Abstract
Infertility affects one in six couples, half of which are caused by a male factor. Male infertility can be caused by both, qualitative and quantitative defects, leading to Oligo- astheno-terato-zoospermia (OAT; impairment in ejaculate sperm cell concentration, motility and morphology). Azoospermia defined as complete absence of sperm cells in the ejaculation. While hundreds of genes are involved in spermatogenesis the genetic etiology of men’s infertility remains incomplete.We identified a hemizygous stop gain pathogenic variation (PV) in the X-linked Germ Cell Nuclear Acidic Peptidase (GCNA), in an Azoospermic patient by exome sequencing. Assessment of the prevalence of pathogenic variations in this gene in infertile males by exome sequence data of 11 additional unrelated patients identified a probable hemizygous causative missense PV in GCNA in a severe OAT patient. Expression of GCNA in the patients’ testes biopsies and the stage of spermatogonial developmental arrest were determined by immunofluorescence and immunohistochemistry. The Azoospermic patient presented spermatogenic maturation arrest with an almost complete absence of early and late primary spermatocytes and thus the complete absence of sperm. GCNA is critical for genome integrity and its loss results in genomic instability and infertility in Drosophila, C. elegans, zebrafish, and mouse. PVs in GCNA appear to be incompatible with male fertility in humans as well: A stop-gain PV caused Azoospermia and a missense PV caused severe OAT with very low fertilization rates and no pregnancy in numerous IVF treatments.
Subject terms: Genetics research, Infertility
Introduction
Around 7% of all men in Western societies experience infertility [1] and the majority of subjects lack a specific etiologic diagnosis [2, 3]. Among many different phenotypes related to sperm pathology, we focus on two phenotypes relevant to our current study. Qualitative and quantitative defects leading to Oligo- astheno-terato-zoospermia (OAT) [impairment in the concentration, motility and morphology (respectively)] of the sperm cells in the ejaculate or azoospermia. Azoospermia is diagnosed when sperm is completely absent in the ejaculation even after centrifugation and is found in more than 10% of infertile men [2]. It is classified into obstructive azoospermia (OA) with normal spermatogenesis and non-obstructive azoospermia (NOA) due to severely impaired or absent spermatogenesis. NOA represents the most severe form of male infertility, with limited treatment options and with low fertility and pregnancy rates depending on the presence or absence of sperm in Testicular Sperm Extraction (TESE) [4]. The consensus view is that male infertility is a complex condition with a significant genetic component [1]. Thus, understanding the underlying genetic basis of spermatogenic failure is of great significance for clinical diagnosis, treatment of male infertility and prognosis for the infertile couple and their future offspring [5]. The current genetic standards for the diagnosis of severe male infertility include testing for numerical and structural chromosomal aberrations and deletions of the AZF regions on the Y chromosome. However, the success rates of these approaches is only between 10 and 20% [2]. Hundreds of infertility gene candidates were identified in animal models, enabling better diagnosis of pathogenic variations causing male infertility specifically in these genes [6–8] but only a minority of the genes responsible for non-obstructive azoospermia and OAT were validated [9]. Currently, out of the 59 spermatogenic failure genes (SPGFs) involved in both OAT and NOA, reported in the Mendelian Inheritance in Man database (OMIM), ten genes are directly involved in protecting genome stability of the male germ and sperm cells. Pathogenic variations (PVs) affecting the integrity of DNA in germ cells are acutely harmful as these cells are uniquely tasked with passing their genomes to the next generation, and specialized pathways have evolved to protect the genomic integrity of pluripotent cells and germ cells [10]. These include PVs in meiosis 1 associated protein (M1AP), a relatively frequent cause of autosomal recessive severe spermatogenic failure and male infertility with strong clinical validity [11] and in FANCM [12], which is involved in DNA repair specifically in inter-strand crosslinks. PVs in three genes, encoding members of the synaptonemal complex assembly: SYCP2 [5]; SYCP3 [13], and SYCE1 [14]. Meiotic arrest and azoospermia are also caused by PVs in TEX11, which regulates homologous chromosome synapsis and double-strand DNA break repair [15]. Previously, we have shown that severe men infertility is caused by PVs in TEX14 [16], which is required for the formation of intercellular bridges in vertebrate germ cells, and in MEIOB, which is required for double-strand break repair, crossover formation, and promotion of true and complete synapsis during meiosis [16, 17]. We also demonstrated that a deletion PV in TDRD9 which preserves genome integrity by silencing of Line-1 retrotransposon in the male germline causes azoospermia [18].
In the present study, we propose that PVs in Germ Cell Nuclear Acidic Peptidase (GCNA) can cause NOA and severe OAT. GCNA functions in DNA-protein crosslink (DPC) repair and eliminates proteins that are inappropriately crosslinked to DNA [19]. DPCs interfere with the transcription, unwinding, replication, and repair of DNA [20]. GCNA exhibits enriched expression in germ cells across species [19] and is expressed throughout germ cell development, including during all key events of meiosis and spermatogenesis [21]. Loss of Gcna results in genomic instability that leads to infertility in Drosophila, C. elegans, zebrafish, and mouse [19, 22, 23]. Indeed, it was suggested that since GCNA is primarily expressed in the male germline, males carrying GCNA PVs are more likely to have germline than somatic phenotypes and possibly be infertile [23]. The results present herein, using whole exome sequencing (WES) identifying PVs in GCNA in infertile males, support this suggestion and propose GCNA as a gene associated with human male infertility.
Materials and methods
Patients and study participants
All study participants consented to undergo genetic evaluations and signed written informed consent. The local institutional review board committees: Soroka Medical Center and Sourasky Medical Center approved the study in accordance with the Helsinki Declaration of 1975. A detailed history and standardized physical, clinical, and laboratory examinations were carried out to record details of the lifestyle, occupation, family reproduction history, physical examination, hormone levels of the infertile men. Chromosome analysis and Y microdeletions were carried out. Multiple semen analyses (1-month interval) were according to World Health Organization (WHO) and Bjorndahl guidelines [24]. All fertile men included in the study were queried about their parental origin, family ancestry, and fertility history.
Testicular tissues extraction and staining
Testicular sperm extraction technique used in this study by both groups was the multiple testicular biopsies as described [25]. GCNA was detected by immunohistochemical staining of Bouin’s fixed testicular biopsies embedded in paraffin as described before [25], using GCNA primary antibody (Novus Biological, NBP1-85993). Slides were examined using microscope Olympus BX40, camera Olympus digital.
Genetic analysis
Genomic DNA was extracted from white blood cells as detailed [16, 18]. For both patients, exome sequencing was carried at Theragene. SureSelectXT Human All Exon V6 kit, was used for library preparation, target size was 58 Mb and the coverage uniformity at X10 for patient 1 and X50 for patient 2 coverage was ≥ 90%.
Bioinformatics analyses
Variants calling and PVs annotation were done as described previously [16, 17]. Briefly, adapter sequences and low‐quality tails of reads were removed with the software Trimmomatic [26]. Row reads were then aligned to the reference genome (hg19) with BWA-MEM [27]. Then, using Picard-Tools software, we removed duplicate and indexed and compressed the aligned sequence to the BAM format [http://broadinstitute.github.io/picard/]. Variant calling and filtration stages were performed using GATK [28], and the result g.vcf files were joint called with an additional five fertile males of similar ethnicity that we have in our database of exome sequencing [16–18, 29], as a negative control. Variants annotation was done using the Annovar tool kit [30], as described before [16, 17]. Likely causative PVs were identified by filtering the data according to (i) the recessive or X-linked model of inheritance, (ii) the expected population frequency of the PVs (MAF < 1%), and (iii) the functional impact of the PVs (conservation, loss of function, functional prediction). Finally, the relevance for the phenotype was assessed using Gene Ontology terms [31], model organism data [32], and comprehensive expression data: RNAseq transcript per million (TPM) values were retrieved from the GTEx portal [33]. A testis specificity value was given to each gene as previously described [16, 17]. Briefly, a synthetic vector for exclusive expression in testis was created. The Pearson product–moment correlation coefficient (r) was calculated for a testis exclusive expression synthetic vector against all the gene expression vectors. Results are displayed as a list of r-values, indicating testis specificity level, where the closer the r-values to r = 1, the higher the testis specificity. Furthermore, additional supportive evidence were obtained by using the GeneCards database [34] by searching association between the candidates’ genes and the terms: “azoospermia” or “meiosis” or “spermatogenesis”.
Verification of the PVs
The variants identified in GCNA of both patients and the variant in PNLDC1 identified in patient 2, were ascertained by PCR of their genomic regions and direct Sanger sequencing of the PCR product as detailed [16, 18]. The primers and conditions for the PCR amplification are provided in Supplementary Table 1. Restriction fragment length polymorphism (RFLP) for verification of the variations of both patients was performed on the PCR products as provided in Supplementary Table 1. The variants were submitted to GV shared LOVD: patient 1: individual #00376828, variant #0000790710 and patient 2:individual #00376843, variant #0000790716.
Immunofluorescence staining of testicular biopsies
Testicular sections of 5 µm from patient-1 and normal adult man fixed tissue were examined for double immunofluorescence staining of GCNA and MAGE-A or DAZL—germ cell markers as previously detailed [18]. The following primary antibodies were used to detect the GCNA (rabbit polyclonal) as for the immunohistochemistry. Mouse monoclonal anti human MAGE-A (sc-20034, Santa Cruz Biotechnology), goat polyclonal antihuman DAZL (sc-27333, Santa Cruz Biotechnology) and mouse monoclonal anti human H2A.X Phospho (Ser139) (613402, BioLegend). Slides were examined for immunofluorescent staining using Nikon eclipse 50 i microscope (Tokyo, Japan) and NIS-Elements F 4.00.00 software.
Results
Clinical phenotypes
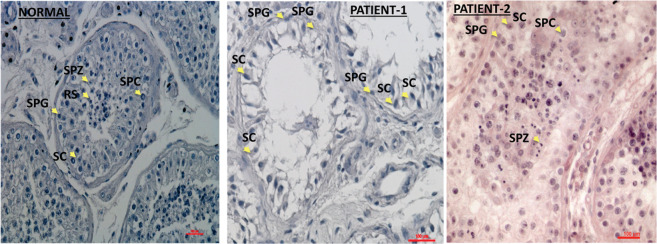
Patient 1: the patient of consanguineous Bedouin descent attended the IVF unit of the Soroka Medical Center due to a history of primary infertility that had lasted for longer than 1 year. There was no history of genetic diseases in the patient family. His medical history was unremarkable, he was in his mid-twenties with BMI 19.2 His karyotype and Y chromosome microdeletion analyses were normal as well as his hormonal profile for LH, testosterone and prolactin. The FSH level was markedly elevated (Table 1) and the testicular volume was small (right ~ 10 mL, left ~ 4.3 mL). Testicular ultrasound did not detect varicoceles. His azoospermia status was defined by two semen analyses in which no sperm was found even after centrifugation. Sperm was not found in TESE. The histology of the testes revealed complete interruption of spermatogenesis uniformly with maturation arrest, presenting only spermatogonial cells (SPG), in all tubules in both left and right testicular biopsies (Fig. 1).
Table 1.
Patients’ hormonal profiles and semen characteristics.
| Hormonal profile | Semen sample 1 | karyotype | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Diagnosis | FSH (mIU/ml) | LH (mIU/ml) | Prolactin (ng/ml) | Testosterone (ng/ml) | Concentration (mil/ml) | Motility (%) | Vitality (%) | |
| 1 | Azoospermia | 40 | 10 | 12 | 6.8 | – | – | – | 46XY |
| 2 | severe OAT | 4.23 | 5.85 | 26.8 | 8.71 | 0.2–5.9*104 | Up to 10 | 31–36 | 46XY |
Normal reference ranges for the hormonal profile were: FSH (0.7–11 mIU\ml), LH (0.8–8 mIU\ml), TSH (0.39–4 mIU\ml), Testosterone Total (2.45–12 ng\ml), Prolactin (1–26 ng\ml).
Fig. 1. Histological presentation of testicular sections of biopsies.
The histological staining by H&E of testicular biopsy with normal spermatogenesis shows seminiferous tubules with complete spermatogenesis that contain all types of spermatogenesis cells such as spermatogonial cells (SPG), spermatocytes (SPC), round spermatid (RS), spermatozoa (SPZ) and Sertoli cells (SC) (according to their location in the seminiferous tubule and the shape of the nucleus). However, the testicular biopsy of the patient 1 showed maturation arrest in the seminiferous tubules that contained only SPG and SC and the biopsy of patient 2 presented complete spermatogenesis with reduced number of RS and SPZ.
Patient 2: The patient attended the Male Fertility Clinic at Tel Aviv Sourasky Medical Center. He was in his mid-thirties; his BMI was 23, with normal size testis and penis. He is of consanguineous Arab descent and reported having a brother, a sister and cousin that may have faced fertility impairments and were treated in another center. His karyotype and Y chromosome microdeletion analyses were normal as well as his FSH, LH, testosterone and prolactin levels (Table 1). Testicular ultrasound did not detect varicocele. He underwent several semen analyses through the period of 3 years and was found to have severe OAT: low sperm cells concentration, low percent of motility mostly with slow motility and low vitality (Table 1). All the sperm cells observed were with abnormal morphology, most with small or rounded head and short or no tail. Cycles of intracytoplasmic sperm injection (ICSI) with ejaculated sperm resulted in very low fertilization rate (8%, 2/35 oocytes), and no pregnancy. The patient returned three years later and asked to try additional IVF\ICSI cycles with testicular sperm. He underwent bilateral multiple location TESE and sperm samples were frozen. Motility was observed for some but all the sperm cells were morphologically pathologic. Few ICSI cycles with thawed testicular sperm were performed and resulted with either low fertilization rate or no fertilization (3/12, 3/12, 0/9). No motile sperm were observed after thawing. The sperm vitality, tested in a sample of one of the biopsies, was 50%. The histology presented spermatocytes and round spermatids in all tubules in both left and right testicular biopsies (Fig. 1). An additional ICSI cycle was performed with ejaculated spermatozoa. None of the 14 oocytes were fertilized. Five years later he underwent an additional IVF treatment with fresh testicular sperm. Six oocytes were injected with motile sperm, but no fertilization resulted. Few months later the last IVF trial was performed with non-motile thawed sperm cells, and resulted in 2 fertilizations out of 5 injected oocytes but no pregnancy obtained.
Identification of the PVs in the infertile patients
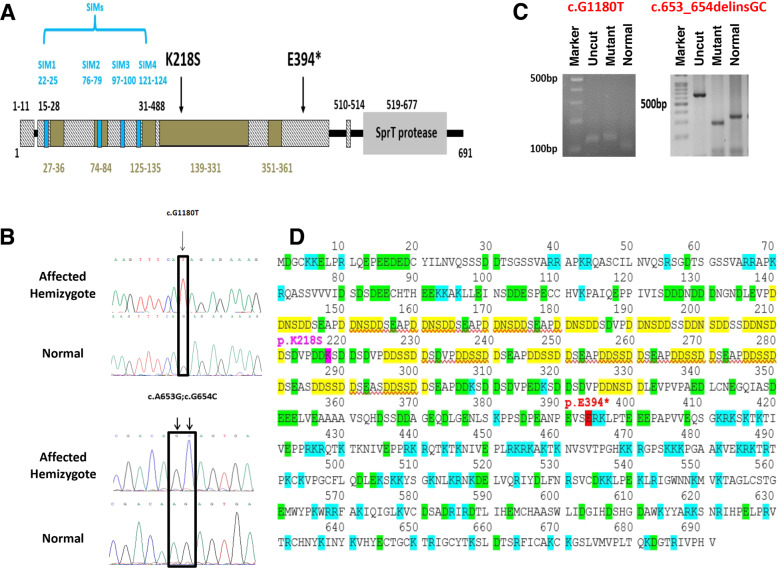
For patient 1, originating from a consanguineous Bedouin family, we assumed homozygosity by descent of a recessive variation or X-linkage model of inheritance as the likely cause of the disorder. Following the filtering steps detailed in the methods section, 14 likely causative PVs in 11 genes were retained. The possible association of these PVs to male infertility was then further assessed as described in the Methods section and shown in Supplementary Tables 2 A and 3 A. The most likely candidate variation was identified in chromosome X: 70824307(GRCh37/ hg19), c.1180 G > T (NM_052957.3), in coding exon 8 of GCNA, causing premature stop codon (p.Glu394*) within the region of the protein defined as an intrinsically disordered region (IDR) (Fig. 2A). The stop gain causing the truncation of the protein is high confidence/likely pathogenic as shown by interVar and MutationTaster. The exome result of this variation was ascertained by PCR of the genomic region and Sanger sequencing of the PCR product (Fig. 2B). The variation was not present in the public databases, nor in any of our collection of Bedouin exomes from 69 individuals (30-female, 39-male). Thus, its prevalence in the Bedouin population is less than 1/99. To verify its contribution to azoospermia in the Arab population, eleven azoospermic Arab patients with similar testicular impairment were tested for the PV by RFLP. None were found to carry the PV (Fig. 2C). In addition, the variation was not present in our in-house genetic variation database (HGVB) obtained from exome sequencing of 500 control individuals from various projects of mixed Israeli populations[16, 17]. The PV of stop codon replacing glutamate at position 394 truncates the protein at the start of the positively charged region of the IDR (Fig. 2D). The loss of the 297 amino acids at the carboxy end will eliminate the conserved metallopeptidase zinc binding like sprT domain (Fig. 2A), which was demonstrated to be required for DNA damage prevention in Drosophila germline [22]. To further assess the role of GCNA in human sperm maturation defects, we re-examined GCNA variations in WES of 11 unrelated patients from the Israeli Arab population, 4 with severe OAT and 7 with azoospermia, who were previously tested but had no conclusive findings.
Fig. 2. The variations in GCNA and the structure of the protein.
A Diagram of the protein domains and the location of the PVs. The disorder region (IDR) is displayed in hatched bars, the low complexity regions within the (IDR) are shown in brown boxes and the SUMO interacting motives (SIM) in blue. The diagram is based on Pfam and SMART embl database, and the SIM repeats are according to Borgermann et al. [46]. B Chromatograms of Sanger sequences presenting the variation in GCNA in the patients in comparison to control. C Verification of the presence of the PVs in NOA patients of the Arab population was done by RFLP on PCR products. c.G1180T was tetsted with the SfcI enzyme. Uncut: 123 bp, normal: 96 bp+27 bp, PV prevents restriction: 123 bp. Verification for the presence of the variation c.653_654delinsGC was done with BstI enzyme. Uncut 675 bp, normal 347 + 118 + (30*7), PV adds a restriction site 317 + 118 + (30*8). D The protein sequence of human GCNA. The location of the stop codon is red. The negatively charged DD(S/N)DD repeats are marked yellow. Positively and negatively charged amino acids are marked blue and green, respectively.
A likely causative PV in GCNA was found only in patient 2: two candidate tandem variations (within the same codon) in GCNA, in exon 8: chrX:g.70823780_70823781 delinsGC; NM_052957.3 (GRCh37/hg19) c.653_654delinsGC that results in conjunction in a change of a basic charged amino acid to a polar amino acid (Lys218Ser) in the largest low complexity region within the IDR domain of the protein (Fig. 2A). This tandem event was not reported in any of the available public database, however, both variants are present in the gnomAD database with MAF = 0.00005 and 0.0001, respectively. It is thus possible that the rarest variant (c.A653G) occurs under the background of c.G654C and the tandem event is present but not reported. We thus inspected this substitution in bam files of 2 individuals obtained from gnomAD and observed that both variations are indeed present together. Therefore the MAF of the tandem event is <=0.00005, all the reports for the pathogenic variation in the gnomAD were heterozygote women [35]. To assess if the GCNA Lys218Ser is the prime candidate substitution we subjected patient 2 to full WES analysis as done for patient 1. The initial bioinformatics filtering results of the WES analysis of patient 2 retained 30 candidate PVs in 30 genes (Supplementary Tables 2B and 3B). The addition filtering for the expression in testis retained only 6 genes that showed sufficient testis expression specificity (Supplementary Table 2B). To relate additional supportive evidences, we use the GeneCards suite [34] and gene expression data, as describe in the methods. This analysis retained three candidate PVs in the genes: PNLDC1, GCNA, and HMOX2 possibly involved in testicular functions. HMOX2 was previously shown to be associated with Leydig cells steroidogenesis but not directly with spermatogenesis [36]. Since patient 2 had a normal hormonal profile, the variation in HMOX2 is less likely to be the causative PV. PNLDC1 shows relatively high testis expression, as well as in several brain tissues [33]. Pnldc1 mutant mice were shown to undergo mainly post meiotic spermatogenesis arrest and also showed that PNLDC1 mediated piRNA trimming throughout mouse spermatogenesis [37]. Both variations in GCNA and in PNLDC1 were not present in our in-house HGVB [16, 17]. The variant in GCNA creates a BstI restriction site which allows RFLP screening (Fig. 1C). This RFLP screening negated the presence of the variations in 80 and 144 fertile and infertile (28 oligozoospermic and 116 azoospermic) Arab men, respectively. To verify whether the variation in PNLDC1 may be the causative PVwe tested its prevalence among Israeli Arabs by RFLP after amplification with a mismatch primer (which creates a FatI restriction site, Supplementary Table 1 and Supplementary Fig. 1). Overall, 17.4% of the tested men (n = 132) have at least one allele with the variant. Two fertile and two infertile men were homozygous for this variant. The allele frequency was similar (10%) among the fertile and infertile men tested. Thus, we discard this PNLDC1 variant as the causative PV. To assess the conservation of the identified variation in GCNA we performed blastp (https://www.ncbi.nlm.nih.gov/) and obtained the multiple sequences alignments of GCNA. The conservation of GCNA is relatively low, specifically, the N’ terminus of the low complexity IDR domain, which includes the mutated 218 amino acid position, was identify only in primates (Supplementary Fig. 2). This limited conservation and the absence of defined motif structure of the IDR domain may explain the failure of in silico predictions to anticipate a deleterious effect (see Supplementary Table 3B). Interestingly, most of the primates that retained the N’ terminus and 218 position, carry the polar amino acid asparagine, while only humans have the charged amino acid lysine, which suggests species-specific nonsynonymous substitutions.
Effect of the PVs on GCNA protein expression in the testis and development of spermatogonial cells
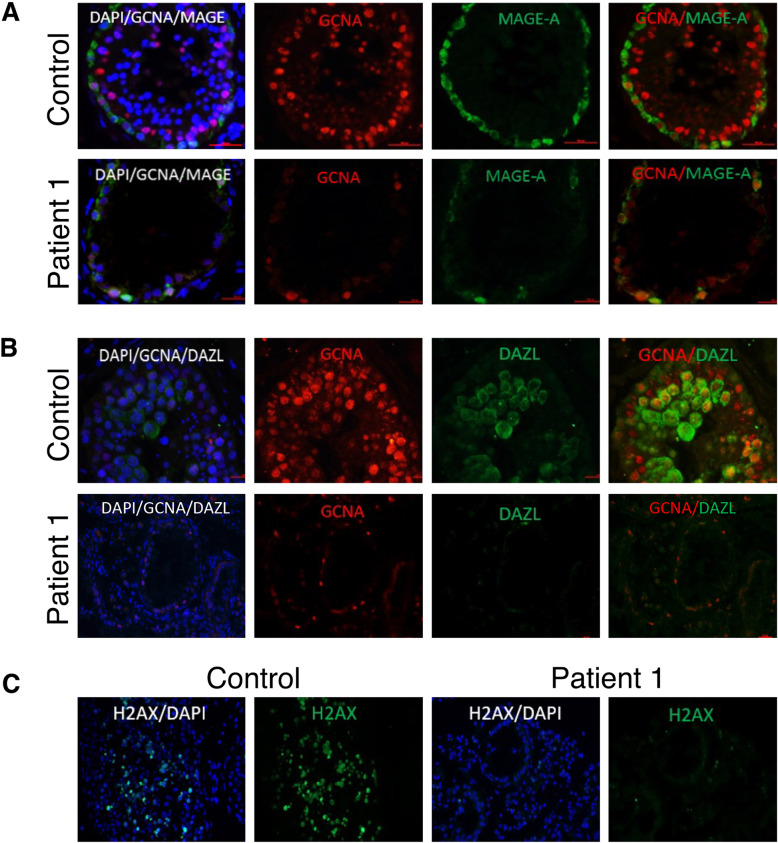
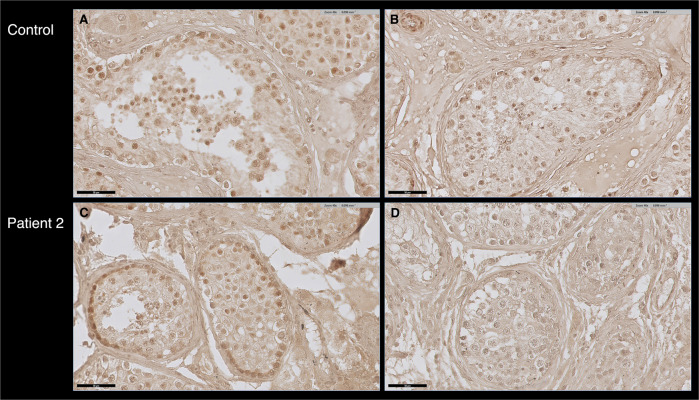
We have verified whether the GCNA protein is expressed in the testicular cells of patient 1 by immunofluorescence analysis on testis biopsies, using a commercial GCNA antibody raised to a peptide within the 50–150 amino acids at the amino terminal of human GCNA. GCNA staining was detected in control testes biopsies and at low levels in spermatogonial cells of patient 1 (Fig. 3). To better define the developmental stages of spermatogenesis in the absence of a functional GCNA, we performed co-immunostaining with the developmental cell markers MAGE and DAZL. MAGE is expressed in all types of spermatogonia and primary spermatocyte and DAZL is expressed in spermatogonial cells until pachytene spermatocytes [38, 39]. In control biopsy we observed that GCNA is co-expressed with both markers. Early and late primary spermatocytes were not observed in the patient testicular biopsy and DAZL expressing cells were not found in the patient testicular biopsy (Fig. 3A, B). Further support for the suspected absence of primary spermatocytes was obtained by immunostaining of patient 1 in comparison to control testis biopsies with phospho-H2AX antibody. PhosphoH2AX is expressed during meiotic prophase 1 [40]. No staining was observed in the patient’s biopsy (Fig. 3C). Staining with the GCNA antibody was observed in the testis biopsy of patient 2, with approximately the same intensity as in the control biopsy (Fig. 4). Presumably, the mutated protein is expressed and the structural change does not affect the antibody epitopes.
Fig. 3. Localization of GCNA protein in different types of spermatogonial cells and effect of the stop PV on the development of spermatogonial cells in human testes.
Testicular biopsies with complete spermatogenesis (control) and from GCNA patient 1 (patient) were examined for: (A) The presence of GCNA (red) in spermatogonial cells which were marked by MAGE-A (specific markers for spermatogonial cells) (green). Immunofluorescence staining was done using specific antibodies for each marker. B The presence of GCNA (red) in germ cells which were marked by DAZL (green) in testicular biopsy from control and from patient. Staining of the patient’s biopsy with DAZL did not show any signal. C The presence of spermatogonial cells expressing phospho-H2AX. Note very few cells expressing phospho-H2AX in the patient’s biopsy.
Fig. 4. Expression of GCNA protein in human testicular tissue.
Expression of GCNA protein in patient 2 OAT testicular tissue in comparison to control. A and C with antibody; B and D without antibody. DAB staining generated a strong brown color at the position of antibody localization.
Discussion
Using exome sequencing, we identified hemizygous likely causative PVs s of GCNA in two unrelated cases of infertile men. No other possible causative PVs were identified in these patients. Notably, the stop gain PV in GCNA identified in patient 1 is novel and absent from human population genome datasets, and the Lys218Ser tandem substitution identified in patient 2 was not reported, although we inspected it in the gnomAD bam files and found it to be extremely rare (MAF < 0.00005), carried solely by females (12 heterozygous). GCNA is extremely intolerant to loss of function (LoF) as reflected by its pLI of 0.99 and observed/expected LoF of 0.06 (gnomAD) [35]. The severity of the pathogenic variation seems to be reflected in the patients’ phenotype. Patient 1, with the truncation PV presents spermatogenic maturation arrest with almost complete absence of early and late primary spermatocytes and thus a complete absence of sperm, whereas patient 2 with the missense PV presents with severe OAT. The differences could reflect variable functionality impairment (or different genetic background), as has been observed in other male infertility genes M1AP (azoospermia and oligozoospermia) [11], KLHL10 (severe oligozoospermia and oligozoospermia) [41], TAF4B (azoospermia and oligozoospermia) [42], TDRD9 (azoospermia and cryptozoospermia) [18], and TEX11 (complete meiotic arrest and mixed testicular atrophy) [15]. Recently a paper describing 7 likely clinically relevant GCNA variants in NOA and cryptozoospermia (no sperm in the ejaculate without evidence of partial reproductive tract obstruction, but with rare sperm seen in centrifuged pellet) [43]. In agreement with our observation, the cryptozoospermic patients had missense PVs in the IDR domain, however no information is available concerning fertilization rates and implantation rates.
GCNA is a multi-functional protein that specifically cleaves DNA-protein crosslinks (DPCs) [22]. DPCs represent particularly intrinsically disordered region (IDR) and SprT domains governing distinct aspects of genome integrity [22]. In most species, but not rodents, GCNA proteins contain a C-terminal SprT- domain insidious lesions that interfere with almost every chromatin-based process including replication, transcription, and chromatin remodeling [44]. Accordingly, SprT enzymatic activity appears necessary for the prevention of DNA damage during Drosophila oogenesis [22]. The IDR of GCNA is essential for its function within germ cells. While the primary amino acid sequence of GCNA’s IDR has diverged, this region has continued to retain a high percentage of aspartic acid and glutamic acid residues. The IDR of GCNA in mice (https://www.uniprot.org/uniprot/A0A1D9BZF0) does not show sequence homology to the IDR of human. However, both structures have two distinct regions: an amino-terminal region containing multiple negatively charged repeats followed by a carboxy-terminal region with many positively charged repeats. The PV Glu394* within the IDR domain truncates the protein just at the start of the positively charged region and thus presumably interferes with the IDR function in addition to eliminating the SprT domain. The replacement of the positively charged lysine at position 218 with polar uncharged serine in the middle of the low complexity region within the IDR domain changes the electrical charge composition in the region, thus presumably interfering with the IDR function, but less than the truncation PV. By blastp and multiple sequence alignment, we observed that human N’ terminus have no homology outside primate (Supplementary Fig. 2), and that the most abundant amino acid in GCNA position 218 is the polar amino acid asparagine. Thus, the human lysine seems to be the result of a species-specific substitution. Interestingly, species-specific amino acid substitutions were previously shown to be a signal for adaptive evolution, specifically in protein-protein interactions [45]. The importance of the IDR distinct composition remains unknown; it may regulate the stability of GCNA or its ability to form condensates. In flies and worms, the GCNA IDR promotes proper chromosome segregation and cell cycle regulation. GCNA physically interacts with topoisomerase II (TOP2), which has germline-specific functions [44], in both mice and worms [23]. Gcna-mutant mice exhibit abnormalities consistent with the inability to process DPCs: including persistent DNA damage, decreased crossovers and crossover interference, and chromatin condensation defects. Thus, suggesting that mouse GCNA may promote germline genome stability through a conserved TOP2 DPC-based mechanism [23].
In conclusion, our genetic and functional analyses in human subjects together with the effect of deletion of Gcna in worm fly and mouse fertility, strongly suggest that PVs of GCNA are a crucial genetic cause of male infertility. Furthermore, the effect of the damaged function of GCNA may indicate that no successful pregnancy outcome could be acquired through ICSI using the spermatozoa of men harboring GCNA PVs. Our study provides new knowledge to clinicians and genetic counselors for understanding the genetic etiology of male infertility.
Supplementary information
Supplementary Figure 2: Assessment of the conservation of the identified PV in GCNA.
Supplementary Table 3: Population frequencies and In-silico predictions for the effect of the candidate variations in patient 1 (A) and patients 2 (B).
Acknowledgements
We are grateful to the affected persons and their families, whose cooperation made this study possible.
Funding
This project was partly supported by The Israeli Ministry of Science, Technology and Space to RP and partly by an internal grant of the Reproductive Hub Faculty of Health Sciences Ben-Gurion University of the Negev, to AZ, EL, EL, MH, and RP, and in part by a grant of the Israeli Science Foundation to MG, and in part by a grant of The Natural Science Foundation of China (NSFC) - Israel Science Foundation (ISF) (NSFC-ISF) (No. 3425/20) to MH.
Data availability
The datasets generated during and/or analysed during the current study are available in the GV shared LOVD repository, https://databases.lovd.nl/shared/genes.
Competing interests
The authors declare no competing interests.
Ethics approval
The local institutional review board committees: Soroka Medical Center and Sourasky Medical Center approved the study in accordance with the Helsinki Declaration of 1975. All study participants consented to undergo genetic evaluations and signed written informed consent.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maram Arafat, Sandra E Kleiman.
These authors jointly supervised this work: Mahmoud Huleihel, Moran Gershoni, Ruti Parvari.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00946-2.
References
- 1.Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol. 2018;15:369–84. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- 2.Tüttelmann F, Ruckert C, Röpke A. Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Medizinische Genetik: Mitteilungsblatt des Berufsverbandes Medizinische Genetik eV. 2018;30:12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, et al. Male infertility. Lancet. 2021;397:319–33. doi: 10.1016/S0140-6736(20)32667-2. [DOI] [PubMed] [Google Scholar]
- 4.Jarvi K, Lo K, Fischer A, Grantmyre J, Zini A, Chow V, et al. CUA guideline: the workup of azoospermic males. Can Urological Assoc J = J de l’Assoc des urologues du Can. 2010;4:163–7. doi: 10.5489/cuaj.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schilit SLP, Menon S, Friedrich C, Kammin T, Wilch E, Hanscom C, et al. SYCP2 translocation-mediated dysregulation and frameshift variants cause human male infertility. Am J Hum Genet. 2020;106:41–57. doi: 10.1016/j.ajhg.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooke HJ, Saunders PT. Mouse models of male infertility. Nat Rev Genet. 2002;3:790–801. doi: 10.1038/nrg911. [DOI] [PubMed] [Google Scholar]
- 7.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci USA. 2003;100:12201–6. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh P, Schimenti JC. The genetics of human infertility by functional interrogation of SNPs in mice. Proc Natl Acad Sci USA. 2015;112:10431–6. doi: 10.1073/pnas.1506974112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oud MS, Volozonoka L, Smits RM, Vissers L, Ramos L, Veltman JA. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod. 2019;34:932–41. doi: 10.1093/humrep/dez022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Wolfswinkel JC. Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integr Comp Biol. 2014;54:700–13. doi: 10.1093/icb/icu084. [DOI] [PubMed] [Google Scholar]
- 11.Wyrwoll MJ, Temel ŞG, Nagirnaja L, Oud MS, Lopes AM, van der Heijden GW, et al. Bi-allelic mutations in M1AP are a frequent cause of meiotic arrest and severely impaired spermatogenesis leading to male infertility. Am J Hum Genet. 2020;107:342–51. doi: 10.1016/j.ajhg.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasak L, Punab M, Nagirnaja L, Grigorova M, Minajeva A, Lopes AM, et al. Bi-allelic recessive loss-of-function variants in FANCM cause non-obstructive Azoospermia. Am J Hum Genet. 2018;103:200–12. doi: 10.1016/j.ajhg.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyamoto T, Hasuike S, Yogev L, Maduro MR, Ishikawa M, Westphal H, et al. Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet. 2003;362:1714–9. doi: 10.1016/S0140-6736(03)14845-3. [DOI] [PubMed] [Google Scholar]
- 14.Maor-Sagie E, Cinnamon Y, Yaacov B, Shaag A, Goldsmidt H, Zenvirt S, et al. Deleterious mutation in SYCE1 is associated with non-obstructive azoospermia. J Assist Reprod Genet. 2015;32:887–91. doi: 10.1007/s10815-015-0445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatsenko AN, Georgiadis AP, Röpke A, Berman AJ, Jaffe T, Olszewska M, et al. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med. 2015;372:2097–107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershoni M, Hauser R, Yogev L, Lehavi O, Azem F, Yavetz H, et al. A familial study of azoospermic men identifies three novel causative mutations in three new human Azoospermia genes. Genet Med. 2017;19:998–1006. doi: 10.1038/gim.2016.225. [DOI] [PubMed] [Google Scholar]
- 17.Gershoni M, Hauser R, Barda S, Lehavi O, Arama E, Pietrokovski S, et al. A new MEIOB mutation is a recurrent cause for azoospermia and testicular meiotic arrest. Hum Reprod. 2019;34:666–71. doi: 10.1093/humrep/dez016. [DOI] [PubMed] [Google Scholar]
- 18.Arafat M, Har-Vardi I, Harlev A, Levitas E, Zeadna A, Abofoul-Azab M, et al. Mutation in TDRD9 causes non-obstructive azoospermia in infertile men. J Med Genet. 2017;54:633–9. doi: 10.1136/jmedgenet-2017-104514. [DOI] [PubMed] [Google Scholar]
- 19.Carmell MA, Dokshin GA, Skaletsky H, Hu YC, van Wolfswinkel JC, Igarashi KJ, et al. A widely employed germ cell marker is an ancient disordered protein with reproductive functions in diverse eukaryotes. Elife. 2016;5:e19993. doi: 10.7554/eLife.19993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yudkina AV, Dvornikova AP, Zharkov DO. Variable termination sites of DNA polymerases encountering a. DNA-protein cross-link. 2018;13:e0198480. doi: 10.1371/journal.pone.0198480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enders GC, May JJ., 2nd Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Developmental Biol. 1994;163:331–40. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- 22.Bhargava V, Goldstein CD, Russell L, Xu L, Ahmed M, Li W, et al. GCNA preserves genome integrity and fertility across species. Dev Cell. 2020;52:38–52.e10. doi: 10.1016/j.devcel.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dokshin GA, Davis GM, Sawle AD, Eldridge MD, Nicholls PK, Gourley TE, et al. GCNA interacts with Spartan and Topoisomerase II to regulate genome stability. Dev Cell. 2020;52:53–68.e6. doi: 10.1016/j.devcel.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Björndahl L, Barratt CL, Mortimer D, Jouannet P. ‘How to count sperm properly’: checklist for acceptability of studies based on human semen analysis. Hum Reprod. 2016;31:227–32. doi: 10.1093/humrep/dev305. [DOI] [PubMed] [Google Scholar]
- 25.Hauser R, Botchan A, Amit A, Ben Yosef D, Gamzu R, Paz G, et al. Multiple testicular sampling in non-obstructive azoospermia-is it necessary? Hum Reprod. 1998;13:3081–5. doi: 10.1093/humrep/13.11.3081. [DOI] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma (Oxf, Engl) 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma (Oxf, Engl) 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arafat M, Harlev A, Har-Vardi I, Levitas E, Priel T, Gershoni M, et al. Mutation in CATIP (C2orf62) causes oligoteratoasthenozoospermia by affecting actin dynamics. J Med Genet. 2020. 10.1136/jmedgenet-2019-106825. [DOI] [PubMed]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–36. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinforma. 2016;54:1.30.1–1.3. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 35.Karczewski KJ, Francioli LC. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. [DOI] [PMC free article] [PubMed]
- 36.Piotrkowski B, Monzón CM, Pagotto RM, Reche CG, Besio M, Cymeryng CB, et al. Effects of heme oxygenase isozymes on Leydig cells steroidogenesis. J Endocrinol. 2009;203:155–65. doi: 10.1677/JOE-09-0061. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura T, Nagamori I, Nakatani T, Izumi N, Tomari Y, Kuramochi-Miyagawa S. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 2018;19:e44957. 10.15252/embr.201744957. [DOI] [PMC free article] [PubMed]
- 38.Di Persio S, Saracino R, Fera S, Muciaccia B, Esposito V, Boitani C. Spermatogonial kinetics in humans. Development. 2017;144:3430–9. [DOI] [PubMed]
- 39.Okada S, Kuroki K, Ruiz CA, Tosi AJ. Imamura M Molecular histology of spermatogenesis in the Japanese macaque monkey (Macaca fuscata) Primates. 2021;62:113–21. doi: 10.1007/s10329-020-00857-8. [DOI] [PubMed] [Google Scholar]
- 40.Okada S, Kuroki K, Ruiz CA, Tosi AJ, Imamura M. Molecular histology of spermatogenesis in the Japanese macaque monkey (Macaca fuscata) Primates. 2021;62:113–21. doi: 10.1007/s10329-020-00857-8. [DOI] [PubMed] [Google Scholar]
- 41.Yatsenko AN, Roy A, Chen R, Ma L, Murthy LJ, Yan W, et al. Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum Mol Genet. 2006;15:3411–9. doi: 10.1093/hmg/ddl417. [DOI] [PubMed] [Google Scholar]
- 42.Ayhan Ö, Balkan M, Guven A, Hazan R, Atar M, Tok A, et al. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet. 2014;51:239–44. doi: 10.1136/jmedgenet-2013-102102. [DOI] [PubMed] [Google Scholar]
- 43.Hardy JJ, Wyrwoll MJ, McFadden W, Malcher A, Rotte N, Pollock NC, et al. Variants in GCNA, X-linked germ-cell genome integrity gene, identified in men with primary spermatogenic failure. Hum Genet. 2021;140:1169–82. doi: 10.1007/s00439-021-02287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, et al. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol Cell. 2016;64:688–703. doi: 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parhad SS, Tu S, Weng Z, Theurkauf WE. Adaptive evolution leads to cross-species incompatibility in the piRNA transposon silencing machinery. Developmental Cell. 2017;43:60–70.e5. doi: 10.1016/j.devcel.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borgermann N, Ackermann L, Schwertman P, Hendriks IA, Thijssen K, Liu JC, et al. SUMOylation promotes protective responses to DNA-protein crosslinks. EMBO J. 2019;38:e101496. doi: 10.15252/embj.2019101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 2: Assessment of the conservation of the identified PV in GCNA.
Supplementary Table 3: Population frequencies and In-silico predictions for the effect of the candidate variations in patient 1 (A) and patients 2 (B).
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the GV shared LOVD repository, https://databases.lovd.nl/shared/genes.