Graphical abstract
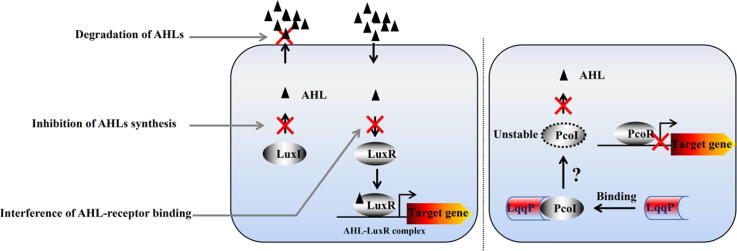
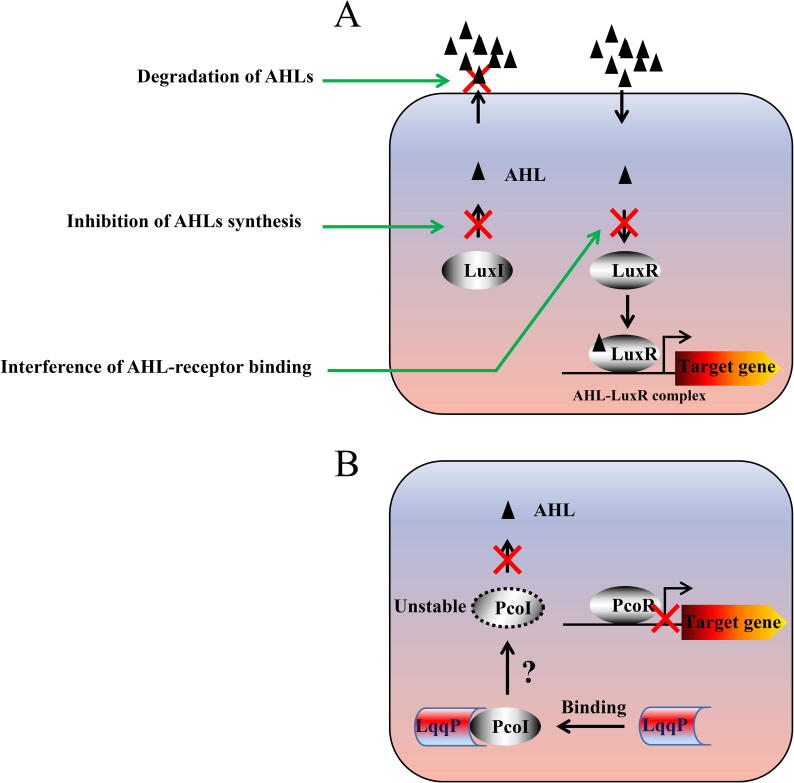
Schematic illustration of AHL quorum quenching strategies. Three known AHL quorum-quenching strategies (left). Quenching of AHL signaling by inhibiting AHL synthesis (middle gray arrow) or AHL-receptor binding (bottom gray arrow) by small molecular chemicals or enzymatically degrading AHL (top gray arrow) are indicated. In the right panel, we proposed a model of LqqP-mediated quenching of PcoI-dependent AHL signaling, representing a new quorum-quenching strategy. The direct binding of LqqP and PcoI reduces the abundance of PcoI (capable of AHL synthesis) in vivo through an unknown mechanism. LuxI and PcoI: AHL synthase, LuxR and PcoR: AHL receptor transcriptional regulator.
Keywords: Quorum sensing, AHL, Quorum quenching, AHL synthase, Autoinducer, Lysobacter, Aminopeptidase
Abstract
Acyl-homoserine lactone (AHL) is the most studied autoinducer in gram-negative bacteria controlling infections of various pathogens. Quenching of AHL signaling by inhibiting AHL synthesis or AHL-receptor binding via small molecular chemicals or enzymatically degrading AHL is commonly used to block bacterial infections. Here, we describe a new quorum-quenching strategy that directly “acquires” bacterial genes/proteins through a defined platform. We artificially expressed a typical AHL synthase gene pcoI from the biocontrol Pseudomonas fluorescens 2P24 in the antifungal bacterium Lysobacter enzymogenes OH11 lacking AHL production. This step led to the discovery of multiple PcoI interacting protein candidates from L. enzymogenes. The individual expression of these candidate genes in 2P24 led to the identification of Le0959, which encodes leucyl aminopeptidase, an effective protein that inhibits AHL synthesis in 2P24. Therefore, we define Le0959 as LqqP (Lysobacterquorum-quenching protein). The expression of pcoI in E. coli could produce AHL, and the introduction of lqqP into E. coli expressing pcoI could prevent the production of AHL. LqqP directly binds to PcoI, and this protein–protein binding reduced the abundance of free PcoI (capable of AHL synthesis) in vivo, thereby blocking PcoI-dependent AHL production. Overall, this study highlights the discovery of LqqP in quenching AHL quorum sensing by binding to AHL synthase via developing a previously-uncharacterized screening technique for bacterial quorum quenching.
1. Introduction
Quorum sensing (QS) is the behavior of communication within and between bacterial species [1], [2], [3]. QS in bacteria produces small signaling molecules, called autoinducers, for cells to communicate with each other. Autoinducers can be released into extracellular environment, with its concentrations increased with cell density. When extracellular autoinducers were accumulated to a certain threshold concentration, they shall activate a number of gene expressions, enabling single-celled bacteria to display group behaviors similar to higher organisms [4], [5]. For example, Vibrio fischeri utilizes quorum sensing to regulate the luminescence activity to achieve mutual symbiosis with squid [6]; Pseudomonas aeruginosa communicates through the quorum sensing to change the state of free-living plankton cells to form community-like biofilms and enable them to become resistant to applied antibiotics [7]. Gram-positive and gram-negative bacteria utilize QS communication to regulate a wide variety of physiological activities, such as virulence, symbiosis, conjugation, motility, competence, antibiotic production, sporogenesis, and biofilm formation [2]. Generally, gram-negative bacteria employ acylated homoserine lactone (AHL, AI-1) for communication, while gram-positive bacteria employ processed oligopeptides as autoinducer peptides (AIP). For instance, Staphylococcus aureus produces 8-amino acid cyclic short peptides as a QS signal to regulate the expression of virulence genes [1], [4], [8], [9]. In particular, AHL is an autoinducer in gram-negative bacteria that has been extensively studied in the past 35 years; they differ in the length and substitution of acyl side chains that confer them the signal specificity [2], [10], [11]. AHL is synthesized by a family of AHL synthases called LuxI through acyl-acyl carrier protein (acyl-ACP) and S-adenosylmethionine (SAM) mediated by MTAN (S-adenosyl homocysteine nucleosidase) and SRH (S-ribosylhomocysteine) [12]. This catalytic reaction is believed to proceed through a two-step mechanism; the intermediate acyl-SAM is formed from the acyl group transfer of acyl-ACP to the amino group of SAM, followed by the lactonization of the methionine moiety and the release of methylthioadenosine [12], [13]. The lowest threshold concentration for detection of AHL and its binding to LuxR-type transcription factors result in the regulation of a diverse array of gene expression, cellular processes, and physiological activities that are regulated by AHL [1]. It has been reported that many plant and animal pathogenic bacteria, i.e. Pseudomonas aeruginosa, can use AHL to control the production of virulence factors and promote host infections [14]. These key observations have motivated microbiologists to use various approaches to block bacterial infections by inhibiting AHL-dependent QS signaling, thereby developing effective strategies for crop protection and drug treatment.
Quorum quenching (QQ) refers to all processes involved in the interference of bacterial QS system [15], [16]. Scientists have developed several efficient screening methods for discovering QS inhibitors for the treatment of bacterial infections [17], [18], [19], and their working mechanisms mainly include (i) inhibition of AHL synthase activity by small-molecule chemicals, such as triclosan (a broad-spectrum antimicrobial agent). Triclosan acts as an enzyme inhibitor of the P. aeruginosa acyl-ACP reductase; it can effectively impair the synthesis of acyl-ACP, resulting in AHL synthase being unable to synthesize N-butyryl-L-homoserine lactone (C4-HSL) in P. aeruginosa [20]; (ii) inhibition of ligand (AHL) receptor interaction to stop AHL signal reception and its transmission. A representative example is the halogenated furanones from the Australian Marine red algae Delisea pulchra, which possess a common structure of a hyperserine lactone ring like AHLs. This structural similarity enables this substance to compete with AHLs for binding to receptors, thereby blocking AHL-induced functional outputs; (iii) disruption of AHL signal molecule by AHL-degrading enzymes. AiiA from Bacillus sp. 240B1, AttM from Agrobacterium tumefaciens C58 (A6), MomL from Muricauda olearia, and AiiD from Ralstonia sp. XJ12B are well-characterized representatives [21], [22], [23], [24]. Moreover, inhibition of protective biofilm formation and efflux pump or blocking autoinducing peptides (AIP)-mediated QS has also been addressed as effective QQ strategies [25], [26], [27]. In this study, we developed a platform to “fish” genes/proteins from bacteria that do not produce AHL to directly bind to and target AHL synthase, thereby generating a new QQ strategy.
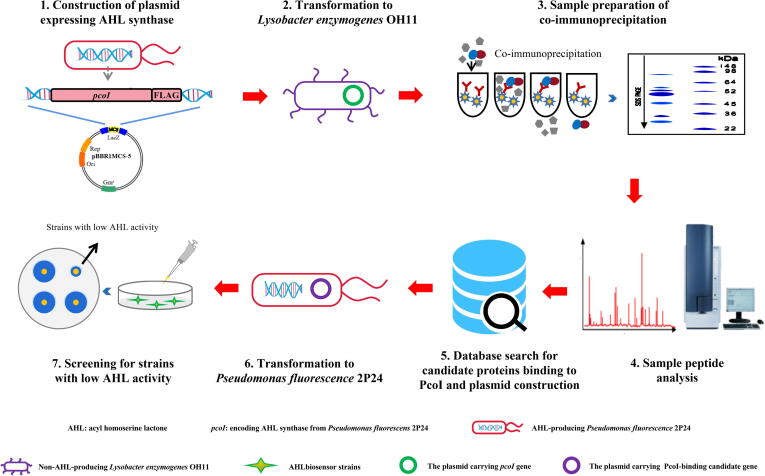
The gammaproteobacterial genus Lysobacter represents a group of antimicrobial biocontrol agents. Bacteria from this genus are common environmental inhabitants; they prey on other microorganisms by using secreted antimicrobial compounds and growth-inhibiting enzymes [28], [29]. Except for L. brunescens OH21 that contains a typical AHL synthase, almost all other reported members of Lysobacter do not produce AHL [30]. This observation led us to hypothesize that the genome of those Lysobacter species that do not produce AHL may carry previously uncharacterized genes/proteins to quench its QS signaling pathway. To discover such naturally occurring, new QQ mechanisms, we aim to identify Lysobacter proteins that directly bind and target AHL synthase by developing a platform involving heterogeneous gene expression and protein binding. To achieve this goal, we selected L. enyzmogenes OH11, which does not produce AHL and is the most studied species in the Lysobacter genus, as the working model. A representative AHL synthase gene known as pcoI from the antifungal Pseudomonas fluorescens 2P24 was fused with the FLAG tag and artificially expressed in L. enzymogenes OH11. The PcoI-FLAG interacting protein candidates from L. enzymogenes OH11 were captured by co-immunoprecipitation (Co-IP) and further expressed in P. fluorescens 2P24 alone to screen for target candidate(s) that can inhibit the production of AHL in P. fluorescens (Fig. 1). Via such screening, we discovered that Le0959 from L. enzymogenes OH11, which encodes an active leucyl aminopeptidase, can function as a new QQ protein. In terms of mechanism, Le0959 failed to directly degrade AHL, but bind with PcoI to reduce the abundance of PcoI (capable of AHL synthesis) as evidenced in E. coli. This study highlights the development of a previously-uncharacterized screen technique for bacterial quorum quenching and the discovery of LqqP in quenching AHL quorum sensing by binding to AHL synthase.
Fig. 1.
A platform for identifying the PcoI-binding proteins from Lysobacter enzymogenes OH11 that inhibit the production of AHL by Pseudomonas fluorescens 2P24. Step 2–3 are proposed by results shown in Fig. S2, while the results of the step 4–7 are provided in Fig. 2A-B.
2. Materials and methods
2.1. Bacterial strains, plasmids and growth conditions
The strains and plasmids used in this study are shown in Table S1. Escherichia coli strains were grown in Lysogenic broth (LB) medium at 37 °C with appropriate antibiotics. Unless otherwise specified, Lysobacter enzymogenes and Pseudomonas fluorescens 2P24 were grown in LB medium at 28 °C using the following antibiotics: kanamycin (Km), 30 μg/mL, gentamicin (Gm), 30 μg/mL, tetracycline (Tc), 30 μg/mL for plasmid maintenance. For growth curve analysis, Pseudomonas fluorescens 2P24 and its derivatives were grown in liquid LB medium at 28 °C, and their growth was monitored by measuring the optical density at 600 nm (OD600) every 2 h. Three replicates were performed for each strain and the bacterial growth curves were drawn based on the average OD600 values.
2.2. Heterogeneous expression of pcoI in L. enzymogenes or E. coli
The coding region of PcoI from P. fluorescens 2P24 was amplified by PCR using the primers listed in Table S2. The PCR product was cloned into the broad-host vector pBBR1-MCS5 and pUCP26 by restriction enzyme digestion (Table S1). The resulting plasmid was transformed into L. enzymogenes OH11 or E. coli strains (DH5α and BL21) by electroporation, and verified by PCR and western blotting.
2.3. Identification of PcoI-binding proteins from L. enzymogenes
The precipitation of the PcoI-FLAG binding proteins and their identification by mass spectrometry were performed as previously described [31]. The plasmid carrying PcoI-FLAG was introduced into L. enzymogenes OH11 by electroporation. The transformant was grown in LB until OD600 reached 1.0. The cells from the 40-mL culture were then harvested, and then resuspend in 2 mL 0.1 M PBS (pH, 7.4), followed by sonication (Sonifier 250; Branson Digital Sonifier, Danbury, USA). The insoluble material was removed by centrifugation at 6000 rpm at 4 °C for 20 min. Then, according to the manufacturer’s instructions, 2 mL of soluble protein was mixed with 50 μL of anti-FLAG magnetic beads (Bimake, Shanghai, China) to capture the PcoI-binding proteins. After incubating overnight at 4 °C, the beads were washed three times with 500 μL of 0.01 M PBS (pH, 7.4) containing 0.5% Tween 20. Proteins bound to the beads were eluted with 90 μL elution buffer (0.2 M glycine, pH 2.5), followed by eluent neutralization with 10 μL neutralization buffer (1.5 M Tris, pH 9.0). The eluted protein samples were identified by mass spectrometry at Beijing Protein Innovation Co., Ltd (Beijing, China).
2.4. AHL bioassays
The AHL plate bioassay was performed as previously described [32]. In brief, the production of AHL was detected by the effective AHL bioassay strain, Agrobacterium tumefaciens JZA1 [32]. The JZA1 strain was inoculated into 100 mL liquid LB medium, with the final concentration of antibiotics being 30 g/mL for gentamicin, 30 g/mL for kanamycin, 30 g/mL for spectinomycin and 12 g/mL for tetracycline. It was incubated at 28 °C for 24 h until the OD600 reached 1.5. 10 mL JZA1 culture was then added into 50 mL LB solid medium at a temperature of 50 °C and a final concentration of 40 g/mL for 5-bromo-4-chloro-3-indolyl-D-galactoside (X-gal) was added at the same time. After mixing, plates containing JZA1 were prepared, followed by inoculation of 3 μL of AHL samples on the plate surfaces. These prepared plates were incubated at 28 °C for 18 h to observe the size of the blue circles.
JZA1-based AHL quantification was carried out by the β-galactosidase method in a previous work [32]. In short, JZA1 and gentamicin (30 g/mL), grammycin (30 g/mL) and tetracycline (12 g/mL) were shaken at 28 °C and 220 rpm for 24 h in 100 mL liquid LB. When OD600 reached 1.5, 1 mL JZA1 culture was added into 100 mL fresh LB medium, followed by adding 2 mL cell-free supernatant of 2P24 and its derivatives. The resulted culture was incubated at 28 °C with shaking at 220 rpm until the OD600 reached to 0.4–0.8. After that, 200 μL culture was added into 800 μL Z-Buffer (8.5 g/L Na2HPO4, 5.5 g/L NaH2PO4·H2O, 0.75 g KCl, 0.246 g/L MgCl2·7H2O, 27 μL β-mercaptoethanol), followed by supplementing 10 μL SDS and 15 μL chloroform to a 2-mL centrifuge tube. After spining for 20 s, 100 μL of 4 mg/mL chromogenic substrate ONPG (2-Nitrophenyl β-D-galactopyranoside, Aladdin, Shanghai, China) was added, followed by inverting and recording the time as T1. When the reaction solution turns yellow, 600 μL 1 mM Na2CO3 was immediately added to stop the reaction and records the time as T2. After centrifuging the terminated solution for 3 min, the optical density of the supernatant at 420 nm (OD420) was measured. AHL activity is quantified by β-galactosidase activity/ (Miller units) and expressed by the formula, 1000 × OD420/OD600 (T2-T1)/0.2.
2.5. Bacterial two–hybrid assay
The BacterioMatch II two-hybrid (B2H) system (Agilent Technologies, USA) was used to detect protein–protein interactions in a previous work [33]. In short, the coding regions of target proteins (LqqP and PcoI) were cloned into pBT and pTGR plasmids and transformed into E. coli blue MRF ´ Kan. Plasmids pBT-GacS and pTRG-GacS (Table S1) were used as positive controls [33], while transformants containing empty pTRG and pBT vectors were used as negative controls. All co-transformants were spotted onto the selective medium and grown at 28 °C for 2 days. If there is a direct physical interaction between PcoI and LqqP, the transformed E. coli strain containing the two vectors are expected to grow well on the reference medium (+3AT + Strr) that is based on the minimal medium (M9) supplemented with 5 mM 3-AT, 2 μg/mL Str, 12.5 μg/mL tetracycline, 34 μg/mL chloramphenicol, and 30 μg/mL kanamycin, as described in a previous work [33]. The LB agar is a non-selective medium (-3AT-Strr) containing 12.5 μg/mL tetracycline, 34 μg/mL chloramphenicol, and 30 μg/mL kanamycin, as described previously [33]. The purpose of this medium is to ensure that both vectors can be successfully transformed into E. coli blue MRF ´ Kan.
2.6. Protein expression and purification
The LqqP and PcoI proteins were expressed as His6-fusions and purified by affinity chromatography. The coding region of pcoI was cloned into plasmid pET30a (Table S1) using primers listed in Table S2. The coding region of lqqP was cloned into plasmid pClod TF (Table S1). The genes were then expressed in E. coli BL21(DE3) (Table S1). The His6-fusion proteins were purified from 400 mL E. coli BL21(DE3) carrying the pET30a or pClod TF plasmid derivatives using Ni-NTA resin (GE Healthcare, Shanghai, China). The strains were grown to OD600 0.4 at 37 °C, and then 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Sigma, USA) was used to induce gene expression at 28 °C for 4 h to obtain PcoI or at 15 °C for 24 h to obtain LqqP. The concentration of purified proteins was determined by BCA protein assay kit (Sangon Biotech, Shanghai, China). Protein purity was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
2.7. Pull-down and Co-IP assays
The pull-down assays were performed as previously described [34]. The reaction mixture contains 5 μM PcoI-His and LqqP-GST or GST in 1.5 mL PBS buffer. Then 50 μL GST resin was added and samples incubated overnight at 4 °C. The resin was collected by centrifugation at 4 °C (500 g, 5 min) and washed 3 times with PBS containing 1% Triton X-100 to remove nonspecifically bound proteins. The proteins captured on the GST-beads were eluted by boiling in 4 × SDS loading dye for 6 min, after which the samples were subjected to SDS-PAGE and Western blotting. Protein detection involved the use of GST- (ab19256), His- (ab18184) and RNA polymerase (ab191598) specific antibodies (Abcam, UK).
The Co-IP assays were performed in a previous work [34]. In brief, the coding regions of the target proteins (LqqP, LqqPC78Yand PcoI) were cloned into pBBR1-MCS5 and pUCP26 plasmids, and co-transformed into E. coli DH5α or BL21(DE3). The transformant was grown in LB until OD600 reached 1.0. The cells from the 40-mL culture were then harvested, then resuspend in 2 mL 0.1 M PBS (pH, 7.4), followed by sonication (Sonifier 250; Branson Digital Sonifier, Danbury, USA). The insoluble material was removed by centrifugation at 6000 rpm for 20 min at 4 °C. Then 50 μL FLAG (Bimake, Shanghai, China) resin was added and samples were incubated overnight at 4 °C. The beads were then washed three times with 500 μL of 0.01 M PBS (pH, 7.4) containing 0.5% Tween 20. Proteins bound to the beads were eluted with 90 μL elution buffer (0.2 M glycine, pH 2.5), followed by neutralization with 10 μL neutralization buffer (1.5 M Tris, pH 9.0). Afterwards, the samples were subjected to SDS-PAGE and western blotting. Protein detection involved the use of FLAG- (M20008S) and His- (ab18184) specific antibodies (Abmart, Shanghai, China).
2.8. Microscale thermophoresis assay, MST
As previously described [33], the protein–protein binding affinities were determined by MST using Monolith NT.115 (NanoTemper Technologies, Germany). In brief, the LqqP-His protein was labeled with the fluorescent dye RED-Tris-NTA (NanoTemper Technologies GmbH, Germany) by amine conjugation. The labeled LqqP protein at a constant concentration (100 nM) in MST buffer was titrated against PcoI-His (concentration range, 0.229 nM-7.5 μM). The MST premium-coated capillaries (Monolith NT.115 MO-K005, Germany) were used to load the samples into the MST instrument at 25 °C with high MST power and 60% LED power. The laser on and off times were set to 30 and 5 s, respectively. All experiments were conducted in triplicate. Data were analyzed using Nanotemper Analysis software v.1.2.101 (NanoTemper Technologies, Germany).
2.9. Bioinformatics analyses
To identify LqqP homologues, LqqP was used as a query to run local BLASTp to identify the corresponding homologues in the selected bacterial genomes. A similar protein was considered present when the E-value was lower than 10-5 with a similarity percentage with the corresponding homologous protein higher than 40%.
2.10. Biofilm formation test
The biofilm formation of P. fluorescens 2P24 was performed as previously described [35]. In short, P. fluorescens 2P24 and its derivative strains were grown in liquid LB until the OD600 reached 1.0. Then 500 μL of bacterial culture was taken and added into a 2-mL tube containing 1 mL of fresh LB. The inoculated culture was cultivated at 28 °C for 36 h. The biofilm was washed three times with sterilized double distilled H2O (ddH2O), and then treated with 0.3% crystal violet (CV) for 15 min. Afterwards, the CV-stained biofilm was washed three times with ddH2O and resuspended in 95% ethanol (1 mL). The biomass was quantified at 570 nm by a spectrophotometer (Biotek, USA).
2.11. LqqP enzymatic activity assay
The leucine aminopeptidase activity was assayed by incubating 3 μg of purified LqqP-His and its derivatives with 100 mM of L-Leucine p-nitroanilide hydrochloride (Aladdin, Shanghai, China) in 1 mL reaction buffer (50 mM Tris-HCl, pH 7.5 and 5 mM NiCl2). The samples were incubated at 28° C for 9 h, and the reaction was stopped by cooling the mixture on ice for 10 min before the optical density at 405 nm (OD450) was measured using a microplate reader to indicate the enzyme activity as previously described [36].
2.12. Homology modeling of LqqP
Modeling of LqqQ was carried out by using the PHYRE2 program (ref?). The Dali program [37] was then used to further search the homologues of the modeled structures in the PDB. Seven structures with a Z score larger than 40 were obtained, and they all have the function of leucine aminopeptidase. We then superimposed the model structure on the highest-scoring analog (PDB: 2EWB) to find its active site residues. We found that five active site residues (K225, D230, D248, D307, and E309) are conserved between the two structures, and they coordinate well with the two zinc ions required for activity [38].
2.13. Data and material availability statement
The sequence data of this study have been submitted to the NCBI GenBank and included in Table S3. All other data required to evaluate the conclusions in the paper are provided in the paper or supporting information.
3. Results
3.1. L. enzymogenes LqqP acts as a new type of quorum-quenching proteins that block production of Pseudomonas AHL
Pseudomonas fluorescens 2P24 encodes a typical AHL synthase called PcoI, which is necessary for AHL production [35], while L. enzymogenes OH11 cannot produce AHL according to an earlier study [39]. To discover whether the L. enzymogenes OH11 genome encodes any genes that can directly target PcoI in P. fluorescens 2P24 to inhibit its AHL production, we established a platform that involves heterogeneous gene expression and protein binding. In this platform (Fig. 1 and Fig. S1), the AHL synthase gene pcoI was fused with the FLAG tag and cloned into a broad-host vector pBBR1-MCS5. The recombinant vector was then transformed into L. enzymogenes OH11 that does not produce AHL, in which the transcription of the pcoI-FLAG fusion gene is driven by a constitutive promoter from the plasmid. This step generated a recombinant strain named OH11-PcoI. The AHL produced by the OH11-PcoI strain was determined and validated by a well-known AHL biosensor strain JZA1 and LC-MS/MS technology (Fig. S1). In the context of OH11-PcoI, proteins that interact with PcoI-FLAG can be captured by Co-IP using anti-FLAG antibody (Fig. S2), and subsequently identified by LC-MS/MS. The identified candidate genes were further expressed separately in the AHL-producing P. fluorescens 2P24 to screen out target candidate(s) that inhibit the production of AHL in P. fluorescens 2P24 (Fig. 1).
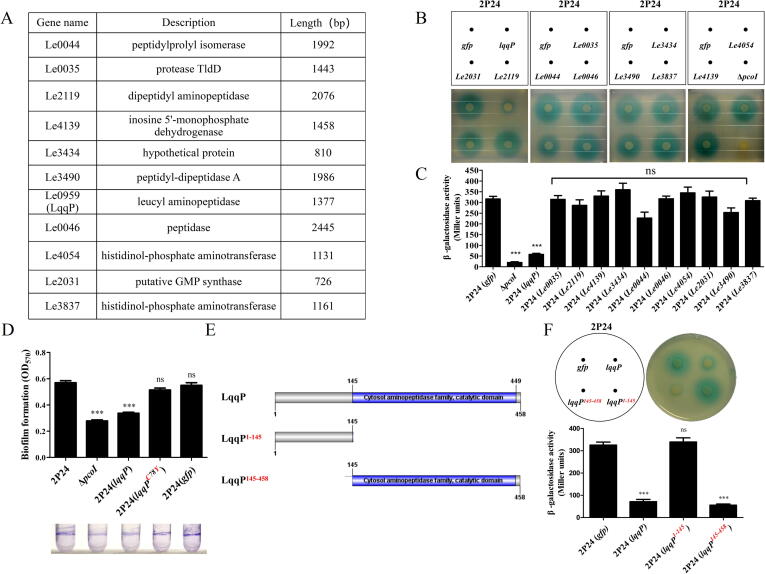
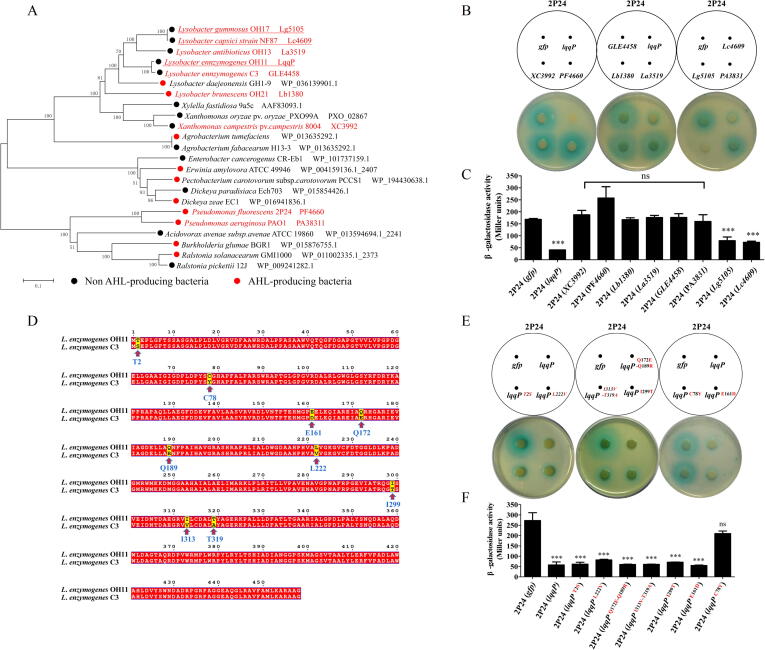
As shown in Fig. 2A, eleven PcoI interaction candidates identified by LC-MS/MS from L. enzymogenes OH11 were randomly selected for further study. Each of the eleven candidate genes was individually cloned into the broad-host vector pBBR1-MCS5 and introduced into 2P24 respectively. The quantitative analysis of AHL recognized by JZA1 revealed that the expression of Le0959 in 2P24 significantly reduced the amount of AHL, while the expression of the remaining ten genes in 2P24 did not show this effect (Fig. 2B & 2C). The pcoI mutant (ΔpcoI) unable to produce AHL was served as a negative control (Fig. 2B & 2C). In the following sections, we define Le0959 as the Lysobacter quorum-quenching protein, LqqP. Since AHL is reported to positively regulate biofilm formation in P. fluorescens 2P24 [35], we investigated whether the expression of lqqP in 2P24 could impair this AHL-controlled phenotype by blocking the AHL production, and results in Fig. 2D supported this hypothesis. We also observed that the expression of lqqP in 2P24 did not affect bacterial growth (Fig. S3). To identify which domain of LqqP functions, we performed protein truncations of LqqP. Results of Fig. 2E & 2F revealed that the expression of the C-terminal domain of LqqP (145–458 aa region) was sufficient to effectively inhibit the amount of AHL produced by 2P24.
Fig. 2.
The expression of lqqP in Pseudomonas fluorescens 2P24 remarkably inhibits the production of AHL. (A) Co-IP reveals a list of potential PcoI-binding proteins from L. enzymogenes OH11; (B-C) The production of AHL in 2P24 carrying each PcoI-binding protein gene shown in Fig. 2A was determined by the AHL bioassay strain JZA1 (B) and the AHL quantification indicated by β-galactosidase assay (C); (D) Biofilm formation of 2P24 carrying lqqP and its variant gene lqqPC78Y. ΔpcoI, the pcoI mutant of P. fluorescens 2P24 lacking the ability to produce AHL; gfp, a negative control; (E-F) Effect of LqqP truncations (E) on blocking AHL production in 2P24 (F). Coding sequences of LqqP truncations were separately cloned into a broad-host vector, pBBR1-MCS5 and transformed into 2P24, followed by AHL plate detection (up panel) and quantification (down panel) as described in panels B-C. In panels B, C, D, F, average data from three experiments are presented, ± SD. ***P < 0.0001 relative to 2P24 or 2P24 expressing gfp. “ns” stands for not statistically significant.
3.2. LqqP encodes an active leucyl aminopeptidase, and its ability to inhibit the production of P. fluorescens AHL does not depend on this enzymatic activity
Since all reported QQ proteins discovered to date function as AHL-degradation enzymes, we tested whether LqqP has any ability to degrade AHL. For this purpose, 100-nM commercial AHL [N-(3-oxodecanoyl)-L-homoserine lactone, 3-oxo-C10-HSL] was added to the culture of the AHL-deficient P. fluorescens mutant ΔpcoI expressing lqqP. The results of Fig. S4 showed that expression of lqqP or the negative control gfp in ΔpcoI failed to degrade the supplemented AHL that is determined by the AHL biosensor strain JZA1. Under similar conditions, expression of the positive control momL encoding a known AHL-degrading enzyme [21] effectively degraded the supplemented AHL in a time-dependent manner (Fig. S4). These results indicate that LqqP does not directly degrade AHL. Moreover, BLASTP searches in the NCBI database reveal that LqqP had no sequence similarity/identify to any reported AHL-degradation enzymes, such as AiiA, AttM, MomL and AiiD.
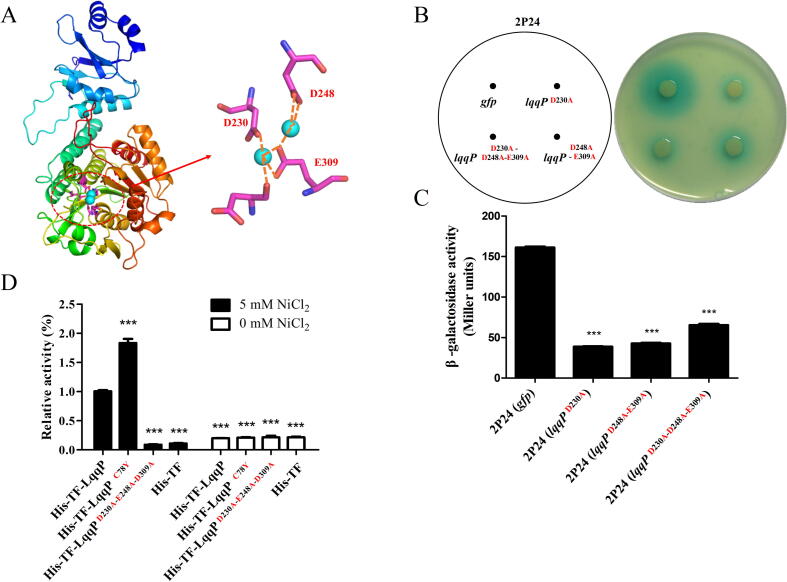
Since LqqP encodes a predicted leucyl aminopeptidase, we are interested to understand whether the enzymatic activity of LqqP is related to its function in P. fluorescens 2P24. A homolog-modeling reveals that LqqP possess three key amino residues –D230, E248, E309 that are expected to be essential for LqqP enzyme activity (Fig. 3A). As a random attempt, we replaced D230 with alanine by site-direct mutagenesis to produce the D230A variant. Expression of the LqqPD230A mutant in the wild-type 2P24 still effectively impaired AHL production (Fig. 3B & 3C). We thus generated a double variant LqqPD248A-E309A by simultaneously replacing D248 and E309 with alanine. Again, the LqqPD248A-E309A variant was still active in inhibiting AHL production when expressed in 2P24 (Fig. 3B & 3C). Finally, we generated a triple variant LqqPD230A-E248A-E309A by replacing D230, D248 and E309 with alanine simultaneously. As the result, introduction of this variant gene into 2P24 still inhibited the production of AHL (Fig. 3B & 3C). These results indicate that the enzyme activity of LqqP does not seem to contribute to its function in blocking AHL production. To validate this finding, we used the pCold-TF vector to express the native LqqP as well as the LqqPD230A-E248A-E309A triple variant, in which both target genes were fused with a His tag and a prokaryotic ribosomal binding partner protein (trigger factor, TF) that can induce co-translational folding of new peptide chains in E. coli BL21. Enzyme activity test showed that the His-TF-LqqP fusion does exhibit aminopeptidase activity (Fig. 3D), while His-TF-LqqPD230A-D248A-E309A triple variant gave no detectable enzyme activity like that of native control, the His-TF fusion protein (Fig. 3D). These results indicate that LqqP encodes an active leucyl aminopeptidase, and its function in blocking AHL production seems to be independent of its enzymatic activity. To support this conclusion, we isolated a variant of LqqP (LqqPC78A) by site mutagenesis and found that this variant LqqP showed higher aminopeptidase activity compared to the native LqqP, but still failed to inhibit the production of the PcoI-dependent AHL in 2P24 (see below).
Fig. 3.
The enzyme activity of LqqP does not seem to be related to its block of AHL production in P. fluorescens 2P24. (A) Homologous modeling of LqqP. The three key residues required for LqqP enzyme activity were predicted and highlighted in red. (B-C) The role of LqqP variants in blocking amount of AHL produced by 2P24. The coding regions of LqqP variants shown in red was expressed in 2P24 and the AHL production was detected in plates (B). (C) AHL quantification shown in panel B. The average data from three experiments is presented, ± SD. ***P < 0.0001 relative to 2P24 expressing gfp. (D) The enzyme activity of LqqP and its derivatives shown in red. LqqP and its variant genes were expressed with N-terminal fused His- and- TF tags. His-TF is served as a negative tag, in which TF is a trigger factor and a prokaryotic ribosomal binding partner protein that can induce co-translational folding of new peptide chains. Average data from three experiments is shown, ± SD. ***P < 0.0001 relative to His-TF-LqqP. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. C78 is required for LqqP function
To explore how LqqP inhibits the production of AHL by P. fluorescens, we set out to identify the key amino residue(s) required for LqqP functioning. Bioinformatics analyses reveal that LqqP is conservatively distributed in many AHL- and non-AHL-producing bacterial species (Fig. 4A). To test whether the LqqP homologs have the ability to block the production of P. fluorescens AHL, we first selected PF4660 and PA3831 from the AHL-producing P. fluorescens 2P24 and P. aeruginosa PAO1 respectively. The introduction of plasmid-borne PF4660 or PA3831 into 2P24 could not inhibit AHL production (Fig. 4B & 4C). We then chose XC3992 from Xanthomonas campestris 8004, which is a phylogenetically-related species of Lysobacter that cannot produce AHL. Similarly, the expression of XC3992 carried by the plasmid in 2P24 failed to impair AHL production (Fig. 4B & 4C). Finally, we chose five LqqP homologous genes from other Lysobacter species, including Lg5105 from L. gummosus OH17, Lc4609 from L. capsici NF87, La3159 from L. antibioticus OH13, GLE4458 from L. enzymogenes C3, and Lb3180 from L. brunescens OH21. The individual expression of these LqqP homologous genes in 2P24 led to the discovery that two of them (Lc4609 and Lg5105), like LqqP itself, prevented the production of AHL, but not for the remaining three (La3159, Lb3180 and GLE4458) (Fig. 4B & 4C). The successful expression of all LqqP homologous genes in 2P24 was confirmed by Western blotting (Fig. S5A). Together, these results indicate that some Lysobacter LqqP homologues are functionally divergent to block the AHL production in P. fluorescens 2P24. Notably, GLE4458 from L. enzymogenes C3 only differs from LqqP with nine residue substitutions (Fig. 4D), but it fails to inhibit AHL production when it is artificially expressed in 2P24 (Fig. 4B & 4C). While there are 9 different residues carried by LqqP compared to GLE4458, only 7 single or combination mutations were made in this study. Briefly, we randomly generated seven LqqP variants by single substitution or combination, which share the same residue(s) of GLE4458, including LqqPT2S, LqqPL222V, LqqPI299T, LqqPC78Y, LqqPE161D, LqqPQ172E-Q189R, and LqqPI313V-T319A (Fig. 4E & 4F). The expression of each LqqP variant in 2P24 by plasmid led to the discovery that only the C78 residue is the key to the LqqP function. The expression of the LqqPC78Y gene in 2P24 could not inhibit the production of AHL (Fig. 4E & 4F). Western blotting revealed that the above single substitution or combination did not affect the abundance of LqqP in 2P24 (Fig. S5B). These results support the conclusion that LqqP seems to require C78 to prevent the production of P. fluorescens AHL.
Fig. 4.
C78 is the key residue for LqqP in inhibiting the production of AHL in P. fluorescens 2P24. (A) The distribution of LqqP homologs in selected bacterial species with or without AHL-producing ability. Nine LqqP homologous genes highlighted in red were selected for heterogeneous expression in 2P24 and the red underline represents their ability to block AHL production; (B) The expression of nine LqqP homologous genes in 2P24 in blocking AHL production. lqqP and gfp were used as positive and negative controls, respectively. (C) AHL quantification corresponding to panel B. (D) The sequence alignment of LqqP from L. enzymogenes OH11 and its homologue, GLE4458 from L. enzymogenes C3. The arrows indicate the nine different amino residues between LqqP and GLE4458. (E-F) The effect of heterogeneous expression of LqqP variants genes with site mutations on the production of AHL in 2P24. Single site mutations or combinations are shown in in red. The AHL production in plates and its quantification are displayed in the upper and lower panels, respectively. lqqP and gfp were used positive and negative controls, respectively. The average data from three experiments is displayed, ± SD. ***P < 0.0001 relative to 2P24 expressing gfp. “ns” stands for not statistically significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. LqqP directly binds to PcoI and this binding weakens the abundance of free PcoI in vivo
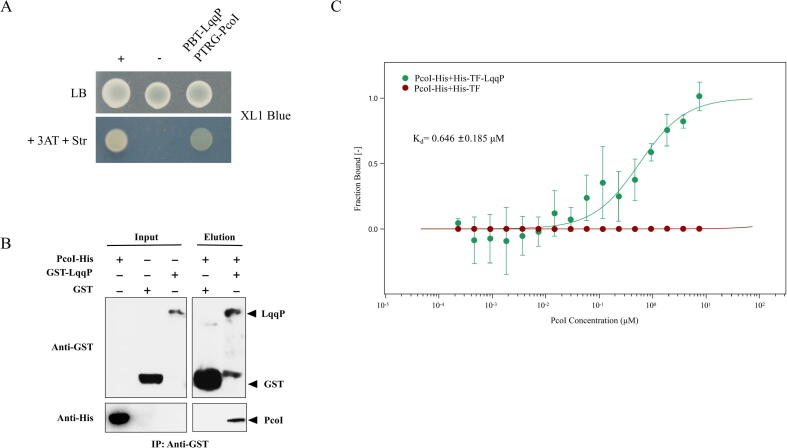
Why LqqP needs residue C78 to prevent the production of P. fluorescens AHL? We first tested whether LqqP directly binds to PcoI, because LqqP is a potent interacting protein of PcoI, as shown in Fig. 2A. To achieve this goal, we first chose the BacterioMatch II two-hybrid (B2H) system, which is a reliable method for evaluating protein–protein interactions based on the transcriptional activation of the HIS3 reporter gene, allowing transformed E. coli strain carrying two target genes to grow in a given media that is supplemented with 3-amino-1, 2, 4-triazole (3-AT), a competitive inhibitor of His3 enzyme [40]. Using this tool, we did detect the LqqP-PcoI interaction (Fig. 5A). To verify this observation, we tested the ability of Pco-His to pull down LqqP-GST, and we did observe a positive signal (Fig. 5B). To further quantify the LqqP-PcoI binding affinity, we adopted the microscale thermophoresis (MST) method, which is a powerful technique for quantifying biomolecular interactions[41]. Through MST, we found that PcoI-His bind to LqqP-TF-His with a moderately strong affinity (Kd, 0.646 µM), while the binding of PcoI-His with TF-His fusion could not be observed by MST (Fig. 5C). These results strongly supported the specific binding of LqqP to PcoI.
Fig. 5.
LqqP-PcoI interactions. (A) B2H assay shows the interaction between LqqP and PcoI. pTRG-PcoI and pBT-LqqP were co-expressed in E. coli XL1-blue. “+”, positive control – co-expression of plasmids pBT-GacS and pTRG-GacS [33]; “-”, negative control – co-expression of empty pTRG and pBT vectors. (B) Pull-down assay to confirm the interaction between PcoI-His and GST-LqqP. Immunoprecipitation was performed using anti-GST antibody. Western blot was performed by using anti-GST and anti-His antibodies. (C) MST measurement of binding affinity: green trace, between PcoI-His and His-TF-LqqP (Kd, 0.64 μM); red trace, between PcoI-His and His-TF (no binding). TF, trigger factor - a prokaryotic ribosomal binding partner protein that can induce co-translational folding of new peptide chains. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
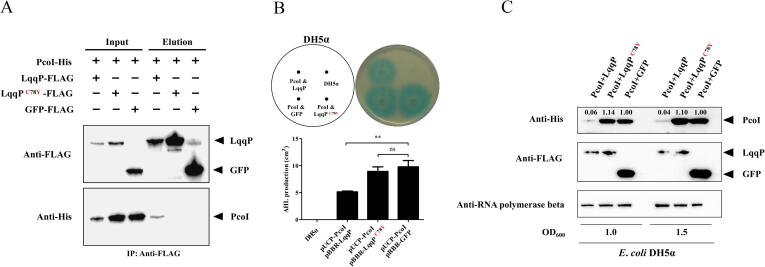
Is C78 essential for the LqqP-PcoI binding? To facilitate the investigation, we used the genetic background based on E. coli for subsequent experiments. We co-transformed the pUCP-PcoI and pBBR-LqqP in E. coli DH5α, and drove the expression of pcoI and lqqP by a constitutive promoter from the corresponding plasmid. The results of the in vivo Co-IP assays clearly reveal that the substitution of C78 by tyrosine (Tyr) prevents the binding of LqqP to PcoI (Fig. 6A), suggesting that C78 is indeed required for the LqqP-PcoI interaction.
Fig. 6.
C78 is essential for both the LqqP-PcoI binding and LqqP functioning in E. coli DH5α. (A) Co-IP reveals that C78 is required for LqqP-PcoI binding. The plasmids pBBR-LqqP and pUCP26-PcoI were co-transformed into E. coli DH5α. Immunoprecipitation was performed using anti-FLAG antibody. Western blot was performed by using anti-FLAG and anti-His antibodies. (B) The effect of the expression of native LqqP and its derivative LqqPC78Y gene on the production of PcoI-dependent AHL in E. coli DH5α. The AHL production and its quantification in the plates are displayed in the upper and lower panels, respectively. AHL production is calculated by dividing the area of the blue circle by the area of the colony. (C) The effect of co-expression of LqqP or its derivative LqqPC78Y gene on the abundance of PcoI in E. coli DH5α. The average data from three experiments is presented, ± SD. **P < 0.01. The band intensities were quantified and analyzed using ImageJ (https://imagej.nih.gov/ij/), with numbers representing the relative intensities of the corresponding bands. The intensity levels of the bands in lane “PcoI-GFP” were set to 1.00. “ns” stands for not statistically significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To understand whether the LqqP-PcoI binding contributes to the function of LqqP by blocking the production of PcoI-dependent AHL, we carried out corresponding experiments. We found that compared with the control E. coli DH5α containing pUCP-PcoI and the GFP vector control (pBBR-GFP), the E. coli DH5α carrying pUCP-PcoI and pBBR-LqqP had significantly lower AHL production (Fig. 6B), indicating that binding of LqqP appears to block the PcoI-dependent AHL production. To support this observation, we further found that the co-expression of PcoI and LqqPC78Y variant in E. coli DH5α failed to inhibit PcoI-mediated AHL production (Fig. 6B). These results together indicate that the association of LqqP with PcoI is essential for LqqP to inhibit the production of PcoI-dependent AHL, and C78 of LqqP seems to play an important role.
How does the binding of LqqP to PcoI inhibit the production of PcoI-dependent AHL? As one of the possibilities, we tested whether the PcoI expression profile would be altered after LqqP binding. To test this hypothesis, the native LqqP and its variant LqqPC78Y were co-expressed with pcoI in E. coli DH5α. We found that, compared to the gfp-pcoI co-expression control, the co-expression of lqqP-pcoI reduced the PcoI expression profile at an OD600 of 1.0 or 1.5 (Fig. 6C), while the co-expression of lqqPC78Y-pcoI resulted in a large abundance of PcoI like the gfp-pcoI co-expression control (Fig. 6C). In these co-expressions, the abundance of GFP or LqqP was not affected (Fig. 6C). As an internal control, the beta subunit of the RNA polymerase showed similar protein levels in all tested samples (Fig. 6C). These findings imply that the binding of LqqP most likely reduces the abundance of free PcoI or makes PcoI unstable in E. coli DH5α. To test whether the LqqP-PcoI binding can directly affect the abundance of PcoI in vitro, we further purified the PcoI-His and His-TF-LqqP fusion proteins, mixed them in a ratio of 1:1, and incubated them for 2 h, 4 h, or 6 h at 28 °C with or without the addition of NiCl2 that is a co-factor required for leucyl aminopeptidase activity. Under both conditions, His-TF-LqqP or His-TF-LqqPC78Y failed to alter the PcoI abundance in vitro (Fig. S6). This outlines a possibility that other unknown factors in E. coli may participate in the processing of LqqP in reducing the abundance of free PcoI.
4. Discussions
In the past decades, thousands of bacterial genomes have been deposited in public database, however, most of them are emphasized for functional genomics study [42]. To expand the usage of bacterial genomes, we have developed a platform (Fig. 1) that involves heterogeneous gene expression and protein interactions to discover bacterial genes for quorum quenching by directly targeting AHL synthase. In this study, we found that LqqP from L. enzymogenes OH11 is a new type of quorum-quenching proteins. LqqP cannot directly degrade AHL, which is a common function of all reported quorum-quenching enzymes, such as AiiA and MomL [21], [23]. The main difference is that LqqP targets PcoI, the AHL synthase of P. fluorescens, by binding to and reducing its free protein abundance, thereby inhibiting the production of PcoI-dependent AHL. Our results thus highlight the discovery of the Lysobacter LqqP in quenching quorum sensing through binding to PcoI and the screening technique developed in this study (Fig. 7).
Fig. 7.
Summary of AHL quorum quenching strategies. (A) Three well-known AHL quorum-quenching strategies. Quenching of AHL signaling by inhibiting AHL synthesis (middle green arrow) or AHL-receptor binding (bottom green arrow) by small molecular chemicals or enzymatically degrading AHL (top green arrow) are indicated. (B) A proposed model of LqqP-mediated quenching of PcoI-dependent AHL signaling, representing a new quorum-quenching mechanism through protein–protein interaction. The direct binding of LqqP and PcoI reduces the abundance of free PcoI in E. coli through an unknown mechanism. LuxI and PcoI: AHL synthase, LuxR and PcoR: AHL receptor transcriptional regulator. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In fact, LqqP encodes an active leucyl aminopeptidase whose function in bacteria is usually to catalyze the release of N-terminal amino acids, preferentially leucine, but not glutamic acid or aspartic acid [43], [44]. In Escherichia coli and Salmonella typhimurium, leucyl aminopeptidase catalyzes the removal of unsubstituted N-terminal amino acids from various peptides, and is involved in the processing and regular turnover of intracellular proteins [45]. Yet, this enzyme also functions as a DNA-binding protein, therefore, as a transcriptional repressor or activator, it controls the expression of virulence-related genes in the operon of pathogenic Escherichia coli, Vibrio cholerae and Pseudomonas aeruginosa [46], [47], [48]. In this study, we further expanded the role of this enzyme family by detecting the ability of LqqP to block the synthesis of PcoI-dependent AHL. Interestingly, although LqqP encodes an active aminopeptidase, its function of blocking the synthesis of PcoI-dependent AHL does not seem to be related to its enzyme activity (Fig. 3B-3D). This mode of action independent of enzyme activity may endow LqqP with unique features as a quorum-quenching protein. This conclusion is partly supported by the following observation: the expression of pcoI in E. coli enables it to produce AHL, in which there is a well-characterized active leucyl aminopeptidase PepA [43]. The failure of multiple LqqP homologues to prevent the production of PcoI-dependent AHL tested in this study can serve as additional evidence. It is also noteworthy that LqqP is discovered in L. enzymogenes OH11 that does not produce AHL, but it is difficult to directly link its function with its genomic distribution in non-AHL-producing bacteria, because we found several LqqP homologues such as XC3992, La3159, and GLE4458 from the phylogenetically-related and non-AHL-producing X. campestris 8004, L. antibioticus OH13 and L. enzymogenes C3, respectively, were all unable to block the PcoI-dependent AHL production (Fig. 4B & 4C). Therefore, how LqqP can function independent of its enzyme activity and specifically bind to PcoI remains unclear. The structural characterization of the LqqP-PcoI complex will allow people to address these interesting questions in the future.
A notable finding of this study is the moderately strong binding affinity between LqqP and PcoI. This specific protein–protein binding appears to be required for LqqP to quench quorum-sensing by targeting PcoI. Although the complex structure of LqqP-PcoI have yet to be determined, we have identified residue C78 of LqqP as an important site for the LqqP-PcoI interaction (Fig. 6A-6C). Interestingly, C78 is not located in the 145–458 region (the C-terminal LqqP truncation) that can efficiently block the PcoI-mediated AHL production (Fig. 2E & 2F). This suggests that the C78 residue may be essential for LqqP to adopt a correct conformation required for PcoI binding or it may be located at the binding interface between LqqP and PcoI. Moreover, C78 did not appear to be a conserved residue signature for LqqP function as a novel AHL quorum-quenching protein, because the LqqP homologues (Lg5105 and Lc4609) that are capable of blocking PcoI-dependent AHL production in 2P24 have a different residue (Y78) at the same position (Fig. S7).
Another notable finding of this study is the demonstration that the co-expression of LqqP and PcoI and their binding seems to significantly reduce the free PcoI abundance as demonstrated in E. coli DH5α (Fig. 6C). Although the underlying mechanism of this observation remains obscure, the targeting of PcoI in E. coli DH5α by LqqP may be associated with the Lon protease, which is an ATP-dependent serine protease that can mediate selective degradation of protein variants or abnormal proteins [49]. According to previous reports, the loss of Lon in Pseudomonas chlororaphis HT66 and Pseudomonas aeruginosa PAO1 significantly increased the abundance of AHL synthase. In vitro degradation assay suggested that the Lon protease can directly degrade AHL synthase [50], [51]. We were surprised to learn that when the LqqP and PcoI genes was co-expressed in E. coli BL21, LqqP could not inhibit the production of PcoI-dependent AHL (Fig. S8A). Compared with DH5α, the Lon protease of E. coli BL21 is mutated. We further observed that in the background of E. coli BL21, the co-expression of lqqP-pcoI resulted in the abundance of PcoI reaching to a level similar to that of the gfp-pcoI co-expression control at OD600 of 1.0 or 1.5 (Fig. S8B). However, other unidentified factors are also possibly present to help the targeting process of PcoI by LqqP in E. coli. Moreover, quenching AHL QS by directly targeting AHL synthase by proteins as presented by the PcoI-LqqP complex is mechanistically different to that of small-molecule inhibitors of AHL synthases, such as the well-studied J8-C8 [52]. It is documented that the small molecule J8-C8 acts as an artificial ligand of the AHL synthase TofI of Burkholderia glumae strain BGR1; it inhibits AHL production by binding to TofI and occupying the binding site of acyl-ACP acyl chain, as reported earlier [52].
The discovery that LqqP can bind to and target PcoI also provides a clue to explain why the PcoI homologues are lost but LqqP remains in L. enzymogenes OH11 during genome evolution. L. enzymogenes OH11 secrets a major antifungal metabolite called heat-stable antifungal factor (HSAF), which enters into the surrounding environment to inhibit the growth of fungal pathogens [53]. We found that the production of AHL in OH11 conferred by the heterogeneous expression of PcoI did not affect HSAF production, while in-frame deletion of lqqP in OH11 caused the strain to overproduce HSAF (Fig. S9). This indicates that LqqP seems to play a vital role in maintaining the HSAF homeostasis, which may help L. enzymogenes to fine-tune the amount of HSAF as an antifungal weapon, because the overproduction of HSAF is more than just an energy-consuming process but also impairs the adaptability of bacteria in nature.
The discovery that LqqP targets PcoI also led us to test whether LqqP also targets other AHL synthases. The result shows that LqqP can also effectively interfere with the RhlI-mediated AHL system in the pathogenic P. aeruginosa PAO1. RhlI in P. aeruginosa is a well-characterized AHL synthase responsible for the synthesis of C4-HSL, which binds to the transcription factor RhlR to activate expression of diverse downstream genes associating with the production of elastase, pyocyanin, hemolysin and rhamnolipid [54], [55]. We found that the expression of lqqP in PAO1 significantly impaired the amount of RhlI-mediated butyl-homoserine lactone (N-butyl homoserine lactones, C4-HSL) and several AHL-controlled functions (Fig. S10A-10E). Furthermore, LqqP directly binds to RhlI (Fig. S11A & 11B). These results indicate that LqqP can target AHL synthases from beneficial and pathogenic Pseudomonas species.
As reported earlier, the AHL-mediated quorum sensing plays key roles not only for the full virulence of plant pathogenic bacteria, but also for promoting antimicrobial activity or colonization capacity of plant beneficial bacteria [35]. Since most reported quorum-quenching enzymes (i.e. AiiA and MomL) have broad-spectrum capacity in degrading AHLs, it seems to be very difficult for them to selectively degrade AHLs produced only by plant pathogenic bacteria but not by plant beneficial bacteria after filing for crop protection. As a unique quorum-quenching protein, LqqP fails to degrade AHLs, but quenches AHL signaling by binding and targeting AHL synthases, as evidenced by the finding of LqqP bound to PcoI and induced PcoI degradation in vivo. This capacity may enable us to design the wild-type LqqP or its engineered versions to recognize only the AHL synthases of plant pathogenic bacteria, but not those of plant beneficial bacteria, by selectively targeting on the AHL signaling of plant pathogenic bacteria to reduce their virulence on hosts without affecting the AHL signaling of plant beneficial bacteria.
Finally, it is also noteworthy that although the heterogeneous expression of LqqP in P. fluorescens 2P24 shows a strong ability in inhibiting the production of PcoI-dependent AHL, our results indicate that LqqP does not seem to have the opportunity to directly enter the P. fluorescens cells because we observed that supplementing the purified LqqP proteins to the culture of P. fluorescens 2P24 did not inhibit AHL production (Fig. S12). Therefore, the delivery of LqqP into AHL-producing bacteria by biotechnology approaches, i.e. nanotechnology may be a promising approach in future.
5. Conclusions
This study highlights a novel quorum-quenching gene through the defined quorum-quenching platform developed in this study. This platform may be universal after minor modifications to uncover genes from bacterial, fungal or plant genomes of interest, to bind and target component(s) of bacterial virulence-associated protein secretion systems and/or signaling pathway(s), by which scientists can develop new strategies to hinder the infection of numerous pathogenic bacteria for crop protection and medical therapy.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Prof. Li-qun Zhang (China Agricultural University) and Prof. Zongtao Zeng (Nanjing Agricultural University, China) for providing P. fluorescens 2P24 and JZA1 as gifts. We also thank Prof. Yongxin He (Lanzhou University, China) for technical support in AHL structural analyses. This study was supported by the National Natural Science Foundation of China [32072470 and 31872016 to G.Q.; 32001955 to L. L.]; the National Key Research and Development Program of China (2018YFE0192600 to T. L.), Shanghai Agriculture Applied Technology Development Program, China (T20200104 to T. L.); the Natural Science Foundation of Jiangsu Province [BK20190026 and BK20181325 to G.Q.]; Fundamental Research Funds for the Central Universities [2020JB05 to T. L.; KJJQ202001, KYT201805, KYZ202106, KYRC2021003 and KYTZ201403 to G.Q.; KJQN202111 to L. L.]; Innovation Team Program for Jiangsu Universities [2017 to G.Q.]. The funders had no role in the study design.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.11.017.
Contributor Information
Jinxing Liao, Email: 2019202019@njau.edu.cn.
Danyu Shen, Email: shendanyu@njau.edu.cn.
Long Lin, Email: linlong@njau.edu.cn.
Hongjun Chen, Email: vetchj@shvri.ac.cn.
Yajie Jin, Email: 1615321647@qq.com.
Shan-Ho Chou, Email: shchou@nchu.edu.tw.
Xiao-Quan Yu, Email: yuxq20@lzu.edu.cn.
Tao Li, Email: litao@shvri.ac.cn.
Guoliang Qian, Email: glqian@njau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fuqua C., Greenberg E.P. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Bio. 2002;3(9):685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 2.Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55(1):165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 3.Ng W.-L., Bassler B.L. Bacterial Quorum-Sensing network architectures. Annu Rev Genet. 2009;43(1):197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papenfort K., Bassler B.L. Quorum sensing signal response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14(9):576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whiteley M., Diggle S.P., Greenberg E.P. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551(7680):313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters C.M., Bassler B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Bi. 2005;21(1):319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 7.Thi M.T.T., Wibowo D., Rehm B.H.A. Pseudomonas aeruginosa biofilms. Int J Mol Sci. 2020;21(22):8671. doi: 10.3390/ijms21228671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua W.C., Winans S.C., Greenberg E.P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176(2):269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji G., Beavis R.C., Novick R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci. 1995;92(26):12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi L.i., Dong X., Grenier D., Wang K., Wang Y. Research progress of bacterial quorum sensing receptors: Classification, structure, function and characteristics. Sci Total Environ. 2021;763:143031. doi: 10.1016/j.scitotenv.2020.143031. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Wu J., Deng Y., Wang J., Wang C., Wang J., et al. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol. 2013;9(5):339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 12.Parsek M.R., Val D.L., Hanzelka B.L., Cronan J.E., Greenberg E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci. 1999;96(8):4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson W.T., Minogue T.D., Val D.L., von Bodman S.B., Churchill M.E.A. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol Cell. 2002;9(3):685–694. doi: 10.1016/S1097-2765(02)00480-X. [DOI] [PubMed] [Google Scholar]
- 14.Pearson J.P., Gray K.M., Passador L., Tucker K.D., Eberhard A., Iglewski B.H., et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci. 1994;91(1):197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandclément C., Tannières M., Moréra S., Dessaux Y., Faure D., Camara M. Quorum quenching: role in nature and applied developments. Fems Microbiol Rev. 2016;40(1):86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 16.Murugayah S.A., Gerth M.L. Engineering quorum quenching enzymes: progress and perspectives. Biochem Soc T. 2019;47(3):793–800. doi: 10.1042/BST20180165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen Q.H., Grove T.L., Booker S.J., Greenberg E.P. A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci. 2013;110(34):13815–13820. doi: 10.1073/pnas.1313098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandi S. Recent Advances in ligand and structure based screening of potent quorum sensing inhibitors against antibiotic resistance induced bacterial virulence. Recent Pat Biotechnol. 2016;10(2):195–216. doi: 10.2174/1872208310666160728104450. [DOI] [PubMed] [Google Scholar]
- 19.Kumar M., Saxena M., Saxena A.K., Nandi S. Recent Breakthroughs in various antimicrobial resistance induced quorum sensing biosynthetic pathway mediated targets and design of their inhibitors. Comb Chem High T Scr. 2020;23(6):458–476. doi: 10.2174/1386207323666200425205808. [DOI] [PubMed] [Google Scholar]
- 20.Hoang T.T., Schweizer H.P. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (fabi): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J Bacteriol. 1999;181(17):5489–5497. doi: 10.1128/JB.181.17.5489-5497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang K., Su Y., Brackman G., Cui F., Zhang Y., Shi X., et al. MomL, a novel marine-derived N -Acyl homoserine lactonase from Muricauda olearia. Appl Environ Microb. 2015;81(2):774–782. doi: 10.1128/AEM.02805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H.B., Wang L.H., Zhang L.H. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci. 2002;99(7):4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Zhang L., Wang L., Zhang H., Xu J., Dong Y. Quenching quorum-sensing dependent bacterial infectionby an N-acyl homoserine lactonase. Nature. 2001;411(6839):813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y., Xu J., Hu J., Wang L., Ong S.L., Leadbetter J.R., et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003;47(3):849–860. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 25.Roy V., Fernandes R., Tsao C., Bentley W.E. Cross species quorum quenching using a native AI-2 processing enzyme. Acs Chem Biol. 2010;5(2):223–232. doi: 10.1021/cb9002738. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Jiao H., Meng J., Qiao M., Du H., He M., et al. Baicalin inhibits biofilm formation and the quorum-sensing system by regulating the msra drug efflux pump in Staphylococcus saprophyticus. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcalde-Rico M., Olivares-Pacheco J., Alvarez-Ortega C., Cámara M., Martínez J.L. Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen P., Cook F.D. Lysobacter a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol. 1978;28(3):367–393. doi: 10.1099/00207713-28-3-367. [DOI] [Google Scholar]
- 29.Puopolo G., Tomada S., Pertot I. The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J Appl Microbiol. 2018;124(1):15–27. doi: 10.1111/jam.13607. [DOI] [PubMed] [Google Scholar]
- 30.Ling J., Zhou L., Wu G., Zhao Y., Jiang T., Liu F. The AHL quorum-sensing system negatively regulates growth and autolysis in Lysobacter brunescens. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian B., Liu X., Jia J., Cai Y., Chen C., Zhang H., et al. MoPpe1 partners with MoSap1 to mediate TOR and cell wall integrity signalling in growth and pathogenicity of the rice blast fungus Magnaporthe oryzae. Environ Microbiol. 2018;20(11):3964–3979. doi: 10.1111/1462-2920.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J., Chai Y., Zhong Z., Li S., Winans S.C. Agrobacterium bioassay strain for ultrasensitive detection of N-Acyl homoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microb. 2003;69(11):6949–6953. doi: 10.1128/AEM.69.11.6949-6953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu G., Han S., Huo C., Chin K.-H., Chou S.-H., Gomelsky M., et al. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res. 2018;46(18):9276–9288. doi: 10.1093/nar/gky803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M., Ren S., Shen D., Yang N., Wang B., Han S., et al. An intrinsic mechanism for coordinated production of the contact-dependent and contact-independent weapon systems in a soil bacterium. Plos Pathog. 2020;16(10):e1008967. doi: 10.1371/journal.ppat.1008967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei H., Zhang L. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek. 2006;89(2):267–280. doi: 10.1007/s10482-005-9028-8. [DOI] [PubMed] [Google Scholar]
- 36.Mun L.S., Tee T.S., Puthucheary S.D. Enzymatic and molecular characterisation of leucine aminopeptidase of Burkholderia pseudomallei. BMC Microbiol. 2013;13(1):110. doi: 10.1186/1471-2180-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L: Using Dali for Protein Structure Comparison. In., vol. 2112. New York, NY: Springer US; 2020: 29-42. [DOI] [PubMed]
- 38.Cappiello M., Alterio V., Amodeo P., Del Corso A., Scaloni A., Pedone C., et al. Metal ion substitution in the catalytic site greatly affects the binding of sulfhydryl-containing compounds to leucyl aminopeptidase. Biochemistry. 2006;45(10):3226–3234. doi: 10.1021/bi052069v. [DOI] [PubMed] [Google Scholar]
- 39.Han Y., Wang Y., Tombosa S., Wright S., Huffman J., Yuen G., et al. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biot. 2015;99(2):801–811. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joung J.K., Ramm E.I., Pabo C.O. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci. 2000;97(13):7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidel S.A.I., Dijkman P.M., Lea W.A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59(3):301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medema M.H., de Rond T., Moore B.S. Mining genomes to illuminate the specialized chemistry of life. Nat Rev Genet. 2021;22(9):553–571. doi: 10.1038/s41576-021-00363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsui M., Fowler J.H., Walling L.L. Leucine aminopeptidases: diversity in structure and function. Biol Chem. 2006;387(12) doi: 10.1515/BC.2006.191. [DOI] [PubMed] [Google Scholar]
- 44.Miller C.G., Mackinnon K. Peptidase Mutants of Salmonella typhimurium. J Bacteriol. 1974;120(1):355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen C., Green L., Miller C.G. Degradation of intracellular protein in Salmonella typhimurium peptidase mutants. J Mol Biol. 1980;143(1):21–33. doi: 10.1016/0022-2836(80)90122-9. [DOI] [PubMed] [Google Scholar]
- 46.Charlier D., Hassanzadeh G.G., Kholti A., Gigot D., Piérard A., Glansdorff N., et al. Involved in pyrimidine regulation of the Escherichia coli carbamoylphosphate synthetase operon encodes a sequence-specific dna-binding protein identical to XerB and PepA, also required for resolution of ColEl multimers. J Mol Biol. 1995;250(4):392–406. doi: 10.1006/jmbi.1995.0385. [DOI] [PubMed] [Google Scholar]
- 47.Behari J., Stagon L., Calderwood S.B. pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J Bacteriol. 2001;183(1):178–188. doi: 10.1128/JB.183.1.178-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolwine S.C., Wozniak D.J. Identification of an Escherichia coli pepA homolog and its involvement in suppression of the algb phenotype in mucoid Pseudomonas aeruginosa. J Bacteriol. 1999;181(1):107–116. doi: 10.1128/JB.181.1.107-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong H., Barbe V., Lee C.H., Vallenet D., Yu D.S., Choi S.-H., et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J Mol Biol. 2009;394(4):644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z., Huang X., Jan M., Kong D., Wang W., Zhang X. Lon protease downregulates phenazine-1-carboxamide biosynthesis by degrading the quorum sensing signal synthase PhzI and exhibits negative feedback regulation of Lon itself in Pseudomonas chlororaphis HT66. Mol Microbiol. 2021;116(2):690–706. doi: 10.1111/mmi.v116.210.1111/mmi.14764. [DOI] [PubMed] [Google Scholar]
- 51.Yang N., Ding S., Chen F., Zhang X., Xia Y., Di H., et al. The Crc protein participates in down-regulation of the Lon gene to promote rhamnolipid production andrhlquorum sensing in Pseudomonas aeruginosa. Mol Microbiol. 2015;96(3):526–547. doi: 10.1111/mmi.2015.96.issue-310.1111/mmi.12954. [DOI] [PubMed] [Google Scholar]
- 52.Chung J., Goo E., Yu S., Choi O., Lee J., Kim J., et al. Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase. Proc Natl Acad Sci. 2011;108(29):12089–12094. doi: 10.1073/pnas.1103165108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lou L., Qian G., Xie Y., Hang J., Chen H., Zaleta-Rivera K., et al. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc. 2011;133(4):643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J., Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6(1):26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brint J.M., Ohman D.E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177(24):7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.