Abstract
During a 1-year survey of Shiga toxin-producing Escherichia coli (STEC) prevalence in central France, 2,143 samples were investigated by PCR for Shiga toxin-encoding genes. A total of 330 (70%) of 471 fecal samples collected from healthy cattle at the Clermont-Ferrand slaughterhouse, 47 (11%) of 411 beef samples, 60 (10%) of 603 cheese samples, and 19 (3%) of 658 stool specimens from hospitalized children with and without diarrhea were positive for the stx gene(s). A STEC strain was isolated from 34% (162 of 471) of bovine feces, 4% (16 of 411) of beef samples, 1% (5 of 603) of cheese samples, and 1.5% (10 of 658) of stool specimens. Of the 220 STEC strains isolated, 34 (15%) harbored the stx1 gene, 116 (53%) harbored the stx2 gene, and 70 (32%) carried both the stx1 and stx2 genes. However, 32 (14.5%) were not cytotoxic for Vero cells. The eae gene, found in 12 (5%) of the 220 strains, was significantly associated with the stx1 gene and with isolates from children. Sequences homologous to ehxA were found in 102 (46%) of the 220 strains. Thirteen serotypes, OX3:H2, O113:H21, O113:H4, OX3:H21, O6:H10, OX178:H19, O171:H2, O46:H38, O172:H21, O22:H16, O91:H10, O91:H21, and O22:H8, accounted for 102 (55%) of 186 typeable isolates, and only one strain (0.5% of the 186 STEC isolates from cattle), belonged to the O157:H7 serotype. We showed that the majority of the STEC isolates from cattle, beef, and cheese were not likely to be pathogenic for humans and that the STEC strains isolated from children in this study were probably not responsible for diarrheal disease. Finally, the strains associated with hemolytic-uremic syndrome in the same geographical area were shown to belong to particular subsets of the STEC population found in the bovine reservoir.
Shiga toxin-producing Escherichia coli (STEC) has been associated with both outbreaks and sporadic cases of human disease, ranging from uncomplicated diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome (HUS). HUS, a life-threatening complication which can occur in 5 to 10% of patients, is characterized by thrombocytopenia, microangiopathic hemolytic anemia, and acute renal failure. The dominant STEC serotype is O157:H7, which is also the most commonly involved in large outbreaks in the United States, Canada, and the United Kingdom. However, strains belonging to more than 100 different O:H types have been associated with human disease. Common STEC serogroups associated with pathogenicity include O26, O91, O103, and O111 (4, 29).
The ability of STEC strains to cause severe disease in humans is undoubtedly related to their capacity to secrete the Stx1 and/or Stx2 Shiga toxins, also called verotoxins (8, 20). The genes encoding these toxins are located in the genomes of temperate bacteriophages. Several other virulence factors that might contribute to the pathogenicity of STEC strains have been described. Among them is intimin, the product of the eae (for E. coli attaching and effacing) gene, a 94-kDa outer membrane protein involved in the intimate attachment of bacteria to enterocytes. The eae gene is part of the locus for enterocyte effacement, required for the formation of the attaching and effacing lesion, initially recognized in enteropathogenic E. coli strains (12, 41). However, many STEC strains involved in severe human disease (including HUS) do not contain the eae gene or do not express a functional intimin as detected by the fluorescent actin-staining test (5, 14, 21, 22, 39). Thus, attaching and effacing lesions might not be essential for the development of STEC-associated severe diseases, and additional factors might be involved. The enterohemolysin, or enterohemorrhagic E. coli Hly, a member of the repeat in toxin (RTX) family of pore-forming cytolysins, has been suspected to have a role in pathogenicity, because it has occurred in the majority of the pathogenic STEC strains tested and was reactive to sera of patients with HUS (36). Two other putative virulence factors have been described: a serine protease (EspP) which can cleave human coagulation factor V and a bifunctional catalase peroxidase, KatP (6, 7). However, there is no experimental proof for the role of these factors in the virulence of STEC (4).
Cattle appear to be the main reservoir of STEC strains, which were recovered from fecal samples of 10 to 20% of healthy animals in the United States and Europe and in as many as 37% of animals in a recent Spanish survey (1, 2, 3, 38). STEC is transmitted to humans through foods contaminated by fecal material, mainly undercooked ground beef and unpasteurized dairy products. Finally, person-to-person transmission and transmission through drinking and recreational water have also been described (16). Interestingly, the epidemiological situation in continental Europe seems to be different from those of North America and the United Kingdom. In continental Europe, human infections are frequently associated with non-O157:H7 serotypes, sporadic cases of infection are more common than cases related to outbreaks, and most cases are not linked to undercooked ground meat, but the means of transmission are often unknown (37).
From August 1996 to May 1997, six STEC strains were isolated from stools of adults suffering from HUS in the Clermont-Ferrand hospital. All the isolates were stx2 positive, but none harbored the eae gene. They belonged to different serotypes: O6:H4, O91:H10, O91:H21, Orough:H16, OX3:H−, and O non-typeable:H− (5). Because of this unusual number of non-O157:H7 STEC infections in the Clermont-Ferrand area, and since very little is known about the prevalence of these organisms in France, a 1-year survey was undertaken. Bovine feces, food samples, and feces from randomly selected hospitalized children were collected and examined for stx1 and stx2 genes by using a PCR technique. Our objectives were (i) to estimate the rate of prevalence of STEC in the environment (cattle and food) in a region of France with both rural and urban populations; (ii) to characterize the strains by means of their biochemical and antimicrobial susceptibility patterns, virulence genes, and serotype; and (iii) to compare them to strains associated with severe disease, isolated in the same area.
MATERIALS AND METHODS
Samples.
Over a 1-year period, from October 1997 to September 1998, a total of 2,143 samples were collected from animals, food, and children. The samples were collected in the same geographical area, the Auvergne region (central France), including the city of Clermont-Ferrand (260,000 inhabitants). The number of samples to be tested monthly, in order to obtain statistically significant data, was estimated according to the literature (3, 16, 32). Bovine feces (about 40 samples per month) were obtained from 471 healthy cattle, 1 to 18 years old, at the city slaughterhouse. Data regarding the age, sex, breed, and geographical origin of each animal were collected. Food samples consisted of 411 beef samples (35/month) and 603 cheeses (50/month) purchased from local retail stores or obtained from the Laboratoire Départemental d'Analyses Vétérinaires et Biologiques, one of the public laboratories involved in controlling food safety. About 55 stool samples per month were collected from randomly selected children (one out of three admissions), with or without diarrhea, hospitalized at the Clermont-Ferrand Teaching Hospital. Data were collected by analysis of medical records and included age, sex, geographical origin, urban or rural origin, length of stay, cause of admission, and presence of diarrhea and of enteric pathogens in the feces. The bacterial pathogens Salmonella spp., Shigella spp., Aeromonas spp., Vibrio cholerae, and Campylobacter, as well as Candida albicans, Entamoeba histolytica, Giardia intestinalis, rotavirus, and enterovirus were assayed by standard methods. Samples were obtained from 658 children (304 female), aged between 1 month and 15 years (mean, 19 months). The age distribution was as follows: 1 to 6 months, 287; 7 to 12 months, 126; 1 to 5 years, 179; 5 to 10 years, 38; 10 to 15 years, 28. Of the 641 children for whom information was available, 611 (92%) came from the Auvergne region and 400 (62%) had an urban way of life. The mean length of stay was 5 days (from 1 to 70 days). Samples were stored for less than 5 days at 4°C before being tested. The reference strains used in this study were E. coli EDL933, an O157:H7 serotype (ATCC 43895) provided by A. O'Brien (26), and E. coli K-12 C600. Six non-O157:H7 STEC isolates associated with HUS, and previously described by Bonnet et al. (5), were also used for comparison with environmental strains from our collection.
Sample preparation and multiplex PCR.
All 2,143 samples were screened for STEC by PCR. Fecal samples (200 mg) were inoculated into 10 ml of MacConkey broth (Difco Laboratories, Detroit, Mich.) and incubated at 37°C for 8 h. Preenriched food samples were prepared by homogenizing a 10-g portion of meat or cheese in 90 ml of MacConkey broth and then incubating the preparations at 37°C overnight. Following incubation, 5 μl of the cultures were streaked out on Drigalski lactose agar plates (Biokar Diagnostics, Beauvais, France) and then incubated at 37°C overnight. A loopful of colonies grown on Drigalski agar was suspended in 200 μl of sterile water and incubated at 100°C for 15 min. Following centrifugation of the lysate, 5 μl of the supernatant was used as a template for the multiplex PCR. Primers specific for stx1 and stx2 are shown in Table 1. The PCR cycle included denaturation for 90 s at 94°C, primer annealing for 90 s at 55°C, and extension for 120 s at 72°C (30 cycles) in a Perkin-Elmer Cetus DNA thermal cycler 2400. Each of the primers was used at 0.5 μM, with 200 μM each deoxynucleotide triphosphate (Boehringer Mannheim, Meyher, France), 1× reaction buffer, 2.5 mM MgCl2, and 2 U of Taq DNA polymerase (Appligène-oncor, Illkrich, France). The reaction products were then analyzed by electrophoresis on 2% agarose gels with 1% ethidium bromide (ProLabo, Strasbourg, France). The expected product sizes are given in Table 1. DNA from the reference strain, E. coli EDL933, and a reagent blank, which contained all components except the template DNA, were included as positive and negative controls, respectively. For each stx PCR-positive sample, and in order to isolate the STEC strain for further characterization, 10 colonies were individually subcultured in 10 ml of Müller-Hinton Broth (Biokar Diagnostics). Lysates were prepared and tested by PCR as described above. The sensitivity of the protocol was assessed using either artificially contaminated (with the O157:H7 strain EDL933) or naturally contaminated samples. The number of STEC organisms in these samples was determined by colony hybridization with the stx1- and stx2-specific probes, with the samples serially diluted and tested following the procedure described above.
TABLE 1.
Primer sequences used in PCR to amplify specific fragments from stx1, stx2, and ehxA genes
| Target | Primer | Oligonucleotide sequence (5′-3′) | Location (bp) within the gene | Size of amplified product (bp) | Reference |
|---|---|---|---|---|---|
| stx1 | VT1c | ACC CTG TAA CGA AGT TTG CG | 31–70 | 140 | 33 |
| VT1d | ATC TCA TGC GAC TAC TTG AC | ||||
| stx2 | LP43 | ATC CTA TTC CCG GGA GTT TAC G | 57–643 | 587 | 9 |
| LP44 | GCG TCA TCG TAT ACA CAG GAG C | ||||
| ehxA | RH35 | CAC ACG GAG CTT ATA ATA TTC TGT CA | 1645–1965 | 321 | 17 |
| RH37 | AAT GTT ATC CCA TTG ACA TCA TTT GAC T |
Hybridization experiments and detection of eae and ehxA.
The identity of the PCR products was confirmed by Southern hybridization after transfer to Hybond N+ nylon membranes (Amersham International, Amersham, United Kingdom), using standard methods and the stx1 and stx2 PCR products of the EDL 933 strain as DNA probes. The PCR products were purified from the agarose gel with a 0.22-μm filter (SPIN-X; Costar, Cambridge, Mass.), and radiolabeled with [32P]dATP (Amersham) using the random-primed DNA labeling kit (Boehringer Mannheim) according to the manufacturer's specifications. The filters were hybridized using a rapid hybridization buffer (Amersham International) as described by the manufacturer, and hybridized filters were exposed to Hyperfilm MP (Amersham International). The eae and ehxA genes were detected by colony blot hybridization following the classic procedure of Maas (23). A 1.4-kb fragment obtained from strain EDL933 and covering the entire eae open reading frame was labeled as described above. The probe used for the detection of ehxA was a 321-bp fragment obtained by PCR, using primers shown in Table 1.
Bacterial strain identification and characterization.
stx PCR-positive isolates were biochemically confirmed to be E. coli by using an API ID32E test (BioMérieux, Marcy-L'Etoile, France). In order to check sorbitol and lactose fermentation, strains were cultured on sorbitol MacConkey agar and on Drigalski lactose agar plates (Oxoid, Hampshire, England, and Biokar Diagnostics, Beauvais, France), respectively. β-d-Glucuronidase activity was assessed using PGUA agar plates (article no. 722; SSI, Copenhagen, Denmark) according to the manufacturer's instructions. The susceptibilities to 32 antimicrobial agents were determined by the disk diffusion method on Müller-Hinton agar (BioMérieux), with disks purchased from Sanofi Diagnostic Pasteur (Marnes La Coquette, France), and interpreted according to the recommended French standards (11). Serotype determination was conducted at The International Escherichia and Klebsiella Reference Centre (WHO) in Copenhagen, Denmark. Serotyping was done by agglutination in microtiter plates and tubes with O and H antisera against O groups O1 to O173 and temporary O antigens OX3 and OX7, plus six putative new O antigens (OX176 through OX181) and 56 H antigens, using the methods described by Orskov and Orskov (27).
Vero cell assay.
The production of Shiga toxin by stx PCR-positive strains was checked by a cytotoxicity assay on Vero cells (20). Vero cells (African green monkey kidney cells; ATCC CRL 1587) were grown at 37°C in EMEM (Seromed, Berlin, Germany) supplemented with 10% fetal calf serum (Seromed), 1% l-glutamine (Life Technologies, Paisley, Scotland), 100,000 U of penicillin per liter, 100 mg of streptomycin per liter, 25 μg of amphotericin B per liter, and 1% minimal essential medium vitamin solution (Life Technologies) in an atmosphere of 5% CO2. The bacterial strains were inoculated into 10 ml of brain heart infusion broth (Biokar Diagnostics) and incubated at 37°C overnight. After centrifugation at 12,000 × g for 5 min, supernatant filtrates were obtained with a 0.45-μm-pore-size filter (PolyLabo, Molsheim, France) and screened for verocytotoxicity. Twofold serial dilutions of bacterial filtrates were done in 96-well flat-bottom microtiter plates (Nunc, Roskilde, Denmark) (100 μl per well; 12 wells per strain; dilutions from 1/2 to 1/2,048). A total of 100 μl of EMEM containing 105 Vero cells in suspension was added to each well. The culture plates were incubated for 24 h at 37°C in a 5% CO2 atmosphere. After 24 h, the cell monolayers were washed with phosphate-buffered saline (pH 7.2) (Seromed) and stained with a crystal violet solution (1.3% crystal violet–5% ethanol in phosphate-buffered saline). The verotoxin titer was expressed as the reciprocal of the highest sample dilution of culture filtrate which caused 50% cell detachment after 24 h of incubation, as judged by the dye intensity and by microscopic observation. E. coli K-12 C600 was used as a negative control.
Statistical methods.
The data were analyzed by the χ2 test, except for the variables needing a two-tailed Fisher exact test. Continuous variables were compared by the Kruskal-Wallis test with Epi-Info version 6.02 software. A P value of <0.05 was considered statistically significant.
RESULTS
Prevalence of STEC in children, healthy cattle, and food.
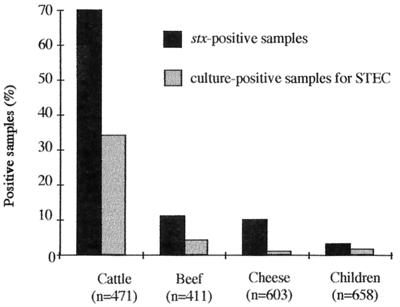
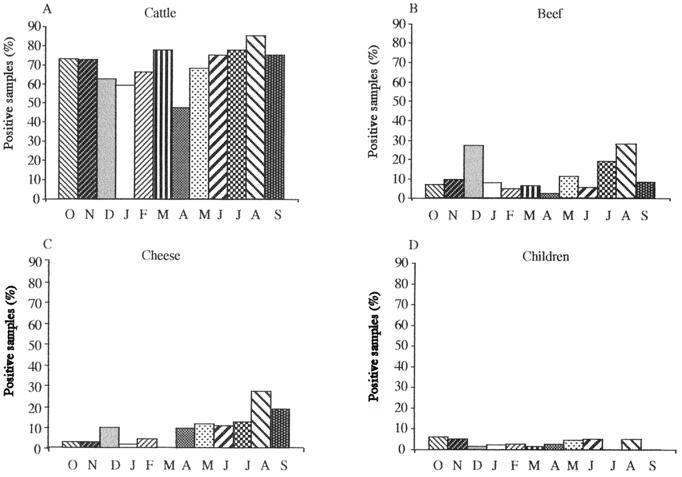
A total of 2,143 samples were investigated for Shiga toxin-encoding genes, using a multiplex PCR method and Southern hybridization with stx-specific probes. Fecal samples were collected from 471 healthy cattle at the Clermont-Ferrand slaughterhouse. The animals originated from different farms located throughout the country, but the majority of them (431 of 471; 91.5%) came from central France. Among the 471 animals tested, 330 (70%) had fecal samples positive for the stx gene(s) (Fig. 1). A significant seasonal shedding of STEC by cattle was observed, with higher rates in August (P < 0.05) (Fig. 2). In contrast, we could not demonstrate any marked variation between the prevalence of stx genes and the geographical origin, age, breed, or sex of the animals. Among the 411 beef samples and 603 cheese samples tested, 11% (47 of 411) and 10% (60 of 603), respectively, were found positive for the presence of the stx gene(s) (Fig. 1). Again, a seasonal pattern was observed in food samples, with higher rates in August for both food types (P < 0.005) and a peak in December for beef samples (P < 0.05) (Fig. 2). Finally, stool samples were obtained from 658 hospitalized children, among whom 255 (39%) had diarrhea. Overall, stx genes were found in 19 (3%) of the 658 stool specimens. Only 8 children out of 19 with stx-positive stools had diarrhea, and in 5 of them, an enteric pathogen other than STEC could be isolated, indicating that only 3 of the cases might be attributed to STEC. No significant association was found between an stx-positive PCR sample and diarrheal disease, and no seasonal variation could be observed (Fig. 2).
FIG. 1.
Prevalence of stx-positive samples and culture-positive samples for STEC, according to the origin. stx-positive samples were determined by using PCR and hybridization with stx-specific probes. The culture-positive samples represent stx-positive samples from which a STEC strain was isolated.
FIG. 2.
Distribution of stx-positive samples relative to the month (represented by initial letters; from October 1997 [O] to September 1998 [S]) and to the origin. (A) Samples collected from cattle (40 samples per month); (B) beef samples (35 per month); (C) cheese samples (50 per month); (D) children's stools (55 per month).
In order to isolate the STEC strains for further characterization, 10 individual colonies from each PCR-positive sample were submitted to a second round of PCR. A STEC isolate was obtained in 34% (162 of 471) of bovine feces, 4% (16 of 411) of beef samples, 1% (5 of 603) of cheese, and 1.5% (10 of 658) of stool specimens from children, which represented from 10 to 50% of the PCR-positive samples (Fig. 1). Two different STEC strains (as determined by genotype and serotype) were isolated from the same sample for 2 meat, 1 cheese and 24 bovine feces samples. We were thus able to establish a collection of 220 STEC strains, including 186 strains isolated from cattle, 18 from beef, 6 from cheese, and 10 from children, all of them isolated in the same geographical area.
Detection of stx1, stx2, eae, and ehxA sequences and significant associations.
Multiplex PCR and Southern blot hybridization demonstrated that 34 (15%) of the 220 STEC strains carried the stx1 gene, 116 (53%) possessed the stx2 gene, and 70 (32%) carried both stx1 and stx2 genes (Table 2). Interestingly, stx1 strains were more prevalent in STEC isolates obtained from children than in those from food or cows (70% versus 37.5 and 10%, respectively) (P < 0.0001), whereas stx2 strains were more commonly recovered from cattle and meat samples than from children and cheese samples (57 and 50% versus 20 and 0%, respectively). Finally, stx1- and stx2-positive strains were more frequently isolated from cheese (67%) than from cows (33%), meat samples (11%), and children (10%). The 220 STEC strains were investigated for the presence of the eae and ehxA genes by colony blot hybridization with specific DNA probes. Only 12 (5%) of the 220 STEC strains were positive for the eae gene. The eae gene was significantly more frequent among STEC strains isolated from children (30%) than in those from cattle (5%) or food samples (0%) (P < 0.05) (Table 2). Furthermore, sequences homologous to eae were significantly more frequent among stx1-positive strains (P < 0.05): 9 of the 34 strains which were positive for the stx1 gene (26%) also reacted with the eae-specific gene probe. Only two of the 116 stx2-positive strains (1.7%) and one of the 70 stx1- and stx2-positive strains (1.4%), which was identified as an O157:H7 isolate, were positive for the eae gene. Among the 220 STEC strains, 102 (46%) harbored sequences homologous to ehxA. Similar proportions of ehxA-positive strains were observed in samples from cattle (48%), beef (33%), cheese (50%), and children (30%) (Table 2). The distribution of the different genotypes is given in Table 3. The predominant genotype among the STEC strains was the stx2 genotype, accounting for 86 of the 220 (39%) isolates tested. The stx1-stx2-ehxA and the stx2-ehxA genotypes were encountered in 54 (24%) and 28 (13%) of the isolates, respectively. As for the global associations between the stx, eae, and ehxA genes, 53% (117 of 220) of the 220 STEC strains were stx positive only, 41% (91 of 220) were stx and ehxA positive, 0.5% (1 of 220) were stx and eae positive, and 5.5% (11 of 220) were stx, eae, and ehxA positive. These data indicated that the eae gene was significantly associated with the ehxA gene (P < 0.005).
TABLE 2.
Distribution of virulence factor-encoding genes in the 220 STEC strains according to the origin of the isolates
| Gene | Distributionc
|
||||
|---|---|---|---|---|---|
| Overall (n = 220) | Cattle (n = 186) | Beef (n = 18) | Cheese (n = 6) | Children (n = 10) | |
| stx1a | 34 (15) | 18 (10) | 7 (39) | 2 (33) | 7 (70) |
| stx2a | 116 (53) | 106 (57) | 9 (50) | 0 (0) | 2 (20) |
| stx1 + stx2a | 70 (32) | 62 (33) | 2 (11) | 4 (67) | 1 (10) |
| eaeb | 12 (5) | 9 (5) | 0 (0) | 0 (0) | 3 (30) |
| ehxAb | 102 (46) | 90 (48) | 6 (33) | 3 (50) | 3 (30) |
Number of stx-positive isolates, by PCR and hybridization.
Number of eae- and ehxA-positive isolates, by colony blot hybridization.
The number in parentheses indicates the percentage of the category positive for the respective genes.
TABLE 3.
Association within virulence factors and distribution according to the origin of the 220 STEC strains collected during this study
| Virulence factor genotypea | Distributionb
|
||||
|---|---|---|---|---|---|
| Overall (n = 220) | Cattle (n = 186) | Beef (n = 18) | Cheese (n = 6) | Children (n = 10) | |
| stx1 | 16 (7) | 4 (2) | 7 (39) | 2 (33) | 3 (30) |
| stx2 | 86 (39) | 79 (42.5) | 5 (28) | 0 (0) | 2 (20) |
| stx1-stx2 | 15 (7) | 13 (7) | 0 (0) | 1 (17) | 1 (10) |
| stx1-ehxA | 9 (4) | 8 (4.5) | 0 (0) | 0 (0) | 1 (10) |
| stx2-ehxA | 28 (13) | 24 (13) | 4 (22) | 0 (0) | 0 (0) |
| stx1-stx2-ehxA | 54 (24) | 49 (26.5) | 2 (11) | 3 (50) | 0 (0) |
| stx1-eae | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) | 1 (10) |
| stx1-eae-ehxA | 8 (4) | 6 (3) | 0 (0) | 0 (0) | 2 (20) |
| stx2-eae-ehxA | 2 (1) | 2 (1) | 0 (0) | 0 (0) | 0 (0) |
| stx1-stx2-eae-ehxA | 1 (0.5) | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) |
The number of stx-positive isolates was obtained by PCR and hybridization, and the numbers of eae- and ehxA-positive isolates were obtained by colony blot hybridization.
The number in parentheses indicates the percentage of positive isolates for the respective association.
Biochemical and antimicrobial susceptibility patterns.
Biochemical profiles were determined for the 220 STEC strains, and a total of 60 different biochemical patterns were distinguished. More than 12% (27 of 220) of the strains did not ferment sorbitol, and 3% (7 of 220) were β-glucuronidase negative. However, only 2 of the 220 strains (1%) were both sorbitol and β-glucuronidase negative: the only O157:H7 isolate of this study and one O49:H− strain (see below). There was no significant association between biochemical characteristics and the origins of the strains. The antibiotic resistance patterns of the 220 isolates were determined. The great majority of them (91%) were sensitive to the 32 antibiotics tested; 20 (9%) were resistant to at least one antibiotic. The antibiotics for which resistance was most frequently observed were sulfonamides (17 isolates), tetracycline (14 isolates), amoxicillin (6 strains, isolated from children and food samples but not from bovine feces), kanamycin (5 isolates), and chloramphenicol (3 isolates). A striking association was established between antibiotic resistance and the origin of the strains: only 4% (8 of 186) of the strains isolated from cows were resistant to at least one antibiotic compared to 33% (8 of 24) of the strains isolated from food samples and 40% (4 of 10) of the strains isolated from children (P < 0.01).
Cytotoxicity on Vero cells.
In order to determine what proportion of the 220 stx-positive strains was indeed producing verotoxin, culture supernatants were tested for cytotoxicity in the Vero cell assay. The verotoxin titer was expressed as the reciprocal of the highest filtrate dilution which caused 50% cell detachment after 24 h of incubation. In our hands, the breakpoint for a positive result was a titer of 4, and all the HUS-associated controls gave titers of >64 (data not shown). On the basis of two independent experiments, the strains were classified into three categories: not cytotoxic for Vero cells (titer, ≤2; 32 isolates [14.5%]), moderately cytotoxic (titer, from 4 to 32; 68 isolates [31%]), and highly cytotoxic (titer, ≥64; 120 isolates [54.5%]). A significant association was observed between highly cytotoxic isolates and the presence of the stx2 gene (P < 0.001) and/or the presence of the enterohemolysin-encoding gene, ehxA (P < 0.001). Finally, the six strains isolated from HUS by Bonnet et al. (5) showed a very high degree of cytotoxicity and were more cytotoxic than their bovine counterparts of identical serotype (data not shown).
Serotyping of STEC isolates.
The O antigen was typeable for 203 of the 220 STEC strains, which could be classified into 58 different serogroups. A total of 97 of the 203 typeable strains (48%) belonged to six serogroups, namely, OX3 (n = 26), O113 (n = 26), O22 (n = 13), O91 (n = 12), O172 (n = 10), and O6 (n = 10) (Table 4). The strains belonging to these six serogroups were isolated from cattle (86 isolates) and food samples (11 isolates), but the strains from children belonged to completely distinct O groups. In terms of flagellar types, 22 isolates were not motile (H−) and 177 isolates were typeable.
TABLE 4.
Distribution within O serogroups and origin of the 198 typeable STEC isolates
| Serogroupa | Total no. of isolates | No. of isolates obtained from:
|
|||
|---|---|---|---|---|---|
| Cattle | Beef | Cheese | Children | ||
| OX3** | 26 | 24 | 1 | 1 | |
| O113* | 26 | 23 | 2 | 1 | |
| O22* | 13 | 12 | 1 | ||
| O91** | 12 | 9 | 2 | 1 | |
| O172 | 10 | 9 | 1 | ||
| O6** | 10 | 9 | 1 | ||
| OX178 | 8 | 8 | |||
| O171* | 7 | 7 | |||
| O46 | 7 | 7 | |||
| O74 | 5 | 5 | |||
| O8* | 4 | 3 | 1 | ||
| O112ac* | 4 | 3 | 1 | ||
| O109 | 4 | 4 | |||
| O105* | 4 | 4 | |||
| O15* | 3 | 2 | 1 | ||
| O39 | 3 | 2 | 1 | ||
| O23 | 3 | 3 | |||
| OX7 | 3 | 3 | |||
| O116 | 3 | 3 | |||
| O96 | 2 | 2 | |||
| O132 | 2 | 2 | |||
| O1* | 2 | 2 | |||
| O2* | 2 | 2 | |||
| O40 | 2 | 2 | |||
| O49* | 2 | 1 | 1 | ||
| O103* | 2 | 1 | 1 | ||
| O141 | 2 | 1 | 1 | ||
| O150 | 2 | 1 | 1 | ||
Other O serogroups isolated once: O20, O77, O84, O87, O88, O98*, O102, O106, O117*, O120, O130, O136, O140, O157*, O159, O163*, “O”K84, and OX177 from cattle; O18ac, O25*, O54, O70, O75*, O128ab*, O153*, and O168 from beef samples; and O26*, O76, O83, and O127 from children's stools. *, serogroups known to be associated with severe disease (4, 21, 40); **, serogroups known to be associated with severe disease and isolated from HUS patients in the same area (5).
A total of 73 different O:H serotypes were identified, accounting for 186 strains, with 34 strains expressing an untypeable O and/or H antigen. A total of 102 out of the 186 typeable isolates (55%) belonged to 13 serotypes, namely, OX3:H2, O113:H21, O113:H4, OX3:H21, O6:H10, OX178:H19, O171:H2, O46:H38, O172:H21, O22:H16, O91:H10, O91:H21, and O22:H8. They were isolated from cattle (93 isolates) and food samples (9 isolates) but not from children. Fifteen serotypes included between two and four isolates each, and 45 serotypes accounted for one isolate each (Table 5). Only one STEC strain, isolated from a cow in September 1998, belonged to the O157:H7 serotype, which represents 0.5% of the STEC organisms isolated from bovine feces. In all, 62 isolates (34%) belonged to Shiga-toxigenic E. coli serotypes previously associated with severe disease in humans (Table 5) (4, 5, 21, 40).
TABLE 5.
Characteristics of the most prevalent serotypes among the STEC strains collected during this study
| Serotypea | Total no. of isolates | No. of isolates from:
|
Characteristics
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cattle | Beef | Cheese | Children | stx1b | stx2b | eaec | ehxAc | Sord | PGUAe | ||
| OX3:H2* | 13 | 11 | 1 | + | + | − | + | + | + | ||
| 1 | + | − | − | − | + | + | |||||
| O113:H21* | 12 | 6 | − | + | − | − | + | + | |||
| 4 | − | + | − | + | + | + | |||||
| 1 | + | + | − | + | + | + | |||||
| 1 | + | + | − | − | + | + | |||||
| O113:H4 | 9 | 6 | 2 | − | + | − | − | + | + | ||
| 1 | − | + | − | − | − | + | |||||
| OX3:H21* | 9 | 9 | − | + | − | − | + | + | |||
| O6:H10 | 8 | 6 | − | + | − | − | + | + | |||
| 1 | − | + | − | − | + | − | |||||
| 1 | + | − | − | − | + | + | |||||
| OX178:H19 | 8 | 6 | + | + | − | + | + | + | |||
| 2 | − | + | − | − | + | + | |||||
| O171:H2 | 7 | 5 | − | + | − | − | + | + | |||
| 2 | − | + | − | − | − | + | |||||
| O46:H38 | 7 | 7 | + | + | − | + | + | + | |||
| O172:H21 | 6 | 5 | + | + | − | − | − | + | |||
| 1 | − | + | − | − | − | + | |||||
| O22:H16 | 6 | 2 | 1 | − | + | − | + | + | + | ||
| 2 | + | + | − | − | + | + | |||||
| 1 | − | + | − | − | + | + | |||||
| O91:H10** | 6 | 6 | − | + | − | − | + | + | |||
| O91:H21** | 6 | 3 | 2 | − | + | − | + | + | + | ||
| 1 | + | + | − | + | + | + | |||||
| O22:H8* | 5 | 4 | − | + | − | − | + | + | |||
| 1 | − | + | − | + | + | + | |||||
| O109:H− | 4 | 2 | + | − | − | + | + | + | |||
| 1 | + | − | − | + | + | − | |||||
| 1 | + | + | − | + | + | + | |||||
| O74:H42 | 4 | 4 | + | + | − | + | + | + | |||
| O113:H− | 3 | 3 | − | + | − | − | + | + | |||
| O105:H18* | 3 | 3 | + | + | − | + | + | + | |||
| O112ac:H19 | 3 | 1 | 1 | − | + | − | − | + | + | ||
| 1 | + | + | − | + | + | + | |||||
| OX7:H16 | 3 | 3 | − | + | − | − | − | + | |||
| O8:H19 | 3 | 3 | + | + | − | + | + | + | |||
| O116:H28 | 2 | 2 | + | − | − | + | + | + | |||
| O132:H18 | 2 | 2 | − | + | − | − | + | + | |||
| O15:H16 | 2 | 2 | − | + | − | − | + | + | |||
| O22:H− | 2 | 1 | − | + | − | − | + | + | |||
| 1 | − | + | − | + | + | + | |||||
| O23:H15 | 2 | 2 | + | + | − | + | + | + | |||
| O40:H21 | 2 | 1 | + | + | − | + | + | + | |||
| 1 | − | + | − | + | + | + | |||||
| O49:H−* | 2 | 1 | − | + | + | + | − | − | |||
| 1 | + | − | + | + | − | + | |||||
| O96:H19 | 2 | 1 | + | + | − | − | − | + | |||
| 1 | − | + | − | + | − | + | |||||
*, serotypes known to be associated with severe disease (4, 21, 40); **, serotypes known to be associated with severe disease and isolated from HUS patients in the same area (5). Other serotypes isolated once: O1:H18, O1:H20, O2:H27, O2:H45, O6:H49, O20:H16, O39:H8, O39:H−, O77:H18, O84:H− O87:H16, O88:H21, O98:H−*, O102:H21, O103:H2*, O106:H42, O116:H21, O117:H7, O130:H43, O136:H12, O140:H32, O141:H−, O150:H−, O157:H7*, O159:H28, O163:H19*, “O”K84:H19, OX3:H43, OX3:H49, OX3:H−**, and OX177:H− from cattle; O25:H21, O54:H2, O75:H8, O128ab:H2*, O150:H8, O153:H8, O168:H8, and O172:H16 from beef samples; O8:H9, O15:H45, and O103:H14 from cheese samples; and O26:H21, O76:H19, and O141:H8 from children's stools.
Multiplex PCR results confirmed by hybridization for the stx genes. −, negative; +, positive.
Colony blot hybridization results for the eae and ehxA genes. −, negative; +, positive.
Sorbitol fermentation. −, negative; +, positive.
β-Glucuronidase production. −, negative; +, positive.
Associations among origin, serotype, and virulence factors (stx1, stx2, eae, and ehxA).
For strains belonging to the 28 serotypes that include more than one isolate (141 strains), the genotypic and biochemical characteristics, according to serotype and origin, are given in Table 5. The stx1 gene was evenly distributed among the 141 strains regardless of their serotypes. The stx2 gene was present in the great majority (133) of the 141 strains. In contrast, the eae gene was absent from the strains belonging to the dominant O:H serotypes, except for two O49:H− isolates. The 10 other strains which harbored eae sequences belonged to serotypes isolated once: O26:H21, O84:H−, O98:H−, O103:H2, O127:H+, O150:H−, O157:H7, OX177:H−, Orough:H+, and Orough:H− (data not shown). As for the biochemical characteristics, the absence of sorbitol fermentation was mainly associated with serotypes O172:H21, OX7:H16, O49:H−, and O96:H19, whereas the absence of β-glucuronidase production was not serotype related. From the data in Table 5, it can be noted that only 9 strains out of 24 from food belonged to the most prevalent serotypes (3 of them differing from bovine strains by virulence gene patterns). Also, the strains from children clearly differ from strains isolated from cattle. Finally, all the strains belonging to serotype O91:H21 except one (n = 6) were stx2 and ehxA positive and all the O91:H10 isolates (n = 6) and the OX3:H− strain were only stx2 positive; none of them harbored sequences homologous to eae (Table 5). These characteristics were identical to those of the strains isolated by Bonnet et al. from HUS in the same geographical area (5).
DISCUSSION
Since very sparse data were available regarding the prevalence of STEC in France, the first aim of this study was to determine the frequency of STEC carriers in healthy cattle and in children, together with the occurrence of STEC in the food supply in central France. We used a preenrichment phase, followed by plating and testing of a set of colonies by PCR, confirmed by Southern hybridization with stx1- and stx2-specific probes. Over 70% of the bovine feces, 11% of the beef samples, and 10% of the cheese tested gave positive results. These figures are rather high compared to the available data from the literature: using PCR, stx genes were found in 24.6% of cattle feces in a recent study in Australia (15), in 46.5% of bovine fecal samples in Canada (34), and in 4.6% of raw meat samples in a survey in Belgium (30). This discrepancy might be due to the high sensitivity of our technique, since we were able to detect as few as 10 STEC organisms in each type of sample (i.e., 10 g of cheese, 10 g of beef, and approximately 200 mg of bovine or human feces). By testing 10 individual colonies, a STEC strain could be isolated in approximately half of the PCR-positive bovine feces samples, beef samples, and stool specimens from hospitalized children (Fig. 1). In several studies based on similar protocols applied to food, bovine feces, or human stools, 40 to 80% of the samples PCR positive for stx genes were culture positive for STEC (10, 24, 28, 30, 31, 34). These results were attributed to the high sensitivity of the PCR technique, which could detect stx genes even in samples where nonpathogenic E. coli was by far dominant (31). In our hands, a STEC strain could be isolated from only 10% of the PCR-positive cheese samples (Fig. 1), indicating that recovering STEC from cheese is even more difficult, either because bacteria occur in very low numbers in cheese or because bacteria are subjected to stress during the cheese-making process.
Overall, stx genes were found in 19 out of 658 (3%) stool specimens from randomly selected hospitalized children, and no significant association could be established between a stx-positive PCR and diarrheal disease. Of the 10 children from whom a STEC was isolated, only three had had symptoms of diarrhea in the 10 days before admission. In each of the three cases, an enteric pathogen (either rotavirus or Salmonella) was isolated from the stool samples. The 10 STEC strains isolated from children might result from prolonged carriage of pathogenic strains after resolution of symptoms, as previously described for O157:H7 strains (16). However, it is also possible that the STEC organisms present in the children in this study were not responsible for diarrheal disease; they might represent healthy carriage, which could be evaluated at 1.5% (10 children of 658 tested) for children under 15. For comparison, stx genes were found in 1.02% of 17,296 fecal samples submitted for routine culture in Belgium (31). This raises questions about the usefulness of PCR assay in identifying diarrheal illness caused by STEC and underlines the necessity to check for Shiga toxin production by STEC isolated from human stools in order to assess their role in pathogenicity (19).
Most of the isolates (66%) belonged to Shiga-toxigenic E. coli serotypes that had not been previously associated with severe disease in humans (Table 5) (4, 5, 21). Very few strains belonging to the major serogroups associated with pathogenicity (i.e., O157, O111, O26, and O103) are present in bovine feces and in food samples in France: only one STEC strain, isolated from a cow in September 1998, belonged to the high-virulence serotype O157:H7, which represents 0.5% of the 186 strains isolated from cattle. For comparison, U.S. surveys indicated that 1.5 to 5.3% of dairy calves and 1.6% of feedlot cattle were positive for E. coli O157:H7 (13, 42). The lower percentage we found (0.5%) is consistent with surveys done using protocols similar to ours in Spain and Germany (2, 25), indicating that E. coli O157:H7 might be less frequently shed by cattle in continental Europe than in England or in North America. However, the O157 prevalence rate in these studies is probably underestimated compared with studies using an immunomagnetic separation procedure with magnetic beads coated with anti-O157 lipopolysaccharide antibodies: in such a survey, Heuvelink et al. (18) isolated E. coli O157 in 57 (10.6%) of 540 adult cattle in The Netherlands.
Recently, STEC strains belonging to serogroups O91, OX3, and O6 were associated with a cluster of HUS cases in adult patients in central France (5). The second major aim of our study was to determine if STEC strains from the environment (bovine feces sampled at the local slaughterhouse or food samples purchased in local stores) had the same characteristics as these HUS isolates. The O:H type and virulence genes were used as markers for clonality, to check whether pathogenic STEC isolates form a population different from those found in the bovine reservoir. Our data indicate that the HUS isolates from 1996 and 1997 do not belong to the serotypes most frequently isolated during the study (OX3:H2, O113:H21, O113:H4, and OX3:H21 [Table 5]). However, 12 STEC strains among the 186 typeable isolates (7%) had characteristics identical to those of these HUS-associated strains: six O91:H10 stx2-positive isolates from cattle, five O91:H21 stx2-ehxA-positive isolates (three from cattle and two from meat), and one OX3:H− stx2-positive isolate from bovine feces. The latter could be a nonmotile variant of an OX3:H21 strain, since they share the stx2 genotype. Further studies at DNA level, involving ribotyping and pulsed-field gel electrophoresis, will be necessary to compare the OX3:H− isolate from HUS and the OX3:H21 isolates. Finally, we did not identify any O6:H4 isolate during this study. From the data obtained here, it can be inferred that the HUS STEC strains characterized by Bonnet et al. (5) form a particular subset of the STEC population found in the bovine reservoir in the same geographical area. Other studies have found the same serotypes from cases of HUS, indicating that common specific virulence properties are present in these strains.
Several authors have underlined the strong association between the carriage of eae and the capacity of STEC strains to cause severe human disease (4, 29). Three (30%) of the strains isolated from children, compared to none of the 24 STEC strains isolated from food and only 9 (5%) of the 186 bovine strains, harbored sequences homologous to eae. It is interesting to note, however, that the three eae strains isolated from children were not associated with diarrhea, perhaps because the intimin is not functional, as previously described for some bovine isolates by Wieler et al. (39), or because the children had been immunized due to previous exposure to eae-positive E. coli infection. Another possibility is that, in some patients, intimin is involved in human gut colonization but not directly in pathogenicity. Indeed, we observed a paradoxical situation with eae-positive nonpathogenic STEC isolates and eae-negative isolates associated with HUS in adults (this study and reference 5). The ehxA gene was evenly distributed among cattle (48%), food (37.5%), and isolates from children (30%). Also, it was found in two (30%) of the six non-O157:H7 strains associated with HUS in the Clermont-Ferrand area (5). Taken together, these data suggest that the enterohemolysin might not be a virulence marker for non-O157:H7 STEC. Our data underlined a significant association between the eae and ehxA genes (P < 0.005). As suggested by Boerlin et al. (4), the association of enterohemolysin with severe disease could be due to confounding effects of the major virulence factor, intimin. Finally, a significant association was observed between highly cytotoxic isolates and the presence of the stx2 gene (P < 0.001), which could explain the capacity of stx2-positive STEC strains to cause severe human disease. In our hands, the pathogenic strains showed a very high degree of cytotoxicity and were more cytotoxic than their bovine counterparts of identical serotype.
Most of the STEC strains isolated from food samples reported in the literature belong to the major O groups found in our study (Table 4). In a survey of foodstuffs in the Seattle area, Samadpour et al. (35) identified eight typeable STEC strains from meat samples, five of which belonged to serogroups OX3 (two isolates), O113, O91, and O6 (one isolate each). In our study, these four O groups accounted for 9 (38%) of 24 isolates from meat or cheese samples and 65 (35%) of the 186 bovine isolates. Thus, some of the STEC serotypes isolated from food indeed correspond to dominant strains from the bovine reservoir and belong to common serogroups regardless of the geographical area. It is interesting to note, however, that 13 (54%) of the 24 food isolates belonged to serogroups infrequently found among bovine isolates. These isolates may have specific properties that allow them to survive in meat or cheese.
In conclusion, although a high proportion of bovine feces and food samples in the Clermont-Ferrand area were PCR positive for stx genes (70 and 10%, respectively), a STEC strain could be isolated in half of them, and 15% of the isolated STEC strains were not cytotoxic for Vero cells. The STEC strains isolated from food represent a particular subset from the bovine reservoir, and very few strains belonged to the serotypes previously reported from cases of HUS or hemorrhagic colitis. Finally, the STEC strains isolated from children in this study were probably not responsible for diarrheal disease. Further studies are needed to identify the properties that distinguish pathogenic STEC isolates from nonpathogenic STEC strains.
ACKNOWLEDGMENTS
We thank Danielle Sirot, from the Clinical Microbiology Laboratory, for help in identifying isolates.
This study was supported in part by the Ministère de l'Enseignement Supérieur et de la Recherche (EA2348), by the Ministère de l'Aménagement du Territoire et de l'Environnement (Programme Environnement Santé EN 98-17), and by a Programme Hospitalier de Recherche Clinique National 1997. N.P. received financial support from the Federation of European Microbiological Societies.
REFERENCES
- 1.Beutin L, Geier D, Steinruck H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco M, Blanco J E, Blanco J, Gonzalez E A, Mora A, Prado C, Fernandez L, Rio M, Ramos J, Alonso M P. Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol Infect. 1996;117:251–257. doi: 10.1017/s0950268800001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco M, Blanco J E, Blanco J, Mora A, Prado C, Alonso M P, Mourino M, Madrid C, Balsalobre C, Juarez A. Distribution and characterization of faecal verotoxin-producing Escherichia coli (STEC) isolated from healthy cattle. Vet Microbiol. 1997;54:309–319. doi: 10.1016/s0378-1135(96)01292-8. [DOI] [PubMed] [Google Scholar]
- 4.Boerlin P, Mcewen S A, Boerlin-Petzold F, Wilson J B, Johnson R P, Gyles C L. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet R, Souweine B, Gauthier G, Rich C, Livrelli V, Sirot J, Joly B, Forestier C. Non-O157 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J Clin Microbiol. 1998;36:1777–1780. doi: 10.1128/jcm.36.6.1777-1780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 7.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 8.Calderwood S B, Acheson D W K, Keusch G T, Barrett T J, Griffin P M, et al. Proposed new nomenclature for SLT (VT) family. ASM News. 1996;62:118–119. [Google Scholar]
- 9.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke R C, Wilson J B, Read S C, Renwick S, Rahn K, et al. Proceedings of the 2nd International Symposium and Workshop on Verocytotoxin (Shiga-like toxin)-Producing Escherichia coli Infections. Amsterdam, The Netherlands: Elsevier Science BV; 1994. Verocytotoxin-producing Escherichia coli (STEC) in the food chain: preharvest and processing perspectives. Recent advances in verocytotoxin-producing Escherichia coli infections; pp. 17–24. [Google Scholar]
- 11.Comité de l'Antibiogramme de la Société Francaise de Microbiologie. Communiqué 1997. Pathol Biol. 1997;45:I–XII. [Google Scholar]
- 12.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 13.Doyle M P, Zhao T, Meng J, Zhao S. Escherichia coli O157:H7. In: Doyle M P, Beuchat L, Montville T, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 171–191. [Google Scholar]
- 14.Dytoc M T, Ismaili A, Philpott D J, Soni R, Brunton J L, Sherman P M. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect Immun. 1994;62:3494–3505. doi: 10.1128/iai.62.8.3494-3505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagan P K, Hornitzky M A, Bettelheim K A, Djordjevic S P. Detection of Shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol. 1999;65:868–872. doi: 10.1128/aem.65.2.868-872.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–758. [Google Scholar]
- 17.Hall, R. H., and J. G. Xu. 21 December 1995. A new and distinctive DNA sequence of E. coli O157:H7 and its use for the rapid, sensitive and specific detection of O157:H7 and other enterohemorrhagic E. coli. International patent WO 95/34682.
- 18.Heuvelink A E, Van Den Biggelaar F L A M, De Boer E, Herbes R G, Melchers W J G, Huis In 'T Veld J H J, Monnens L A H. Isolation and characterisation of verocytotoxin-producing Escherichia coli O157 strains from dutch cattle and sheep. J Clin Microbiol. 1998;36:878–882. doi: 10.1128/jcm.36.4.878-882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karmali M A, Petric M, Bielaszewska M. Evaluation of a microplate latex agglutination method (Verotox-F Assay) for detecting and characterizing verotoxins (Shiga toxins) in Escherichia coli. J Clin Microbiol. 1999;37:396–399. doi: 10.1128/jcm.37.2.396-399.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic-uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 21.Keskimäki M, Ikäheimo R, Kärkkäinen P, Scheutz F, Ratiner Y, Puohiniemi R, Siitonen A. Shiga toxin-producing Escherichia coli serotype OX3:H21 as a cause of hemolytic-uremic syndrome. Clin Infect Dis. 1997;24:1278–1279. doi: 10.1086/513668. [DOI] [PubMed] [Google Scholar]
- 22.Lindgren S L, Melton A R, O'Brien A D. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect Immun. 1993;61:3832–3842. doi: 10.1128/iai.61.9.3832-3842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maas R. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid. 1983;10:296–298. doi: 10.1016/0147-619x(83)90045-8. [DOI] [PubMed] [Google Scholar]
- 24.Mariani-Kurkdjian P, Cave H, Elion J, Loirat C, Bingen E. Direct detection of verotoxin genes in stool samples by polymerase chain reaction in hemolytic uremic syndrome patients in France. Clin Microbiol Infect. 1997;3:117–119. doi: 10.1111/j.1469-0691.1997.tb00260.x. [DOI] [PubMed] [Google Scholar]
- 25.Montenegro M A, Bülte M, Trumpf T, Aleksic S, Reuter G, Bulling E, Helmuth R. Detection and characterization of fecal verotoxin-producing Escherichia coli from healthy cattle. J Clin Microbiol. 1990;28:1417–1421. doi: 10.1128/jcm.28.6.1417-1421.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien A O, Lively T A, Chen M E, Rothman S W, Formal S B. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet. 1983;1:702. doi: 10.1016/s0140-6736(83)91987-6. [DOI] [PubMed] [Google Scholar]
- 27.Orskov F, Orskov I. Serotyping of Escherichia coli. In: Bergan T, editor. Methods in microbiology. Vol. 14. London, United Kingdom: Academic Press; 1984. pp. 43–112. [Google Scholar]
- 28.Paton A W, Paton J C, Goldwater P N, Manning P A. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J Clin Microbiol. 1993;31:3063–3067. doi: 10.1128/jcm.31.11.3063-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paton J C, Paton A W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierard D, Damme L V, Moriau L, Stevens D, Lauwers S. Virulence factors of verocytotoxin-producing Escherichia coli isolated from raw meats. Appl Environ Microbiol. 1997;63:4585–4587. doi: 10.1128/aem.63.11.4585-4587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierard D, Stevens D, Moriau L, Lior H, Lawers S. Isolation and virulence factors of verocytotoxin-producing Escherichia coli in human stool samples. Clin Microbiol Infect. 1997;3:531–540. doi: 10.1111/j.1469-0691.1997.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 32.Pierard P, Vandamme L, Stevens D, Morian L, Lanwers S. Detection of verocytotoxin-producing Escherichia coli in meat in Belgium. In: Karmali M, Goglio A G, editors. Recent advances in verocytotoxin-producing Escherichia coli infections. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 78–80. [Google Scholar]
- 33.Pollard D R, Johnson W M, Loir H, Tyler S D, Rozee K D. Differentiation of Shiga toxin and Vero cytotoxin type 1 genes by polymerase chain reaction. J Infect Dis. 1990;162:1195–1198. doi: 10.1093/infdis/162.5.1195. [DOI] [PubMed] [Google Scholar]
- 34.Rahn K, Wilson J B, Mcfadden K A, Read S C, Ellis A G, Renwick S A, Clarke R C, Johnson R P. Comparison of Vero cell assay and PCR as indicators of the presence of verotoxigenic Escherichia coli in bovine and human fecal samples. Appl Environ Microbiol. 1996;62:4314–4317. doi: 10.1128/aem.62.12.4314-4317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samadpour M, Ongerth J E, Liston J, Tran N, Nguyen D, Whittam T, Wilson R A, Tarr P I. Occurrence of shiga-like toxin-producing Escherichia coli in retail fresh seafood, beef, lamb, pork, and poultry from grocery stores in Seattle, Washington. Appl Environ Microbiol. 1994;60:1038–1040. doi: 10.1128/aem.60.3.1038-1040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tozzi A E, Fisher I S T. Epidemiology of STEC infections in Europe: ENTER-NET and other data. Second International Symposium of the European Study Group on Enterohemorrhagic Escherichia coli, Brussels. Acta Clin Belg. 1999;54:34. [PubMed] [Google Scholar]
- 38.Wells J G, Shipman L D, Greene K D, Sowers E G, Green J H, et al. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J Clin Microbiol. 1991;29:985–989. doi: 10.1128/jcm.29.5.985-989.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wieler L H, Vieler E, Erpenstein C, Schlapp T, Steinrück H, Bauerfeind R, Byomi A, Baljer G. Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J Clin Microbiol. 1996;34:2980–2984. doi: 10.1128/jcm.34.12.2980-2984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. 6 September 1999, revision date. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). Report of a WHO Scientific Working Group meeting, 23–26 June 1998, Berlin, Germany. [Online.] http://www.who.int/emc-documents/zoonoses/whocsraph988c.html. [24 January 2000, last date accessed].
- 41.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhao T, Doyle M P, Shere J, Garber L. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl Environ Microbiol. 1995;61:1290–1293. doi: 10.1128/aem.61.4.1290-1293.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]