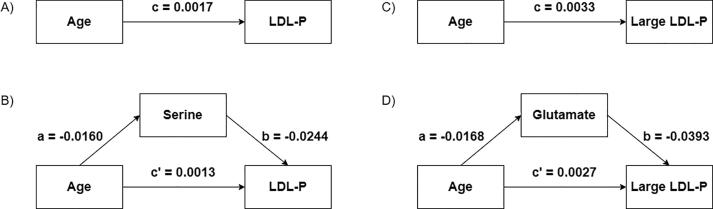
Statistical mediation analysis. Subsequently, we examined to what extent LMWMs explained the associations between age and lipoprotein profiles (Fig. 3). Although the association between age and LDL-P was attenuated (β = 0.0013, SE = 0.0006) after the inclusion of serine (and BMI) in the model, the associations between age and serine levels with LDL-P remained statistically significant (p-values of 0.0425 and 0.0301, respectively) (Fig. 3A-B). In addition, the inclusion of glutamate (together with SBP and BMI) in the model studying the relationship between age and large LDL-P also attenuated this latter association (β = 0.0027, SE = 0.0012), and age and glutamate were still statistically associated with large LDL-P (p-values of 0.0264 and 0.0268, respectively) (Fig. 3C-D). These two mediation effects were statistically significant as tested using the bootstrapping technique. Briefly, the estimated mediation effect (a*b) of the relationship between age and LDL-P by serine was 4.06x10-4 (95% CI: 3.93x10-4 – 4.19x10-4). The estimated mediation effect of the relationship between age and large LDL-P was 5.92x10-4 (95% CI: 5.77x10-4 – 6.07x10-4). Proportionally, serine mediated 24.1% (95% CI: 22.9 – 24.7) of the effect of age on LDL-P, and glutamate mediated 17.9% (95% CI: 17.6 – 18.5) of the effect of age on large LDL-P. The remaining associations between LMWMs and lipoprotein profiles shown in Table 3 disappeared after adjustment for age and BMI (and SBP for the association between isobutyrate and LDL-P). However, in all these models, the association between age and the corresponding lipoprotein parameter remained statistically significant after the inclusion of the corresponding age-related LMWMs in the statistical model.
Fig. 3.
Mediation analysis of A-B) the relationship between age and LDL-P by serine and C-D) the relationship between age and large LDL-P by glutamate. a, c and c’ represent the effects (regression coefficient) of age on the corresponding LMWM or lipoprotein parameter, and b represents the effect (regression coefficient) of a particular LMWM on the corresponding lipoprotein parameter. The covariates have been omitted for visualization purposes.
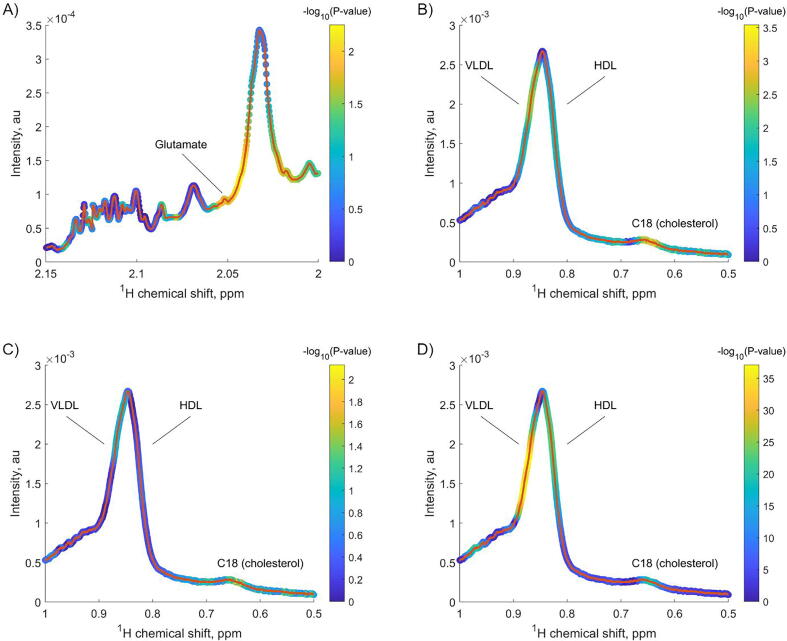
Validation of the identified mediation effects. Afterwards, we explored whether the identified mediation effects could be replicated in a second dataset obtained from the Metabolomics Workbench repository. In this dataset, quantitative lipoprotein data was not available. However, we leveraged the diffusion NMR spectra as a surrogate of an NMR-based lipoprotein profile. As shown in Fig. 4 and Table 6, we could replicate the mediation effect of the relationship between age and a surrogate of cholesterol, namely the integral intensity (from 0.63 to 0.66 ppm) of the NMR signal arising from the C18 cholesterol moiety, by the integral intensity (from 2.048 to 2.056) of the glutamate NMR signal. Briefly, Fig. 4A shows statistically significant associations (colormap) between chronological age and the glutamate NMR signals around 2.05 ppm (path a). Also, Fig. 4B shows statistically significant associations between chronological age and the C18 NMR signals around 0.65 ppm (path c). Finally, Fig. 4C-D show statistically significant associations between the C18 NMR signals with chronological age and the glutamate NMR signals (paths c’ and b, respectively). Table 6 shows the regression coefficients (β), standard errors (SE) and 95% confidence intervals for the different paths as determined by the bootstrapping method. Importantly, all the regression coefficients were statistically significant, and of the same direction as in the first dataset. All regression models included smoking status, BMI, disease status and the disease status*age interaction term.
Fig. 4.
Colour-coded NMR envelopes of A) LMWM (CPMG pulse) and B-D) LIPO (diffusion-based pulse) windows. Each NMR feature was considered in the different regression models. The red line represents the experimental NMR spectrum of a reference sample. The colour coding represents the statistical significance of the corresponding associations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 6.
Regression coefficients (β), standard errors (SE) and 95% confidence intervals for the different paths as determined by the bootstrapping method.
| Path | β | SE | 95% Confidence interval |
|---|---|---|---|
| a (Age -> Glutamate) | −0.0667 | 8.03x10-4 | −0.0683, −0.0651 |
| b (Glutamate -> C18) | −0.0012 | 4.27x10-6 | −0.0013, −0.0012 |
| c’ (Age -> C18) | 8.84x10-5 | 1.20x10-6 | 8.60x10-5, 9.07x10-5 |
| a * b | 8.29x10-5 | 1.02x10-6 | 8.09x10-5, 8.49x10-5 |
Fine mapping of the glutamate NMR signal. To conclude, we performed a more finned mapping of the NMR region containing the glutamate signal to further validate this particular annotation. To note, we further validated this annotation because only glutamate could be validated in the second sample set. For this validation, we took the NMR signal (2.047 ppm) that was annotated as glutamate and performed a correlation analysis with all the signals within the [2.6–1.9] ppm range (Supplementary Fig. S5A). Notably, this correlation analysis was performed considering the raw NMR data, not the peaks from the peak picking step. Supplementary Fig. S5A depicts a fingerprint composed of a number of “peaks” (correlation is present with the reference signal) that deviate from 0 (no correlation is present with the reference signal). Since Gowda (18) and other authors [23], [24] have identified glutamate, glutamine, and proline in this region, Supplementary Fig. S5B-C show the NMR spectra of these metabolites obtained from the Human Metabolome Database to guide the matching of the different “peaks” with the corresponding signals from these pure compounds. Importantly, this correlation analysis showed high correlation coefficients (r > 0.8) within the [2.3–2.35] ppm range, a region that matches another glutamate signal (see Supplementary Fig. S3A) that was nominally associated with age (with a similar effect size an p-value). On the other hand, although proline also shows an NMR signal within the same region, the fact that the correlation coefficients within the [2–1.95] ppm range are lower (r ∼ 0.4), and that this region matches another proline signal, allow us to confidentially annotate the reference signal as glutamate.
4. Discussion
4.1. Effects of age on lipoprotein profiles
We identified positive and significant associations between age and the concentrations of 1) small VLDL, 2) total LDL, 3) large LDL, 4) total HDL, and 5) small HDL particles. Importantly, these associations remained significant after adjustment for age-related metabolomic features. As we will discuss later, these results may indicate that although lipoproteins increase with age among healthy men, they present a beneficial lipoprotein profile that may be associated with healthy aging. Briefly, small VLDL and large LDL particles do not predispose to an atherogenic profile as large VLDL and small LDL particles do [25], [26]. Moreover, the positive and significant associations between age and HDL parameters include the small, atheroprotective, and antioxidant HDL subclass [27]. When comparing our results with previous findings (Table 7), the direct relationship between age and the 1) concentration of small VLDL and 2) total LDL particle concentrations follows a common pattern across previous studies. However, the direct relationship between age and the other lipoprotein parameters (large LDL, total HDL, and small HDL) shows certain degree of discordant results when comparing them with the results of previous literature. Nevertheless, this discordance must be taken cautiously since all the studies have very different study designs. First, the study reported by Freedman et al. involved participants from the general population, and thus was not focused on a healthy population as was our case. Second, the study reported by Heijmans et al. involved the study of families, and only total LDL-P and HDL-P were available. Finally, we found a total agreement with the study reported by Rajalahti and co-authors.
Table 7.
Comparison of the direction of the relationship between age and different lipoprotein parameters across studies.
| This study | Freedman et al. (9) | Heijmans et al. (28) | Rajalahti et al. (29) | |
|---|---|---|---|---|
| Small VLDL | + | n.s. | n.a. | + |
| Total LDL | + | + | – | + |
| Large LDL | + | n.s. | n.a. | + |
| Total HDL | + | n.s. | n.s. | + |
| Small HDL | + | n.s. | n.a. | + |
n.s.: nonsignificant; n.a.: not available.
Briefly, in the seminal work by Freedman et al., large and intermediate VLDL particle concentrations, but not small VLDL particle concentrations, were positively associated with age in men. In contrast, and more in line with our results, total and intermediate LDL particle concentrations, but not small LDL particle concentrations, were positively associated with age in men [9]. Of note, Freedman et al. did not find correlations between HDL subclasses and age in men. In addition, Heijmans et al. reported that long-lived siblings had lower LDL particle concentrations and larger mean LDL particle size compared with their offspring [28]. The authors argued that this effect could be associated with a decrease in the concentration of small LDL particles in long-lived individuals. Another study that reported changes in lipoprotein patterns during aging in men found increases in small VLDL concentrations as well as in total, large and medium LDL classes and subclasses [29]. They also found increases in concentrations of “healthy” lipoproteins such as HDL and almost no changes in concentrations of the atherogenic small and very small LDL particles. Notably, in this study the lipoprotein profiles were determined using high-performance liquid chromatography. Finally, Slade et al. found a negative and significant association between age and VLDL size in men [13]. This result is in line with an increase in small VLDL concentrations.
4.2. Effects of age on serine and glutamate levels
Of the different age-related metabolic features found to be associated with age in our study, we focus our discussion on those features that remained significant when included in the statistical model modeling the effect of age on lipoprotein profiles. Specifically, we identified negative and significant associations between age and the metabolic features annotated as serine and glutamate. There have been previous attempts to study the effects of age on metabolite concentrations under a similar study design (e.g., linear regression analysis of metabolomic profiles on chronological age). Serine and glutamate were previously reported to be modulated by age in a study investigating the effects of age, sex and race on the relative concentrations of metabolites in the blood of healthy (and adult) men and women [11]. However, they reported a direct relationship between these same metabolites and age. In another study, Menni et al. reported associations between age and both glutamate and serine levels; whereas glutamate showed a direct relationship with age, serine was inversely related to age, agreeing with our results [12].
Other studies have investigated age-related metabolic changes using different study designs. For example, Collino et al. showed higher serine concentrations in centenarian’s offspring compared to offspring of non-long-lived parents [15]. In another study, Chak et al. found a direct relationship between age and serine levels in women but not in men. However, the relationship between age and serine levels was significant in men under FDR-adjusted p-values in both the discovery and replication studies [30]. Finally, in a longitudinal study investigating longitudinal changes of the plasma metabolome, the authors found that serine decreased with age [31].
4.3. Roles of lipoproteins, serine, and glutamate in aging processes: Focus on oxidative stress
Aging is a process characterized by the progressive loss of tissue and organ function [32]. The oxidative stress theory of aging is based on the hypothesis that age-associated functional losses are due to the accumulated damage induced by reactive oxygen and nitrogen species [33]. In parallel, oxidative stress is involved in several age-related conditions such as cardiovascular diseases [34]. In this line, current evidence links atherosclerosis with oxidized LDL-cholesterol [35]. In our study, we found a positive and significant statistical association between age and small HDL particle concentrations. Current knowledge about the role of HDL subclasses in atherogenesis acknowledges the atheroprotective and antioxidative role of the small HDL subclass, which protects LDL particles from oxidation [36], [37], [38]. Indeed, the antioxidative activity of HDL subclasses increases with the density [36]. Importantly, these properties may be impaired in disease states such as atherogenic dyslipidemia or familial hypercholesterolemia [38], [39].
Serine is a nonessential amino acid and a predominant source of one-carbon groups for the de novo synthesis of purine nucleotides that are essential for cell proliferation. From a metabolic point of view, oxidative stress resistance has been noted as a potential biological pathway of serine related to biological aging [30]. Importantly, the antiatherogenic and antioxidant properties of serine have already been demonstrated [40]. Serine has also been linked with decreased lifespan in yeast models through sensitization to oxidative stress and activation of the TOR-S6 signaling pathway [41]. Moreover, serine is necessary for the metabolism of fats and fatty acids, for muscle growth and for maintaining a healthy immune system [15]. In this line, serine levels were previously found to be lower in plasma under inflammatory conditions [42].
Finally, prior literature has already shown the interconnection between lipoproteins and metabolites such as glutamate in relation to aging and disease processes like atherosclerosis and cardiovascular diseases. In the case of glutamate, systemic plasma glutamate levels have been shown to be elevated in several diseases characterized by chronic oxidative stress and inflammation [43], and thus the link between glutamate, oxidative stress and neurological disorders has been extensively described [44]. For example, Wang et al. applied a targeted metabolomics approach to investigate the mechanisms of hyperlipidemia and discover potential biomarkers in control and hyperlipidemic rats [45]. They found that arginine levels reflected oxidative stress behavior, and that hyperlipidemia was also closely related to oxidative stress. Importantly, glutamate plays an important role in decreasing arginine as an oxidative stress messenger and regulator. In addition, Zheng et al. reported that baseline glutamate levels were associated with increased cardiovascular disease risk in the PREvención con DIeta MEDiterránea (PREDIMED) trial [46]. More recently, an association between the single nucleotide polymorphism (SNP) rs10911021 and oxidative stress biomarkers in coronary artery disease patients was reported [47]. Importantly, this SNP is present upstream of the GLUL gene, which affects glutamate metabolism.
5. Study strengths and limitations
Age-related changes in metabolites, lipids and lipoproteins are well described in the general population, but to the best of our knowledge, no study has described the statistical mediation effects in healthy subjects recruited following our strict criteria. Here, we took advantage of the technical possibility to simultaneously assess a very diverse and informative set of markers to study how they associate with age in the absence of any pathological condition in healthy men ranging from 19 to 75 years. Indeed, our study is probably the one that has used the strictest inclusion criteria in order to recruit a healthy population. Furthermore, we were able to validate the statistical mediation effect of glutamate on the relationship between chronological age and LDL on a second dataset. Besides, as a cross-sectional study, we could only explore the correlations among variables. In this regard, a follow-up study will be of great value to determine the longitudinal changes. In addition, the statistical focus of this study does not allow for the experimental confirmation of the identified mediation effects or for the identification of the signaling or metabolic pathways that would be affected by age.
6. Conclusions
There is already a large body of research studying the relationships between aging and different molecular species with the ultimate goal of better understanding aging mechanisms and age-related diseases. Considering the large number of metabolites and other molecular entities that can be profiled in metabolomics studies and the possible biological interrelations between them, a more complex statistical approach can contribute to this current research topic. Here, we hypothesized that the relationship between chronological age and lipoprotein concentrations was statistically mediated by LMWMs. Indeed, we found that a proportion of the effect of age on LDL-P levels was mediated by serine levels, and a proportion of the effect of age on large LDL-P levels was mediated by glutamate levels. Thus, our results suggest that the age-related increase in LDL particles may be mediated by a decrease in the nonessential amino acids serine and glutamate. Further research is needed to investigate the potential biological roles of serine, glutamate and LDL particles in aging mechanisms. Overall, since similar studies continue to appear in the literature, we encourage researchers to consider these and/or other statistical scenarios (e.g., statistical interactions) when evaluating the interrelations between age factors and the metabolome.
Funding
EU FP5 Program ‘‘Quality of Life and Management of Living Resources,’’ Key Action 1, ‘‘Food, Nutrition, and Health,’’ entitled ‘‘Vitamin A, Vitamin E, and Carotenoid Status and Metabolism during Ageing: Functional and Nutritional Consequences,’’ acronym VITAGE (contract QLK1-CT-1999-00830). Project PI08/1579, Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación; CIBER in Diabetes and Associated Metabolic Diseases, Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the staff and volunteers who participated in the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.11.022.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Costantino S., Paneni F., Cosentino F. Ageing, metabolism and cardiovascular disease. J Physiol. 2016;594(8):2061–2073. doi: 10.1113/JP270538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy B.K., Berger S.L., Brunet A., Campisi J., Cuervo A.M., Epel E.S., et al. Geroscience: Linking Aging to Chronic Disease. Cell. 2014;159(4):709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma R., Ramanathan A. The Aging Metabolome—Biomarkers to Hub Metabolites. Proteomics. 2020;20(5-6):1800407. doi: 10.1002/pmic.v20.5-610.1002/pmic.201800407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newgard C. Interplay between Lipids and Branched-Chain Amino Acids in Development of Insulin Resistance. Cell Metab. 2012;15(5):606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R., Li B., Lam S.M., Shui G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression. J Genet Genomics. 2020;47(2):69–83. doi: 10.1016/j.jgg.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Dettmer K., Aronov P.A., Hammock B.D. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Wang G., Jing R., Hu T., Likhodii S., Sun G., et al. Metabolomics analysis of human plasma metabolites reveals the age- and sex-specific associations. J Liq Chromatogr Relat Technol. 2020;43(5-6):185–194. [Google Scholar]
- 8.Emwas A.-H., Roy R., McKay R.T., Tenori L., Saccenti E., Gowda G.A.N., et al. NMR Spectroscopy for Metabolomics Research. Metabolites. 2019;9(7):123. doi: 10.3390/metabo9070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman D.S., Otvos J.D., Jeyarajah E.J., Shalaurova I., Cupples L.A., Parise H., et al. Sex and Age Differences in Lipoprotein Subclasses Measured by Nuclear Magnetic Resonance Spectroscopy: The Framingham Study. Clin Chem. 2004;50:1189–1200. doi: 10.1373/clinchem.2004.032763. [DOI] [PubMed] [Google Scholar]
- 10.Jové M., Maté I., Naudí A., Mota-Martorell N., Portero-Otín M., De la Fuente M., et al. Human Aging Is a Metabolome-related Matter of Gender. J Gerontol. 2016;71(5):578–585. doi: 10.1093/gerona/glv074. [DOI] [PubMed] [Google Scholar]
- 11.Lawton K.A., Berger A., Mitchell M., Milgram K.E., Evans A.M., Guo L., et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9(4):383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 12.Menni C., Kastenmuller G., Petersen A.K., Bell J.T., Psatha M., Tsai P.-C., et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int J Epidemiol. 2013;42:1111–1119. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slade E., Irvin M.R., Xie K., Arnett D.K., Claas S.A., Kind T., et al. Age and sex are associated with the plasma lipidome: findings from the GOLDN study. Lipids Health Dis. 2021;20(1) doi: 10.1186/s12944-021-01456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Z., Zhai G., Singmann P., He Y., Xu T., Prehn C., et al. Human serum metabolic profiles are age dependent. Aging Cell. 2012;11(6):960–967. doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collino S., Montoliu I., Martin F.-P.J., Scherer M., Mari D., Salvioli S., et al. Metabolic Signatures of Extreme Longevity in Northern Italian Centenarians Reveal a Complex Remodeling of Lipids, Amino Acids, and Gut Microbiota Metabolism. Wertheimer A, editor. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock E., Winklhofer-Roob B.M., Ribalta J., Scotter M., Vasson M.P., Brtko J., et al. Vitamin A, vitamin E and carotenoid status and metabolism during ageing: functional and nutritional consequences (VITAGE PROJECT) Nutr Metab Cardiovasc Dis NMCD. 2001;11:70–73. [PubMed] [Google Scholar]
- 17.Alonso A., Rodríguez M.A., Vinaixa M., Tortosa R., Correig X., Julià A., et al. Focus: A Robust Workflow for One-Dimensional NMR Spectral Analysis. Anal Chem. 2014;86(2):1160–1169. doi: 10.1021/ac403110u. [DOI] [PubMed] [Google Scholar]
- 18.Nagana Gowda G.A., Gowda Y.N., Raftery D. Expanding the Limits of Human Blood Metabolite Quantitation Using NMR Spectroscopy. Anal Chem. 2015;87(1):706–715. doi: 10.1021/ac503651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Karu N., et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero P.R., Kobayashi N., Wedell J.R., Baskaran K., Iwata T., Yokochi M., et al. BioMagResBank (BMRB) as a Resource for Structural Biology. Methods Mol Biol. 2020;2112:187–218. doi: 10.1007/978-1-0716-0270-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeyarajah E.J., Cromwell W.C., Otvos J.D. Lipoprotein Particle Analysis by Nuclear Magnetic Resonance Spectroscopy. Clin Lab Med. 2006;26(4):847–870. doi: 10.1016/j.cll.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Laíns I., Duarte D., Barros A.S., Martins A.S., Gil J., Miller J.B., et al. Human plasma metabolomics in age-related macular degeneration (AMD) using nuclear magnetic resonance spectroscopy. Clark SJ, editor. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson JK, Foxall PJD, Spraul M. 750 MHz IH and IH-l3C NMR Spectroscopy of Human Blood Plasma. :19.
- 24.Vaarhorst A.A.M., Verhoeven A., Weller C.M., Böhringer S., Göraler S., Meissner A., et al. A metabolomic profile is associated with the risk of incident coronary heart disease. Am Heart J. 2014;168(1):45–52.e7. doi: 10.1016/j.ahj.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Guardiola M., Solà R., Vallvé J.C., Girona J., Godàs G., Heras M., et al. Body mass index correlates with atherogenic lipoprotein profile even in nonobese, normoglycemic, and normolipidemic healthy men. J Clin Lipidol. 2015;9(6):824–831.e1. doi: 10.1016/j.jacl.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Austin M.A., Breslow L., Hennekens C.H., Willett W.C., Krauss R.M. Low-Density Lipoprotein Subclass Patterns and Risk of Myocardial Infarction. JAMA. 1988;260:1917–1921. [PubMed] [Google Scholar]
- 27.Asztalos B.F., Tani M., Schaefer E.J. Metabolic and functional relevance of HDL subspecies. Curr Opin Lipidol. 2011;22:176–185. doi: 10.1097/MOL.0b013e3283468061. [DOI] [PubMed] [Google Scholar]
- 28.Heijmans B.T., Beekman M., Houwing-Duistermaat J.J., Cobain M.R., Powell J., Blauw G.J., et al. Lipoprotein Particle Profiles Mark Familial and Sporadic Human Longevity. PLoS Med. 2006;3(12):e495. doi: 10.1371/journal.pmed.0030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajalahti T., Lin C., Mjøs S.A., Kvalheim O.M. Changes in serum fatty acid and lipoprotein subclass concentrations from prepuberty to adulthood and during aging. Metabolomics. 2016 Mar;12(3):51. doi: 10.1007/s11306-016-0968-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chak C., Lacruz M., Adam J., Brandmaier S., Covic M., Huang J., et al. Ageing Investigation Using Two-Time-Point Metabolomics Data from KORA and CARLA Studies. Metabolites. 2019;9(3):44. doi: 10.3390/metabo9030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darst B.F., Koscik R.L., Hogan K.J., Johnson S.C., Engelman C.D. Longitudinal plasma metabolomics of aging and sex. Aging. 2019;11(4):1262–1282. doi: 10.18632/aging.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flatt T. A New Definition of Aging? Front Genet. 2012;3 doi: 10.3389/fgene.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckman K.B., Ames B.N. The Free Radical Theory of Aging Matures. Physiol Rev. 1998;78(2):547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 34.Liguori I., Russo G., Curcio F., Bulli G., Aran L., Della-Morte D., et al. Oxidative stress, aging, and diseases. Clin Interv Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gradinaru D., Borsa C., Ionescu C., Prada G.I. Oxidized LDL and NO synthesis—Biomarkers of endothelial dysfunction and ageing. Mech Ageing Dev. 2015;151:101–113. doi: 10.1016/j.mad.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Kontush A., Chantepie S., Chapman M.J. Small, Dense HDL Particles Exert Potent Protection of Atherogenic LDL Against Oxidative Stress. Arterioscler Thromb Vasc Biol. 2003;23(10):1881–1888. doi: 10.1161/01.ATV.0000091338.93223.E8. [DOI] [PubMed] [Google Scholar]
- 37.Kontush A., Chapman M.J. Antiatherogenic function of HDL particle subpopulations: focus on antioxidative activities. Curr Opin Lipidol. 2010;21:312–318. doi: 10.1097/MOL.0b013e32833bcdc1. [DOI] [PubMed] [Google Scholar]
- 38.Camont L., Chapman M.J., Kontush A. Biological activities of HDL subpopulations and their relevance to cardiovascular disease. Trends Mol Med. 2011;17(10):594–603. doi: 10.1016/j.molmed.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Hussein H., Saheb S., Couturier M., Atassi M., Orsoni A., Carrié A., et al. Small, dense high-density lipoprotein 3 particles exhibit defective antioxidative and anti-inflammatory function in familial hypercholesterolemia: Partial correction by low-density lipoprotein apheresis. J Clin Lipidol. 2016;10(1):124–133. doi: 10.1016/j.jacl.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Maralani M.N., Movahedian A., Javanmard S.H. Antioxidant and cytoprotective effects of L-Serine on human endothelial cells. Res Pharm Sci. 2012;7:209–215. [PMC free article] [PubMed] [Google Scholar]
- 41.Mirisola M.G., Taormina G., Fabrizio P., Wei M., Hu J., Longo V.D., et al. Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging. PLoS Genet. 2014;10(2):e1004113. doi: 10.1371/journal.pgen.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suliman M.E., Qureshi A.R., Stenvinkel P., Pecoits-Filho R., Bárány P., Heimbürger O., et al. Inflammation contributes to low plasma amino acid concentrations in patients with chronic kidney disease. Am J Clin Nutr. 2005;82:342–349. doi: 10.1093/ajcn.82.2.342. [DOI] [PubMed] [Google Scholar]
- 43.Davalli A.M., Perego C., Folli F.B. The potential role of glutamate in the current diabetes epidemic. Acta Diabetol. 2012;49(3):167–183. doi: 10.1007/s00592-011-0364-z. [DOI] [PubMed] [Google Scholar]
- 44.Coyle J.T., Puttfarcken P. Oxidative stress, glutamate, and neurodegenerative disorders. Science. 1993;262(5134):689–695. doi: 10.1126/science.7901908. [DOI] [PubMed] [Google Scholar]
- 45.Wang X.-F., Zhang Y.-x., Ma H.-Y. Targeted profiling of amino acid metabolome in serum by a liquid chromatography-mass spectrometry method: application to identify potential markers for diet-induced hyperlipidemia. Anal Methods. 2020;12(18):2355–2362. doi: 10.1039/d0ay00305k. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y., Hu F.B., Ruiz‐Canela M., Clish C.B., Dennis C., Salas‐Salvado J., et al. Metabolites of Glutamate Metabolism Are Associated With Incident Cardiovascular Events in the PREDIMED PREvención con DIeta MEDiterránea (PREDIMED) Trial. J Am Heart Assoc. 2016;5(9) doi: 10.1161/JAHA.116.003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shahid S.U., Shabana, Humphries S. The SNP rs10911021 is associated with oxidative stress in coronary heart disease patients from Pakistan. Lipids Health Dis. 2018;17(1) doi: 10.1186/s12944-017-0654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.