Abstract
Asymptomatic infection of COVID-19 is a global threat for public health. Unfortunately, the study about metabolic dysregulation of asymptomatic infection is barely investigated. Here, we performed carboxylic submetabolome profiling of serum from 62 asymptomatic and 122 control individuals, by a highly sensitive chemical isotope labelling method. Twenty-one discriminative carboxylic features, including 12-hydroxyeicosatetraenoic acid, cholic acid, glycoursodeoxycholic acid and 15,16-dihydroxyoctadeca-9,12-dienoic acid were discovered to be dysregulated in asymptomatic patients. This panel containing 21 carboxylic features could accurately identify asymptomatic patients based on a random forest model, providing an accuracy of 85.7% with only 3.6% false positive rate and 7.1% false negative rate. The dysregulated metabolites found in asymptomatic patients covered several important pathways, such as arachidonic acid metabolism, synthesis of bile acid, β-oxidation of fatty acids, activation of macrophage and platelet aggregation. This work provided valuable knowledge about serum biomarkers and molecular clues associated with asymptomatic COVID-19 patients.
Keywords: COVID-19 asymptomatic infection, Carboxylic metabolic dysregulation, Chemical isotope labelling, Machine learning
Graphical abstract
1. Introduction
COVID-19 is an unprecedented global threat for its tremendous infection velocity and awful effect on production and life [1]. Most confirmed patients had respiratory symptoms such as cough, expectoration and respiratory distress. Conversely, asymptomatic patients were diagnosed positive of nucleic acid test but exhibited no typical clinical symptoms [2]. As most asymptomatic patients were not aware of being infected and did not seek medical assistance, a large number of these individuals may have been undetected. Increasing studies showed clear epidemiological evidence of human-to-human spread from COVID-19 asymptomatic patients [3]. Therefore, asymptomatic infection has become a serious public health issue and the key point to totally control the spread of COVID-19.
Currently, nucleic acid test is the gold standard method for diagnosing COVID-19 [4,5]. Unfortunately, increasing reports have shown that false negative results are almost inevitable in nucleic acid test when virus load is low [6]. In this case, detecting serum antibodies was came up with as an effective supplement for enhanced accuracy [4]. Nevertheless, the median response time of IgM and IgG in serum against virus infection is 5 days and 14 days, respectively. Thus, serological test seems unlikely to play a timely diagnostic role in the early stage of infection [7]. New complementary method should be developed for more sensitive and specific risk assessment for asymptomatic infection.
Studies on genome and proteomic have discovered that COVID-19 is associated with complement activation, dysregulated lipid transport, and neutrophil activation [8]. Compared with genome and proteome, metabolome is more likely to reflect the immediate changes in patients while viral invasion. Arachidonic acid (AA) has been suggested to play an important role in susceptibility to COVID-19 [9]. Biosynthesis of bile acid was a key clinical manifestation of liver damage by SARS-CoV-2 infection [10]. Significantly altered lipid metabolism of short- and medium-chain saturated fatty acids was observed, too [11]. Above examples confirmed the metabolomic dysregulation in COVID-19 symptomatic patients. Nevertheless, the dysregulated metabolome signature characterization about COVID-19 asymptomatic infection is still barely studied.
Compared with traditional untargeted metabolome, submetabolome focusing on specific functional groups can offer more precise and sensitive signature characterization of specific metabolites, usually performed with chemical isotope labelling method (CIL-LC-MS) [12]. In previous reports, carboxylic metabolites alterations were proved to play a vital role in viral infections and acute respiratory distress syndrome [13,14]. Hence, we are curious whether SARS-CoV-2 can induce characteristic carboxylic metabolites alterations that can be detected in the serum of asymptomatic patients.
In this study, we conducted profiling of carboxylic submetabolome in asymptomatic COVID-19 patients’ serum by a modified CIL-LC-MS method based on our previous work [15]. The workflow was presented in Fig. 1 a. We identified 21 dysregulated carboxylic metabolites, 6 of which were confirmed using standards validation or homemade database. A random forest model based on above discriminative carboxylic biomarkers was established for risk assessment, with mean accuracy of 85.7% in independent validation set. We further discussed the possible infection mechanism in COVID-19 asymptomatic based on these dysregulated carboxylic metabolites. These dysregulated metabolites covered several important pathways, including AA metabolism, β-oxidation of fatty acids, bile acid synthesis, activation of macrophage and platelet aggregation, and these disrupted pathways may account for the occult infection of SARS-Cov-2 virus. Our results will offer a promising tool for prevention and treatment of asymptomatic infection and provide carboxylic metabolites signature in COVID-19 asymptomatic patients for better understanding the covert infection mechanism.
Fig. 1.
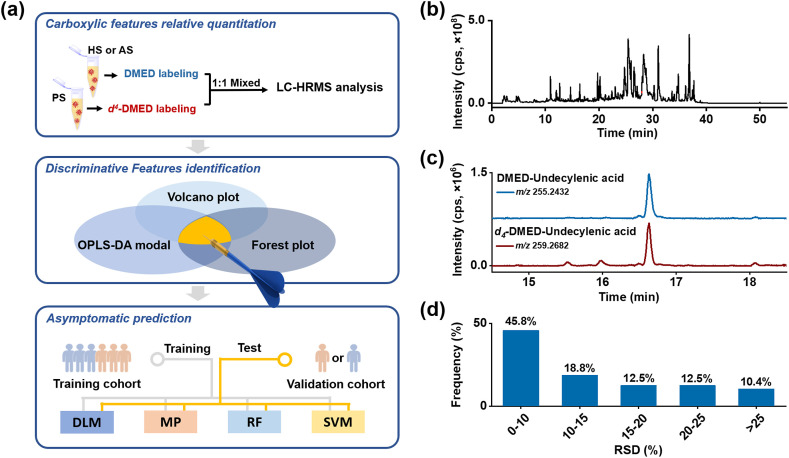
(a) The workflow of carboxylic submetabolome-driven signature characterization. Abbreviations: PS, pooled serum; HS, healthy serum; AS, asymptomatic serum; DMED, 2-dimethylaminoethylamine; DLM, Decision linear model; MP, Multi-layer perceptron; RF, Random forest; SVM, Support vector machine. (b) Total ion chromatography of DMED labelled serum detected in positive mode. (c) Extracted ion chromatograms at m/z 255.2432 and 259.2682 from DMED- and d4-DMED-labelled undecylenic acid in serum sample. (d) Frequency histogram of RSDs of potential carboxyl metabolites in serum.
2. Materials and methods
2.1. Sample collection
COVID-19 asymptomatic patients were diagnosed according to the Chinese Government Diagnosis and Treatment Guideline (6th version) (NHCPRC, 2020): (1) no clinical symptoms, (2) nucleic acid or serological IgM test was positive. Another 122 healthy individuals from physical examination clinic at the same period, were selected to match the asymptomatic patients with a ratio of 1:2, according to the basic characteristics including age and gender. This study was approved by the Ethics Review Commission of Wuhan Prevention and Treatment Centre for Occupational Diseases (reference no. 202002).
2.2. Sample pretreatment and LC-Orbitrap MS analysis
The sample pretreatment and LC-Orbitrap MS analysis method was based on our previous method with modification [15]. The inactivation of viruses was operated in a biosafety hood. 300 μL ethanol was added into 100 μL serum, following with vigorously shaking. Proteins were denatured and precipitated by centrifugation at 13,000 g for 15 min at 4 °C. 10 μL supernatant was evaporated to dryness under nitrogen stream. The residue was dissolved in 100 μL aqueous hydrochloric acid solution (pH 2). Then, extraction of the aqueous solution was performed three times (3 × 300 μL) with ethyl acetate. All supernatant was combined after centrifugation at 13,000 g for 5 min at 4 °C. Next, concentration with nitrogen stream at room temperature was performed for following chemical labelling.
For DMED and d 4-DMED labelling, 200 μL ACN, 30 μL 20 μmol/L TEA, 15 μL 20 μmol/L CMPI and 15 μL 20 μmol/L DMED or d 4-DMED was added into above residue. The mixture was incubated at 40 °C for 1 h. Resulting solution was dried under nitrogen and reconstruction in 50 μL ACN/water (v/v, 1/9). Finally, 50 μL sample was injected for LC-Orbitrap MS analysis.
Conventional untargeted metabolome was profiled for comparison of sensitivity and accuracy with modified CIL-LC-MS method. Briefly, 300 μL ethanol was added into 100 μL serum. The mixture was shaken vigorously for 2 min. Proteins were denatured and precipitated by centrifugation. The supernatant was dried under nitrogen gas. The reconstituted solution and injection volume for LC-MS kept the same as that in CIL-LC-MS.
LC-MS analysis for CIL-LC-MS was performed on an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, USA) coupled with an UltiMate 3000 UHPLC System (Thermo Fisher Scientific, USA). LC separation was performed on an Acquity UPLC BEH C18 column (50 × 2.1 mm i. d., 1.7 μm; Waters, Milford, USA) with a flow rate of 0.4 mL/min at 40 °C. FA in water (0.1%, v/v, solvent A) and ACN (solvent B) were employed as mobile phases for analysis. A gradient of 0–5 min 5% B, 5–45 min 5%–95% B, 45–55 min 95% B, 55–55.1 min 95%–5% B and 55.1–60 min 5% B was used. Carboxylic metabolites were detected in a positive ion mode using the full scan mode (MS1, m/z 180–650) at a resolution of 60,000. The source and ion transfer parameters applied were as follows: heater temperature, 300 °C; capillary temperature, 350 °C; sheath gas flow, 35 arbitrary; auxiliary gas flow, 15 arbitrary; spray voltage, 3.5 kV; capillary voltage, 35 V; S-lens RF level, 60%; automatic gain control, 1 × 106; the maximum injection time, 100 ms. MS2 spectra were acquired based on the mass list of preselected precursor ions (paired peaks) in collision-induced dissociation (CID).
Conventional untargeted metabolome LC-MS analysis was performed under the same instrumental conditions as CIL-LC-MS, except carboxylic metabolites were detected in a negative ion mode using the full scan mode (MS1, m/z 100–800) at a resolution of 60,000.
2.3. Carboxylic metabolite identification
For the discriminative carboxylic feature identification, we annotated them using the five-level system advocated by the Metabolomics Standards Initiative (MSI) as follows [16]: the metabolites were identified as MSI level 1, when their accurate m/z, retention index (RI), and MSn spectra were coincident with that of the standard or the record in our homemade database CLMD (https://www.clmdb.com). Metabolites were defined as MSI level 2 if they had interpretable MSn spectra and could be matched to a consistent structure in metabolome databases, such as HMDB (https://hmdb.ca) or METLIN (https://metlin. Scripps. edu/); metabolites were categorized as MSI level 3 if the formula were matched to that in above public databases, but lack of high-quality MSn spectra and commercially available standards; metabolites not matched in public database were classified as MSI level 4; metabolites without a defined molecular formula were annotated as MSI level 5. More detailed information on data extraction and formula calculation was elucidated in Supporting Information.
2.4. Relative quantification of carboxylic features
A pooled serum sample labelled with d 4-DMED served as internal standard (IS) and quality control (QC) sample to correct the MS fluctuations. Generally, individual samples of COVID-19 asymptomatic and healthy control were labelled with DMED. Then, the samples labelled with DMED and the IS were mixed at a ratio of 1:1 (v/v) for analysis. Relative concentrations of carboxylic metabolites in individual serum samples were calculated by peak intensity ratios (DMED-labelled peak to d 4-DMED labelled peak).
2.5. Statistical analysis
Missing values were imputed with the 1/2 minimal value. Kruskal-Wallis test and logistic regression model were performed with IBM SPSS 26.0 (IBM SPSS Inc., New York, USA) software. Orthogonal projections to latent structures discriminative analysis (OPLS-DA) was performed with SIMCA 14.1 (Umetrics AB, Umea, Sweden).
2.6. Machine learning
PyCharm Community Edition 2020.1.3 was employed for developing machine learning models to differentiate and predict the healthy and asymptomatic cases. The investigated classification models included decision linear model (DLM), multi-layer perceptron (MP), random forest (RF) and support vector machine (SVM). The in-built “sklearn” module was employed for investigating and comparing the model performances in fitting and generalization.
3. Results and discussion
3.1. Clinical characteristics of asymptomatic COVID-19 serum
A cohort of 184 subjects including 62 asymptomatic COVID-19 patients and healthy controls (n = 122), matched according to age and gender, was procured in this study. The detailed patient descriptions including gender, age, IgM and IgG level, nucleic acid detection results were shown in Table S1 and Table S2. 11.3% (7/62) was tested seropositive for IgM in the asymptomatic group, and 90.3% (56/62) appeared to be seropositive for IgG. During the process of SARS-CoV-2 infection, IgM is produced in the early stage, while IgG is likely to exist for a longer period [4]. The high positive rate of IgG and relatively low positive rate of IgM may indicate that most patients in this study were in the middle or late stage of infection.
3.2. CIL-LC-MS method optimization and validation
We made a modification of our reported CIL-LC-MS method for compatibility of virus-infected serum samples [15]. Additional viral inactivation step was necessary. In that case, 75% ethanol, rather than ethyl acetate in our reported CIL method, was utilized for viral inactivation and metabolite extraction in our initial protocol. Nevertheless, we found the polar metabolites extracted in 75% ethanol interfered derivatization process and resulted to decreased derivatization efficiency (data not shown). Therefore, we suppled a liquid-liquid extraction (LLE) with ethyl acetate to remove polar metabolites. The labelling efficiency of modified protocol can reach over 93% (data not shown).
The validation of the modified CIL-LC-MS method was performed. The total ion chromatography (TIC) showed abundant peaks were detected under positive ion mode after CIL (Fig. 1b). In our proposed screening rule, DMED and d 4-DMED labelled peak pairs exhibited 4.025 Da mass difference, with RI difference <1% and ratio of peak intensity between 0.5 and 2, taking undecylenic acid as an example (Fig. 1c). Most RSDs of the detected peak pairs in QC were below 10% (Fig. 1d), indicating good stability and reproducibility of the modified method.
We further compared quantitative sensitivity and accuracy of the modified CIL-LC-MS method with conventional untargeted metabolomic method. As results, the ratio of signal to noise of 9-decenoic acid, undecylenic acid, 3-hydroxydecanoic acid and 12-HETE got 3-100-fold improved compared to that of unlabelled method (Fig. S1). Besides, the quantitative accuracy was also improved (Fig. 1d and Fig. S2), for the special peak matching principle in CIL-LC-MS method can correct signal fluctuations with the help of isotope internal standard.
Thereafter, the paired peaks satisfied with RSDs<30% across QC samples, and missing values < 20% in each group were screened for further analysis. A total of 96 potential carboxylic features were filtered out (Table S3), and corresponding quantitative result in participants was presented in Table S4.
3.3. Discriminative carboxylic features screening and identification in asymptomatic COVID-19 serum
To find discriminative carboxylic metabolites in asymptomatic patients, the above 96 candidate carboxylic features were classified by two criteria: (1) p < 0.05 and |log2FC (fold change)|>0.25; (2) variable importance for projection (VIP) value > 1. Features meeting both two criteria were defined as rank 1, satisfied with p < 0.05 and |log2FC|>0.25 were rank 2, and only living up to VIP value > 1 were rank 3 (Table S5).
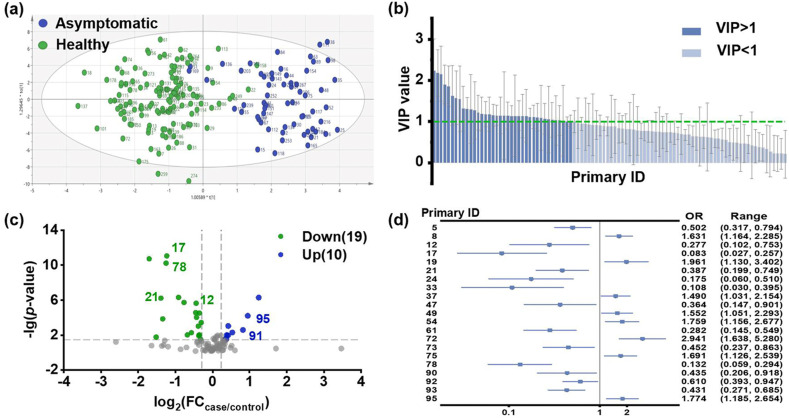
OPLS-DA model was constructed utilizing above 96 features for visualizing the classification performance of the metabolic peaks. As seen in Fig. 2 a, healthy and asymptomatic group can be distinguished in the optimized OPLS-DA model. The validation plot demonstrated that the model was not over-fitting (Fig. S3). A total of 38 potential carboxylic features with VIP value > 1 were filtrated out for further discriminative features screening (Fig. 2b).
Fig. 2.
Discovery of discriminative features by statistical analysis. (a) OPLS-DA score plot of overall metabolic peaks. (b) VIP list of carboxyl metabolites in optimized OPLS-DA model. (c)Volcano plots showing changed contents of carboxyl metabolites in asymptomatic patients. (d) Logistic regression model with covariates age and sex. Forest plots illustrate the magnitude of odds ratios with indicator of significance of the estimate in the model.
Data analysis of the Kruskal-Wallis test and FC were presented in volcano plot (Fig. 2c). Based on the criterion of p < 0.05 and |log2FC|>0.25, 29 features with significant changes were obtained between healthy and asymptomatic groups. 19 features got down-regulated in the asymptomatic group, while 10 features got up-regulated.
Based on above criteria, a total of 44 discriminative carboxylic features were picked out, and 23 of them were listed as rank 1 (Table S5). Among the 23 features, 14 got down-regulated and 9 got up-regulated in asymptomatic group.
Furthermore, to exclude the effect of age and sex, logistic regression model with covariates age and sex was set up to search for truly significant variables from above features. Features with overall false discovery rate <0.05 were short-listed and grouped for visual clarity in a forest plot (Fig. 2d). Our result illustrated that 21 features were significant with asymptomatic infection (Table S5, marked as red), including 8 features consisted with alleviation and 13 features representing deterioration. Most features significant correlated with asymptomatic infection got down-regulated, while features with ID 72 and 95 got upregulated (marked with *** in Fig. S4). Although there was no significant difference regarding the clinic pathological features between asymptomatic patients and healthy control, the metabolomic profiles effectively categorized them into two distinct groups.
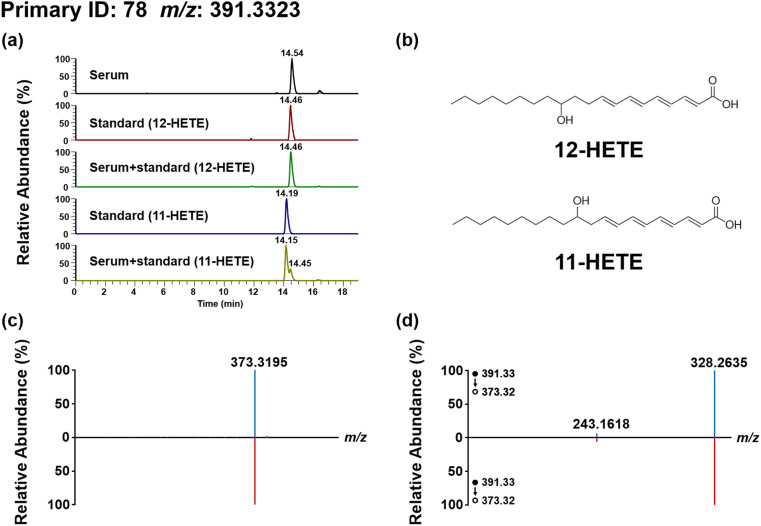
We then performed structure annotation of the discriminant features according to MSI [16]. Taking 12-hydroxyeicosatetraenoic acid (12-HETE, MSI level 1) as an example (Fig. 3 ), the m/z and MSn of candidate feature in serum matched well with both 11-HETE and 12-HETE standards; the RI of candidate feature matched well with 12-HETE standard while mismatched with 11-HETE. Therefore, we identified it as 12-HETE. For the structural annotation of 15,16-dihydroxyoctadeca-9,12-dienoic acid (15, 16-DiHODE, MSI level 2, Fig. S10), the terminal aldehyde-containing ion of 278.2115 and terminal alkenyl-containing ion of 262.2165 with a m/z difference of 15.995 (-O), were generated via fragmentation of the three-membered epoxide ring of the fragment ion m/z 320.2572. According to our previous report [17], it was a fragmentation characteristic of DMED labelled epoxy/dihydroxy-oxylipins. Furthermore, above diagnostic ion pairs revealed the epoxy groups are located at C-15 and C-16. Moreover, the product ions (m/z 338.2690, m/z 320.2572 and m/z 302.2467) observed in the MS2 spectra suggested the loss of two H2O from the parent ion, indicating the primary structure contained two hydroxy. The two adjacent hydroxy converted to above three-membered epoxide ring by loss of two H2O in MS2. Thus, it was identified as 15, 16-DiHODE.
Fig. 3.
Identification of the arachidonic acid derivative in serum sample. (a) The XIC of 12-HETE or 11-HETE in serum sample, standard solution and standard addition serum samples; (b)The chemical structure of 12-HETE and 11-HETE; (c) The MS2 spectrum of 12-HETE in serum sample and in 12-HETE standard solution; (d) The MS3 spectrum of fragment ion m/z 373.32 (391.33–373.32) in serum sample and in 12-HETE standard solution. LC separation was performed on an Acquity UPLC BEH Phenyl column (50 × 2.1 mm i. d., 1.7 μm; Waters, Milford, USA) with a flow rate of 0.4 mL/min at 40 °C. FA in water (0.1%, v/v, solvent A) and ACN/MeOH (7/3, v/v, with 0.1% FA, solvent B) were employed as mobile phases for analysis. A gradient of 0–5 min 20–30% B, 5–17 min 30–40% B, 17–19 min 40-20% B was used.
Overall, 6 metabolites were identified as MSI level 1 (Fig. 3, Figures S5-S9), 3 metabolites were identified as MSI level 2 (Figures S10-S12), 19 metabolites were assigned to MSI level 3 and 16 metabolites were assigned to MSI level 4. Specifically, 9-decenoic acid, undecylenic acid, 3-hydroxydecanoic acid and 12-HETE were downregulated in asymptomatic patients, while cholic acid and glycoursodeoxycholic acid (GUDCA) were upregulated.
3.4. Association of serum carboxylic features with IgG/IgM levels
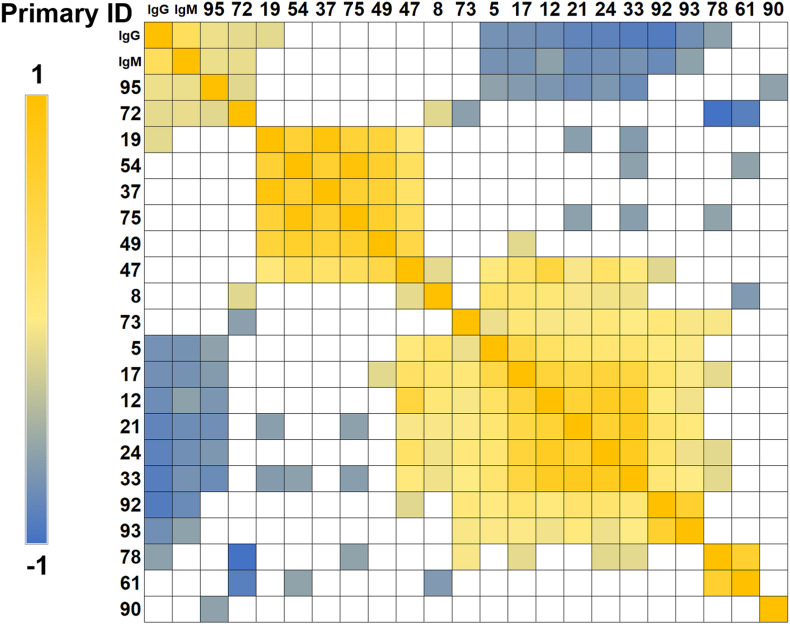
Spearman correlations were performed to investigate the correlation between above dysregulated carboxylic features and IgG/IgM levels in COVID-19 asymptomatic group (Fig. 4 ). IgG and IgM are known as the signal of inflammation. It can be observed that feature 95 and feature 72, identified as GUDCA and 15,16-DiHODE, displayed significantly positive correlations with IgG and IgM. This suggested that the increased GUDCA and 15,16-DiHODE level were associated with aggravated systemic inflammation. Besides, feature 5, 17, 12, 21, 24, 33, 92 and 93 presented negative correlations with IgG and IgM. Among them, feature 5 (C10H12O2), 17 (undecylenic acid), 12 (9-decenoic acid), 24 (C12H22O2), 33 (C14H24O2) were identified by standards or postulated by their formulas as unsaturated fatty acids. Down-regulation of these fatty acid metabolites and their products in COVID-19 patients has been reported [18]. The reduction was regarded as a feedback to declining lung functions and limiting blood oxygen, reducing reliance on oxygen for energy production [18]. Meanwhile, these fatty acids may serve as precursor of PUFA-PCs, which displayed significant negative correlations with clinical indices of systemic inflammation [18].
Fig. 4.
Clustered Spearman correlation matrix for the distinguished metabolites. Color indicates the correlation coefficient between the two orthogonal factors. Only correlations with p < 0.05 were indicated as colored squares. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Machine learning for risk assessment of asymptomatic patient
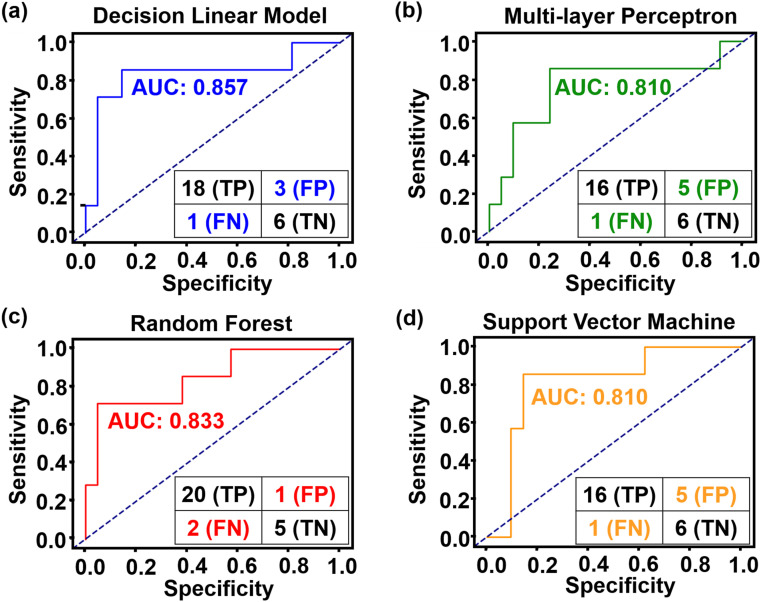
Based on above 21 dysregulated carboxylic features, a machine learning trial was made for discrimination of asymptomatic patients from healthy control using different machine learning methods, including DLM, MP, RF and SVM. Confusion matrix presented the relationship of true value and predicted value. 110 samples (55 asymptomatic patients and 55 healthy control) were randomly selected as training cohorts to establish the prediction model for asymptomatic infection, and other 7 asymptomatic patients and 21 healthy controls were used as independent validation cohort to evaluate the model. As shown in Fig. 5 , the AUCs of optimized DLM, MP, RF and SVM models were 0.857, 0.810, 0.833 and 0.810, respectively. RF model exhibited better diagnose performance with accuracy of 85.7%, specificity of 90.9% and sensitivity of 83.3%, with only one false positive and two false negative case. The results suggested the RF model can play an auxiliary role in the risk assessment of asymptomatic patients.
Fig. 5.
Four machine learning models for asymptomatic infection risk assessment. (a) Decision linear model, (b) Multilayer perceptron, (c) Random forest, (d) Support vector classifier. In each model, confusion matrix at bottom right represented performance. AUC: area under the curve; TP: true positive; FP: false positive; FN: false negative; TN: true negative. Sensitivity = TP/(TP + FN). Specificity = TN/(TN + FP).
3.6. Pathological discussion of COVID-19 asymptomatic infection
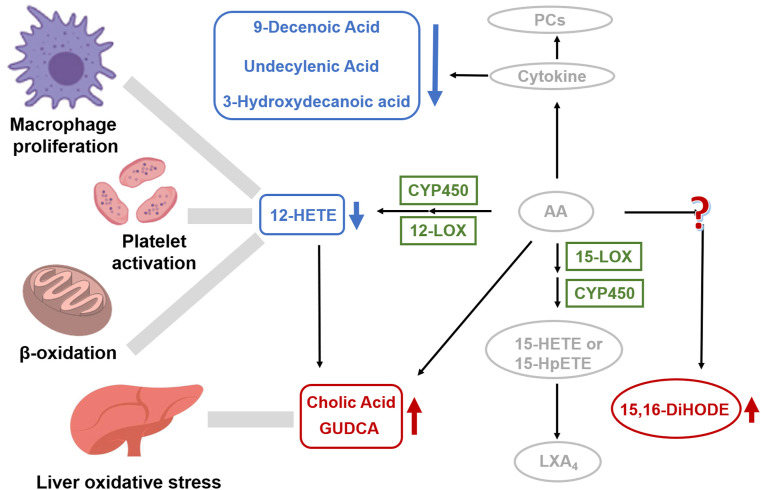
The identified 6 metabolites can provide deeper understanding on the pathogenesis metabolism of asymptomatic infection on molecular level. The identified 12-HETE is known as a metabolic product of AA [19], which was reported to be down-regulated in some respiratory infectious diseases [20]. Similar down-regulation of 12-HETE was found in asymptomatic patients (Fig. 6 ). In previous reports, concentration of AA was directly correlated with the severity of COVID-19 [18,21]. Besides, AA was reported to induce cytokine storm, dysregulated unsaturated fatty acids and activated immune response in COVID-19 patients [22]. As metabolic product of AA, we speculated that the reduced 12-HETE might be an early signal of AA metabolic dysregulation in asymptomatic patients. In addition, the known physiological functions of 12-HETE was related to the regulation of platelet aggregation [23]. Thrombocytopenia was observed in symptomatic COVID-19 patients [24]. It suggested the down-regulation of 12-HETE might lead to thrombocytopenia. Moreover, 12-HETE can be converted into (ω-1)-hydroxylation and β-oxidation products by active M2 macrophages [21]. Coincidentally, Liao et al. revealed macrophage infiltration and activation in COVID-19 patients [25]. Therefore, the reduction of 12-HETE might be associated with M2 macrophage activation.
Fig. 6.
Dysregulated metabolites and their associated pathway. A hypothetical-systems view of the responses to SARS-CoV-2 infection. Blue cycle and arrow, downregulated metabolites; Red cycle and arrow, upregulated metabolites; Grey font and arrow, known pathway and reported clinical manifestation; Green font, known enzymes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Cytokine storm in COVID-19 was reported to influence the concentrations of unsaturated fatty acids [22]. The observed downregulation of 9-decenoic acid and undecylenic acid in our results provided auxiliary proof for dysregulation of unsaturated fatty acids induced by cytokine disorder. 9-decenoic acid was regarded as the main contributors of β-oxidation [20]. Salaun et al. found that 9-decenoic acid was a weak competitive inhibitor of the in-chain hydroxylase, which can inactivate the P450 enzyme [26]. we hypothesized 9-decenoic acid played a similar role in asymptomatic patients. As for undecylenic acid, it was reported to be associated with the oxidation pathway of P450 enzyme [27]. Meanwhile, Lin et al. proved that undecylenic acid can directly bind with ZIKV WT-RdRp (a kind of flavivirus) and had a relatively weak interaction with D535A-RdRp, which could serve as a promising lead compound against ZIKV [28]. Therefore, undecylenic acid may work in a similar way against COVID-19. Besides, we found 3-hydroxydecanoic acid got down-regulated in asymptomatic patients (Fig. 6), while upregulated in symptomatic [29]. 3-hydroxydecanoic acid is a medium-chain fatty acid, whose sensing receptor GPR84 is a pro-inflammatory receptor [30]. Peters et al. declared that 3-hydroxydecanoic acid can stimulate GPR84 and cause a sustained ERK activation [31]. The ERK pathway was closely related to the human immunodeficiency regulatory protein, which can enhance the replication and infectivity of viruses such as HIV by regulating T-cell activity [32]. Therefore, we speculated that 3-hydroxydecanoic acid may play a similar role in COVID-19 asymptomatic patients.
Unlike the downregulation tendency of fatty acids, cholic acid and GUDCA got up-regulated in asymptomatic patients (Fig. 6). Their physiological functions were related to the conjugation with glycine or taurine by bile acid-CoA synthetase and bile acid-CoA [33], P450 enzyme metabolism [34]. In previous studies on symptomatic COVID-19, increased synthesis of bile acid [35], abnormally high levels of ketone bodies and 2-hydroxybutyric acid were found, indicating enhanced hepatic glutathione synthesis and oxidative stress [36]. Meanwhile, biliverdin, the primary cytoprotective antioxidant [37], was found up-regulated in COVID-19, also suggesting enhanced oxidative stress in disease state on the other hand [18]. Moreover, AA was proved to have an influence on the metabolism of bile acid [38]. As metabolic product of AA, we supposed 12-HETE may act as a messenger of AA for enhanced synthesis of bile acids and GUDCA, as well as regulation of oxidative stress in the liver.
More interesting result in this study was the upregulation of a lipoxin, 15,16-DiHODE (Fig. 6), which was seldom reported in other COVID-19 related studies. Generally, lipoxin metabolites play anti-inflammatory or pro-inflammatory roles in physiology, such as anti-inflammatory LXA4, producing by 15-HETE or 15-HPETE under the catalysis of 15-LOX enzyme or P450 enzyme. We speculated that 15, 16-DiHODE may have corresponding anti-inflammatory or pro-inflammatory function in COVID-19 asymptomatic patients, which was worth further study.
4. Conclusion
In summary, we reported a serum carboxylic submetabolome profiling of asymptomatic COVID-19 patients by a CIL-LC-MS method. Twenty-one discriminative carboxylic biomarkers were filtered out, 6 of which were confirmed using standards validation. The confirmed 6 carboxylic acids may disorder the following pathways: metabolic disturbance of AA and M2 activation of macrophages may have an adverse influence on the synthesis of 12-HETE; Down-regulation of unsaturated fatty acid may be associated with the inflammatory response induced by deregulated AA; 12-HETE may be an omen of dysregulated AA, thus affecting bile acid synthesis and several amino acid metabolisms; The upregulation of a lipoxin, 15,16-DiHODE, may have anti-inflammatory or pro-inflammatory function in COVID-19 asymptomatic patients, which was worth further study. Furthermore, we built four machine models for risk assessment of asymptomatic patients based on these potential carboxylic biomarkers. RF model provided the best performance with accuracy of 85.7%, sensitivity of 90.9% and specificity of 83.3%. To sum up, our results offered the missing puzzles of carboxylic metabolites signature characterization in asymptomatic COVID-19 patients. It was also conducive for the study of pathogenic mechanism of COVID-19, promoting the prevention and control of the crisis.
Credit author statement
Jing Xu: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. Yu Yuan: Investigation, Resources. Yao-Yu Chen: Investigation. Cai-Feng Xiong: Investigation. Zheng Zhang: Conceptualization, Investigation, Resources, Methodology, Software, Writing – review & editing. Yu-Qi Feng: Conceptualization, Resources, Writing – review & editing, Funding acquisition, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the National Key Research and Development Program of China (2018YFA0900400), National Natural Science Foundation of China (21635006, 31670373, 21721005).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.123086.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Inizan T.J., Célerse F., Adjoua O., El Ahdab D., Jolly L.-H., Liu C., Ren P., Montes M., Lagarde N., Lagardère L., Monmarché P., Piquemal J.-P. High-resolution mining of the SARS-CoV-2 main protease conformational space: supercomputer-driven unsupervised adaptive sampling. Chem. Sci. 2021;12:4889–4907. doi: 10.1039/d1sc00145k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng J., Su D., Zhang Z., Wang M. Identification and management of asymptomatic carriers of coronavirus disease 2019 (COVID-19) in China, Influenza. Other. RESP (Rev. Epidemiol. Sante Publique) 2020;14(5):599–600. doi: 10.1111/irv.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X., Sun J., Nie S., Li H., Kong Y., Liang M., Hou J., Huang X., Li D., Ma T., Peng J., Gao S., Shao Y., Zhu H., Lau J.Y., Wang G., Xie C., Jiang L., Huang A., Yang Z., Zhang K., Hou F.F. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. 2020;26(8):1193–1195. doi: 10.1038/s41591-020-0949-6. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q., He Z., Mao F., Pei H., Cao H., Liu X. Diagnostic technologies for COVID-19: a review. RSC Adv. 2020;10(58):35257–35264. doi: 10.1039/d0ra06445a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wikramaratna P.S., Paton R.S., Ghafari M., Lourenço J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Euro Surveill. 2020;25(50) doi: 10.2807/1560-7917.ES.2020.25.50.2000568. pii=2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overmyer K.A., Shishkova E., Miller I.J., Balnis J., Bernstein M.N., Peters-Clarke T.M., Meyer J.G., Quan Q., Muehlbauer L.K., Trujillo E.A., He Y., Chopra A., Chieng H.C., Tiwari A., Judson M.A., Paulson B., Brademan D.R., Zhu Y., Serrano L.R., Linke V., Drake L.A., Adam A.P., Schwartz B.S., Singer H.A., Swanson S., Mosher D.F., Stewart R., Coon J.J., Jaitovich A. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12(1):23–40. doi: 10.1016/j.cels.2020.10.003. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoxha M. What about COVID-19 and arachidonic acid pathway? Eur. J. Clin. Pharmacol. 2020;76(11):1501–1504. doi: 10.1007/s00228-020-02941-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D., Shu T., Yang X., Song J.-X., Zhang M., Yao C., Liu W., Huang M., Yu Y., Yang Q., Zhu T., Xu J., Mu J., Wang Y., Wang H., Tang T., Ren Y., Wu Y., Lin S.-H., Qiu Y., Zhang D.-Y., Shang Y., Zhou X. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020;7(7):1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas T., Stefanoni D., Dzieciatkowska M., Issaian A., Nemkov T., Hill R.C., Francis R.O., Hudson K.E., Buehler P.W., Zimring J.C., Hod E.A., Hansen K.C., Spitalnik S.L., D'Alessandro A. Evidence of structural protein damage and membrane lipid remodeling in red blood cells from COVID-19 patients. J. Proteome Res. 2020;19(11):4455–4469. doi: 10.1021/acs.jproteome.0c00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S., Luo X., Li L. Chemical isotope labeling LC-MS for high coverage and quantitative profiling of the hydroxyl submetabolome in metabolomics. Anal. Chem. 2016;88(21):10617–10623. doi: 10.1021/acs.analchem.6b02967. [DOI] [PubMed] [Google Scholar]
- 13.Shan J., Qian W., Shen C., Lin L., Xie T., Peng L., Xu J., Yang R., Ji J., Zhao X. High-resolution lipidomics reveals dysregulation of lipid metabolism in respiratory syncytial virus pneumonia mice. RSC Adv. 2018;8(51):29368–29377. doi: 10.1039/c8ra05640d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam Vincent C., Quehenberger O., Oshansky Christine M., Suen R., Armando Aaron M., Treuting Piper M., Thomas Paul G., Dennis Edward A., Aderem A. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154:213–227. doi: 10.1016/j.cell.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan B.-F., Zhu Q.-F., Guo N., Zheng S.-J., Wang Y.-L., Wang J., Xu J., Liu S.-J., He K., Hu T., Zheng Y.-W., Xu F.-Q., Feng Y.-Q. Comprehensive profiling of fecal metabolome of mice by integrated chemical isotope labeling-mass spectrometry analysis. Anal. Chem. 2018;90(5):3512–3520. doi: 10.1021/acs.analchem.7b05355. [DOI] [PubMed] [Google Scholar]
- 16.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A., Fan T.W., Fiehn O., Goodacre R., Griffin J.L., Hankemeier T., Hardy N., Harnly J., Higashi R., Kopka J., Lane A.N., Lindon J.C., Marriott P., Nicholls A.W., Reily M.D., Thaden J.J., Viant M.R. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong C.-F., Zhu Q.-F., Chen Y.-Y., He D.-X., Feng Y.-Q. Screening and identification of epoxy/dihydroxy-oxylipins by chemical labeling-assisted ultrahigh-performance liquid chromatography coupled with high-resolution mass spectrometry. Anal. Chem. 2021;93(28):9904–9911. doi: 10.1021/acs.analchem.1c02058. [DOI] [PubMed] [Google Scholar]
- 18.Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., Chua G.H., Zhang C., Meng F.P., Xu Z., Fu J.L., Huang L., Xia P., Yang T., Zhang S., Li B., Jiang T.J., Wang R., Wang Z., Shi M., Zhang J.Y., Wang F.S., Shui G. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metabol. 2020;32(2):188–202. doi: 10.1016/j.cmet.2020.06.016. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.J., Cheng X., Yan Z.Z., Fang J., Wang X., Wang W., Liu Z.Y., Shen L.J., Zhang P., Wang P.X., Liao R., Ji Y.X., Wang J.Y., Tian S., Zhu X.Y., Zhang Y., Tian R.F., Wang L., Ma X.L., Huang Z., She Z.G., Li H. An ALOX12-12-HETE-GPR31 signaling axis is a key mediator of hepatic ischemia-reperfusion injury. Nat. Med. 2018;24(1):73–83. doi: 10.1038/nm.4451. [DOI] [PubMed] [Google Scholar]
- 20.Luzardo-Ocampo I., Campos-Vega R., Gonzalez de Mejia E., Loarca-Piña G. Consumption of a baked corn and bean snack reduced chronic colitis inflammation in CD-1 mice via downregulation of IL-1 receptor, TLR, and TNF-α associated pathways. Food Res. Int. 2020;132:109097. doi: 10.1016/j.foodres.2020.109097. [DOI] [PubMed] [Google Scholar]
- 21.Kriska T., Thomas M.J., Falck J.R., Campbell W.B. Deactivation of 12(S)-HETE through (ω-1)-hydroxylation and β-oxidation in alternatively activated macrophages. J. Lipid Res. 2018;59(4):615–624. doi: 10.1194/jlr.M081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takagi J., Usui T., Kawajiri K., Kobayashi Y., Sato F., Saito Y. 12S-Hydroxyeicosatetraenoic acid plays a central role in the regulation of platelet activation. Biochem. Bioph. Res. Co. 1991;179(1):345–351. doi: 10.1016/0006-291x(91)91376-n. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y.-F., Zhang Y., Chi H.-B., Chen S.-Y., Peng M.-F., Luo L.-F., Chen L.-P., Li J., Shen B., Wang D.-L. The hemocyte counts as a potential biomarker for predicting disease progression in COVID-19: a retrospective study. Clin. Chem. Lab. Med. 2020;58(7):1106–1115. doi: 10.1515/cclm-2020-0377. [DOI] [PubMed] [Google Scholar]
- 25.Liao M., Liu Y., Yuan J., Wen Y., Zhang Z. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 doi: 10.1101/2020.1102.1123.20026690. [DOI] [Google Scholar]
- 26.Salaun J.P., Reichhart D., Simon A., Durst F., Reich N.O., Ortiz de Montellano P.R. Autocatalytic inactivation of plant cytochrome P-450 enzymes: selective inactivation of the lauric acid in-chain hydroxylase from Helianthus tuberosus L. by unsaturated substrate analogs. Arch. Biochem. Biophys. 1984;232(1):1–7. doi: 10.1016/0003-9861(84)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Chen X., Su Z., Horner J.H., Newcomb M. Oxidation of 10-undecenoic acid by cytochrome P450(BM-3) and its Compound I transient. Org. Biomol. Chem. 2011;9(21):7427–7433. doi: 10.1039/c1ob06035j. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y., Zhang H., Song W., Si S., Han Y., Jiang J. Identification and characterization of Zika virus NS5 RNA-dependent RNA polymerase inhibitors. Int. J. Antimicrob. Agents. 2019;54(4):502–506. doi: 10.1016/j.ijantimicag.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 29.De Silva I.W., Nayek S., Singh V., Reddy J., Granger J.K., Verbeck G.F. Paper spray mass spectrometry utilizing Teslin (R) substrate for rapid detection of lipid metabolite changes during COVID-19 infection. Analyst. 2020;145(17):5725–5732. doi: 10.1039/d0an01074j. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki M., Takaishi S., Nagasaki M., Onozawa Y., Iino I., Maeda H., Komai T., Oda T. Medium-chain fatty acid-sensing receptor, GPR84, is a proinflammatory receptor. J. Biol. Chem. 2013;288(15):10684–10691. doi: 10.1074/jbc.M112.420042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters A., Rabe P., Krumbholz P., Kalwa H., Kraft R., Schöneberg T., Stäubert C. Natural biased signaling of hydroxycarboxylic acid receptor 3 and G protein-coupled receptor 84. Cell Commun. Signal. 2020;18(1):31. doi: 10.1186/s12964-020-0516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrager J.A., Der Minassian V., Marsh J.W. HIV Nef increases T cell ERK MAP kinase activity. J. Biol. Chem. 2002;277(8):6137–6142. doi: 10.1074/jbc.M107322200. [DOI] [PubMed] [Google Scholar]
- 33.Vaz F.M., Ferdinandusse S. Bile acid analysis in human disorders of bile acid biosynthesis. Mol. Aspect. Med. 2017;56:10–24. doi: 10.1016/j.mam.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Deo A.K., Bandiera S.M. Identification of human hepatic cytochrome P450 enzymes involved in the biotransformation of cholic and chenodeoxycholic acid. Drug Metab. Dispos. 2008;36(10):1983–1991. doi: 10.1124/dmd.108.022194. [DOI] [PubMed] [Google Scholar]
- 35.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Chen H., Guo T. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruzzone C., Bizkarguenaga M., Gil-Redondo R., Diercks T., Arana E., Garcia de Vicuna A., Seco M., Bosch A., Palazon A., San Juan I., Lain A., Gil-Martinez J., Bernardo-Seisdedos G., Fernandez-Ramos D., Lopitz-Otsoa F., Embade N., Lu S., Mato J.M., Millet O. SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. Iscience. 2020;23:101645. doi: 10.1016/j.isci.2020.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barañano D.E., Rao M., Ferris C.D., Snyder S.H. Biliverdin reductase: a major physiologic cytoprotectant. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99(25):16093–16098. doi: 10.1073/pnas.252626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barberis E., Timo S., Amede E., Vanella V.V., Puricelli C., Cappellano G., Raineri D., Cittone M.G., Rizzi E., Pedrinelli A.R., Vassia V., Casciaro F.G., Priora S., Nerici I., Galbiati A., Hayden E., Falasca M., Vaschetto R., Sainaghi P.P., Dianzani U., Rolla R., Chiocchetti A., Baldanzi G., Marengo E., Manfredi M. Large-scale plasma analysis revealed New mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 2020;21(22):8623. doi: 10.3390/ijms21228623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.