Abstract
In the rapidly expanding field of peptide therapeutics, the short in vivo half-life of peptides represents a considerable limitation for drug action. D-peptides, consisting entirely of the dextrorotatory enantiomers of naturally occurring levorotatory amino acids (AAs), do not suffer from these shortcomings as they are intrinsically resistant to proteolytic degradation, resulting in a favourable pharmacokinetic profile. To experimentally identify D-peptide binders to interesting therapeutic targets, so-called mirror-image phage display is typically performed, whereby the target is synthesized in D-form and L-peptide binders are screened as in conventional phage display. This technique is extremely powerful, but it requires the synthesis of the target in D-form, which is challenging for large proteins. Here we present finDr, a novel web server for the computational identification and optimization of D-peptide ligands to any protein structure (https://findr.biologie.uni-freiburg.de/). finDr performs molecular docking to virtually screen a library of helical 12-mer peptides extracted from the RCSB Protein Data Bank (PDB) for their ability to bind to the target. In a separate, heuristic approach to search the chemical space of 12-mer peptides, finDr executes a customizable evolutionary algorithm (EA) for the de novo identification or optimization of D-peptide ligands. As a proof of principle, we demonstrate the validity of our approach to predict optimal binders to the pharmacologically relevant target phenol soluble modulin alpha 3 (PSMα3), a toxin of methicillin-resistant Staphylococcus aureus (MRSA). We validate the predictions using in vitro binding assays, supporting the success of this approach. Compared to conventional methods, finDr provides a low cost and easy-to-use alternative for the identification of D-peptide ligands against protein targets of choice without size limitation. We believe finDr will facilitate D-peptide discovery with implications in biotechnology and biomedicine.
Abbreviations: MIPD, mirror-image phage display; PPI, protein-protein interaction; D-AA, dextrorotatory amino acid; L-AA, levorotatory amino acid; PSMα3, phenol soluble modulin alpha 3; MRSA, methicillin-resistant Staphylococcus aureus; PD-1, receptor programmed death 1; MD, molecular dynamics; EA, evolutionary algorithm; SPPS, solid phase peptide synthesis; NCL, native chemical ligation; MIVS, mirror-image virtual screening; MIEA, mirror-image evolutionary algorithm
Keywords: D-peptide, Web server, Evolutionary algorithm, Peptide design, Molecular docking, Mirror-image phage display
1. Introduction
Advances in peptide synthesis and peptide functional screening methods in the last decades have brought about a renaissance in peptide drug discovery [1]. The potential of peptides and peptide-based compounds as diagnostic, theragnostic and therapeutic tools is increasingly recognized in academia and industry, leading to a steady increase in peptide drugs in clinical trials and among FDA approvals, like Setmelanotide™ for the treatment of genetically determined obesity [2,3]. Next to hormone analogues such as insulin substitutes, peptide therapeutics are increasingly used to target protein-protein interactions (PPIs) to interfere with a variety of pathology-related cellular processes [4]. Especially as personalized medicine is advancing, the need for nontoxic and high-affinity ligands to target PPIs increases. The major advantages of peptides as drug candidates are their specificity and affinity towards their target, which exceed that of often used small molecule drugs and which are, in some cases, even comparable to those of antibodies [5]. Due to their small size, peptide drugs are also considerably less costly to manufacture and have an increased tissue penetration, which is instead a major limitation of antibodies. In fact, peptides have even been suggested as an alternative to antibodies. However, a considerable shortcoming of conventional peptide therapeutics is their short in vivo half-life due to proteolytic degradation [4]. This was successfully overcome by the development of so-called D-peptides, which are made exclusively of dextrorotatory amino acids (D-AAs) – the enantiomers of the canonical levorotatory amino acids (L-AAs) [6]. Since all known naturally occurring proteases are exclusively specific for the naturally occurring peptide bonds between L-AAs and sterically incompatible with the peptide bond between two D-AAs, D-peptides are intrinsically protease-resistant, which dramatically increases their metabolic stability [7]. As a consequence, D-peptides are also less immunogenic [8] and have an increased oral bioavailability as compared to conventional L-peptides [9].
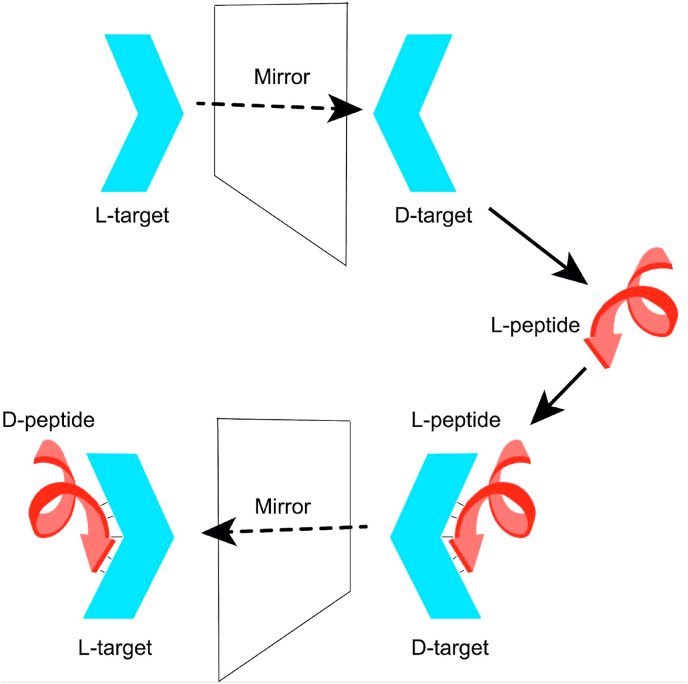
To identify a D-peptide ligand to a target protein of interest, a method called mirror-image phage display (MIPD) was developed, as an extension of the powerful conventional phage display technology [10,11]. Analogous to conventional phage display, MIPD is based on the screening of a random peptide library, which is expressed on the surface of bacteriophages, against an immobilized target in multiple cycles of selection and amplification [12,13]. Since the generation of D-peptide libraries in biological protein expression systems such as phages is not yet possible, MIPD takes advantage of the principle that the interaction of two molecules is analogous to that of their mirror-image counterparts [14]. Thus, MIPD is performed by screening an L-peptide phage library against the mirror-image version of the target protein – a protein of the same amino acid sequence made completely of D-AAs. The interaction between L-peptide ligands and the D-protein target will also exist between the mirror-image D-peptide ligand and the naturally occurring L-configuration of the target protein [11] (Fig. 1).
Fig. 1.
Schematic representation of the stereochemistry of a protein - peptide interaction. If an L-peptide ligand binds a protein's mirror-image (D-protein), then their corresponding mirror-images (D-peptide ligand and L-protein) will also bind to each other in exactly the same fashion.
Several groups have applied MIPD, identifying D-peptide ligands of clinical potential, such as inhibitors of the immune checkpoint receptor programmed death 1 (PD-1) pathway [15], inhibitors of glycoprotein 41 to block HIV entry [16] or inhibitors of toxic amyloid beta plaque in Alzheimer's disease [17]. In the latter two cases, D-peptide therapeutics reached the clinical trial phase (ClinicalTrials.gov identifiers: NCT04672083 and NCT04711486).
Despite its potential, MIPD suffers from some limitations that impede its broader adoption. The current technical means of chemical D-protein synthesis limit the choice of target proteins in terms of size and sequence [18]. Furthermore, the correct in vitro folding of chemically synthesized proteins is not always successful and becomes more challenging the longer the protein. The longest D-protein to be synthesized and proven to be correctly folded to date is the mirror-image version of the E. coli enzyme DapA. This 312 D-AA-long protein was synthesized by Weinstock and colleagues using solid phase peptide synthesis (SPPS) and native chemical ligation (NCL) [19]. Apart from target-related restrictions, MIPD is highly time consuming and costly, and requires specialized know-how and equipment for chemical protein synthesis and subsequent purification of D-protein targets [20]. Furthermore, MIPD often yields off-target polystyrene binding peptides as an artefact [21].
An alternative way to identify L- and D-peptide ligands, which does not suffer from the abovementioned restrictions, is computational modelling of PPIs [22]. Recent publications have shown the feasibility of identifying biologically active D-peptide ligands by modelling the structure of short helical D-peptide segments with molecular dynamics (MD) simulations and by screening peptide libraries generated this way for ligand-target interactions via molecular docking [23,24]. These programs enable the high-throughput in silico screening of peptide libraries; however, with increasing peptide length, the size of peptide libraries and, consequently, the search space and computational cost for docking-based screenings increase exponentially to a point where they are simply too large to be systematically explored. For example, a peptide library of all possible 12-mer peptides made of canonical L-AAs would contain 2012 (4.096 × 1015) sequences. The chemical space would further be vastly expanded by the integration of noncanonical L- or D-AAs. In such cases, where deterministic approaches reach their limits, evolutionary algorithms (EA) represent a viable alternative [25,26]. EA-based methods apply the principles of Darwinian evolution to identify and optimize peptide ligands in a directed selection process. An initial population of randomly selected peptides is evaluated for evolutionary fitness, which is, in most cases, defined as their binding affinity towards a given target. After scoring, the peptides with the highest fitness are recombined by crossing over their sequences at a randomly chosen position to give rise to a second generation of peptides - similar to chromosome crossover during meiosis. Furthermore, the algorithm can introduce point mutations with a certain probability to avoid sequence convergence. The population will then again be subjected to the scoring function for evolutionary fitness and be further optimized in multiple cycles (“generations”) of the algorithm [27,28]. This approach has been successfully employed for peptide design by utilizing in vitro binding assays as a scoring function to determine peptide fitness [29]. Circumventing the need for extensive peptide synthesis for the validation of each generation, an in silico docking step has been integrated into the framework of an EA for de novo identification of peptide ligands [25,30].
Most of the aforementioned computational tools require either commercial software or a strong expertise in bioinformatics. While there are plenty of well-established, open-source software tools available for performing MD and molecular docking, for example GROMACS [31] and AutoDock Vina [32], handling these programs and integrating them into an effective pipeline for the purpose of screening requires extensive bioinformatics know-how. Recent studies report that the majority of wet lab researchers do not feel confident with the use of bioinformatic tools, programming languages or handling big data, revealing a gap between the rapidly increasing knowledge in the field of bioinformatics and university education [33]. This often underappreciated factor restricts the accessibility of these highly useful tools for peptide drug discovery. The need for computational power for extensive screening presents another obstacle.
To make computational identification and optimization of D-peptide ligands available to the scientific community we developed finDr, an easy-to-use web server based on established MD and docking software, which allows for the identification of L- and D-peptide ligands for any given target protein structure. As an extension of mere library screening, finDr also allows for the optimization of the ligands using a target-based EA. As a proof of principle, we identified D-peptide inhibitors against phenol soluble modulin alpha 3 (PSMα3), a protein toxin of the medically relevant methicillin-resistant Staphylococcus aureus (MRSA) [34]. Alongside our computational approach, we chemically synthesized D-PSMα3 and performed MIPD. Using in vitro binding assays, we show that the peptide optimized by the EA is an even better binder to PSMα3.
2. Methods
2.1. MIPD and phage ELISA
MIPD was performed using the Ph.D.™-12 Phage Display Peptide Library from New England Biolabs Inc (NEB) following the manufacturer's instructions (for MIPD principle, see Fig. 3). Briefly, 96-well plates were coated overnight at 4 °C; first with streptavidin (100 mg/mL in 0.1 M NaHCO₃ pH = 8.6), then with the target protein D-PSMα3, attached to a Lys-PEG4-Biotin Linker (obtained from GeneCust) (100 μg/mL in 0.5% TBS-Tween20). The target solution was removed, and the wells were blocked with 5 mg/mL bovine serum albumin (BSA) in 0.1 M NaHCO₃ for 1 h. Afterwards the plate was washed 6 times with TBS-T before a solution of 1011 phages (containing approximately 109 unique phage clones) were added to the wells and incubated for 30 min. Non-binding phages were removed by washing 15 times with TBS-T before eluting the target-binding phages with a 0.2 M glycine-HCL solution (pH 2.2). The eluted phages were added to a log phase (OD = 0.5) culture of K12 ER2738 E. coli (obtained from NEB) and amplified for 3.5 h at 37 °C, shaking. The amplified phages were purified by PEG precipitation according to the manufacturer's instructions and the titer of the phage solution was determined by adding serial dilutions of phages to K12 ER2738 E. coli and plating them on X-Gal/isopropyl β-d-1-thiogalactopyranoside agar plates. The number of plaque forming units (pfu) was determined by counting blue plaques in the bacterial lawn after overnight incubation at 37 °C. At least 109 pfu of the amplified target binding phages were used for the subsequent round of panning. To eliminate nonspecific binders, the blocking step of the second panning was performed with 0.1% gelatine in TBS-T. After three rounds of panning, the eluted phages were plated with K12 ER2738 E. coli as before and single plaques containing individual phage clones were picked for DNA isolation and sequencing. Further, the phage clones were amplified and purified for verification of their D-PSMα3 binding in a phage ELISA. 96-well plates were coated as before and blocked with BSA, then 3.5 × 109 pfu of the single phage clones were added and incubated for 90 min at 37 °C, shaking. The plates were washed 6 times with 0.1% TBS-T and then incubated with Horseradish peroxidase (HRP)-conjugated anti-M13 antibody (1:5000, Sino Biological, Inc., Beijing) for 2 h, washed another three times and incubated with TMB. After 3–6 min the reaction was stopped with 2 M sulfuric acid and the absorbance was detected at 450 nm using a standard plate reader. D-PSMα3 specific binding was determined in comparison to BSA/streptavidin binding in wells without D-PSMα3 coating. Phages obtained from a mock panning without D-PSMα3 coating served as negative control.
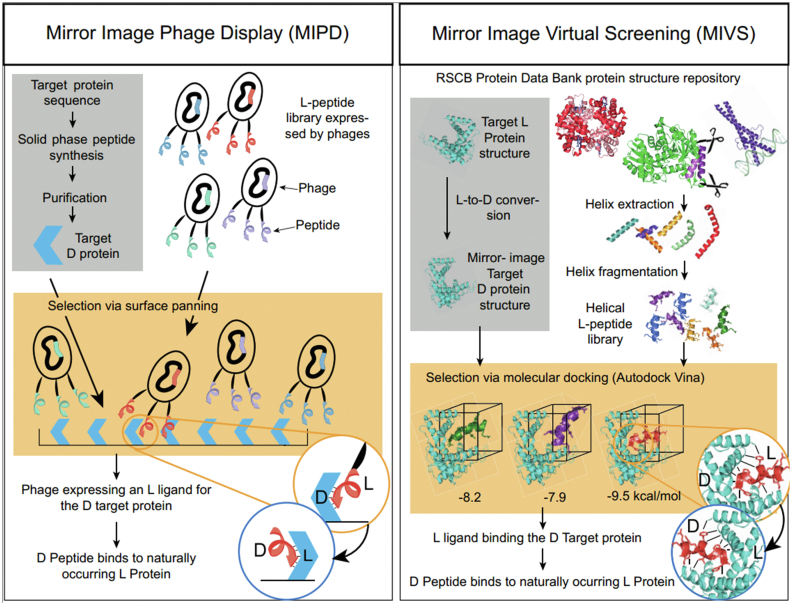
Fig. 3.
Schematic representation of the MIPD and MIVS workflow. MIPD: A library of random L-peptides expressed on the surface of bacteriophages is selected via surface panning against a chemically synthesized D-analogue of the target L-protein. MIVS: A structural library of helical L-peptide segments extracted from the PDB is screened for binding affinity towards an in silico mirrored D-version of the target L-protein structure via molecular docking. Both methods yield L-peptide ligands to D-protein targets. Consequently, the corresponding D-peptides bind to the naturally occurring L-protein target (see Fig. 1).
2.2. Solid phase peptide synthesis of L-peptide ligands
L-Peptide ligands were synthesized in a 50 μmol scale using 9-fluorenylmethyloxycarbonyl (Fmoc) solid-phase peptide synthesis (SPPS) using a MultiPep RSi peptide synthesizer (Intavis Bioanalytical Instruments). All reagents and protected AAs were purchased from Merck. Wang resins pre-coupled with the C-terminal amino acid of the respective peptide were swollen for 20 min in N,N-dimethylformamide (DMF) before synthesis. The subsequent elongation of the peptide chain was performed in sequential cycles of coupling to Fmoc-protected amino acids (0.5 M). The coupling step was carried out two times for each AA. The Fmoc group was removed using 20% piperidine in DMF. For the coupling reaction itself the activator O-benzotriazole-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate (HBTU, 4 eq) dissolved in DMF and the bases N-methylmorpholine (NMM, 0.1 eq), were added and reacted for 30 min. Capping was performed using 2,6-lutidine (5%) and acetic anhydride (5%) in DMF for 5 min. This cycle was repeated until the full length of the desired peptide was reached. Then the remaining Fmoc protection groups were cleaved by 20% piperidine and the side chain-protecting groups (tert-butyl ether, tert-butyl, triphenylmethyl (Trt) or tert-butyloxycarbonyl (Boc)) removed using trifluoroacetic acid (TFA, 95% TFA + 2.5% water, 2.5% Triisopropyl silane (TIPS)). After cleavage, the peptides were precipitated in diethyl ether at −20 °C and subsequent centrifugation. The fully synthesized ligands were then purified by preparative HPLC (Knauer) over an acetonitrile water gradient and their identity was validated using HPLC-MS.
2.3. SCORE real-time binding assays
The binding of the peptide ligands to PSMα3 was measured by single colour reflectometry (SCORE), formerly known as 1λ-imaging reflectometric interferometry (iRIf), which was described in detail previously [35]. In short, an array of L-peptide ligands (10 μg/mL in PBS) was spotted on 3D-N-hydroxysuccinimide (3D-NHS) coated custom SCORE glass slides purchased from Biametrics GmbH (Tübingen, Germany) now BioCopy GmbH (Emmendingen, Germany). BSA, a scrambled 12-mer peptide and biotinylated BSA (all 10 μg/mL in PBS) were used as negative controls. PSMα3 (10 μg/mL in PBS) was flushed over 3D-NHS slides at a constant flow rate (60 μL/min) and temperature (22 °C) through the microfluidic flow cell of the SCORE device. Flushing of PSMα3 was preceded and followed by a base- and endlining step with PBS 0.5% BSA. The association of D-PSMα3 to the peptide-coated spots was quantified in real-time by measuring the change of the optical properties of the biolayer. Binding of PSMα3 increases the pathlength for monochromatic light, leading to an interference-change of the light beams that are reflected in different planes of the biolayer resulting in a change in intensity. Drift correction of the binding kinetics was performed using the software ANABEL [36] (anabel.skscience.org).
2.4. Generating model structures of mirror-image D-protein analogues of L-proteins
To obtain the mirror-image of a protein structure, the atoms' X-coordinates were mirrored along the Y/Z plane (Supplementary Fig. 1). As a result of this process, all AAs change their conformation to the d-enantiomer and the protein backbone forms left-handed helices. The accuracy of this D-protein structural prediction method was verified with the natural sweetener protein monellin, which is one of the few proteins that have been synthesized in their mirror-image version and for which crystal structure information is available for both L- and D-protein [37]. The crystal structure of D-monellin and our D-monellin model, created by inversion of the L-monellin crystal structure, were aligned with a negligible root mean square deviation (RMSD) of 0.052 Å, indicating that both structures are indistinguishable (Supplementary Fig. 2).
2.5. Creating a library of 3D helical peptide structures
A library of helical peptides was generated by extracting the sequence and the structural information of all ≥12 AA-long alpha helical segments within the 175.434 protein structures within the PDB (as of May 2021) [22]. The peptide length of 12 AAs was chosen based on results from Ding and colleagues indicating that peptides <12 AAs are more likely to assume unstructured conformations in solution [38]. Following this step, the structure of every occurring 12 AA-long sequence within helices of ≥12 AAs was extracted using a sliding window approach. Upon obtaining these helical segments, all duplicates were removed and, in case of alternative rotameric conformations of single AAs, only the ‘A’ conformation was kept. To restore the peptides' truncated C-terminus resulting from computational breakage of the peptide bond to a biologically correct carboxyl moiety, each PDB file was modified. This was achieved by adding a second oxygen atom to the C-terminal CA using the two vectors C → O and C → CA, inverting this sum and scaling it to 1.262 Å. In this way the bond angles of the OXT to CA and O are nearest to 120°. The bond length was obtained from the C-Terminal OXT - C bond of the randomly chosen PDB structure 1N9F [39].
2.6. Energy minimization and MD simulations
The structures of the target protein (PSMα3, PDB ID: 5KGY) and the ligand peptides were preprocessed by removing any present atoms that are not part of the protein, such as crystal waters. Simulations were performed with GROMACS v2019.3 [31] using the Amber99sb-ildn force field [40,41]. To support the formylated N-terminus of our target protein PSMα3, formyl group parameters were generated using antechamber [42] and acpype [43] with formyl atom charges generated as BCC-AM1 charges [44,45]. The formyl parameters are listed in Supplementary Table 1. The peptides and proteins were simulated in a rhombic dodecahedron box with dimensions of 7.63 × 7.64 × 5.44 nm3 and periodic boundary conditions (PBC) including 10045 SPC-E water molecules as solvent [46]. To obtain a simulation box without net charges, a suitable number of counter ions was added (2 Cl-ions in the case of PSMα3). To prevent steric clashes or aberrant geometry within the structure, a steepest descent energy minimization (EM) was performed, followed by two equilibration steps (0.1 ns NVT and 0.1 ns NPT). Van der Waals interactions were treated via a 1.0 nm cut-off, electrostatic interactions were calculated using PME [47] with a minimal real space cut-off of 1.0 nm. A temperature of 300 K was used with a modified Berendsen thermostat [48] (coupling time constant of 0.1 ps). The pressure of 1.0 bar was held using a Parinello-Rahman barostat [49] (coupling time constant of 2.0 ps). This was followed by 50 ns MD simulations with a time step of 2 fs using a leapfrog integrator and constraints on all covalent bonds involving hydrogen atoms. Afterwards, the trajectories of the atoms were clustered using an algorithm as described in Daura et al. [50] with a cut-off of 0.12 Å. Based on this clustering, the most populated conformation of the target protein was extracted for docking.
2.7. Molecular docking with AutoDock Vina
To predict the free energy of binding the protein-ligand complex, the docking program AutoDock Vina [32] together with MGLTools [51,52] was used. AutoDock scores a ligand binding mode via an empirical function. The search space for AutoDock Vina's search function was defined as a cubus with an edge 30 Å larger than the target protein in order to avoid steric restrictions in the ligand's possible binding positions to the target. The exhaustiveness of the search function for the ligand's optimal binding position to the target protein was set to 10 for mirror-image virtual screening (MIVS) and to 1 for the mirror-image evolutionary algorithm (MIEA) due to different requirements in terms of computational cost.
2.8. Mirror-image evolutionary algorithm (MIEA)
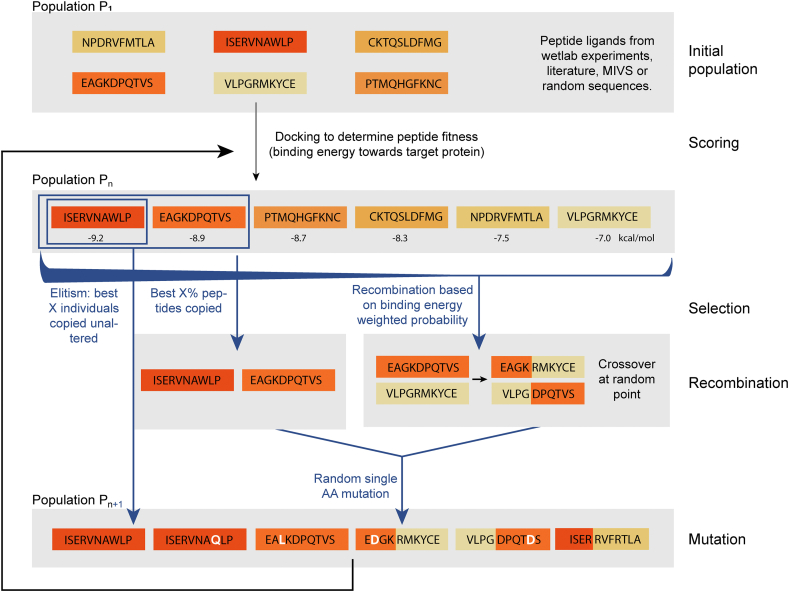
An initial population of helical L-peptides was docked against the three most occurring conformations of the target protein D-PSMα3, taken from 3 respective 50 ns MD simulations. The peptides were then ranked according to their binding affinity, which corresponds to evolutionary fitness. For evaluation of the peptides’ best binding affinity, three conformations of the target protein were used. A second-generation peptide population of the same size was generated in four steps (Fig. 5): First, the 30% of peptide sequences with the best binding energy were copied to the new population (copy rate). Next, crossover recombination of all peptide sequences from the initial population was performed at a randomly chosen point in the sequence. Peptides were chosen for recombination using a weighted probability distribution based on their binding energies. The copied and newly recombined peptide sequences of this new population were then subjected to random mutations by exchanging single AAs at randomly chosen positions with other AAs (mutation rate = 0.7). Finally, adhering to the principle of elitism, the two peptide sequences with the highest binding energy were copied without any alteration from the previous population to avoid losing favourable peptide sequences during recombination. Then, helical structures for the amino acid sequences in this new population were modeled using the peptide building tool of PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) and energy minimization of these structures was performed using GROMACS (version 2019.3) [31]. The population was then again subjected to evaluation by docking and recombination.
Fig. 5.
Diagram of MIEA. The fitness of all peptides in a population Pn is evaluated by molecular docking. Based on this, a population Pn+1 is newly generated by copying the peptides with the lowest binding energy, with and without crossover recombination of their sequences, and introducing random mutations. Further, a number of X individual peptides with the best binding energy are directly copied into the population Pn+1 without alteration. This population Pn+1 is then again evaluated via docking to complete the MIEA cycle.
2.9. Web server development
finDr uses HTTPS for networking, AMQP for job queueing, MySQL for job data storage, TypeScript (https://www.typescriptlang.org/), React (https://reactjs.org/) and Blueprint (https://blueprintjs.com/) for the user interface, JavaScript and Express (https://expressjs.com/) for the backend and finally Python for the orchestration of the heavy computation carried out by AutoDock Vina (version 1.1.2) [32] and GROMACS (version 2019.3) [31]. finDr is served from a virtual server running Ubuntu 20.04.2 and can be accessed via https://findr.biologie.uni-freiburg.de/.
Submitted PDB files (e.g. demo file ErbB2, PDB id: 1S78) [53] are processed with PDBfixer (https://github.com/caiyingchun/pdbfixer). Next, EM is applied by using GROMACS (see Section 2.6) [31]. The output is passed to Autodock Vina [32] to proceed in one of the two different modalities chosen by the user as MIVS or MIEA.
2.10. Statistical analysis
Statistical analysis of all datasets was performed using GraphPad PRISM 5.0.
3. Results and discussion
3.1. Identifying D-peptide ligands for PSMα3 using mirror-image phage display (MIPD)
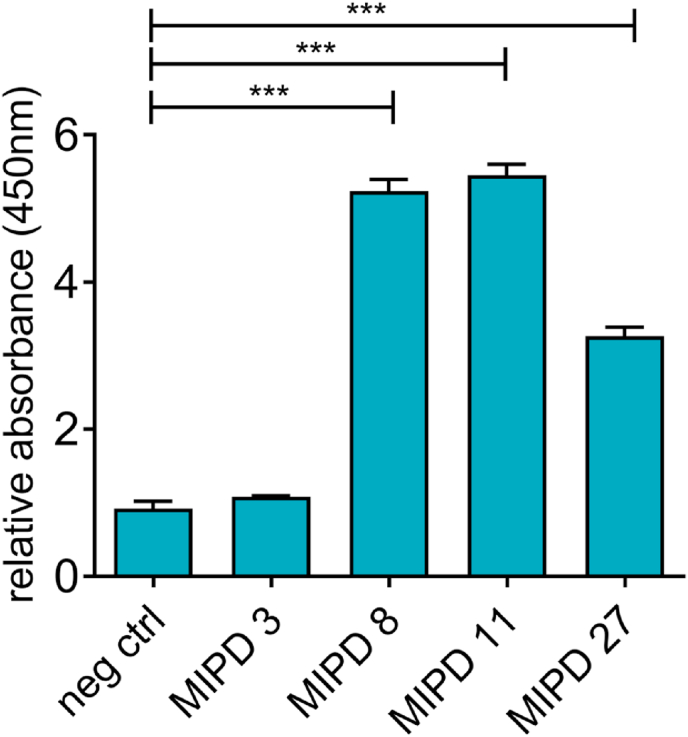
We selected PSMα3 as target protein for the identification of D-peptide ligands. PSMα3 is a toxin secreted predominantly by multiresistant strains of Staphylococcus aureus [54]. It is a helical peptide of 22 AAs, with a hydrophobic and a hydrophilic face opposing each other. Due to this amphipathic nature, PSMα3 can insert itself into biological membranes and form pores that induce cytolysis [54]. Blocking PSMα3 function would reduce the virulence of an MRSA infection. Beside its biological relevance, PSMα3 was chosen as a model target due to its small size, which facilitates chemical synthesis. For D-peptide ligand identification by MIPD, a library of M13 phages expressing 10⁹ random 12-mer L-peptides fused to the N-terminus of the minor coat phage protein pIII was screened for binding to D-PSMα3 via surface panning (Fig. 3). D-PSMα3-binding phages were amplified in K12 ER2738 E. coli cells and subjected to two further rounds of selection by surface panning. The phage clones that survived the selection process were then sequenced to obtain the amino acid sequences of the peptide ligands. To verify the binding of the phage-bound peptides to D-PSMα3 and to exclude nonspecific binders we performed phage ELISA. Three phage clones (MIPD 8, 11 and 27) bound selectively to D-PSMα3 as compared to a randomly chosen clone of control phages and one unspecific binder (MIPD 3) (Fig. 2). There were no obvious similarities in net charge, hydrophobicity or sequence in the different ligands (Supplementary Table 1). Considering that phage-bound L-peptides might display different binding behaviours than the isolated peptides in solution, phage ELISA is not the method of choice to measure binding affinity. To verify their binding to D-PSMα3 under more physiological conditions, the L-peptide ligands were synthesized by SPPS and validated by single colour reflectometry (SCORE), which is a technique that enables the label-free assessment of real-time binding kinetics (see Section 2.3 Fig. 7).
Fig. 2.
Selective binding of MIPD-derived phage clones to D-PSMα3. Results of a phage ELISA performed with isolated phages from MIPD displaying the indicated peptides on their surfaces. The absorbance resulting from phage binding to D-PSMα3 coated wells was normalized to that resulting from unspecific phage binding to D-PSMα3 free control wells. As negative control, a phage clone was randomly chosen from the phage library. Data represent mean + SEM of triplicates. P values were calculated by a two sided, unpaired Student's t-test ***: P ≤ 0.0001.
Fig. 7.
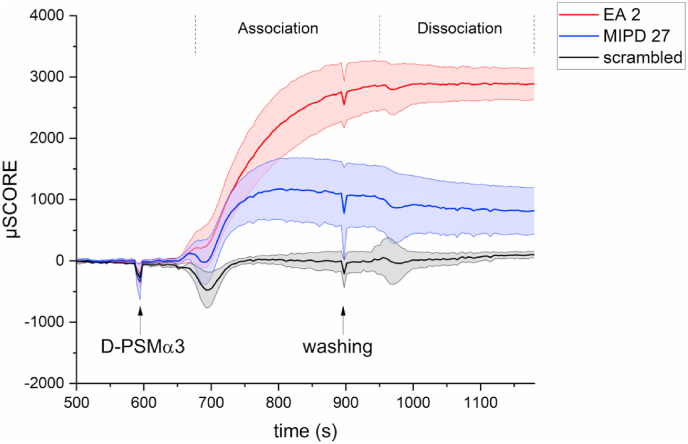
SCORE binding assay of MIPD- and MIEA-derived peptide ligands to PSMα3. Real-time binding kinetics of L-MIPD27 and L-EA2 to D-PSMα3 as measured by SCORE. The mean intensities of four spots (EA2, MIPD27) and 2 spots (scrambled) with standard deviation are depicted. The association and dissociation phases are indicated; the dissociation kinetic starts shortly after induction of the washing step due to methodological reasons.
3.2. Mirror-image virtual screening (MIVS) as an in silico alternative to MIPD
Despite the success of MIPD, which led to the identification of three candidate L-peptides able to bind to D-PSMα3, its costly and labour-intensive nature motivated us to create an affordable and accessible high-throughput in silico alternative (Fig. 3). Just like MIPD, the workflow we established is also based on screening L-peptide ligands for their ability to bind to the D-version of the target, with the major difference being that the binding is assessed in silico and the mirroring of the target to its D-form also occurs only virtually.
As for MIPD, a target D-protein is required for the MIVS protocol (Fig. 3). Similarly to >99% of all crystallized proteins, our target protein PSMα3 has only been subjected to structural analysis in its naturally occurring L-enantiomeric version. Hence, we used a structural model of D-PSMα3 based on X-coordinate inversion of the first model of the NMR structure of L-PSMα3 (PDB ID: 5KGY) [55] (see Section 2.4, Supplementary Fig. 2A).
To generate a large in silico peptide library that could be used to find binders to our target of choice, we extracted the sequence and the structural information of all alpha helical segments within the 175434 protein structures deposited in the PDB [22] and fragmented them into a structural library of 1458278 12-mer peptide segments (Fig. 3; for details on the extraction and fragmentation procedures see Section 2.5). We provide this structural L-peptide library for free download as well as for usage on our web server. It has been shown in various circular dichroism-based analyses of peptides in solution that the propensity of an amino acid sequence to fold as a helix is maintained even after that sequence is isolated from the full protein it belongs to Refs. [56,57]. Therefore, it is likely that the helical structural models of the peptides in our library are representative of their actual secondary structure in solution.
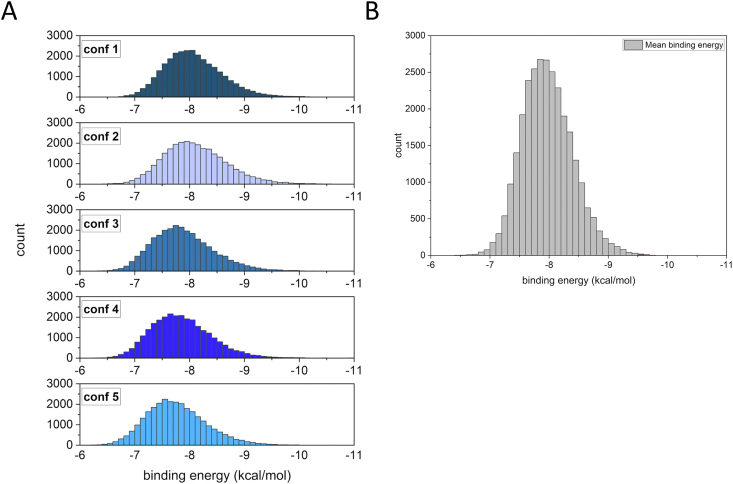
To identify potential ligands binding to our model target protein PSMα3, we performed MIVS using the molecular docking program AutoDock Vina [32] for the evaluation of the binding of L-peptides from our library to the structural model of D-PSMα3. In order to obtain a more realistic model of D-PSMα3, taking into account its conformational freedom, we performed five molecular dynamics simulations with GROMACS [31]. We extracted the 5 most representative conformations of the 50-ns MD simulations. Then, a random sample of 28647 helical 12-mer peptides from our structural L-peptide library was docked against these 5 different D-PSMα3 conformations (due to the computational cost, not the whole library could be docked). On account of the variation of the binding surfaces of the 5 different D-PSMα3 conformations, the average binding energies of the peptides to D-PSMα3 differ by 0.1–0.3 kcal between conformations (Fig. 4A). To determine the peptides that are most likely to bind to D-PSMα3 under physiological conditions we ranked the docking results according to their average binding energy to all 5 most often occurring conformations (Fig. 4B). The most promising peptide ligand predicted by MIVS has an average binding affinity of −10.1 kcal/mol and was derived from helix 13 of human DNA polymerase beta (PDB id: 1BPY).
Fig. 4.
Mirror-image virtual screening oftheL-peptide library to D-PSMα3. A: Distribution of binding energies of 28.647 L-peptides, each docked to 5 different conformations of D-PSMα3. B: Histogram of the L-peptides’ mean binding energy to the 5 different D-PSMα3 conformers.
As mentioned above, the MIVS approach for the identification of D-peptide ligands to PSMα3 was limited due to restrictions in computational capacity. The peptides that were screened represent only a small fraction (2%) of the whole peptide library. Hence it can be assumed that there would exist peptides with an even better binding affinity to PSMα3 than the ones that were identified by MIVS. In order to further enhance sampling, without exorbitantly increasing the computational cost, we decided to turn to a heuristic approach to further explore the enormous chemical space of 12-mer peptide ligands.
3.3. A mirror-image evolutionary algorithm (MIEA) for optimization of D-peptide ligands
To overcome the above mentioned inherent limitations of MIVS we employed a Darwinian evolutionary algorithm to find D-peptide ligands to PSMα3. In the MIEA an initial population of 88 helical L-peptides was docked against three conformations of D-PSMα3 and then ranked according to binding affinity, which corresponds to evolutionary fitness in our setup. A second-generation peptide population of the same size was then generated by selection of the fittest individual peptides, as well as recombination among peptides, in a process similar to chromosomal crossover during meiosis, and random introduction of point mutations. The AA sequences of this new population were then modeled with a helical structure template using PyMOL and energy minimization of this structure using GROMACS. The newly generated population was then again subjected to evaluation by docking, selection and recombination (see Section 2.8 and Fig. 5).
For D-peptide ligand identification to our model target PSMα3 we customized the MIEA parameters. First, we incorporated the sequences of the peptides, that we obtained by MIPD and that bound to D-PSMα3 in the phage ELISA, into the initial population. We also included seven 12-mer peptides extracted from L-PSMα3, which is known to form complexes with its mirror-image D-protein analogue [58], and 60 peptides that were identified as the best binders by MIVS using a randomly chosen subset of 30034 L-peptides in our helical library. To maintain sufficient diversity in the population we added 16 randomly chosen peptides from our helical peptide library as well as Glu12 / Asp12 polypeptides. Furthermore, during the mutation step, new AAs were chosen using a weighted probability distribution, where Glu and Asp had a slightly higher chance of being chosen based on the assumption that negatively charged AAs would increase the probability of a peptide to bind to the functionally relevant, positively charged lysine residues of D-PSMα3 [59].
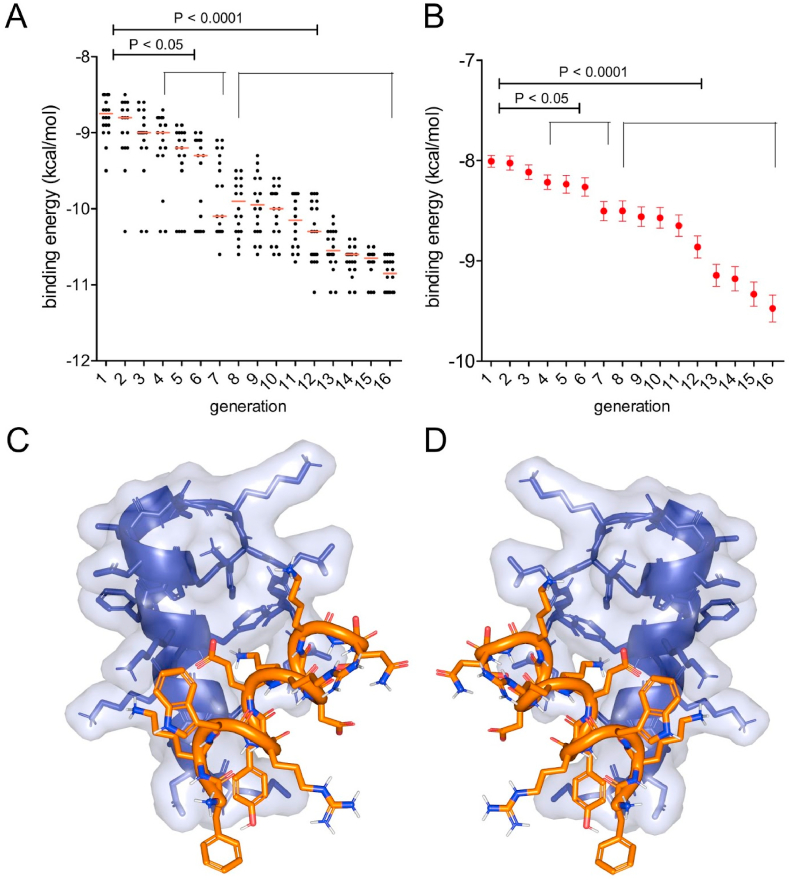
After 15 generations of MIEA with the above mentioned parameters, a peptide ligand “EA2” emerged that had a 1.6 kcal/mol (17%) better binding energy than the best binder in the initial population (Fig. 6A) (−9.5 kcal/mol vs −11.1 kcal/mol). Also, there was an overall improvement of the average binding energy of the peptide populations towards D-PSMα3 of 1.5 kcal/mol (18%, from −8 to −9.5 kcal/mol on average) (Fig. 6B). This demonstrates that not only individual peptides with a high binding affinity emerge from the MIEA, but that the whole pool of peptides is continually improved during the process. Depending on the individual target protein, the choice of initial population and parameters, the number of generations necessary to achieve a significant improvement of binding energy with MIEA might vary. The binding of the L-peptide EA2 to D-PSMα3 is shown in Fig. 6C. To complete the cycle, we reverted the structures of L-EA2 and D-PSMα3 to obtain their mirror-images: the naturally occurring L-PSMα3 and the D-peptide ligand EA2. In order to prove that D-EA2 and L-PSMα3 interact in the same manner as their mirror-image analogues, we conducted docking simulations. The result confirms that the binding modes for L-EA2/D-PSMα3 (Fig. 6C) and D-EA2/L-PSMα3 (Fig. 6D) are identical (RMSD = 0). Hence, we conclude that MIEA successfully identified a D-peptide ligand for the Staphylococcus aureus toxin L-PSMα3.
Fig. 6.
Improvement of binding affinity of L-peptides to D-PSMα3 over 15 generations of MIEA. A: Binding energies of L-peptides to D-PSMα3 per generation of an MIEA, assessed by molecular docking using AutoDock Vina. Only the 20 best binding peptides of each generation are shown. B: Binding energy of all 88 peptides to D-PSMα3 in each generation. Mean binding energy of each peptide population and SEM are shown. Statistical significance of the difference from the initial population was determined by an unpaired, two-tailed t-test. C: Association of the L-peptide ligand L-EA2 (sequence FKWRYERDKKQS, shown in orange) to D-PSMα3 (shown in blue). D: Association of the D-peptide ligand D-EA2 (sequence all D-FKWRYERDKKQS, shown in orange) to L-PSMα3 (shown in blue). Bound states in C and D were obtained by Autodock Vina.
3.4. Experimental verification of MIEA-derived ligands
To experimentally validate our in silico predictions we synthesized the L-peptide EA2, which was the best binding peptide predicted by MIEA, via SPPS. We did not synthesize any of the peptides predicted by MIVS, since MIEA covers a larger chemical space as compared to the limited MIVS approach. Next, we assessed the binding kinetics of L-EA2 to D-PSMα3 using SCORE [35]. For comparison we also included in this analysis one L-peptide ligand obtained by MIPD, namely MIPD 27. We chose to validate the interaction between the L-peptide and the target D-protein rather than between the D-peptide and the L-target out of convenience, considering that, following the principle of mirror-image stereochemistry, the two ligand-protein pairs would interact in exactly the same manner. The L-peptide ligands were immobilized on an 3D-NHS SCORE slide, which was flushed with D-PSMα3. As a further control to exclude nonspecific protein binding by the ligands, the spots were also flushed with BSA (data not shown). Measuring the real-time binding kinetics at each spot revealed that both peptides bound to D-PSMα3, with EA2 having higher affinity than MIPD 27 (Fig. 7A). As expected, our scrambled 12-mer control peptide did not bind to D-PSMα3, demonstrating that the binding of our MIPD- and MIEA-derived peptide ligands to D-PSMα3 is specific. Interestingly, SCORE also revealed that there is only a low stereoselectivity of the EA2/PSMα3 and MIPD27/PSMα3 complexes. The L-peptide ligands bind not only to D-PSMα3 but also to L-PSMα3 (Supplementary Fig. 2). The lack of complete stereoselectivity in this case is most likely due to the small size and amphiphilic structure of PSMα3, where all residues are exposed to the solvent and easily accessible. However, for larger biomolecules with well-defined binding pockets, which would sterically restrict access to most peptides, a very high degree of stereoselectivity was reported [14,60]. However, in the physiological context, stereoselectivity is not relevant since the D-form of the target protein is not present and does not compete with the L-form for binding to the ligand. Further experiments are required to evaluate whether the binding of the D-peptide ligands to L-PSMα3 has a functional impact on its toxicity. Nevertheless, the experimental verification of the L-ligands’ ability to bind to D-PSMα3 demonstrates that our docking-based MIEA approach for ligand identification is successful.
3.5. finDr: a web server for in silico D-peptide identification and optimization
The computational process for in silico D-peptide identification described in this work requires bioinformatics know-how and high computing power. To facilitate the adoption of this approach for D-peptide ligand identification and optimization we developed the web server finDr. finDr offers a tool for easy conversion of any L-protein structure to its mirror-image D-protein analogue and allows the user to perform MIVS and MIEA.
As its first modality, finDr can perform MIVS for the identification of D-peptide ligands to any protein target of choice. For this purpose, finDr generates a mirror-image D-analogue of the target protein and screens L-peptides from our structural peptide library via molecular docking. The identified L-peptide ligands for the D-version of the target are subsequently inverted to D-form; these are expected to bind to the naturally occurring L-protein target (Fig. 1). Importantly, finDr's peptide library screening modality could also be used to identify L-peptide ligands to an L-protein target, whenever the superior stability and pharmacokinetic profile of D-peptides are not required.
The user only has to provide a file containing the structural information of the protein of interest in PDB format, which will be used for the conversion to D-form whenever requested by the user to perform MIVS. To increase the accuracy of the structural model, it is recommended to perform an MD of the target molecule beforehand (for example by using web servers such as MDWeb [61] or FG-MD [62]) in order to obtain the most frequently occurring conformation of the target protein [63]. Then, random peptides from our in silico library of short helices will be docked against the target. To make the docking process more efficient and specific, the user can also choose a target region of interest, such as the active site of an enzyme or the ligand binding pocket of a receptor. This helps to reduce the chances of predicting on-target off-site binders that do not influence the target protein's biological activity or the PPI of interest, which can always arise from MIPD [64]. finDr provides as output a complete list of peptide sequences ranked according to their binding affinity to the target. The structures of the ligand/target protein complexes resulting from docking can be also downloaded.
In its second modality, finDr allows users to perform MIEA to any chosen target protein and to customize parameters such as the size of the initial population, the degree of elitism, the recombination and mutation rate (see Section 2.8). Furthermore, the user can upload individual peptide sequences to be included in the initial population to adjust the starting point of the MIEA. While EA has often been successfully used as a tool for de novo ligand identification [25], target-based choice of the EA initial population and above mentioned parameters can increase the performance of an EA as it integrates existing knowledge about the binding properties into the process of evolutionary optimization [30]. This knowledge can easily be obtained by performing MIVS prior to MIEA. Based on the work of Garton and colleagues on retro-inverso peptides, it would also be possible to integrate information about existing L-peptide ligands from the literature into the MIEA process for further optimization of D-peptide ligands with finDr [22]. Alternatively, finDr's EA modality could also be used to optimize conventional L-peptide ligands without L-to-D conversion. As output, finDr provides a list of all peptide sequences and their corresponding binding energies to the target protein conformations, as well as charts visualizing the improvement of binding energy over all MIEA generations (Fig. 8).
Fig. 8.
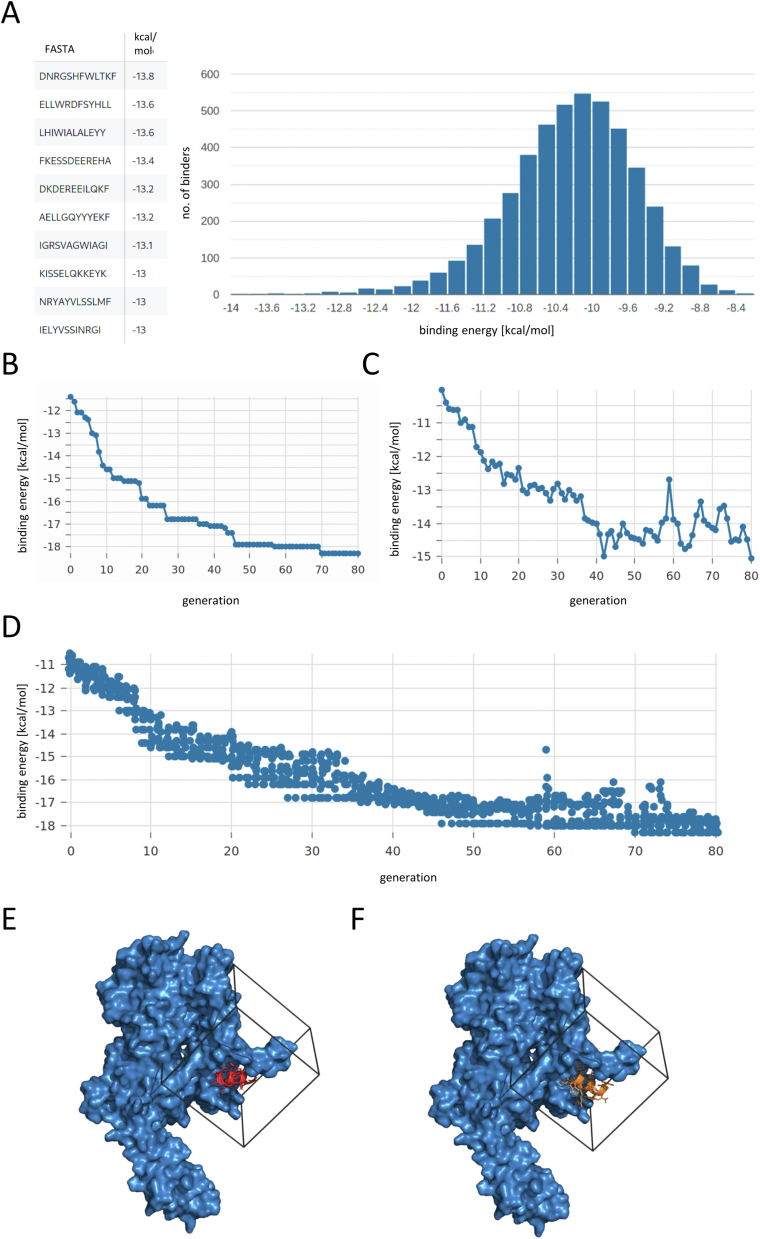
D-peptide ligand identification for ErbB2 with finDr. A: MIVS - Histogram of the binding energies of all docked library peptides, screenshot from finDr results webpage. B: MIEA - Binding energy of L-peptides to D-ErbB2 per generation of the MIEA. Only the 20 best binding peptides are shown. C: MIEA - Mean binding energy per generation D: Binding energy of the best binding peptide in each generation of MIEA. E, F: L-ErbB2 in complex with its D-peptide ligand derived from MIVS (E) and MIEA (F) (visualized by PyMOL, Gridbox for docking is shown in black).
3.6. Testing MIVS and MIEA on another target to evaluate finDr's performance
To test the performance of finDr we conducted MIVS and MIEA to identify D-peptide ligands for the human growth factor receptor 2 (ErbB2) (PDB ID: 1S78), which is overexpressed in several types of human cancer and is of critical importance for their pathology [53]. This target was also chosen to demonstrate that finDr is capable of handling large protein targets. We decided to target the extracellular dimerization domain of ErbB2 with the aim to find a D-peptide inhibitor that sterically inhibits its dimerization and thus the associated downstream signalling, similarly to the mechanism of action of the ErbB2 targeting monoclonal antibody Pertuzumab [53]. Guided by the well-described dimerization interface of ErbB2 and the knowledge of the residues involved in antibody binding, we defined a gridbox for docking, in order to exclude on-target off-site binders and to reduce the computational cost for docking.
Within 6 h and 40 min, 4573 12-mer helical peptides were docked to the target protein by MIVS (AutoDock Vina exhaustiveness = 1). This equals approximately 685 peptides per hour; however, this number may vary depending on the size of the target and the chosen gridbox. The best binding ligand in this case has the sequence DNRGSHFWLTKF and binds with a binding energy of −13.8 kcal/mol (Fig. 8A, E).
For the same target protein we performed MIEA with an initial population of 55 helical peptides randomly chosen from our library, a copyrate of 0.3, mutation rate of 0.7 and an elitism rate of 2 individuals. Within 7 h and 10 min, 80 generations of optimization were completed and a D-peptide ligand with a binding energy of −18.3 kcal/mol emerged (Fig. 8B-D, F). This represents an improvement of 6.9 kcal/mol (60.5%) as compared to the best binder present in the initial population (−11.4 kcal/mol). This demonstrates that, regardless of the target size, finDr's MIEA modality succeeds in identifying D-peptides with high affinity for the target protein.
4. Conclusion and outlook
We presented a computational method for the identification of D-peptide ligands, both in a deterministic approach via MIVS of a helical peptide library and by exploring the immensely large chemical space of 12-mer peptides with an EA in a heuristic manner. In a proof-of-concept example, we demonstrated the successful transfer of our in silico predictions to in vitro conditions by verifying the binding of an MIEA-derived peptide ligand to our model target PSMα3. With our web server finDr we provide a novel openly accessible, easy-to-use interface for the identification of peptide ligands to any chosen target structure, using either MIVS, MIEA or a combination of the two (https://findr.biologie.uni-freiburg.de/). finDr allows the user to either apply default settings or customize the screening taking into account specific features of the target of choice. Depending on individual settings, such as the exhaustiveness of docking or the size of the MIEA populations, the time required to apply finDr can vary and is currently limited by the available computational capacity (maximum 6 h). The typical limitations of molecular docking simulations apply to finDr too, especially considering that it uses a rigid model of proteins for docking to reduce computational cost. Currently almost all computational models of PPIs that take into account the proteins’ flexibility are not suitable for large-scale screening due to their computational cost. Nevertheless, we were able to experimentally confirm the predictions obtained with the finDr approach. As for any virtual screening approach, we propose to test a large number of peptides in a subsequent in vitro experiment to validate the predictions and to analyze the biological activity of the D-peptide ligands in a physiological context.
In conclusion, we provided a new tool for D-peptide ligand identification via MIVS and MIEA. We envisage that finDr will facilitate the identification of D-peptides and help promote their application in biotechnology and biomedicine.
Author contributions
All authors conceived and designed the study and discussed the results as iGEM Team Freiburg 2019. MIPD experiments were designed, conducted and analyzed by HE, SH and CR under the supervision of NG. Chemical synthesis of peptides was done by FG and JN with the help of NH and under the supervision of MK. SCORE measurements were done by JN and NK. All computational pipelines were conceptualized, created, applied and analyzed by HE, JN, FG, FK, with the suggestions of SW and supervision of MAÖ. All finDr web page implementation was done by FK and set up by KV with optimization feedback from MAÖ. Conceptualization figures were generated by PK. HE, JN, FG, FK, MAÖ and BDV wrote the manuscript with feedback from all authors. NG, BDV and MAÖ were in charge of overall direction and planning of the project.
Declaration of competing interest
None.
Acknowledgements
We thank, Günter Roth and BioCopy GmbH for their help with SCORE measurements, Dominik Bezold for his support with peptide synthesis and purification, Hanna Wagner for her guidance on working with phages, Michael Kay for providing D-proteins to establish the MIPD protocol, the Centre for Integrative Biological Signalling Studies (CIBSS) for equipment and laboratory support, Dieter Willbold for helpful discussions on D-peptides and their pharmacokinetic properties, Frank Rosenau for giving an example of MIPD, Jeffrey Bode and Stephen Kent for the discussions on chemical synthesis and ligation methods. We also acknowledge support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC and the German Research Foundation (Deutsche Forschungsgemeinschaft - DFG) through grant no INST 37/935-1 FUGG. This work was funded by the following people and organizations: CIBSS - Centre for Integrative Biological Signalling Studies / BIOSS - Centre for Biological Signalling Studies (University of Freiburg), Student deanery (Studiendekanat) Molecular Medicine (University of Freiburg), Integrated DNA Technologies (IDT), Maria Ladenburger Stiftung (Freiburg, Germany), Friends of the University of Freiburg, Dr. Falk Pharma GmbH (Freiburg, Germany), OFAMED Open Student Council Medicine (Offene Fachschaft Medizin e.V.) - University of Freiburg, Faculty of Biology, Neue Universitätsstiftung Freiburg, Abcam, Geneious, Institute for Biochemistry (University of Freiburg), Thorsten Hugel Group (University of Freiburg), BioCopy GmbH (Emmendingen, Germany), Hahn-Schickard Institute (Freiburg, Germany), Zahnarztpraxis Thomas Ruckes (Germany), Merck Millipore, New England Biolabs GmbH, Zymo Research, Carl Roth GmbH + Co. KG (Germany), Eurofins Scientific, Faust Lab Science GmbH (Germany), Macherey-Nagel (Germany), Greiner Bio-One, Jena Bioscience (Germany), NIPPON Genetics Europe (Germany), GENEWIZ Europe - A Brooks Life Sciences Company (Germany), Institute for Biology III (University of Freiburg), La Luna Baila, Sparkasse Freiburg - Nördlicher Breisgau (Germany).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.11.004.
Contributor Information
Nicole Gensch, Email: nicole.gensch@bioss.uni-freiburg.de.
Barbara Di Ventura, Email: barbara.diventura@biologie.uni-freiburg.de.
Mehmet Ali Öztürk, Email: mehmet.oeztuerk@bioss.uni-freiburg.de.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Angell Y., Holford M., Moos W.H. Building on success: a bright future for peptide therapeutics. Protein Pept. Lett. 2018;25:1044–1050. doi: 10.2174/0929866525666181114155542. [DOI] [PubMed] [Google Scholar]
- 2.Al Shaer D., Al Musaimi O., Albericio F., de La Torre B.G. FDA TIDES (peptides and Oligonucleotides) harvest. Pharmaceuticals. 2019;13 doi: 10.3390/ph13030040. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clément K., van den Akker E., Argente J., Bahm A., Chung W.K., Connors H., de Waele K., Farooqi I.S., Gonneau-Lejeune J., Gordon G., Kohlsdorf K., Poitou C., Puder L., Swain J., Stewart M., Yuan G., Wabitsch M., Kühnen P., Pigeon-Kherchiche P., Flaus-Furmaniuk A., Bald M., Denzer C., von Schnurbein J., Abawi O., Blume-Peytavi U., Krabusch P., Mai K., Schnabel D., Wiegand S., Flück C.E., Schulz E., Voss E., Bratina N., Weiss K., Martos-Moreno G.Á., Danset A., Gougis P., Dubern B., van den Akker E.L., Bahm A.L., Connors H.S., Farooqi I.S., Gordon G.A., Swain J.M., Stewart M.W., Yuan G. Efficacy and safety of setmelanotide, an MC4R agonist, in individuals with severe obesity due to LEPR or POMC deficiency: single-arm, open-label, multicentre, phase 3 trials. Lancet Diabetes Endocrinol. 2020;8:960–970. doi: 10.1016/S2213-8587(20)30364-8. [DOI] [PubMed] [Google Scholar]
- 4.Lee A.C.-L., Harris J.L., Khanna K.K., Hong J.-H. A comprehensive review on current advances in peptide drug development and design. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20102383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ladner R.C., Sato A.K., Gorzelany J., de Souza M. Phage display-derived peptides as therapeutic alternatives to antibodies. Drug Discov Today. 2004;9:525–529. doi: 10.1016/S1359-6446(04)03104-6. [DOI] [PubMed] [Google Scholar]
- 6.Le Zhao, Lu W. Mirror image proteins. Curr Opin Chem Biol. 2014;22:56–61. doi: 10.1016/j.cbpa.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uppalapati M., Lee D.J., Mandal K., Li H., Miranda L.P., Lowitz J., Kenney J., Adams J.J., Ault-Riché D., Kent S.B.H., Sidhu S.S. A potent d-protein antagonist of VEGF-A is nonimmunogenic, metabolically stable, and longer-circulating in vivo. ACS Chem Biol. 2016;11:1058–1065. doi: 10.1021/acschembio.5b01006. [DOI] [PubMed] [Google Scholar]
- 8.Dintzis H.M., Symer D.E., Dintzis R.Z., Zawadzke L.E., Berg J.M. A comparison of the immunogenicity of a pair of enantiomeric proteins. Proteins. 1993;16:306–308. doi: 10.1002/prot.340160309. [DOI] [PubMed] [Google Scholar]
- 9.Pappenheimer J.R., Dahl C.E., Karnovsky M.L., Maggio J.E. Intestinal absorption and excretion of octapeptides composed of D amino acids. Proc Natl Acad Sci USA. 1994;91:1942–1945. doi: 10.1073/pnas.91.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun N., Funke S.A., Willbold D. Mirror image phage display--generating stable therapeutically and diagnostically active peptides with biotechnological means. J Biotechnol. 2012;161:121–125. doi: 10.1016/j.jbiotec.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher T.N., Mayr L.M., Minor D.L., Milhollen M.A., Burgess M.W., Kim P.S. Identification of D-peptide ligands through mirror-image phage display. Science. 1996;271:1854–1857. doi: 10.1126/science.271.5257.1854. [DOI] [PubMed] [Google Scholar]
- 12.Smith G.P., Petrenko V.A. Phage display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 13.Smith G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 14.Klussmann S., Nolte A., Bald R., Erdmann V.A., Fürste J.P. Mirror-image RNA that binds D-adenosine. Nat Biotechnol. 1996;14:1112–1115. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- 15.Chang H.-N., Liu B.-Y., Qi Y.-K., Zhou Y., Chen Y.-P., Pan K.-M., Li W.-W., Zhou X.-M., Ma W.-W., Fu C.-Y., Qi Y.-M., Liu L., Gao Y.-F. Blocking of the PD-1/PD-L1 interaction by a D-peptide antagonist for cancer immunotherapy. Angew Chem Int Ed Engl. 2015;54:11760–11764. doi: 10.1002/anie.201506225. [DOI] [PubMed] [Google Scholar]
- 16.Welch B.D., VanDemark A.P., Heroux A., Hill C.P., Kay M.S. Potent D-peptide inhibitors of HIV-1 entry. Proc Natl Acad Sci USA. 2007;104:16828–16833. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aileen Funke S., van Groen T., Kadish I., Bartnik D., Nagel-Steger L., Brener O., Sehl T., Batra-Safferling R., Moriscot C., Schoehn G., Horn A.H.C., Müller-Schiffmann A., Korth C., Sticht H., Willbold D. Oral treatment with the d-enantiomeric peptide D3 improves the pathology and behavior of Alzheimer's Disease transgenic mice. ACS Chem Neurosci. 2010;1:639–648. doi: 10.1021/cn100057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mäde V., Els-Heindl S., Beck-Sickinger A.G. Automated solid-phase peptide synthesis to obtain therapeutic peptides. Beilstein J Org Chem. 2014;10:1197–1212. doi: 10.3762/bjoc.10.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstock M.T., Jacobsen M.T., Kay M.S. Synthesis and folding of a mirror-image enzyme reveals ambidextrous chaperone activity. Proc Natl Acad Sci USA. 2014;111:11679–11684. doi: 10.1073/pnas.1410900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coin I., Beyermann M., Bienert M. Solid-phase peptide synthesis: from standard procedures to the synthesis of difficult sequences. Nat Protoc. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]
- 21.Vodnik M., Strukelj B., Lunder M. HWGMWSY, an unanticipated polystyrene binding peptide from random phage display libraries. Anal Biochem. 2012;424:83–86. doi: 10.1016/j.ab.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Garton M., Nim S., Stone T.A., Wang K.E., Deber C.M., Kim P.M. Method to generate highly stable D-amino acid analogs of bioactive helical peptides using a mirror image of the entire PDB. Proc Natl Acad Sci USA. 2018;115:1505–1510. doi: 10.1073/pnas.1711837115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y., Chen Y., Tan Y., Hu R., Niu M.-M. An NRP1/MDM2-targeted D-peptide supramolecular nanomedicine for high-efficacy and low-toxic liver cancer therapy. Adv Healthc Mater. 2021 doi: 10.1002/adhm.202002197. [DOI] [PubMed] [Google Scholar]
- 24.Yang W., Zhang Q., Zhang C., Guo A., Wang Y., You H., Zhang X., Lai L. Computational design and optimization of novel d-peptide TNFα inhibitors. FEBS Lett. 2019;593:1292–1302. doi: 10.1002/1873-3468.13444. [DOI] [PubMed] [Google Scholar]
- 25.Belda I., Madurga S., Llorà X., Martinell M., Tarragó T., Piqueras M.G., Nicolás E., Giralt E. ENPDA: an evolutionary structure-based de novo peptide design algorithm. J Comput Aided Mol Des. 2005;19:585–601. doi: 10.1007/s10822-005-9015-1. [DOI] [PubMed] [Google Scholar]
- 26.Holland J.H. In: Adaptive control of Ill-defined systems. Selfridge O.G., Rissland E.L., Arbib M.A., editors. Springer; Boston, MA: 1984. Genetic algorithms and adaptation; pp. 317–333. [Google Scholar]
- 27.Hiss J.A., Hartenfeller M., Schneider G. Concepts and applications of "natural computing" techniques in de novo drug and peptide design. Curr Pharmaceut Des. 2010;16:1656–1665. doi: 10.2174/138161210791164009. [DOI] [PubMed] [Google Scholar]
- 28.Belda I., Llorà X., Martinell M., Tarragó T., Giralt E. In: Genetic and evolutionary computation – GECCO 2004. Kanade T., Kittler J., Kleinberg J.M., Mattern F., Mitchell J.C., Naor M., Nierstrasz O., Pandu Rangan C., Steffen B., Sudan M., Terzopoulos D., Tygar D., Vardi M.Y., Weikum G., Deb K., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2004. Computer-aided peptide evolution for virtual drug design; pp. 321–332. [Google Scholar]
- 29.Röckendorf N., Borschbach M., Frey A. Molecular evolution of peptide ligands with custom-tailored characteristics for targeting of glycostructures. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe K., Kobayashi N., Sode K., Ikebukuro K. Peptide ligand screening of alpha-synuclein aggregation modulators by in silico panning. BMC Bioinf. 2007;8:451. doi: 10.1186/1471-2105-8-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berendsen H., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 32.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attwood T.K., Blackford S., Brazas M.D., Davies A., Schneider M.V. A global perspective on evolving bioinformatics and data science training needs. Briefings Bioinf. 2019;20:398–404. doi: 10.1093/bib/bbx100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vestergaard M., Frees D., Ingmer H. Antibiotic resistance and the MRSA problem. Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.GPP3-0057-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burger J., Rath C., Woehrle J., Meyer P.A., Ben Ammar N., Kilb N., Brandstetter T., Pröll F., Proll G., Urban G., Roth G. Low-volume label-free detection of molecule-protein interactions on microarrays by imaging reflectometric interferometry. SLAS Technol. 2017;22:437–446. doi: 10.1177/2211068216657512. [DOI] [PubMed] [Google Scholar]
- 36.Krämer S.D., Wöhrle J., Rath C., Roth G. Anabel: an online tool for the real-time kinetic analysis of binding events. Bioinf Biol Insights. 2019;13 doi: 10.1177/1177932218821383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung L.W., Kohmura M., Ariyoshi Y., Kim S.H. Structure of an enantiomeric protein, D-monellin at 1.8 A resolution. Acta Crystallogr D Biol Crystallogr. 1998;54:494–500. doi: 10.1107/S0907444997012225. [DOI] [PubMed] [Google Scholar]
- 38.Ding F.-X., Schreiber D., VerBerkmoes N.C., Becker J.M., Naider F. The chain length dependence of helix formation of the second transmembrane domain of a G protein-coupled receptor of Saccharomyces cerevisiae. J Biol Chem. 2002;277:14483–14492. doi: 10.1074/jbc.M111382200. [DOI] [PubMed] [Google Scholar]
- 39.Miele A.E., Federici L., Sciara G., Draghi F., Brunori M., Vallone B. Analysis of the effect of microgravity on protein crystal quality: the case of a myoglobin triple mutant. Acta Crystallogr D Biol Crystallogr. 2003;59:982–988. doi: 10.1107/S0907444903005924. [DOI] [PubMed] [Google Scholar]
- 40.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case D.A., Aktulga H.M., Belfon K., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden T.A., Duke R.E., Giambasu G., Gilson M.K., Gohlke H., Goetz A.W., Harris R., Izadi S., Izmailov S.A., Jin C., Kasavajhala K., Kaymak M.C., King E., Kovalenko A., Kurtzman T., Lee T.S., LeGrand S., Li P., Lin C., Liu J., Luchko T., Luo R., Machado M., Man V., Manathunga M., Merz K.M., Miao Y., Mikhailovskii O., Monard G., Nguyen H., O'Hearn K.A., Onufriev A., Pan F., Pantano S., Qi R., Rahnamoun A., Roe D.R., Roitberg A., Sagui C., Schott-Verdugo S., Shen J., Simmerling C.L., Skrynnikov N.R., Smith J., Swails J., Walker R.C., Wang J., Wei H., Wolf R.M., Wu X., Xue Y., York D.M., Zhao S., Kollman P.A. Amber. 2021;2021 [Google Scholar]
- 42.Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Da Sousa Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon parser interfacE. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 45.Jakalian A., Bush B.L., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J Comput Chem. 2000;21:132–146. doi: 10.1002/(SICI)1096-987X(20000130)21:2<132:AID-JCC5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 46.Bryk T., Haymet A.D.J. Ice 1h/water interface of the SPC/E model: molecular dynamics simulations of the equilibrium basal and prism interfaces. J Biol Chem. 2002;117:10258–10268. doi: 10.1063/1.1519538. [DOI] [Google Scholar]
- 47.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 48.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 49.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 50.Daura X., Gademann K., Jaun B., Seebach D., van Gunsteren W.F., Mark A.E. Peptide folding: when simulation meets experiment, angew. Chem Int Ed. 1999;38:236–240. doi: 10.1002/(SICI)1521-3773(19990115)38:1/2<236:AID-ANIE236>3.0.CO;2-M. [DOI] [Google Scholar]
- 51.Sanner M.F. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 52.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franklin M.C., Carey K.D., Vajdos F.F., Leahy D.J., de Vos A.M., Sliwkowski M.X. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–328. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 54.Wang R., Braughton K.R., Kretschmer D., Bach T.-H.L., Queck S.Y., Li M., Kennedy A.D., Dorward D.W., Klebanoff S.J., Peschel A., DeLeo F.R., Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 55.Towle K.M., Lohans C.T., Miskolzie M., Acedo J.Z., van Belkum M.J., Vederas J.C. Solution structures of phenol-soluble modulins α1, α3, and β2, virulence factors from Staphylococcus aureus. Biochemistry. 2016;55:4798–4806. doi: 10.1021/acs.biochem.6b00615. [DOI] [PubMed] [Google Scholar]
- 56.Myers J.K., Pace C.N., Scholtz J.M. Helix propensities are identical in proteins and peptides. Biochemistry. 1997;36:10923–10929. doi: 10.1021/bi9707180. [DOI] [PubMed] [Google Scholar]
- 57.Muñoz V., Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. II. Helix macrodipole effects and rational modification of the helical content of natural peptides. J Mol Biol. 1995;245:275–296. doi: 10.1006/jmbi.1994.0023. [DOI] [PubMed] [Google Scholar]
- 58.Yao Z., Cary B.P., Bingman C.A., Wang C., Kreitler D.F., Satyshur K.A., Forest K.T., Gellman S.H. Use of a stereochemical strategy to probe the mechanism of phenol-soluble modulin α3 toxicity. J Am Chem Soc. 2019;141:7660–7664. doi: 10.1021/jacs.9b00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheung G.Y.C., Kretschmer D., Queck S.Y., Joo H.-S., Wang R., Duong A.C., Nguyen T.H., Bach T.-H.L., Porter A.R., DeLeo F.R., Peschel A., Otto M. Insight into structure-function relationship in phenol-soluble modulins using an alanine screen of the phenol-soluble modulin (PSM) α3 peptide. Faseb J. 2014;28:153–161. doi: 10.1096/fj.13-232041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milton R.C., Milton S.C., Kent S.B. Total chemical synthesis of a D-enzyme: the enantiomers of HIV-1 protease show reciprocal chiral substrate specificity corrected. Science. 1992;256:1445–1448. doi: 10.1126/science.1604320. [DOI] [PubMed] [Google Scholar]
- 61.Hospital A., Andrio P., Fenollosa C., Cicin-Sain D., Orozco M., Gelpí J.L. MDWeb and MDMoby: an integrated web-based platform for molecular dynamics simulations. Bioinformatics. 2012;28:1278–1279. doi: 10.1093/bioinformatics/bts139. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J., Liang Y., Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumari I., Sandhu P., Ahmed M., Akhter Y. Molecular dynamics simulations, challenges and opportunities: a biologist's prospective. Curr Protein Pept Sci. 2017;18:1163–1179. doi: 10.2174/1389203718666170622074741. [DOI] [PubMed] [Google Scholar]
- 64.Castel G., Chtéoui M., Heyd B., Tordo N. Phage display of combinatorial peptide libraries: application to antiviral Research. Molecules. 2011;16:3499–3518. doi: 10.3390/molecules16053499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.