Abstract
The Cockayne syndrome B protein (CSB) is required for coupling DNA excision repair to transcription in a process known as transcription-coupled repair (TCR). Cockayne syndrome patients show UV sensitivity and severe neurodevelopmental abnormalities. CSB is a DNA-dependent ATPase of the SWI2/SNF2 family. SWI2/SNF2-like proteins are implicated in chromatin remodeling during transcription. Since chromatin structure also affects DNA repair efficiency, chromatin remodeling activities within repair are expected. Here we used purified recombinant CSB protein to investigate whether it can remodel chromatin in vitro. We show that binding of CSB to DNA results in an alteration of the DNA double-helix conformation. In addition, we find that CSB is able to remodel chromatin structure at the expense of ATP hydrolysis. Specifically, CSB can alter DNase I accessibility to reconstituted mononucleosome cores and disarrange an array of nucleosomes regularly spaced on plasmid DNA. In addition, we show that CSB interacts not only with double-stranded DNA but also directly with core histones. Finally, intact histone tails play an important role in CSB remodeling. CSB is the first repair protein found to play a direct role in modulating nucleosome structure. The relevance of this finding to the interplay between transcription and repair is discussed.
Nucleotide excison repair (NER) is a versatile DNA repair pathway that removes a wide range of DNA lesions, including the main UV light-induced lesions, i.e., cyclobutane pyrimidine dimers and 6-4 photoproducts. Approximately 30 proteins are involved in mammalian NER, which proceeds via a well-characterized, multistep reaction (for a recent review, see reference 13). One of the immediate consequences of DNA damage is blockage of transcription. To allow a rapid resumption of this vital process, a specialized repair mode that preferentially recognizes and repairs transcription-blocking lesions has evolved (3, 37). This process is known as transcription-coupled repair (TCR). Some aspects of TCR are highly conserved among Escherichia coli, yeasts, and mammals (57). Oxidative damage, mainly processed by the base excision repair pathway, is also removed in a transcription-coupled manner (9, 30). Thus, the importance of TCR is not limited to NER.
Inherited defects in TCR constitute the molecular basis of Cockayne syndrome (CS). CS patients are UV light sensitive and show severe developmental and neurological dysfunctions (4, 39). Two human genes are specifically required for TCR, CSA and CSB. These genes are defective in CS complementation groups A and B (21, 54). Several recent observations suggest that the CS proteins may have a subtle, additional role in transcription (1, 8, 44, 53, 56).
CSA specifies a five-WD-40-repeat-containing protein (21). The CSB gene encodes a nuclear protein of 168 kDa, and its central part is highly homologous to the helicase domain present in the SWI2/SNF2 family of proteins (15, 54). The recombinant CSB protein is a double-stranded DNA-dependent ATPase (activated by both naked and nucleosomal DNA) but, like other members of the SWI2/SNF2 family, is not a classical helicase (7, 11, 40, 45). Changing the invariant lysine to an arginine residue within the Walker A motif, responsible for NTP-binding and hydrolysis, abolished CSB ATPase activity but only partially affected its biological function in living cells (7). Both in vivo and in vitro it was found that CSB interacts with RNA polymerase II (RNAP II) (45, 53, 56). However, the role of CSB both in TCR and in RNAP II transcription is unknown.
Several SWI2/SNF2-related proteins are part of large multiprotein complexes involved in transcription regulation (64). In vitro studies have well documented the ability of these remodeling machines to alter chromatin structure in an ATP-dependent manner (27). Recently, three ATPases, the human Brg1 and hBrm together with the Drosophila ISWI, have been reported to exhibit chromatin remodeling activities on their own (10, 41). Brg1 and hBrm are the ATPase components of two distinct human SWI/SNF complexes (28, 61), while ISWI is part of three Drosophila remodeling complexes, ACF, CHRAC, and NURF (6). These experiments have led to the view that the ATPase subunits are the catalytic core of the remodeling machines, while the other associated proteins may have regulatory or auxiliary functions. Interestingly, the SWI2/SNF2 family also includes proteins involved in various DNA repair pathways, such as RAD 16 (global genome NER), RAD 5 (postreplication repair), RAD 54 (recombination repair), and CSB (TCR) (15). The fact that chromatin structure hinders the repair process (47) suggests that these SWI2/SNF2-related DNA repair proteins may possess a similar remodeling activity in order to provide access to damaged DNA. However, very little is known about the proteins and the molecular mechanism involved in chromatin transitions in DNA repair. In this study, we examined the involvement of CSB, as an isolated recombinant protein, in chromatin remodeling by in vitro accessibility assays on nucleosomal DNA fragments. Our results show that CSB can alter DNA conformation and is able to induce changes in chromatin structure in an ATP-dependent fashion. In addition, we demonstrate that CSB directly interacts with the core histones. We provide evidence that CSB exhibits chromatin remodeling activity, thus linking repair to chromatin remodeling.
MATERIALS AND METHODS
Proteins.
Recombinant, epitope-tagged (N-terminal hemagglutinin antigen epitope [HA] and C-terminal histidine stretch [His6]) CSB and CSBK538R mutant proteins were overexpressed using the baculovirus system and purified as described previously (7), except that a Mono Q column was substituted for the final purification step. The eluate from the Ni2+-nitrilotriacetic acid-agarose column was loaded on a Mono Q column equilibrated with buffer A (25 mM HEPES–KOH [pH 7.9], 0.05% Nonidet P-40, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride) in 0.1 M KCl, and the adsorbed proteins were eluted by a 0.1 to 1 M KCl gradient. The elution profile and purity of the CSB fractions were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining. Protein concentration was ∼20 ng/μl.
The hSWI/SNF complex was purified from HeLa cells by affinity chromatography using a FLAG epitope tag on the Ini1 subunit, and its functional characterization was performed as described previously (43). HeLa core histone octamers were purified as described previously (66). H1-depleted HeLa polynucleosomes were isolated and quantified as described previously (43).
ATPase assay.
Standard reactions (10-μl volumes) were performed as described previously (7). Plasmid DNA (300 ng), HeLa polynucleosomes (300 ng) (untreated or trypsinized) and HeLa core histones (40 ng to 400 ng) were tested as cofactors. No ATPase activity intrinsic to HeLa polynucleosomes or core histones was detected. Incubation was for 30 min at 30°C, and it was followed with separation by thin-layer chromatography. ATP hydrolysis was determined by image analysis on a PhosphorImager (Molecular Dynamics).
Topological assay on plasmid DNA.
pBlueScript II KS was singly nicked by treatment with bovine pancreatic DNase I (Boehringer Mannheim) (12) and purified by phenol-chloroform extraction and ethanol precipitation. Reactions (60-μl volumes) contained 100 ng of nicked plasmid and the indicated amounts of CSB or CSBK538R and were performed as described previously (52) except that the KCl concentration was adjusted to 60 mM. After 10 min, 1 U of E. coli DNA ligase was added and the incubation was continued for 50 min. Where not specified, reaction mixtures contained not ATP but NAD as the cofactor of the E. coli DNA ligase. Topoisomers were resolved by electrophoresis on 1% agarose gels containing 0.5 μg of chloroquine per ml. Gels were run in 1× Tris-borate-EDTA for 20 h at 70 V, followed by Southern blotting, hybridization with a pBluescript probe, and autoradiography. Two-dimensional gel electrophoresis was performed as described previously (49, 52). In a control experiment, purified CSB was incubated with closed, supercoiled plasmid DNA under the above-described experimental conditions. Reactions were analyzed on a 0.8% agarose gel. No formation of relaxed DNA was observed.
Mononucleosome assembly and DNase I accessibility assay.
Mononucleosome cores were assembled by stepwise salt dilution (23) on MluI-EcoRI DNA fragments obtained by digestion of the TPT plasmid (43). The DNA sequence includes two GT-phasing sequences (46) and was 155 bp after labeling with the Klenow fragment and [32P]dCTP. After assembly, mononucleosomes were purified on a 5 to 30% glycerol gradient as described previously (23). Ten nanograms of these tailed mononucleosomes was diluted in 100 μl of buffer V (25 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and trypsinized for 10 min at room temperature with trypsin (5 ng/μl) as described by Guyon et al. (19). Reactions were stopped with a 15-fold (wt/wt) excess of soybean trypsin inhibitor. Trypsinization was monitored by electrophoresis on a 5% nondenaturing acrylamide gel.
DNase I footprinting reactions (with 25-μl volumes) were performed as described previously (23, 43), except that 0.5 mg of bovine serum albumin (BSA) per ml was added. Where indicated, the reaction mixtures contained the nonhydrolyzable ATP analog ATPγS (Sigma) or 1 U of apyrase (Sigma) (dissolved in 20 mM HEPES [pH 7.9], 1 mM MgCl2, 1 mM DTT, 1 mM EDTA, 1 mg of BSA per ml. The KCl concentration was adjusted to 60 mM in all reaction mixtures. After incubation at 30°C for 45 min, followed by incubation for 5 min at room temperature, the reaction mixtures were subjected to digestion with DNase I. Tailed mononucleosomes were digested with 0.1 U of DNase I; tailless monunucleosomes were digested with 0.025 U of DNase I. Processing and denaturing PAGE were performed as described previously (23).
Gel-shift and histone octamer transfer reactions.
For gel-shift analysis, remodeling reaction mixtures (containing 0.3 ng of either labeled nucleosome particles or labeled naked DNA and the indicated amount of CSB) were directly loaded onto 4% polyacrylamide gels (80:1 acrylamide-to-bisacrylamide ratio) containing 12.5 mM Tris, 100 mM glycine, and 0.5 mM EDTA. Gels were run at ∼150 V for ∼2.5 h at 4°C. Where indicated, reactions were stopped by the addition of KCl (final concentration, 160 mM) and competitor DNA (2 μg of plasmid DNA, 0.5 μg of polynucleosomes) prior to loading on the gel. Octamer transfer reaction mixtures contained unlabeled HeLa polynucleosomes in excess (10 ng), the naked labeled TPT MluI-EcoRI fragment (1 ng) as a possible acceptor of core particles, and CSB (40 ng) or hSWI/SNF (200 ng) and were performed under the same conditions as for the DNase I footprinting. After 60 min at 30°C, KCl and 2 μg of plasmid DNA (as described above) were added to stop the reactions. Samples were further incubated at 30°C for 10 min and analyzed by electrophoretic mobility shift assay using 5% polyacrylamide gels (22). Product analysis was performed using a PhosphorImager. In the presence or absence of ATP, no de novo formation of core particles on the naked DNA could be detected in the CSB reactions, whereas octamer transfer was catalyzed by hSWI/SNF in an ATP-dependent manner (data not shown).
Plasmid chromatin remodeling assays.
Chromatin was assembled on the 3.35-kb pG5HC2AT plasmid (65) in Drosophila embryo extracts and subsequently treated with Sarkosyl to disrupt endogenous remodeling activities according to published protocols (58, 59). Sarkosyl and ATP were removed by gel filtration on Micro Bio-spin columns (Bio-Gel polyacrylamide P-6; Bio-Rad). Sarkosyl-treated chromatin (∼40 ng of DNA) was incubated with CSB or CSBK538R (estimated amount, 160 ng) or with hSWI/SNF complex (300 ng) for 90 min at 30°C in 70 μl of EX buffer (58) containing 60 mM KCl. Micrococcal nuclease (MNase) digestion and agarose gel electrophoresis were performed as described previously (58). Southern blotting, hybridization with 32P-labeled total plasmid DNA, and autoradiography were used to visualize the DNA fragments.
The supercoiling assay on reconstituted plasmid chromatin was performed as described previously (23). The pG5HC2AT plasmid was internally labeled and assembled into nucleosomes using purified HeLa core histones and a heat-treated Xenopus egg extract (23). A glycerol gradient-purified template (1 ng of DNA) was incubated with the indicated amounts of CSB and hSWI/SNF, and remodeling reactions were carried out in 12.5- or 25-μl volumes for 90 min at 30°C, as described previously (23). Reaction mixtures contained 60 mM KCl. Similar results were obtained when the pG5HC2AT plasmid was assembled with Drosophila embryo extracts (58) and treated with Sarkosyl and the reactions were carried out under experimental conditions that were the same as those described above for the MNase analysis.
Antibodies and IPs.
Purified CSB (500 ng) and HeLa core histones (2 μg) were incubated in vitro in buffer A containing 0.01% Nonidet P-40 and 0.1 M KCl for 2 h at 4°C with rotation. Immunoprecipitation (IP) was performed overnight at 4°C either with rabbit polyclonal antibodies raised against CSB (3 μl of crude serum) (56) or with 2 μg of mouse monoclonal antibodies that recognize an epitope present on all four histones (anti-histone, pan H11-4; Boehringer Mannheim). Mock IPs were carried out either in the absence of CSB or in the absence of histones. Binding of the antibody-protein complexes to protein A-Sepharose beads, equilibrated in buffer A, was for 2 h at 4°C. The beads were washed three times with 20 volumes of buffer A containing 0.1% Nonidet P-40 and 0.35 mM KCl and three times with buffer A containing 0.01% Nonidet P-40 and 0.1 M KCl. After boiling of the beads, the bound proteins were analyzed by SDS-PAGE using either 8% acrylamide gels (to visualize CSB) or 16.5% acrylamide gels (to visualize the histones), followed by silver staining.
Far-Western analysis.
Samples (3.6 μg) of H1-depleted HeLa polynucleosomes (untreated or trypsinized) and samples of two control proteins, cytochrome c (0.9 μg; Sigma) and bovine serum albumin (DNase free, 0.9 μg; Pharmacia Biotech), were used. HeLa polynucleosomes were trypsinized by following a previously described protocol (19). The trypsinization reaction was for 30 min at room temperature, and it was stopped by the addition of a 20-fold excess (wt/wt) of soybean trypsin inhibitor (Sigma) and monitored by SDS-PAGE and Coomassie blue staining. All protein samples were separated on SDS–16.5% polyacrylamide gels and stained with Coomassie blue or transferred to a nitrocellulose membrane for far-Western analysis. The membrane was blocked with skimmed milk containing 0.05% Tween 50 and incubated with purified CSB (100 ng/ml) overnight at 4°C. Bound protein complexes were visualized by incubation with affinity-purified anti-CSB antibodies (56) and goat anti-rabbit secondary antibodies (BIOSOURCE) and by the alkaline phosphatase detection method.
RESULTS
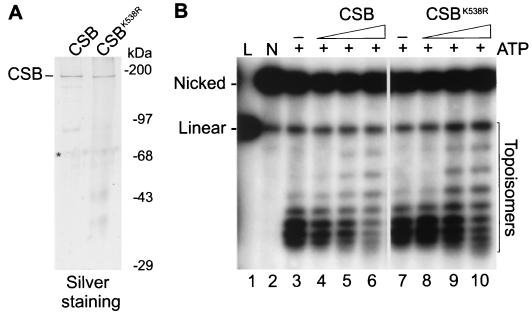
In this study, we utilized highly purified, recombinant CSB protein. Wild-type and ATPase-deficient (CSBK538R) mutant CSB proteins, both epitope tagged (HA-CSB-his6), were isolated from baculovirus-infected Sf21 insect cells as described previously. The tags do not interfere with the biological function of CSB in vivo (7). Both proteins were purified to near homogeneity, as determined by silver staining (Fig. 1A). As previously described, CSB ATPase activity was stimulated by double-stranded but not by single-stranded DNA. Both naked and nucleosomal DNAs, with or without histone tails, exhibited similar levels of stimulation (reference 7 and data not shown). When HeLa core histones were tested as cofactors, no increase of ATP hydrolysis by CSB was detected (data not shown; see Materials and Methods). This emphasizes that CSB ATPase activity is strictly dependent on double-stranded DNA. The hSWI/SNF complex was isolated from HeLa cells and functionally characterized as described previously (43).
FIG. 1.
CSB introduces negative supercoils in plasmid DNA upon binding. (A) Purified recombinant CSB and CSBK538R proteins. Aliquots of the Mono Q fractions of both proteins were separated by SDS-PAGE (8% acrylamide) and visualized by silver staining. In addition to the major CSB band, two degradation products (as determined by Western blotting) were visible. ∗, keratins which were also present in empty lanes. Molecular size markers are indicated. (B) Shift in the topoisomers distribution upon CSB binding. L, linear DNA (lane 1). Singly nicked (N) plasmid DNA (100 ng) (lane 2) was incubated in the presence of ATP alone (lanes 3 and 7) or with increasing amounts (20, 40, and 80 ng) of CSB (lanes 4, 5, and 6, respectively) or CSBK538R (lanes 8, 9, and 10, respectively). The DNA molecules were closed by the addition of E. coli DNA ligase, and the topoisomers were resolved by electrophoresis on a 1% agarose gel containing 0.5 μg of chloroquine per ml.
CSB influences DNA topology by inducing negative supercoiling.
CSB contains seven conserved motifs characteristic of the SNF2/SWI2 family of putative helicases (15, 54). Nevertheless, CSB, as well as most of the SWI2/SNF2-related proteins tested to date, fails to show classical helicase activity as assayed by oligonucleotide strand displacement (7, 11, 45, 51). However, the lack of overt helicase activity does not exclude the possibility that these proteins may work by altering the DNA conformation, thereby inducing a local separation of the DNA strands. To address this possibility, we tested CSB activity in a topological assay (52). Singly nicked DNA plasmid was incubated with increasing amounts of purified CSB or CSBK538R (Fig. 1B). Subsequently, E. coli DNA ligase was added to covalently close the nick in order to preserve any protein-induced change in linking number (ΔLk) in the duplex DNA. To determine the extent and the direction of induced ΔLk, the topoisomer population was analyzed by agarose gel electrophoresis in the absence or presence of the intercalating agent chloroquine (Fig. 1B and data not shown). As shown in Fig. 1B, lane 5, addition of 40 ng of CSB shifted the topoisomers' distribution. The number of slowly migrating DNA topoisomers in the gel increased with increased CSB concentration (Fig. 1B, lane 6, and data not shown). We calculate that the presence of CSB at an approximate molar ratio of 10:1 per 2.96 kb of plasmid DNA molecule was sufficient to induce the topological effect apparent in Fig. 1B, lane 6.
To examine whether ATP hydrolysis was required for this reaction, two experiments were performed. The ATPase-deficient CSBK538R mutant was used, and since the E. coli ligase requires only NAD, we could perform the reaction in the absence of ATP. In both cases near wild-type ΔLk values were obtained (Fig. 1B, lanes 8 to 10, and results not shown). Furthermore, in control experiments we did not detect any significant contaminating topoisomerase activity in the CSB preparations (data not shown; see Materials and Methods). The chloroquine included in the gels shown in Fig. 1B intercalates in the DNA double helix and introduces positive supercoiling in the covalently closed relaxed plasmid. CSB counteracted chloroquine action, causing an electrophoretic retardation of the DNA topoisomers (Fig. 1B, lanes 4 to 6). We conclude that binding of CSB causes a change in DNA conformation detected as negative supercoiling in our topological assay. The direction of CSB-induced ΔLk was confirmed by analysis of the topoisomers on two-dimensional gels (data not shown) (52).
CSB remodels a nucleosome core in an ATP-dependent manner.
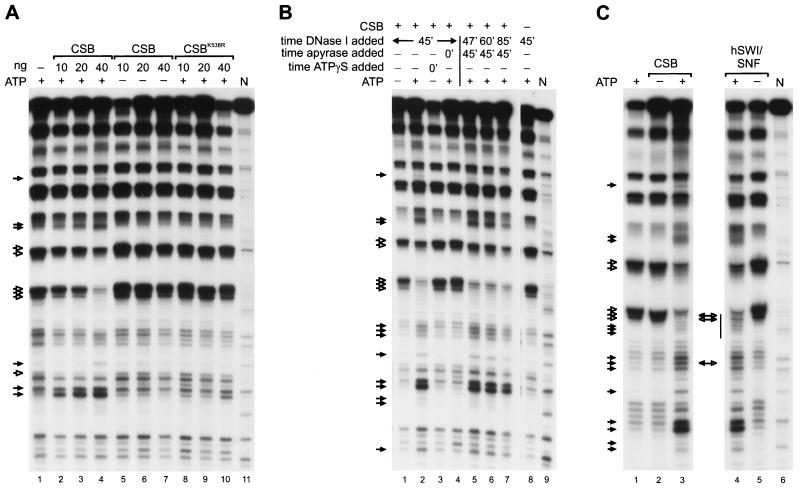
The high homology shared with the SWI2/SNF2-related proteins suggested that CSB might play a role in chromatin remodeling. However, such a property has not been established for any of the DNA repair members of this family. We first investigated this possibility at the basic level of chromatin organization, the nucleosome. For this purpose, we reconstituted rotationally phased mononucleosomes with purified histones on a radioactively labeled 155-bp DNA fragment using a salt dilution procedure. Alterations of the nucleosomal structure were assessed by DNase I footprinting (23) (see Materials and Methods). In the absence of remodeling activities, DNase I digestion of nucleosomes gave rise to the characteristic pattern of DNA fragments with a periodicity of 10 bp, which is clearly distinct from the pattern generated on naked DNA (Fig. 2A, compare lanes 1 and 11). Mononucleosomes were incubated with increasing amounts of purified CSB protein. In the presence of ATP, CSB visibly altered the DNase I accessibility to nucleosomal DNA. Remodeling is particularly evident from the change in intensity of some specific DNA bands (Fig. 2A, lanes 2 to 4, note bands marked by arrows). The changes in the digestion pattern were proportional to the amount of CSB added. When the naked 155-bp DNA fragment was incubated with CSB, no specific DNA footprint by CSB was observed, as expected for a non-sequence-specific DNA binding protein (data not shown). We estimate that a ratio of approximately 4:1 for CSB versus core particles promoted alterations of the nucleosomal structure (Fig. 2A, lane 2). No alterations were observed in the absence of ATP (Fig. 2A, lanes 5 to 7) or with the addition of the nonhydrolyzable ATP analogue [γ-S]ATP (Fig. 2B, lane 3), nor was the ATPase-deficient CSBK538R able to promote significant remodeling (Fig. 2A, lanes 8 to 10). However, gel-shift experiments have shown that binding of CSB to DNA and to nucleosomes is independent from the presence of ATP and is displayed with wild-type efficiency by the CSBK538R mutant (data not shown). This indicates that sole binding of CSB to DNA does not support changes in DNase I accessibility. In addition, the fact that CSB was not able to remodel mononucleosomes in the presence of the nonhydrolyzable ATP analogue [γ-S]ATP (Fig. 2B, lane 3) indicates that ATP hydrolysis, rather than ATP binding, is required for remodeling by CSB.
FIG. 2.
Mononucleosome remodeling by CSB. (A) Mononucleosome remodeling by CSB is ATP dependent. End-labeled nucleosome particles (approximately 3 ng of total nucleosomes) were incubated in the presence of ATP alone (lane 1) or with increasing amounts (10, 20, and 40 ng) of CSB in the presence (lanes 2, 3, and 4, respectively) or absence (lanes 5, 6, and 7, respectively) of ATP. Similar reactions were performed with CSBK538R in the presence of ATP (lanes 8 to 10). Remodeling was assessed by DNase I digestion. Filled arrows, sites of enhanced cutting due to the presence of CSB; open arrows, sites of reduced cleavage. N, naked control DNA (lane 11). (B) Nucleosome remodeling by CSB is stable upon removal of ATP by apyrase. Reactions were performed as described for panel A, and reaction mixtures contained 40 ng of CSB alone (lane 1) or with ATP (lane 2); where indicated, [γ-S]ATP (ATPγS) (2 mM) or apyrase (1 U) was added. [γ-S]ATP did not support remodeling (lane 3). Similarly, addition of apyrase (1 U) prior to CSB inhibited nucleosome remodeling (lane 4). However, addition of apyrase (1 U) after CSB had been present for 45 min did not inhibit or reverse nucleosome disruption, as shown by DNase I digestion after 2 min (lane 5), 15 min (lane 6), and 40 min (lane 7). As a control, nucleosomes were incubated in the presence of ATP alone (lane 8). N, naked DNA (lane 9). Arrows and filled arrows are as defined for panel A. (C) CSB nucleosome remodeling pattern is very similar but not identical to the one generated by the hSWI/SNF complex. Reactions were performed as described for panel A and reaction mixtures contained ATP alone (lane 1), 60 ng of CSB alone (lane 2) or with ATP (lane 3), or 300 ng of the isolated hSWI/SNF complex with ATP (lane 4) or alone (lane 5). Sites of enhanced (filled arrows) and of decreased (open arrows) DNase I digestion are indicated. Double-headed arrows between lanes 3 and 4 represent cleavage sites that distinguish the two nucleosome disruption activities. Bar, approximate position of the nucleosomal dyad axis. N, naked control DNA.
Consistent with these findings, when apyrase, which hydrolyzes ATP, was added to a reaction mixture containing nucleosomes before addition of CSB, ATP-dependent nucleosome remodeling was inhibited (Fig. 2B, compare lanes 2 and 4). To analyze the stability of the altered nucleosome, we first incubated identical samples containing ATP, nucleosomes, and CSB for 45 min, after which we exposed them to apyrase and subsequently subjected them to digestion with DNase I at time points ranging from 2 min to 40 min (Fig. 2B, lanes 5 to 7). The specific DNase I digestion pattern was conserved until 40 min past the addition of apyrase (Fig. 2B, lanes 5 to 7). These data suggest that the CSB-induced structural changes in the nucleosome are stable and that maintenance of the remodeled state does not require continuous ATP hydrolysis.
In order to qualitatively compare CSB activity with those of other chromatin-remodeling factors, we assayed CSB and the purified hSWI/SNF multiprotein complex (43) in identical disruption reactions. The remodeling activities of the entire hSWI/SNF complex and of its two separate ATPase-subunits were recently shown to be qualitatively, although not quantitatively, similar (41). The results presented in Fig. 2C reveal that the DNase I digestion patterns mediated by CSB and by hSWI/SNF are very similar. However, subtle differences in DNase accessibility were visible in the central portion of the DNA fragment (Fig. 2C, compare lanes 3 and 4), as was consistently observed in independent experiments (data not shown).
Plasmid chromatin remodeling by CSB.
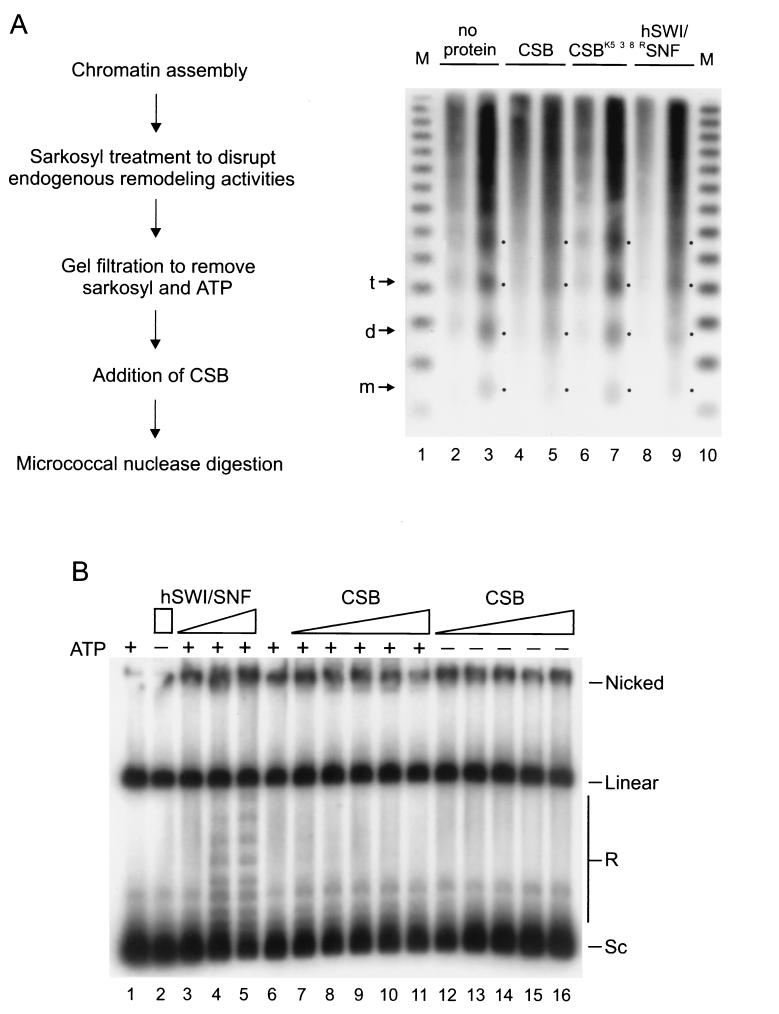
To examine whether CSB could reorganize an array of nucleosomes, a situation which more closely resembles that observed in vivo, we used Drosophila embryo extracts to reconstitute chromatin on a DNA plasmid in vitro (58). The chromatin assembly and spacing complexes (24, 59) active in these extracts catalyze the deposition of nucleosomes with uniform spacing, as visualized by MNase digestion (see Fig. 3A and Materials and Methods). Before the chromatin templates were incubated with CSB, the endogenous Drosophila remodeling activities were disrupted by Sarkosyl treatment (59). Sarkosyl and ATP were subsequently removed by gel filtration (Fig. 3A) (see Materials and Methods). The regularity of the nucleosomal array, which was maintained after the Sarkosyl treatment, was significantly perturbed by the addition of CSB in the presence of ATP (Fig. 3A, compare lanes 3 and 5). The loss of periodic spacing between the nucleosomes was visible when CSB was present in an ∼20-fold molar excess compared to the level of the 3.35-kb plasmid. Since this plasmid contains an average of 16 nucleosomes, we estimate that CSB is active at an approximate equimolar ratio with nucleosome particles. A similar irregular MNase pattern was induced by the hSWI/SNF complex in an ATP-dependent reaction (Fig. 3A, compare lanes 9 and 5, and data not shown). In contrast, in the presence of the CSBK538R ATPase-deficient mutant, the nucleosomes were found in the original regular array (Fig. 3A, compare lanes 7 and 3). Similar results were obtained in control reactions from which ATP was omitted (not shown). These data indicate that CSB uses the energy from ATP-hydrolysis to remodel nucleosomes on large DNA molecules. This change could result from repositioning of nucleosome octamers and/or generation of an altered nucleosome that is more accessible to nucleases.
FIG. 3.
Remodeling of plasmid chromatin by CSB. (A) Remodeling of nucleosome arrays detected by MNase digestion. CSB induces loss of the regular nucleosome repeat characteristic of Drosophila-reconstituted chromatin in an ATP-dependent manner. A schematic outline of the assay is shown at left. Sarkosyl-stripped chromatin (40 ng) was incubated either in the absence of proteins (lanes 2 and 3) or with CSB (lanes 4 and 5), CSBK538R (lanes 6 and 7), or hSWI/SNF (lanes 8 and 9) in the presence of ATP (as described in Materials and Methods). Nucleosome organization was studied by MNase digestion (8 U), performed for 60 and 120 s, respectively. The positions of mononucleosomes (m), dinucleosomes (d), and trinucleosomes (t) are indicated by arrows and by dots. The DNA size marker (M) represents a ladder of 123-bp repeats (lane 1). (B) Remodeling of nucleosome arrays visualized as ATP-dependent changes in supercoiling. CSB, unlike hSWI/SNF, does not induce visible changes in the topology of nucleosomal plasmid DNA. Nucleosomal template was incubated with topoisomerase I alone (lanes 1 and 6) or with either hSWI/SNF (200 ng in lane 2; 20, 60 and 200 ng, respectively, in lanes 3, 4, and 5) or increasing amounts of CSB (threefold increments starting from 2.9 ng in lanes 7 and 12), in the presence (lanes 7 to 11) or absence (lanes 12 to 16) of ATP. The molar ratio of CSB (in lane 11) to the ATPase subunits of hSWI/SNF (in lane 5) was approximately 20:1, as determined by silver staining (not shown). Sc, supercoiled; R, relaxed or partially supercoiled.
One hallmark of the SWI2/SNF2 type complexes is the ability to change the topology of nucleosomal plasmid DNA in an ATP-dependent manner (23, 28, 63). This particular property (to date reported only for SWI2/SNF2-related complexes) is also displayed by the isolated human SNF2 homologs Brg1 and hBrm (41). To assay CSB for this activity, closed circular plasmid chromatin was reconstituted with Xenopus oocyte heat-treated extracts (23) (see Materials and Methods). Nucleosomes introduce negative supercoils in plasmid DNA, which can be visualized after deproteinization as a rapid migration (relative to that of relaxed DNA) in agarose gel electrophoresis (Fig. 3B, lane 1). As previously characterized, addition of hSWI/SNF, in the presence of ATP and topoisomerase I, reduces the supercoiling of the reconstituted plasmid. This is detected as the appearance of topoisomers that have a reduced mobility (Fig. 3B, lanes 3 to 5) (23). In contrast, CSB failed to show detectable activity in this assay (Fig. 3B, lanes 7 to 11) even when present in an ∼20-fold molar excess compared to amounts of the hSWI/SNF ATPase subunits, as determined by silver staining (Fig. 3B, compare lane 11 with lane 5; data not shown). Similar results were obtained on chromatin templates assembled with Drosophila embryo extracts (58) (data not shown; see Materials and Methods). These data indicate mechanistic differences in chromatin remodeling by CSB and hSWI/SNF.
Nucleosome remodeling by CSB does not result in complete disruption of DNA-histone contacts or in octamer transfer to free DNA in trans.
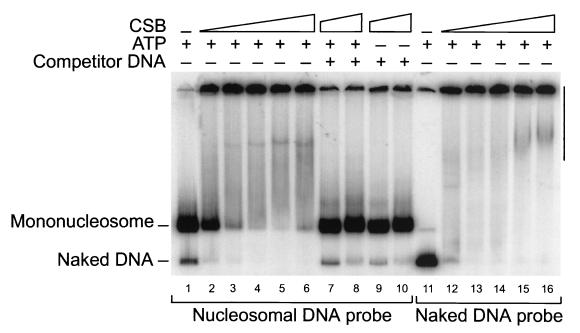
The results described above establish the ability of CSB to influence chromatin structure. To gain insight into the mechanism of remodeling by CSB, we used gel mobility shift experiments to determine whether CSB dissociates histones from DNA. As with other remodeling complexes, mixing CSB with labeled nucleosomes (or naked DNA) creates a mixture that does not enter the gel (Fig. 4, lanes 2 to 6 and 12 to 16) (see Materials and Methods). To determine whether histones have dissociated from DNA, we performed the remodeling reactions and then we added an excess of unlabeled competitor DNA and analyzed the reaction products on native gels. Importantly, addition of naked DNA (see Materials and Methods) to the samples after remodeling but prior to electrophoresis reverted the nucleosome-CSB complexes and regenerated the two DNA species in the original ratio (Fig. 4, lanes 7 to 10). A comparison of Fig. 4, lanes 7 and 8, with Fig. 4, lane 1, indicates that the remodeling reaction did not release appreciable amounts of labeled nonnucleosomal DNA. Thus, under the conditions used, nucleosome remodeling by CSB does not involve a complete dissociation of the core histones from the DNA.
FIG. 4.
Gel-shift analysis of CSB mononucleosome remodeling reactions. CSB does not catalyze the complete dissociation of histone octamers from DNA upon nucleosome binding and remodeling. Mononucleosomes (lanes 1 to 10) or free DNA (lanes 11 to 16) were incubated with increasing amounts of CSB (10, 20, 30, 60, and 100 ng for lanes 2, 3, 4, 5, and 6, respectively, and for lanes 12, 13, 14, 15, and 16, respectively; 60 and 100 ng, respectively, in lanes 7 and 8 and 9 and 10) and reactions were performed as described for those whose results are shown in Fig. 2, with (+) or without (−) ATP. Reaction mixtures were analyzed directly on native polyacrylamide gels or, where indicated, were treated with excess of cold competitor plasmid DNA before being loaded (lanes 7–10) (see Materials and Methods). Multiple DNA-protein complexes are visible (verticle bar).
Yeast SWI/SNF, yeast RSC, and human SWI/SNF (33, 40a, 62) can transfer histone octamers from excess donor nucleosomes to labeled free DNA to form nucleosomes. In contrast, the ISWI ATPase and the ISWI-based complexes CHRAC and NURF cannot perform this activity (20, 29). We failed to detect octamer transfer activity by CSB under experimental conditions in which transfer was readily detected by SWI/SNF (data not shown; see Materials and Methods).
CSB interacts with histone proteins.
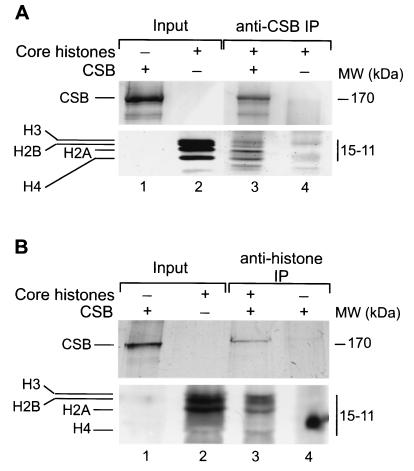
The findings described above indicate that CSB binds to nucleosomes and alters their structure upon ATP hydrolysis. To test how CSB is targeted to the core particles, we investigated whether CSB binds the histones by IP analysis. Isolated HeLa core histones [present as H2A-H2B dimers and a heterogeneous population of H3-H4 dimers and (H3-H4)2 tetramers] (2, 14, 26) were incubated with purified CSB, and IP was carried out either with an anti-CSB antibody (56) or with an antibody that recognizes an epitope present on all histones (H11-4) (see Materials and Methods). After extensive washing of the protein A beads, the bound fraction was analyzed by SDS-PAGE followed by silver staining. The results presented in Fig. 5A show that the anti-CSB antibody was able to immunoprecipitate all four histones only when CSB was present (compare Fig. 5, lanes 3 and 4, noting that the weak bands visible in lane 4 do not correspond with histones and are derived from the serum used). Conversely, using anti-histone antibodies, specific co-IP of CSB with histones was consistently observed (Fig. 5B, compare lanes 3 and 4). These data point to a specific direct interaction between CSB and the core histones.
FIG. 5.
CSB directly interacts with purified HeLa core histones. IP analysis was performed on reaction mixtures containing purified CSB and core histones (lanes 1 and 2, respectively, in panels A and B) (see Materials and Methods). After extensive washing, the antibody-bound protein complexes were separated on SDS–16.5% and –8% polyacrylamide gels to visualize the histones and CSB, respectively. Gels were stained with silver. (A) IP was carried out with anti-CSB antibodies. Analysis of the beads by SDS-PAGE showed that all four core histones specifically coimmunoprecipitate with CSB (lane 3) and not in a mock IP (lane 4). (B) IP with anti-histone, pan antibodies is shown. CSB is present in the bound fraction together with the histones (lane 3). No nonspecific binding is detected in the mock IP (lane 4).
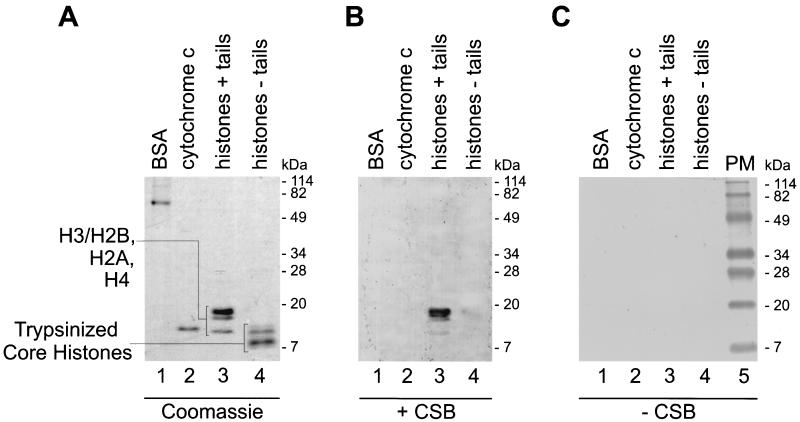
The interaction of purified CSB with histones was further examined by far-Western analysis. Human H1-depleted polynucleosomes and several control proteins (see Materials and Methods) were separated by SDS-PAGE. The purity of the protein samples used is shown on the Coomassie blue-stained gel (Fig. 6A). All four core histones were recognized by CSB, as shown by far-Western analysis (Fig. 6B, lane 3). In addition, CSB was able to bind to each of the separate core histones (data not shown). Under identical conditions, no association with BSA or with the highly basic cytochrome c protein was detected (Fig. 6B, lanes 1 and 2), suggesting that the net positive charge of the histones does not (solely) account for the binding. In the absence of the CSB protein, no background staining due to a specific antibody binding was detected (Fig. 6C).
FIG. 6.
CSB binds histone proteins in far-Western analysis. Samples of BSA (lanes 1), cytochrome c (lanes 2), and H1-depleted HeLa polynucleosomes (untreated [+ tails] [lanes 3] or trypsinized [− tails] [lanes 4]) were separated by SDS-PAGE (16.5%) and stained with Coomassie blue (A) or subjected to immunoblotting, whereupon the membrane was probed with purified CSB (B) or mock incubated (C). Anti-CSB antibodies detected binding of CSB to the immobilized histones (see Materials and Methods for details). No binding of CSB to the tailless histones was detected, suggesting that histone N-terminal tails are important for CSB-core histone interaction. The positions of prestained molecular mass markers (PM) are shown.
When DNA is wrapped around a histone octamer to form a nucleosome, only the 15 to 30 residues at the amino termini of the histones, commonly named tails, are protruding from the core structure (34). Histone tails have been demonstrated to play a crucial role in the regulation of chromatin accessibility (35). To analyze whether histone amino termini are important for CSB targeting, we followed an established procedure to remove them from all four core histones (19) (see Materials and Methods). HeLa polynucleosomes were digested with trypsin and analyzed by SDS-PAGE. After trypsin cleavage the histones ran with the characteristic size of tailless histones (Fig. 6A, lane 4). In the far-Western analysis, as shown in Fig. 6B, lane 4, we could not detect binding of CSB to the tailless histones. This further confirms the specificity of the interaction and provides evidence that CSB associates with the exposed histone tails.
CSB remodeling activity on trypsinized mononucleosomes.
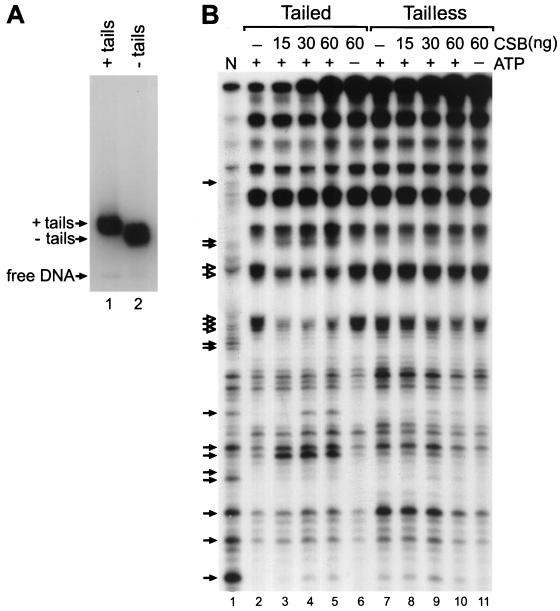
To investigate the functional significance of the presence of histone tails on CSB remodeling activity, we used the DNase I accessibility assay to determine whether CSB was able to remodel a tailless mononucleosome substrate. Tailless mononucleosomes were prepared by digesting glycerol gradient-purified tailed mononucleosomes with trypsin (19) (see Materials and Methods). The removal of the tails was confirmed by the electrophoretic mobility of tailless nucleosomes, which was faster than that of their tailed counterparts (Fig. 7A). As expected, the tailless nucleosomes have a sensitivity to DNAse I digestion that is slightly altered compared to that of intact nucleosomes (Fig. 7B, lanes 2 and 7). We then incubated CSB with either tailed or tailless nucleosomes and compared the remodeling activities. CSB induced ATP-dependent changes in the DNase I cleavage pattern on tailed nucleosomes (Fig. 7, lanes 3 to 5) in a fashion similar to that shown in Fig. 2. In contrast, at equivalent protein concentrations, CSB remodeling activity on the tailless substrates was strongly reduced (Fig. 7B, compare lanes 8 to 10 and 3 to 5). These results further support the significance of the histone tails for proper CSB functioning.
FIG. 7.
Remodeling of tailed (+ tails) and tailless (− tails) mononucleosomes by CSB. (A) Reconstituted mononucleosomes were analyzed by electrophoretic mobility shift assay. Nucleosomes with tails are shown (lane 1). Nucleosomes without tails (lane 2) have been generated by trypsinization of labeled tailed nucleosomes after glycerol gradient purification (Materials and Methods). (B) DNase I accessibility assay. Tailed (lanes 2 to 6) and trypsinized (lanes 7 to 11) mononucleosomes were incubated alone (lanes 2 and 7) or with increasing amounts (15, 30 and 60 ng) of CSB in the presence of ATP (lanes 3, 4, and 5, respectively, and 8, 9, and 10, respectively) or with 60 ng of CSB in the absence of ATP (lanes 6 and 11). Remodeling was assessed by DNase I digestion and denaturing gel electrophoresis. Filled arrows, sites of enhanced cutting due to the presence of CSB; open arrows, sites of reduced cleavage. N, naked control DNA (lane 1).
DISCUSSION
In this study, we further characterize the biochemical activities of CSB by analyzing its interaction with DNA and chromatin.
CSB alters DNA double helix conformation upon binding.
Our results show that CSB binds to double-stranded DNA and that this binding causes a significant change in linking number as evidenced by the appearance of negatively supercoiled DNA in a topological assay (Fig. 1B). This activity is reminiscent of that of the hRad54 recombination-repair protein, another member of the SWI2/SNF2 family (52). It also resembles the effect of binding of HMG-box-containing polypeptides, such as HMG-1, mtTF1, and LEF-1, to DNA (16, 18, 50). In contrast, DNA binding of the yeast SWI/SNF complex induces positive supercoils. However, CSB and ySWI/SNF share the property that ATP hydrolysis is not required to alter DNA topology (42). Relevant to this point is our observation that ATPase-deficient CSB (CSBK538R) still retains partial activity when microinjected into living CS-B cells (7). In theory, CSB binding can induce negative supercoiling in two ways. One possibility is that CSB wraps the DNA around its surface in a left-handed manner, which will cause a change in writhe. Alternatively, CSB DNA binding could induce a change in twist, which may result in local unwinding of the DNA double helix. At present, we cannot discriminate between these two possibilities.
CSB shares chromatin remodeling properties with both SWI2/SNF2 and ISWI-containing complexes.
Purified, recombinant CSB has intrinsic chromatin remodeling activities in vitro. It catalyzes the remodeling of both reconstituted mononucleosomes and nucleosomes uniformly spaced on plasmid DNA. Both activities require the energy of ATP hydrolysis. As demonstrated for hBrm and Brg1, additional proteins may be needed to increase the rate of remodeling by CSB (41). Our findings are consistent with the present view that SWI2/SNF2-related ATPases form the functional core of chromatin remodeling machines (10, 41).
Although ATP-dependent chromatin remodeling complexes perform similar activities, mechanistic differences distinguish the SWI2/SNF2 family from the ISWI-based complexes (27). Additionally, the three ISWI family remodeling complexes display different activities in vitro (6). An interesting question is whether the CSB remodeling activity resembles more that of the SWI2/SNF2-type proteins or that of ISWI.
CSB activity on mononucleosomes is similar to that of the SWI2/SNF2-containing complexes (11, 28). In fact, CSB induces significant changes in the DNase I cleavage pattern on reconstituted mononucleosomes (Fig. 2). In contrast, the ISWI-based CHRAC complex does not support nucleosome remodeling in similar experiments (29). The DNase I digestion patterns induced by CSB and by the hSWI/SNF complex are very similar (Fig. 2C). Only subtle differences are observed near the nucleosomal dyad axis, the suggested location of SWI/SNF binding (42, 43), possibly because CSB and hSWI/SNF bind slightly different sites on the nucleosome. During the mononucleosome remodeling reaction, both CSB and hSWI/SNF generate an altered nucleosome structure that is stable after removal of ATP (Fig. 2B) (23). This activity has not been tested for ISWI-based complexes. CSB activity on reconstituted plasmid chromatin presents two aspects. On one hand, CSB shows the same behavior of hSWI/SNF and the Drosophila NURF, as they all disorganize the regular repeat of nucleosome arrays (Fig. 3) (55; G. Schnitzler, unpublished observations). This activity distinguishes CSB from ISWI (10) and from the ISWI-based remodeling complexes CHRAC and ACF (24, 59). On the other hand, CSB does not induce detectable loss of superhelical density of nucleosomal plasmid, a characteristic activity displayed by SWI2/SNF2 complexes (Fig. 3B) (23). This might reflect differences in the details of the remodeling reactions. Generally, it is argued that chromatin-remodeling complexes work without removal of histone octamers from the DNA. CSB results are consistent with this idea (Fig. 4). However, SWI2/SNF2-containing complexes (RSC, hSWI/SNF, and yeast SWI/SNF) can transfer histone octamers to an acceptor DNA molecule in trans (33, 40a, 62). We were not able to detect this type of activity for CSB (data not shown). In this respect, CSB is more like the ISWI-based complexes NURF and CHRAC (20, 29). In conclusion, the data presented here indicate that CSB remodeling activity has some properties in common with both the SWI2/SNF2-based and the ISWI-based family. This may be due to the fact that CSB was tested as isolated polypeptide and not as a complex as in the case of the others. An additional interesting possibility is that CSB may be distinct in its capabilities from both classes of remodeling complexes.
Intact histone tails are important for efficient nucleosome remodeling by CSB.
To get insight into the mechanism of how CSB destabilizes the nucleosomal structure, it is important to identify its target sites. Does CSB recognize the DNA surface or can CSB also contact the histone proteins directly, or can it do both? Our in vitro data indicate that CSB can do both. Figure 1B reveals its interaction with double-stranded DNA. The results presented in Fig. 5 and 6 show that CSB interacts with the core histones, suggesting multiple contacts between CSB and the histone proteins. Interestingly, trypsinized core histones completely failed to be recognized by CSB (Fig. 6B). Histone tails have been shown to differentially influence the activity of ATP-driven chromatin remodeling complexes. They are essential for the Drosophila NURF action (17), while hSWI/SNF and yeast SWI/SNF complexes can remodel tailless nucleosomes (19, 32). Here, we present evidence that CSB remodeling activity is severely reduced on tailless mononucleosomes (Fig. 7B), providing a first indication that interaction with the histone tails is important for CSB function.
Implications for transcription-coupled repair.
A dual functionality has been proposed for CSB. First, CSB is specifically required for the transcription-repair coupling reaction. Second, several observations support an additional, nonessential role of CSB in transcription itself (1, 57). Very recently, specific fragile sites in metaphase chromosomes have been detected at abundantly transcribed chromosomal loci in CSB-deficient cells, suggesting an involvement of CSB in transcription elongation of highly transcribed (structured) genes (67). The capacity of CSB to remodel nucleosome structure may be relevant for both the repair and the transcription function, as discussed below.
In relation to the role of CSB in repair, CSB may be involved in chromatin rearrangements at repair sites. These rearrangements may include both opening of chromatin, to facilitate displacement of the stalled polymerase complex and/or favor accessibility of NER enzymes to the damage, and the rapid refolding of nucleosomes following repair synthesis (36, 38). In addition, the capability of CSB to modulate DNA double helix conformation may directly facilitate TCR. In a mechanism similar to nucleosome disruption, CSB could use ATP hydrolysis to weaken the RNAP II-DNA contacts at the site of a DNA lesion, thereby accomplishing displacement or removal of the blocked polymerase, which is an obligatory step for repair (30). The observed interaction of CSB with RNAP II, both in vivo and in vitro (44, 53, 56), might specifically target CSB to sites of blocked transcription.
With respect to the role of CSB in transcription, it is well documented that nucleosomes constitute a strong barrier to transcription elongation (5, 25). A mild stimulation of transcription elongation by CSB on naked DNA templates in vitro has recently been reported (44; our unpublished observations). The data presented here open the possibility that CSB may play a role in facilitating transcription by RNAP II through pause sites on natural chromatin templates in vivo.
The experiments presented here extend CSB function to chromatin remodeling and place the protein at the crossroad between DNA repair, transcription, and chromatin structure. It is possible that defective chromatin rearrangements during DNA repair or transcription may contribute to the severe clinical symptoms of CS patients, which cannot be explained solely by a DNA repair defect. Other examples of severe biological consequences of alterations of chromatin-modifying activities have been recently reported, e.g., the hSNF5 component of the hSWI/SNF complex (60) and proteins that regulate histone acetylation (31, 48). Finally, CSB is the first of the class of repair proteins of the SWI/SNF family for which a chromatin-remodeling activity is documented. This suggests that also the other repair members of this family mediate the cross talk between repair and chromatin structure.
ACKNOWLEDGMENTS
We are very grateful to the entire Kingston laboratory for indispensable help, especially to J. Guyon and L. Corey for sharing reagents and protocols and M. Phelan and G. Narlikar for fruitful suggestions. We are also indebted to P. Becker for Drosophila facilities. We thank the members of our laboratory for constant support and helpful discussion and D. Bootsma for his continued interest. We acknowledge S. van Baal for his assistance in computer work and M. Kuit for photography.
This work was supported by EC grant no. 7010-212 and the Dutch Science Foundation (NWO 901-501-151). R.K. is a fellow of the Royal Netherlands Academy of Arts and Sciences; J.H.J.H. is recipient of a SPINOZA award from NWO.
REFERENCES
- 1.Balajee A S, May A, Dianov G L, Friedberg E C, Bohr V A. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxevanis A D, Godfrey J E, Moudrianakis E N. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- 3.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 4.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. In: Scriver C R, Beaudet A L, Sly W S, Valle D, editors. The metabolic basis of inherited disease. 8th ed. New York, N.Y: McGraw-Hill Book Co.; 1997. [Google Scholar]
- 5.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 7.Citterio E, Rademakers S, van der Horst G T, van Gool A J, Hoeijmakers J H, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 8.Citterio E, Vermeulen W, Hoeijmakers J H J. Transcriptional healing. Cell. 2000;101:447–450. doi: 10.1016/s0092-8674(00)80854-5. [DOI] [PubMed] [Google Scholar]
- 9.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 10.Corona D F, Langst G, Clapier C R, Bonte E J, Ferrari S, Tamkun J W, Becker P B. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 11.Cote J, Quinn J, Workman J L, Peterson C I. Stimulation of GAL derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 12.Crisona N J, Kanaar R, Gonzalez T N, Zechiedrich E L, Klippel A, Cozzarelli N R. Processive recombination by wild-type gin and an enhancer-independent mutant. Insight into the mechanisms of recombination selectivity and strand exchange. J Mol Biol. 1994;243:437–457. doi: 10.1006/jmbi.1994.1671. [DOI] [PubMed] [Google Scholar]
- 13.de Laat W L, Jaspers N G, Hoeijmakers J H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 14.Eickbush T H, Moudrianakis E N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978;17:4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 15.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher R P, Lisowsky T, Parisi M A, Clayton D A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 17.Georgel P T, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci USA. 1997;94:12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guyon J R, Narlikar G J, Sif S, Kingston R E. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol Cell Biol. 1999;19:2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamiche A, Sandaltzopoulos R, Gdula D A, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 21.Henning K A, Li L, Iyer N, McDaniel L, Reagan M S, Legerski R, Schultz R A, Stefanini M, Lehmann A R, Mayne L V, Friedberg E C. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 22.Huang S Y, Garrard W T. Electrophoretic analyses of nucleosomes and other protein-DNA complexes. Methods Enzymol. 1989;170:116–142. doi: 10.1016/0076-6879(89)70044-6. [DOI] [PubMed] [Google Scholar]
- 23.Imbalzano A N, Schnitzler G R, Kingston R E. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 25.Izban M G, Luse D S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 26.Karantza V, Baxevanis A D, Freire E, Moudrianakis E N. Thermodynamic studies of the core histones: ionic strength and pH dependence of H2A-H2B dimer stability. Biochemistry. 1995;34:5988–5996. doi: 10.1021/bi00017a028. [DOI] [PubMed] [Google Scholar]
- 27.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 28.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 29.Langst G, Bonte E J, Corona D F, Becker P B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 30.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 31.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 32.Logie C, Tse C, Hansen J C, Peterson C L. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry. 1999;38:2514–2522. doi: 10.1021/bi982109d. [DOI] [PubMed] [Google Scholar]
- 33.Lorch Y, Zhang M, Kornberg R D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 34.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 35.Luger K, Richmond T J. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 36.Meijer M, Smerdon M J. Accessing DNA damage in chromatin: insights from transcription. Bioessays. 1999;21:596–603. doi: 10.1002/(SICI)1521-1878(199907)21:7<596::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 38.Moggs J G, Almouzni G. Chromatin rearrangements during nucleotide excision repair. Biochimie. 1999;81:45–52. doi: 10.1016/s0300-9084(99)80037-6. [DOI] [PubMed] [Google Scholar]
- 39.Nance M A, Berry S A. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 40.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 40a.Phelan M L, Schnitzler G R, Kingston R E. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol Cell Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 42.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 43.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 44.Selby C P, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selby C P, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 46.Shrader T E, Crothers D M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smerdon M J, Conconi A. Modulation of DNA damage and DNA repair in chromatin. Prog Nucleic Acid Res Mol Biol. 1999;62:227–255. doi: 10.1016/s0079-6603(08)60509-7. [DOI] [PubMed] [Google Scholar]
- 48.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark W M, Sherratt D J, Boocock M R. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989;58:779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- 50.Stros M, Stokrova J, Thomas J O. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994;22:1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swagemakers S M, Essers J, de Wit J, Hoeijmakers J H, Kanaar R. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J Biol Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 52.Tan T L, Essers J, Citterio E, Swagemakers S M, de Wit J, Benson F E, Hoeijmakers J H, Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 53.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Troelstra C, Van Gool A, De Wit J, Vermeulen W, Bootsma D, Hoeijmakers J H J. ERCC6, a member of a subfamily of putative helicases, is involved in Cockaynes syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 55.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 56.van Gool A J, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly J M, Bootsma D, Hoeijmakers J H J. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in a RNA polymerase II containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Gool A J, Van der Horst G T J, Citterio E, Hoeijmakers J H J. Cockayne syndrome: defective repair of transcription? EMBO J. 1997;16:4155–4162. doi: 10.1093/emboj/16.14.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varga-Weisz P D, Bonte E J, Becker P B. Analysis of modulators of chromatin structure in Drosophila. Methods Enzymol. 1999;304:742–757. doi: 10.1016/s0076-6879(99)04045-8. [DOI] [PubMed] [Google Scholar]
- 59.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 60.Versteege I, Sevenet N, Lange J, Rousseau-Merck M F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 61.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 62.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 63.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 64.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 65.Workman J L, Taylor I C, Kingston R E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991;64:533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- 66.Workman J L, Taylor I C, Kingston R E, Roeder R G. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 1991;35:419–447. doi: 10.1016/s0091-679x(08)60582-8. [DOI] [PubMed] [Google Scholar]
- 67.Yu A, Fan H-Y, Liao D, Bailey A D, Weiner A M. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]