Abstract
It has been shown that phages have evolved anti-CRISPR (Acr) proteins to inhibit host CRISPR-Cas systems. Most acr genes are located upstream of anti-CRISPR-associated (aca) genes, which is instrumental for identifying these acr genes. Thus far, eight Aca families (Aca1–Aca8) have been identified, all proteins of which share low sequence homology and bind to different target DNA sequences. Recently, Aca1 and Aca2 proteins were discovered to function as repressors by binding to acr-aca promoters, thus implying a potential anti-anti-CRISPR mechanism. However, the structural basis for the repression roles of Aca proteins is still unknown. Here, we elucidated apo-structures of Aca1 and Aca2 proteins and their complex structures with their cognate operator DNA in two model systems, the Pseudomonas phage JBD30 and the Pectobacterium carotovorum template phage ZF40. In combination with biochemical and cellular assays, our study unveils dimerization and DNA-recognition mechanisms of Aca1 and Aca2 family proteins, thus revealing the molecular basis for Aca1-and Aca2-mediated anti-CRISPR repression. Our results also shed light on understanding the repression roles of other Aca family proteins and autoregulation roles of acr-aca operons.
Keywords: anti-CRISPR-associated, Aca1, Aca2, anti-CRISPR, X-ray crystallography, protein–DNA complex
Abbreviations: aca, anti-CRISPR-associated; acr, anti-CRISPR; CRIPSR, clustered regularly interspaced short palindromic repeat; HTH, helix-turn-helix; IPTG, isopropyl-β-D-thiogalactoside; IR, inverted repeat; ITC, isothermal titration calorimetry; LB, Luria–Bertani; MGE, mobile genetic element; SAD, single-wavelength anomalous diffraction
Bacteria and archaea have evolved several immune mechanisms, including CRISPR-Cas systems, to defend against invaders such as bacterial viruses (phages) and other mobile genetic elements (MGEs) (1, 2). CRISPR-Cas systems are remarkably diverse, with RNA-guided adaptive immune systems composed of two classes and six types (I–VI), each with various subtypes (3, 4, 5). CRISPR-Cas systems carry out their defense activities in three stages (6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16): (i) During the adaptation stage, hosts capture invader-derived DNA and integrate them as spacers into the clustered regularly interspaced short palindromic repeats (CRISPRs) in their chromosome; (ii) during the expression stage, the CRISPR arrays are transcribed and processed into mature CRISPR RNAs, which then associate with CRISPR-associated (Cas) effectors to form surveillance complexes; (iii) during the interference stage, surveillance complexes can identify foreign genetic elements that are complementary to the CRISPR RNAs, thus leading to target interference or cleavage through intrinsic or recruiting nuclease activity. Like an arms race, phages and other MGEs have coevolved anti-CRISPR (Acr) proteins to counteract the hosts’ CRISPR-Cas systems (17, 18, 19). More than 40 Acr proteins have been identified as having inhibitory effects on distinct CRISPR-Cas systems since it was first reported in 2013 (17, 20). The Acr proteins are very diverse and display no common features in sequences and structures; however, most anti-CRISPR (acr) genes are found upstream of a conserved gene, which is referred to as an anti-CRISPR-associated (aca) gene (17). The conservation of aca genes has enabled the development of guilt-by-association bioinformatic methods to identify potential Acr proteins. So far, eight families of aca genes (aca1–aca8) have been reported (https://tinyurl.com/anti-CRISPR) (21, 22, 23, 24).
Although Aca proteins are predicted to possess a helix-turn-helix (HTH) DNA-binding motif that would potentially help regulate adjacent acr genes (21, 22, 23, 24), the exact functions of Aca proteins have just started to be elucidated (25, 26). The Aca1 protein of Pseudomonas aeruginosa phage JBD30, which appears to function as a repressor by binding to inverted repeat 2 (IR2) in the promoter of the acrIF1-aca1 operon to downregulate the expression of the AcrIF1 protein, is conserved among different anti-CRISPR-encoding phages and prophages from Pseudomonas species (20, 22). This repressive effect is essential, as without it, strong constitutive transcription from acr promoters would be detrimental for phages survival (26). Likewise, the Aca2 protein of Pectobacterium carotovorum temperate phage ZF40 was demonstrated to function as an autoregulator through dimerization and binding to inverted repeat 1 (IR1) in the promoter of the aca2-acrIF8 operon to downregulate the AcrIF8 protein level (25). Interestingly, this autoregulation mechanism of Aca proteins is also employed by some fusion anti-CRISPR proteins, including AcrIIA1 of Listeria monocytogenes temperate phages and its orthologs (27), as well as AcrIIA13, AcrIIA14, and AcrIIA15 of Staphylococcus species (28), all of which contain an N-terminal HTH motif (NTD) that acts as a repressor in the acr promoter and a C-terminal domain that functions as an anti-CRISPR. Interestingly, heterologous overexpression of Aca1 and AcrIIA1NTD, or the host hijacked version of the AcrIIA1NTD homolog, can inhibit phages, thus indicating that Aca proteins and AcrIIA1NTD homologs are engineered into or naturally co-opted by hosts as anti-anti-CRISPR mechanisms against phages and other MGEs (27). More remarkably, the anti-anti-CRISPR mechanism of Aca1 has been utilized recently to overcome the Acr protein-mediating impediment of a Cascade-Cas3 bacterial genome editing tool, which further highlights the potential application of Aca families in genome editing (29). The repressor and potential anti-anti-CRISPR functions of the Aca families depend on their associations with inverted repeats in the acr promoters; however, the structural basis for how Aca proteins recognize their cognate DNA targets is unknown.
Here, we focused on the Aca1 protein from P. aeruginosa phage JBD30 and the Aca2 protein from P. carotovorum template phage ZF40. We determined the apo-structure of JBD30 Aca1 and its complex structure with inverted repeat 2 (IR2) DNA from the promoter of the acrIF1-aca1 operon, as well as the apo-structure of ZF40 Aca2 and its complex structure with IR1 DNA from the promoter of the acrIF8-aca2 operon. Our structural studies demonstrate that both the JBD30 Aca1 and ZF40 Aca2 form rigid homodimers and that the ZF40 Aca2 has a distinct dimerization mechanism as compared with the JBD30 Aca1. The sequence conservations further support that dimerization is an intrinsic feature of Aca1 and Aca2 families. JBD30 Aca1 and ZF40 Aca2 each contain an HTH DNA-binding motif and utilize the recognition helix α3 of the HTH domain to recognize its cognate operator DNA. Binding with the JBD30 Aca1 dimer bends the IR2 DNA into an arch-like structure with an overall bending of 42°, while binding with ZF40 Aca2 does not bend IR1 DNA. Residues of JBD30 Aca1 and ZF40 Aca2 involved in DNA recognition were verified by isothermal titration calorimetry (ITC) assays. The eGFP-fluorescence reporter assays further determined the conserved residues of JBD30 Aca1 and ZF40 Aca2 that are involved in transcription repression. In summary, our structural, biochemical, and cellular analysis uncovered the conserved recognition and repression modes of the Aca1 and Aca2 families and provides a structural basis that establishes the potential value of Aca proteins in future applications.
Results
Overall structure of Aca1 homodimer
To dissect the molecular basis for Aca-mediating anti-CRISPR repression, we first solved a 1.80 Å-resolution apo-structure of the full-length Aca1 (a.a. 1–79) from P. aeruginosa phage JBD30, for which the initial phase had been determined by a single-wavelength anomalous diffraction (SAD) method with a selenomethionine-derived Aca1 crystal diffracted to 1.85 Å resolution (Table 1).
Table 1.
Data collection and refinement statistics
| Crystallographic statistics | Native–Aca1 | SeMet–Aca1 | Aca1–IR2 | Native–Aca2 | SeMet–Aca2 | Aca2–DNA |
|---|---|---|---|---|---|---|
| Accession code | 7VJN | 7FA3 | 7VJM | 7VJO | 7VJP | 7VJQ |
| Data collection | ||||||
| Space group | P212121 | P212121 | P4122 | C2 | P21 | C2 |
| Cell dimensions | ||||||
| a, b, c (Å) | 45.70, 50.44, 64.92 | 45.43, 50.44, 64.74 | 42.71, 42.71, 362.37 | 96.20, 34.57, 81.30 | 39.65, 67.15, 42.60 | 129.31, 168.47, 41.49 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 124.58, 90 | 90, 106.92, 90 | 90, 90.06, 90 |
| Resolution (Å) | 40.00–1.80 (1.86–1.80)a | 40.00–1.85 (1.92–1.85) | 30.00–3.00 (3.16–3.00) | 50.00–1.23 (1.25–1.23) | 67.15–1.59 (1.68–1.59) | 40.00–2.80 (2.90–2.80) |
| Rmerge | 0.069 | 0.084 | 0.092 | 0.171 | 0.094 | 0.060 |
| I/σI | 29.4 (3.0) | 28.4 (2.7) | 20.1 (4.6) | 18.28 (2.7) | 8.8 (3.5) | 16.9 (3.0) |
| CC1/2 of the highest-resolution shell | 0.944 | 0.917 | 0.980 | 0.816 | 0.774 | 0.918 |
| Completeness (%) | 99.7 (99.4) | 99.3 (94.6) | 87.0 (85.1) | 98.9 (99.8) | 99.4 (99.7) | 95.6 (78.8) |
| Redundancy | 10.2 | 11.9 | 11.3 | 6.3 | 5.1 | 3.4 |
| Refinement | ||||||
| Resolution (Å) | 39.83–1.80 | 22.05–1.85 | 29.79–3.00 | 39.60–1.31 | 32.96–1.59 | 38.48–2.79 |
| Completeness (%) | 99.54 | 87.39b | 85.71 | 97.27b | 99.25 | 79.71b |
| No. reflections | 14,401 | 11,611 | 6470 | 51,858 | 28,304 | 17,557 |
| Rwork/Rfree (%) | 19.04/21.45 (24.00/27.88)a | 19.86/22.03 (19.79/25.61) | 19.49/25.36 (23.10/33.03) | 15.56/18.48 (20.87/22.54) | 15.20/18.80 (18.58/22.41) | 19.01/21.85 (26.52/34.00) |
| No. atoms | 1235 | 1220 | 1948 | 4212 | 2189 | 3103 |
| Protein | 1104 | 1133 | 1151 | 3739 | 1907 | 1928 |
| Ligand/Water | 131 | 87 | 473 | 282 | 74 | |
| DNA | 797 | 1101 | ||||
| B–factors | 37.23 | 27.40 | 58.40 | 20.08 | 19.53 | 55.35 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.013 | 0.008 | 0.004 | 0.007 | 0.006 | 0.003 |
| Bond angles (°) | 1.211 | 0.969 | 0.688 | 0.954 | 0.767 | 0.548 |
| Ramachandran plot | ||||||
| Outliers (%) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Favored/allowed (%) | 100.00/0.00 | 100.00/0.00 | 96.45/3.55 | 100.0/0.00 | 99.15/0.85 | 98.73/1.27 |
Values in parentheses are for the highest-resolution shell. One crystal was used for each data set.
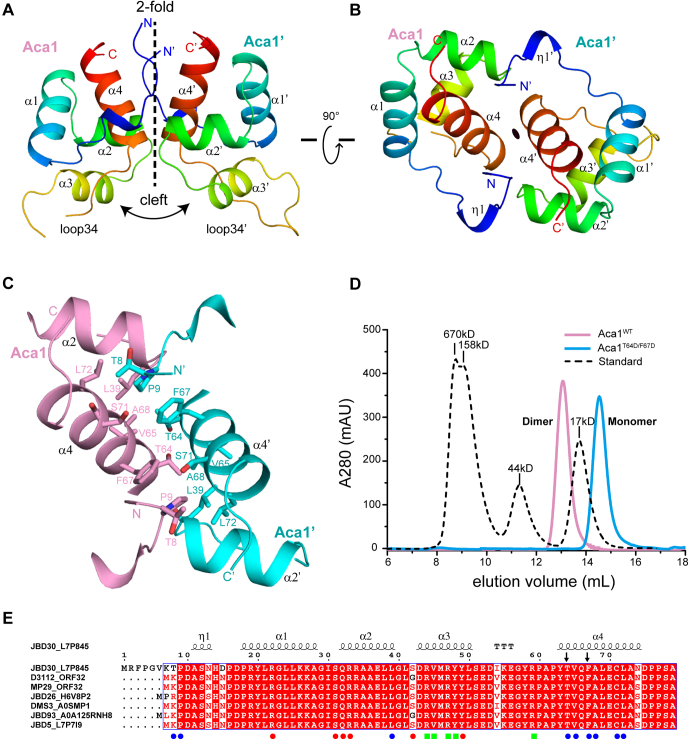
The space group of the native structure is P212121, and two molecules of Aca1 are located in an asymmetric unit where they form a homodimer with twofold symmetry (Fig. 1, A and B). Due to the good quality of the electron density map, amino acids (a.a.) 6 to 54 and 58 to 77 of protomer A and 7 to 77 protomer B could be modeled. The overall structure of the Aca1 protomer contains four α-helices, α1 (a.a. 18–28), α2 (a.a. 32–39), α3 (a.a. 43–50), and α4 (a.a. 63–74), which form a tetra-helical bundle and a small 310-helix (a.a. 11–13; η1 helix) located at the extended N-terminus (Fig. 1, A and B). The α2 and α3 helices are nearly perpendicular to one another and are connected by a sharp turn, which corresponds to the predicted HTH, with α3 being the recognition helix (26). Another interesting feature is that loop34 (a.a. 51–62) connecting the α3 and α4 helices protruded from the globular tetra-helical bundle, which is also involved in DNA recognition (see below).
Figure 1.
Overall structure of apo-Aca1.A and B, two overall views of the Aca1 homodimer structure 90° apart in the illustration. The two protomers Aca1 and Aca1′ are colored by rainbow from N terminus (blue) to C terminus (red), respectively. The twofold symmetry, cleft, and long loops are highlighted. C, intermolecular contacts of the dimer interface. The two protomers Aca1 and Aca1′ are colored in pink and cyan, respectively. The residues involved in interactions are shown in sticks representation. D, size-exclusion chromatography analysis of the oligomeric statuses of wild-type Aca1 and double-mutant T64D/F67D. The black dashed line is a molecular weight standard. E, sequence alignment of Aca1 from phage JBD30 and other phages. Under alignment, the residues involved in dimer formation are indicated with blue circles, while the residues interact with the DNA backbone and bases are indicated with red circles and green rectangles, respectively. Secondary structures of Aca1 from Pseudomonas phage JBD30 are shown on top of the alignment. The two residues Thr64 and Phe67 selected for the double mutant T64D/F67D in panel D are indicated with arrows.
Intermolecular contacts of the Aca1 homodimer
Regarding the quaternary structure, hydrophobic contacts between α4 of protomer A and α4′ of protomer B constitute the major interactions of the dimer interface, which is further stabilized by hydrophobic contacts from the two N-termini and the α2 and α2′ helices (Fig. 1C). Residues Thr64, Val65, Phe67, Ala68, and Ser71 in α4 interact with their equivalent residues in α4′ to form a hydrophobic core with a twofold axis going through the center of the core. Residues Thr8 and Pro9 in the N-terminus and Leu39 at the end of α2 of protomer A interact with the hydrophobic core from opposite sides, whereas the equivalent residues of promoter B form similar interactions (Fig. 1C). Concordantly, our size-exclusion chromatography analysis revealed that wild-type Aca1 formed a dimer conformation in solution and that a double mutant of Aca1 involving substitutions of Thr64 and Phe67 with aspartic acids (T64D/F67D) in the dimer interface could disrupt dimer formation (Fig. 1D). Interestingly, sequence alignments revealed that Aca1 had homologs in JBD30-related phages and bacteria, with respective sequence identities of 90 to 96% and 47 to 72% (Figs. 1E and S1A). Most of the residues involved in dimerization, except Thr8, are strictly conserved in the phage homologs and type-conserved in the bacterial homologs, which indicates that dimerization is an intrinsic feature of the Aca1 family. It should be noted that the residue Thr64 in the dimer interface is substituted by an arginine residue in the Pseudomonas syringae homolog (Fig. S1A), which may locally disturb the dimer interface. Thus, this bacterial homolog might form monomer, but not dimer, in solution. Moreover, the dimerization process of Aca1 places the recognition helices α3 and α3′ in an antiparallel position that is suitable for recognition of operator DNA containing an inverted repeat, and forms an open cleft between α3 and α3′, which is expected to accommodate the operator DNA (Fig. 1A).
A structure-based similarity search with the Dali server (30) revealed that the Aca1 protomer structure displayed significant similarities to the HTH domains of many transcription regulators with low sequence identities, including the well-studied Vibrio cholerae antitoxin and autorepressor HigA2 (PDB ID: 5JAA; Z-score 7.5; 2.20 Å RMSD for 61 Cα atoms; sequence identity 11%) and Escherichia coli antitoxin and transcription repressor MqsA (PDB ID: 3GN5; Z-score 6.9; 2.50 Å RMSD for 63 Cα atoms; sequence identity 17%) (Fig. S2, A and B) (31, 32). As both HigA2 and MqsA can form dimers in solution, we thus extended the structural comparisons to the level of quaternary structure and revealed that the quaternary structure of the Aca1 dimer was also similar to that of the two transcription regulators (Fig. S2, C and D).
Structure of the Aca1 homodimer in complex with IR2 DNA
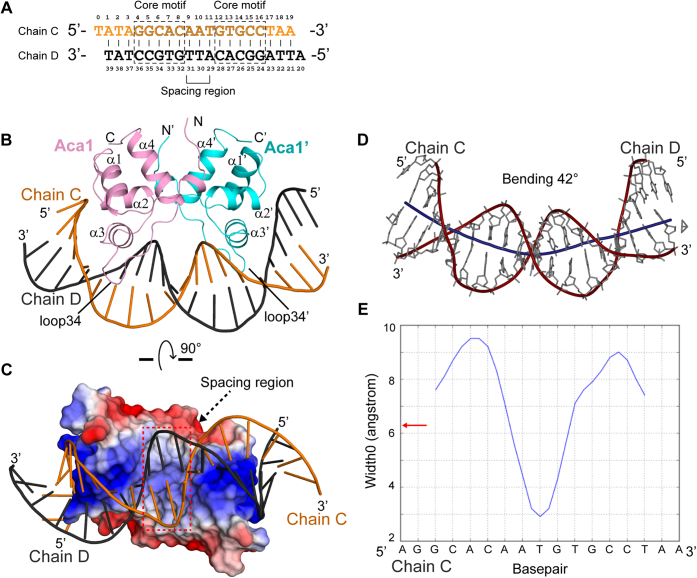
To further understand the molecular basis for Aca1-mediating anti-CRISPR repression, we designed a 19-bp operator DNA (Fig. 2A, Table S1) that corresponded to the inverted repeat 2 (IR2) in the acrIF1-aca1 promoter of phage JBD30 (Fig. S3A) (26), for cocrystallization with Aca1. The designed 19-bp IR2 DNA contained 5′ overhangs that would facilitate crystal packing during crystallization. We successfully solved the crystal structure of Aca1 in complex with the 19-bp IR2 DNA (hereafter referred to as Aca1–IR2 complex), which was refined to 3.0 Å resolution (Table 1). The whole complex structure exhibited a similar twofold symmetry as the apo-structure, with each Aca1 protomer binding to a half site of the IR2 DNA (Fig. 2B), and the 5′ overhangs indeed facilitate crystal packing (Fig. S3B). Consistently, two halves of the complex structure were superimposed with a small RMSD of 0.20 Å (for 62 Cα atoms) (Fig. S3C). The IR2 DNA adopts an arch-like conformation, with the 5′ part of both DNA strands in the flanking region being grasped by α2 and α2′ of the Aca1 dimer, respectively (Fig. 2B). The recognition helices α3 and α3′ are inserted into the major groove to recognize the core motifs specifically, and the two conserved loops, loop34 and loop34′, also interact with the major groove (Fig. 2B). Moreover, the minor groove is accommodated in the cleft of the Aca1 dimer, thus, the protein and DNA were complementary at the binding interface (Fig. 2C). Further analysis using the program Curve+ (33) revealed that the IR2 DNA bound by Aca1 curved by 42° and the minor groove width narrowed around the spacing region (Fig. 2, D and E).
Figure 2.
Structure of Aca1 in complex with IR2 DNA.A, the sequence of the 19-bp 5′-overhang IR2 DNA used for crystallization. The core motifs and spacer region are highlighted. B, overall structure of Aca1 in complex with 19-bp-DNA. The Aca1 dimer is colored as in Figure 1C. C, surface view of the Aca1–IR2 complex color-coded with the electrostatic potential (blue, positively charged; red, negatively charged). The cleft enclosing the spacer region is highlighted. D, bent conformation of the IR2 DNA. Helical axis of curvature is highlighted with a dark blue line through the center of DNA. E, the blue curve represents variation of the minor groove widths (Å) along the Aca1-bound IR2 DNA (Chain C 5′-AGGCACAATGTGCCTAA-3′). The red arrow denotes the widths of minor groove in a canonical B-form DNA. Both panels D and E are generated using the program Curves+ (33).
The structural comparison between the apo-structure and complex structure further revealed that the overall structure of the Aca1 dimer did not show any significant conformational change upon IR2 DNA binding, with a small RMSD of 0.45 Å (for 124 Cα atoms) that indicated a rigid dimer of Aca1 (Fig. S3D). However, the conformations of loop34 and loop34′ were adjusted to bind the major groove of the IR2 DNA. The rigid and twofold symmetric dimer separated the two antiparallel recognition helices α3 and α3′ in a distance of approximately 29 Å (measured between the Cα atoms of Arg47 and Arg47′), while the distance between adjacent major grooves in B-form DNA was approximately 34 Å. This was consistent with the observation that binding of the Aca1 dimer induced the IR2 DNA to bend by approximately 42°.
Recognitions of IR2 DNA by the Aca1 homodimer
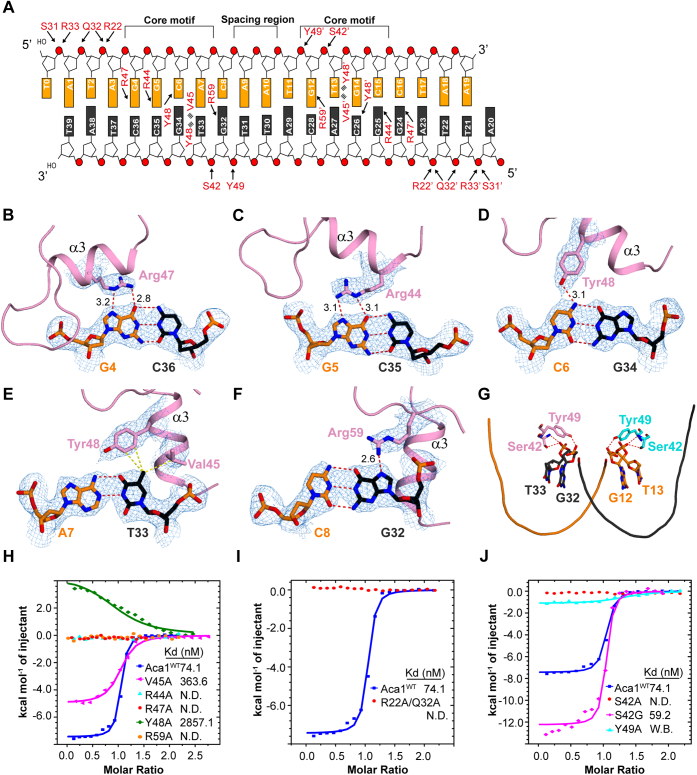
Due to the twofold symmetrical feature of the Aca1–IR2 complex, we selected one-half of the structure to further analyze the intermolecular contacts. The side chains of Ser31 and Gln32 established hydrogen bonds in the phosphate groups of A1 and A3, respectively, in the flanking region (Fig. 3A), whereas the main chain of Gln32 is hydrogen-bonded to the phosphate group of T2 in the flanking region. Moreover, Arg33 and Arg22 each form two salt-bridges with the phosphate groups of A1 and A3, respectively. Ser31, Gln32, and Arg33 are located at the N-terminal end of Aca1 α2, and Arg22 is from α1. Thus, these residues in the Aca1 protomer and their equivalents in the other protomer grasp the phosphate backbone in the 5′ part of both DNA strands.
Figure 3.
Detailed interactions and ITC results between Aca1 and IR2 DNA.A, schematic representation of the detailed interactions between Aca1 and IR2 DNA. Hydrogen bonds and salt-bridges are indicated by black arrows and hydrophobic interactions are indicated with multi-lines. A 3D view of the overall interactions between Aca1 and IR2 DNA is shown in Figure S4, A and B. B–F, detailed interactions in the core motifs. Hydrogen bonds and distance are shown in red dashed lines, and hydrophobic interactions are shown in yellow dashed lines. The 2|Fo|–|Fc| σ-weighted map is contoured at 1.0 σ. G, Aca1 Ser42 and Tyr49 bind to the minor groove around the spacer region. H, ITC results of Aca1-wt and mutants with 21-bp blunt-end IR2 DNA. I, ITC results of Aca1-wt and R22A/Q32A mutant with 21-bp IR2 DNA. J, ITC results of wild-type Aca1, S42A, S42G, and Y49A with 21-bp IR2 DNA. All raw data of the ITC experiments are shown in Table S2, Figures S5 and S6. N.D., not detected. W.B., week binding.
The specificity between Aca1 and the IR2 DNA comes primarily from the recognitions between Aca1 α3 and the core motifs (half-site 5′-GGCAC-3′): In the G4:C36 pair, the O6 and N7 atoms of G4 were hydrogen-bonded to the side chain of Arg47 (Fig. 3B), which were bidentate-bonding interactions that conferred specific recognition of guanine base by arginine residue (34); in the G5:C35 pair, the O6 and N7 atoms of G5 were hydrogen-bonded to the side chain of Arg44 (Fig. 3C), which were also conferred specific recognition of G5; in the C6:G34 pair, the N4 atom of C6 established a hydrogen bond with the side chain of Tyr48 (Fig. 3D); in the A7:T33 pair, the C7 methyl group of T33 formed hydrophobic interactions with the side chains of Val45 and Tyr48 (Fig. 3E), which seemed to confer specific recognition of A7 through its pairing base T33; and in the C8:G32 pair, the N7 atom of G32 formed a hydrogen bond with the side chain of Arg59 (Fig. 3F). Taken together, the two guanine and adenine bases in the core motifs (half-site 5′-GGCAC-3′) were specifically recognized by Aca1, while the two cytosine bases may contribute less to the specific recognitions. Thus, the residues Arg44, Val45, Arg47, and Tyr48 in α3 and Arg59 in loop34, together with their equivalents in the other Aca1 protomer, materialized the specific recognitions of the core motifs in IR2 DNA.
The minor groove around the spacing region is accommodated in the cleft of the Aca1 dimer. The spacing nucleotides (A9-A10-T11 or A29-T30-T31) had little contact with the proteins, but the phosphate groups of the adjacent nucleotides (G12-T13 or G32-T33) were bound by proteins (Fig. 3, A and G): both the side chain and main chain of Ser42 were hydrogen-bonded to the phosphate group of T33, whereas the side chain of Tyr49 formed a hydrogen bond with the phosphate group of G32. Similar interactions were observed for Ser42 and Tyr49 of the other Aca1 protomer with the phosphate groups G12 and T13. Thus, these interactions were expected to further stabilize the bent conformation of IR2 DNA.
Considered together, these analyses elucidated the molecular basis for specific recognition of IR2 DNA by the Aca1 homodimer.
Mutagenesis studies on Aca1
To validate the essential roles of Aca1 residues in the recognition of IR2 DNA, we performed mutagenesis studies with ITC experiments. At first, the Aca1 residues Arg44, Val45, Arg47, Tyr48, and Arg59, most of which are involved in specific interactions with the core motifs within the IR2 DNA, were each substituted with alanine. The ITC results revealed that (Fig. 3H; raw data in Table S2 and Fig. S5, A-F), compared with the wild-type Aca1 (Kd of 74.1 nM), the Aca1 R44A, R47A, and R59A mutants disrupted interactions with the 21-bp blunt-end IR2 DNA (5′-ATTAGGCACAATGTGCCTAAT-3′), while Y48A greatly reduced binding (Kd of 2857.1 nM). These results were consistent with the structural observations that these residues established prominent hydrogen bonds with the bases of the core motifs (Fig. 3, B–F). In contrast, the V45A mutant seemed to just partially influence binding (Kd of 363.6 nM), which was reasonable, because Aca1 Val45 and Tyr48 contributed to hydrophobic interactions in the recognition of T33. The contribution from Tyr48 was preserved in the V45A mutant, or the alanine substitution complemented a portion of the hydrophobic interactions (Fig. 3, E and H). Please note that the importance of the Aca1 residue Arg44 had been well studied in vitro and in vivo by others (26). Interestingly, the aforementioned residues of the JBD30 Aca1, including Arg44, Val45, Arg47, Tyr48, and Arg59, are strictly conserved in the phage homologs (Fig. 1E), while some of these residues are not conserved in the bacterial homologs, with Arg47 and Arg59 being strictly conserved, Tyr48 being type-conserved, and Arg44 and Val45 not conserved (Fig. S1A), implicated that these bacterial homologs may be able to bind to different DNA targets. Consistent with these observations, the phage Aca1 homologs were found to be involved in forming acr-aca1 operons, and their target DNAs (IR2 and IR1) were highly conserved (Fig. S4C). In contrast, at present we could not confirm that the bacterial Aca1 homologs were involved in forming acr-aca1 operons in their genomic contexts. Thus, the interactions of the bacterial Aca1 homologs and their potential target DNAs need to be studied in future.
Stabilization of the complex is achieved in part by contribution from nonspecific interactions. The Aca1 double-mutant R22A/Q32A disrupted binding to the 21-bp blunt-end IR2 DNA as verified by ITC (Figs. 3, A and I, Table S2 and S6A). Moreover, 23-bp and 21-bp blunt-end IR2 DNA, which were expected to preserve interactions in the flanking regions, showed similar binding affinities to Aca1 (Kd of 32.2 and 74.1 nM, respectively), whereas the interactions of 19-bp and 17-bp blunt-end IR2 DNA, which were reduced in the flanking regions, disrupted or weakened binding to Aca1 under the ITC conditions (Kd of 740.7 nM and N.D., respectively) (Figs. 3A and S6, B–D). These results verified the essential functions of these residues in recognition of the flanking regions in DNA. Furthermore, phosphate groups of the adjacent nucleotides (G12-T13 or G32-T33) surrounding the spacing region in DNA were held by Ser42 and Tyr49 (Fig. 3, A and G), which were expected to facilitate DNA bending. Consistently, the S42A and Y49A mutants almost disrupted interactions with IR2 DNA in vitro (Figs. 3J ; Table S2; S6, E and G). It was also noted that most of the residues involved in nonspecific interactions with DNA were strictly conserved and type-conserved in phage and bacterial homologues, respectively (Figs. 1E and S1A). Intriguingly, the residue Ser42, important for Aca1-DNA complex formation, was substituted by a glycine residue in the homologs from the phages D3112 and JBD93 (Fig. 1E). In contrast to the JBD30 Aca1 S42A mutant, the S42G mutant retained a similar DNA binding affinity as the wild-type Aca1 (Kd of 59.2 and 74.1 nM, respectively; Figs. 3J and S6F). It seemed that the substitution of a serine by an alanine at position 42 of JBD30 Aac1 would cause loss of the side-chain O–H···O hydrogen-bonding interactions, while the substitution by a glycine might, in our speculation, retain the interactions through the Cα–H···O hydrogen bond.
Interestingly, there were two inverted repeats, IR1 and IR2, in the acrIF1-aca1 promoter of P. aeruginosa phage JBD30, among which only IR2 was responsive for repression while IR1 titrated Aca1 away from IR2, thus fine-tuning repression by Aca1 in vivo (Fig. S3A) (26). There is a slight difference between 21-bp IR1 DNA (5′-AAGCGGCACACTGTGCCTATT-3′) and 21-bp IR2 DNA (5′-ATTAGGCACAATGTGCCTAAT-3′). Consistent with these observations, our ITC results revealed that 21-bp IR1 DNA bound to Aca1 with similar affinity as 21-bp IR2 DNA bound to Aca1 in vitro (Kd of 53.2 and 74.1 nM, respectively; Figs. S6H and S5A), which indicated that differences in the sequences of the flanking and spacing regions did not significantly influence interactions. Similar affinities between IR2 and IR1 to Aca1 suggested that these sites compete for Aca1 binding, which was observed in vivo in a previous study (26).
Considered together, these results support specific and nonspecific interactions between Aca1 and the IR2 DNA as essential for complex formation.
Aca1 residues involved in transcriptional repression
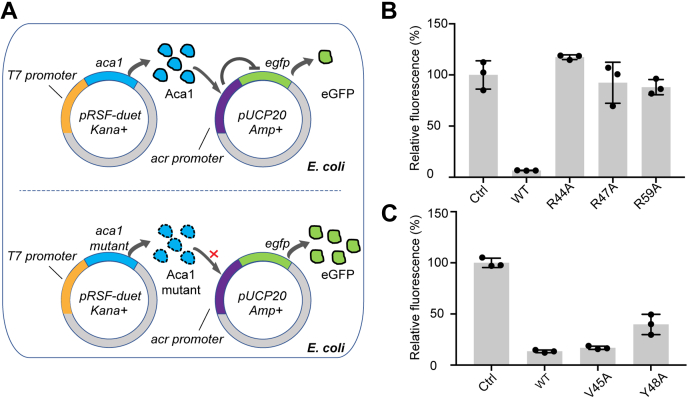
Given that all the mutagenesis studies were performed in vitro, we performed an in vivo eGFP-fluorescence reporter assay (25, 26) to evaluate the importance of the key residues for transcriptional repression of the acrIF1-aca1 operon. The promoter of the pUCP20 plasmid carrying the egfp gene was substituted with the sequence of the acrIF-aca1 promoter. The wild-type and mutant Aca1 genes were cloned into the pRSFDuet1 vector, which was controlled by the T7 promoter for isopropyl-β-D-thiogalactoside (IPTG)-induced protein production. The pUCP20 and pRSFDuet1 (wild-type or mutant Aca1) plasmids were cotransformed into the BL21 (DE3)-RIL E. coli strain for reporter assays (Fig. 4A). The fluorescence intensity revealed that a low level of eGFP underexpressed the wild-type Aca1 protein, thus indicating the robust repression of acrIF1-aca1 operon by Aca1. However, when the pRSFDuet1 plasmids carried the Aca1 mutant of R44A, R47A, or R59A, the fluorescence intensity was comparable to that of the control (Fig. 4B). The Aca1 Y48A mutant showed moderate repressive activity, while the V45A mutant retained its strong repressive effect on eGFP expression, which was similar to that of the wild-type Aca1 (Fig. 4C). These results were highly consistent with our structural observations and ITC results and revealed the involvement of more key residues of Aca1, as well as the well-studied Arg44 (26), in transcriptional repression of the acrIF1-aca1 operon.
Figure 4.
Aca1 can repress the expression of acrIF1-aca1 operon-controlled eGFP.A, schematic of the reporter assays in vivo. B and C, the repression activity of Aca1 and mutations on the acrIF1-aca1 promoter, which was determined as the median percentage of eGFP fluorescence intensity compared with the control. In (B) and (C), data are presented as the mean ± standard deviation of three biological replicates.
Molecular basis of Aca2 homodimers
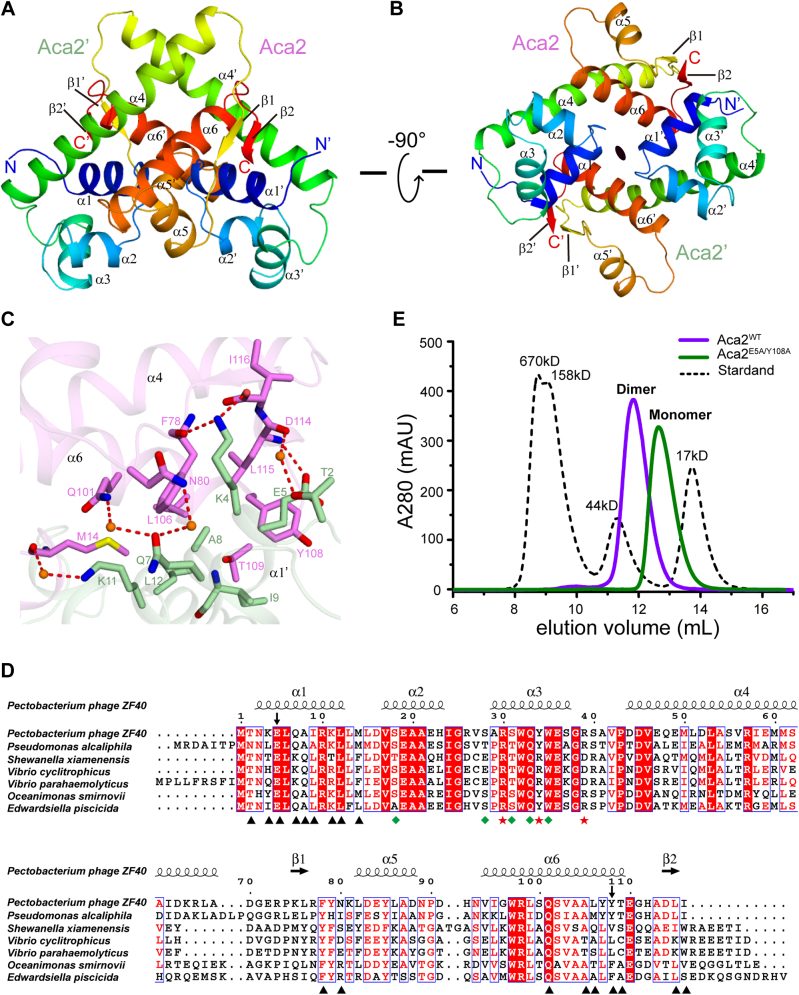
The proteins of different Aca1 families contain approximately 70 residues; for example, the Aca1 of P. aeruginosa phage JBD30 has 79 residues (Fig. 1E). Proteins of the Aca2 family have approximately 120 residues; for example, the Aca2 of P. carotovorum temperate phage ZF40 has 116 residues. The ZF40 Aca2 had been reported to form dimers in solution (25). We hypothesized that the extra C-terminal region of Aca2 is involved in quaternary-structure organization. To test this hypothesis, we solved the crystal structure of full-length ZF40 Aca2 refined to the 1.31 Å resolution, of which the phase was determined using the SAD method with selenomethionine-derived Aca2 crystal diffracted to the 1.59 Å resolution (Table 1). The overall structure of Aca2 also formed a dimer, with each protomer containing six α-helices and two parallel β-strands (Fig. 5, A and B). The N-terminal part of Aca2 (a.a. 1–70) constituted by the α1-α2-α3-α4 elements forms a structure that is largely similar to that of Aca1 with an RMSD of 0.93 Å (for 32 Cα atoms) (Fig. S7A), while the C-terminal extension (a.a. 71–116) constituting the β1-α5-α6-β2 elements formed a distinct structure that is involved in dimerization (Fig. 5, A and B). Intermolecular contacts within the Aca2 dimer mainly form between the C-terminal extension β1-α5-α6-β2 of one protomer and the α1′ helix of the other protomer, and vice versa: the residues Met14 in the loop connecting α1 and α2, the residues Phe78 and Asn80 in the loop connecting β1 and α5, the residues Gln101, Ala105, Leu106, Tyr108, and Thr109 in α6, and the residues Asp114, Leu115, and Ile116 in β2 of one protomer formed extensive hydrogen-bonding and hydrophobic interactions with the residues Thr2′, Lys4′, Glu5′, Gln7′, Ala8′, Ile9′, Lys11′, and Leu12′ in α1′ of the other promoter (Fig. 5, B and C). Moreover, most of the residues involved in dimerization, especially those residues located in the central region of the interface, were conserved or type-conserved in Aca2 homologs (Fig. 5, C and D), thus indicating that dimerization is also an intrinsic feature of the Aca2 family. Consistently, a double mutant E5A/Y108A of Aca2 disrupted dimerization, as was revealed by gel filtration assays (Fig. 5E).
Figure 5.
Structural insight of Aca2 homodimer.A and B, two overall views of the Aca2 homodimer structure 90° apart in the illustration. The two protomers Aca2 and Aca2′ are colored by rainbow from N terminus (blue) to C terminus (red), respectively. C, intermolecular contacts of the Aca2 homodimer. The two protomers Aca2 and Aca2′ are colored in violet and pale green, respectively. The residues involved in interactions are shown in sticks representation. Water molecules are shown as bright orange spheres. Direct and water-mediated hydrogen bonds are shown in red dashed lines. D, sequence alignment of Aca2 from different species. Under alignment, the residues involved in dimer formation are indicated with a black triangle, while the residues interact with the DNA backbone and bases are indicated with green rhombus and red star, respectively. Secondary structures of Aca2 from Pectobacterium phage ZF40 are shown on top of the alignment. The two residues Glu5 and Tyr108 selected for the double mutant E5A/Y108A in panel E are indicated with arrows. E, size-exclusion chromatography analysis of the oligomeric statuses of wild-type Aca2 and double mutant E5A/Y108A. The black dashed line is a molecular weight standard.
The structural elements involved in dimerization of Aca2 are totally different from those involved in the dimerization of Aca1 (Figs. 1C and 5C), which makes it impossible for the quaternary structure of the Aca2 dimer to be superimposed onto the quaternary structure of the Aca1 dimer. Although we could superimpose the structure of the N-terminal part of one Aca2 protomer with that of one Aca1 protomer (Fig. S7A), it would place other protomers of Aca2 and Aca1 on opposite sides (Fig. S7B). Thus, Aca2 and Aca1 families should adopt different dimerization mechanisms.
Recognitions of IR1 DNA by Aca2 homodimers
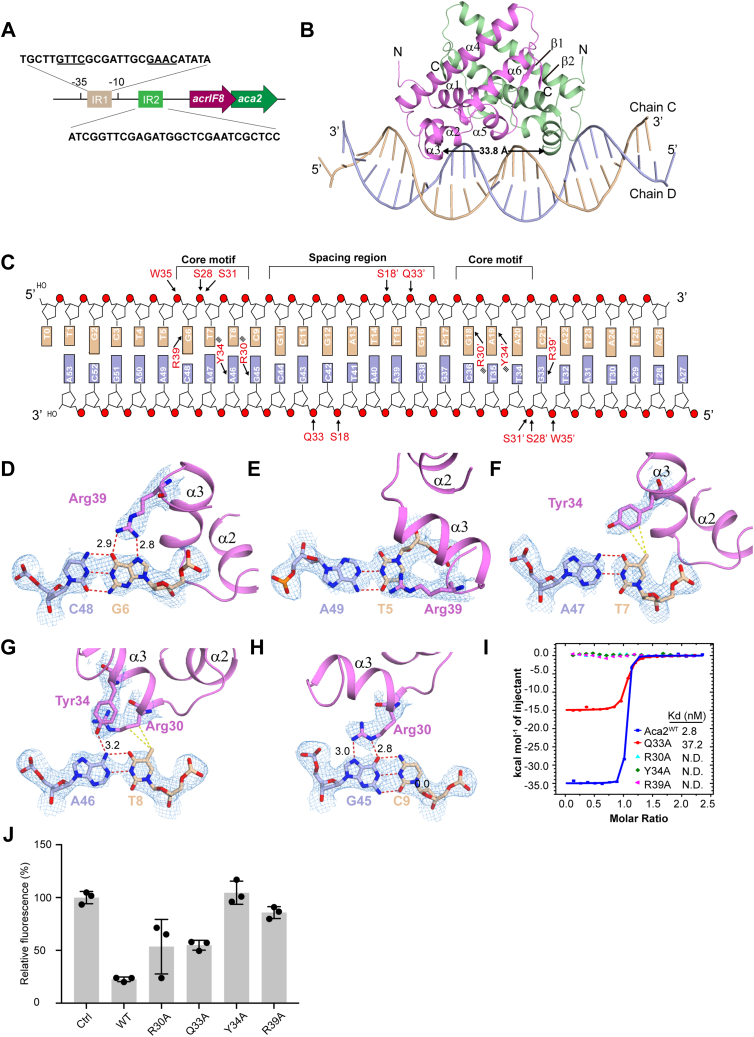
As ZF40 Aca2 bound tightly to the IR1 region from the acrIF8-aca2 operon in vitro (25) (Figs. 6A and S8A), we designed a 5′-overhanged 26-bp DNA derived from the IR1 region and solved a complex structure of the Aca2 dimer with this IR1 DNA (Figs. 6B and S7C). The 5′ overhangs indeed facilitate crystal packing in the complex structure (Fig. S7D). A small RMSD of 0.63 Å (for 205 Cα atoms) was revealed when compared with the Aca2 dimer in the apo- and complex structures, which suggested that Aca2 adopted a rigid dimer, just like Aca1 (Fig. S7E). The rigid Aca2 dimer separates the recognition helices α3 and α3′ in a distance of approximately 33.8 Å (measured between the Cα atoms of Tyr34 and Tyr34′), while the distance between adjacent major grooves of B-form DNA was just approximately 34 Å (Fig. 6B). Consistently, IR1 DNA in the complex structure was just slightly bent upon binding by the Aca2 dimer (Figs. 6B and S7, F and G).
Figure 6.
DNA-binding properties of Aca2.A, schematic representation of the acrIF8-aca2 operon. B, complex structure of Aca2 with 26-bp DNA substrates. The distance between the two α3 is indicated by a black arrow. C, schematic representation of the detailed interactions between Aca2 and IR1 DNA. The hydrogen bonds and salt-bridges are indicated by a black arrow, and the hydrophobic interactions are indicated with multi-lines. A 3D view of the overall interactions between Aca2 and IR1 DNA is shown in Figure S9, A and B. D–H, detailed interactions in the core motifs. Hydrogen bonds and distance are shown in red dashed lines, and hydrophobic interactions are shown in yellow dashed lines. The 2|Fo|–|Fc| σ-weighted map is contoured at 1.0 σ. I, ITC results of the wild-type and mutant Aca2 to 26-bp DNA substrates. All raw data of the ITC experiments are shown in Table S2 and Figure S8. J, the repression activity of Aca2 and mutants on the acrIF8-aca2 promoter, which was determined as the median percentage of eGFP fluorescence intensity compared with the control. Data are presented as the mean ± standard deviation of three biological replicates. N.D., not detected.
The core motifs (half-site 5′-GTTC-3′) of the IR1 DNA were specifically recognized by the Aca2 dimer (Fig. 6, B and C): In the G6:C48 pair, the O6 and N7 atoms of G6 were hydrogen-bonded to the side chain of Arg39, which were bidentate-bonding interactions that conferred specific recognition of G6 (Fig. 6D); interestingly, Arg39 was also extensively stacked with the ring and methyl group of T5, however, which seemed to not contribute to specific recognitions (Fig. 6E); in the T7:A47 pair, the methyl group of T7 formed hydrophobic contacts with the side chain of Tyr34 (Fig. 6F), which may confer certain extent of specificity for recognition of T7; in the T8:A46 pair, the methyl group of T8 formed hydrophobic contacts with the side chain of Arg30, and the N6 atom of A46 was hydrogen-bonded to the side chain of Tyr34 (Fig. 6G), which may also confer certain extent of specificity for recognition of T8; and in the C9:G45 pair, the O6 and N7 atoms of G45 were hydrogen-bonded to the side chain of Arg30 (Fig. 6H), which seemed to confer specific recognition of C9 through its pairing base G45. Taken together, each base of the core motifs (half-site 5′-GTTC-3′) was specifically recognized by the Aca2 dimer. Moreover, the sequence alignment of Aca2 homologs (sequence identities about 42–58%) revealed that the key residues Arg30 and Tyr34 of ZF40 Aca2 were strictly conserved and typed-conserved, respectively, while the residue Arg39 was not conserved in some homologs including Aca2 from Shewanella and Vibrio, with a substitution of an aspartic acid in this position (Fig. 5D). A further examination of the alignment uncovered that the Aca2 homologs from Shewanella and Vibrio had a nearby arginine residue in position 40 (Fig. 5D), which may complement for the specific interactions with DNA, as was modeled in the Coot program (data not shown). Consistent with the variation in the protein sequences, the core motifs within the IR1 DNAs are slightly different in the Shewanella and Vibrio species, with a half-site 5′-GGTC-3′ (Fig. S9C). In addition to these base-specific interactions, some backbone phosphate groups were also bound by the Aca2 dimer (Fig. 6C): The phosphate group of G6 formed a hydrogen bond with the side chain of Trp35; the phosphate group of T7 was hydrogen-bonded to the side chains of Ser28 and Ser31; and the phosphate groups of G43 (C11:G43 pair) and C42 (G12:C42 pair) were hydrogen-bonded to Gln33 and Ser18, respectively (Fig. S9D). Consistent with the structural observations, the ITC results revealed that single mutants of Aca2 R30A, Y34A, or R39A could disrupt binding to the IR1 DNA, while the mutant of Aca2 Q33A just slightly influenced binding (Fig. 6I; raw data in Table S2 and Fig. S8, A–E).
As done for Aca1, we also performed an in vivo eGFP-fluorescence reporter assay for Aca2 (25, 26) to evaluate the importance of the key residues in transcriptional repression of the acrIF8-aca2 operon. The results revealed that all the Aca2 mutants, including R30A, Q33A, Y34A and R39A, reduced the repressive effect on eGFP expression, which further corroborated our structural observations and ITC results and unraveled the involvement of more key residues of Aca2, as well as the well-studied Arg30 (26), in transcriptional repression of the acrIF8-aca2 operon (Fig. 6J).
Considered together, the above analyses unraveled the molecular basis of the specific recognitions between the IR1 DNA and the Aca2 homodimer.
Discussion
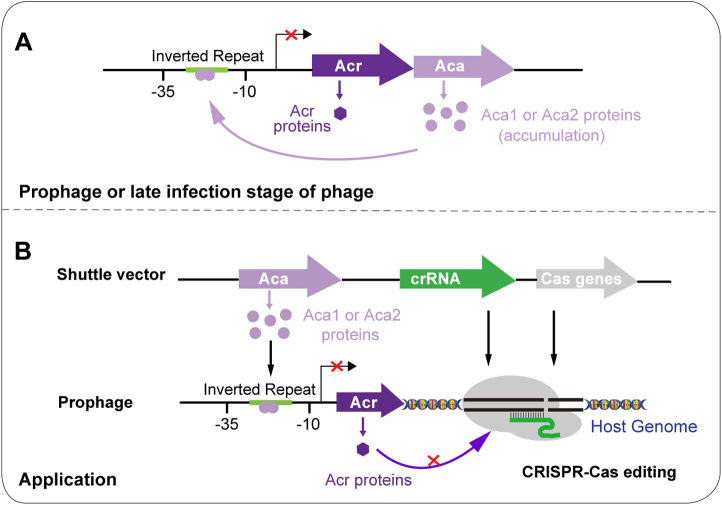
Our studies revealed the structural basis for Aca1-and Aca2-mediating anti-CRISPR repression. Both JBD30 Aca1 and ZF40 Aca2 adopt rigid dimer conformations. The structural and sequence conservations support that dimerization is an intrinsic feature of Aca1 and Aca2 families, although Aca1 and Aca2 have different dimerization mechanisms. The JBD30 Aca1 dimer binds and bends 19-bp operator DNA into an arch-like structure with an overall bending of 42°. This kind of bending mode is usually seen for the HTH-type transcriptional repressors (35). As compared with Aca1, ZF40 Aca2 binds to a roughly unbent 26-bp operator DNA, which may be due to the fact that Aca2 has little, if any, interaction with the flanking regions of its target DNA, while Aca1 has several interactions with the flanking regions of its target DNA. ZF40 Aca2 had been reported to bend its operator DNA by approximately 37° using a DNA-bending assay (25). This contrast might be due to the fact that much longer DNA fragments (∼200 bp) were used in their experiments. Our results have further revealed that the JBD30 Aca1 and ZF40 Aca2 each specifically recognize their cognate operator DNA with the recognition helix α3 of the HTH domain. The residues of JBD30 Aca1 involved in specific interactions with DNA (including Arg44, Val45, Arg47, Tyr48 and Arg59) are strictly conserved in its phage homologs and less conserved in its bacterial homologs, while the residues of ZF40 Aca2 involved in specific interactions with DNA (including Arg30, Tyr34, and Arg39) are conserved within all the homologs. The exact function of the bacterial Aca1 homologs needs to be further studied in future. Moreover, our eGFP-fluorescence reporter assay revealed that all these conserved residues of JBD30 Aca1 and ZF40 Aca2 were involved in transcription repressions. Thus, our study unveils the conserved recognition and repression modes of the Aca1 and Aca2 families (Fig. 7A). There are eight Aca families, Aca1–Aca8. Although they are all anti-CRISPR-associated proteins, they share low sequence homology and bind to different target DNA. Thus, more structural and biochemical studies are needed to characterize the repression mechanisms of the remaining Aca3–Aca8 families.
Figure 7.
Schematic diagram showing the repressor roles of Aca1 and Aca2 and their potential value in genome editing.A, in prophage or late infection stage of phage, the accumulated Aca1 or Aca2 proteins can function as repressor through binding to the inverted repeat (IR) in the promoter of the acr-aca operon, which then alleviates the deleterious effects on phages caused by the strong and constitutive transcription from the promoter of the operon. We have presented the structural basis for Aca1-and Aca2-mediating anti-CRISPR repression. B, a prophage within a bacterial host may have inhibitory effects on genome editing in the bacterial host, as inhibited by Acr proteins produced from the prophage. The epitopic expression of the Aca1 or Aca2 proteins can potentially repress the transcription from the acr promoter, which thus releases the inhibitory effects of the Acr proteins on CRISPR-Cas-mediated genome editing in the host, as proved by Csörgő et al. (29). Our structures can be used as a template/start to design stronger Aca1 and Aca2 repressors (higher binding affinity to the target DNA), which would potentially provide a more powerful anti-anti-CRISPR mechanism for different applications including genome editing.
Moreover, the Aca proteins could be engineered into or naturally co-opted by hosts as an anti-anti-CRISPR mechanism against phages and other MGEs. For example, the potential anti-anti-CRISPR mechanism of Aca1 was recently utilized to overcome the Acr protein-mediating impediment to enhance the efficiency of a Cascade-Cas3 bacterial genome editing tool (29). Thus, our study provides a structural basis for this anti-anti-CRISPR mechanism. The structural analyses reported here can be used as a template/start to design stronger Aca repressors (higher binding affinity to the target DNA), which would potentially provide a more powerful anti-anti-CRISPR mechanism for different applications including genome editing (Fig. 7B).
A structure-based similarity search with the Dali server (30) revealed that the ternary and quaternary structures of Aca1 displayed similarities to the antitoxins and repressors of V. cholerae HigA2 (PDB ID: 5JAA; Z-score 7.5; 2.20 Å RMSD for 61 Cα atoms) and E. coli MqsA (PDB ID: 3GN5; Z-score 6.9; 2.50 Å RMSD for 63 Cα atoms), which implicates that the Aca1 family might originate from antitoxin (Fig. S2, A–D). This evolutionary relationship is also suggested by a recent bioinformatic report (36). The Dali search also revealed that both the overall ternary and quaternary structures of Aca2 only showed high similarities to two bacterial proteins: Salmonella typhimurium YdiL and Shewanella oneidensis SO_3848 (Fig. S10, A and B). The PDBs of these two bacterial proteins were deposited by the Midwest Center for Structural Genomics, which should be acknowledged. Though the function of the two bacterial proteins is unknown, the unexpectedly high similarities of structures to Aca2 may implicate that they may function as transcription repressors just as Aca2 did.
In conclusion, our studies present series of high-resolution crystal structures of the Aca1 and Aca2 families, reveal the structural basis for Aca1-and Aca2-mediating anti-CRISPR repression, and provided a structural basis that establishes the potential value of the Aca proteins in future applications (e.g., genome editing).
Experimental procedures
Cloning and protein expression and purification
The genes that encoded the full-length Aca1 protein (1–79) (Uniprot ID: L7P845) of P. aeruginosa phage JBD30 and the full-length Aca2 protein (1–116) (Uniprot ID: H9C180) of P. carotovorum were synthesized by GENEWIZ and then cloned, by recombinational method, into a modified pGEX-6P-1 plasmid with an N-terminal GST-tag followed by a 3C protease cleavage site, respectively. Mutations were generated by standard polymerase chain reaction using Takara MutanBEST Kit. Recombinant plasmids were verified by DNA sequencing.
For the protein expression, all plasmids including wild-type Aca proteins and mutants were transformed into BL21 (DE3)-RIL strain and then cultured in Luria–Bertani (LB) medium supplemented with 34 mg/l chloramphenicol and 100 mg/l ampicillin at 37 °C to an optical density OD600 of 0.6 to 0.8. The protein expression was induced with 0.5 mM IPTG, and the cells were further incubated overnight at 20 °C. To induce the expression of selenomethionine (SeMet)-derived Aca1 protein, cells were first cultured in LB medium; when the OD600 reached 1.0 to 1.2, the cells were collected by centrifugation at 1000g and then transferred to M9 medium supplemented with the seven amino acids, Val, Ile, Leu, Phe, Thr, Trp, and Lys each at a final concentration of 50 mg/l. After 1-h incubation at 37 °C, SeMet at a final concentration of 60 mg/l was added to the culture, and cells were induced for expression with 0.5 mM IPTG and further incubated overnight at 20 °C. The collected cells were resuspended and lysed by sonication in a buffer containing 20 mM Tris-HCl pH 7.5, 1 M NaCl. The cell extracts were centrifuged at 38,000g for 1 h at 4 °C. Supernatants were collected and incubated with Glutathione Sepharose 4B Beads (GE Healthcare) for 4 to 6 h, then centrifuged at 500g for 2 min to remove the supernatants, the beads bound with the GST-tagged protein were washed three times with washing buffer containing 20 mM Tris-HCl pH 7.5, 1 M NaCl, 0.5% Triton X-100. The protein-bound beads were suspended with a buffer of 20 mM Tris-HCl pH 7.5, 1 M NaCl, then 3C protease (cleavage site: LEVLFQ/GPLGS. Home-made protease) was added and incubated at 4 °C overnight to cut the GST-tag and release the protein of interest from the beads. The eluted fractions were further purified on a HiLoad 16/600 Superdex 75 column (GE Healthcare), at a flow rate of 1 ml/min, with the buffer containing 20 mM Tris-HCl pH 7.5, 1 M NaCl. Peak fractions were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (15%, w/v) and stained with Coomassie brilliant blue R-250. Pure fractions were pooled together and concentrated by centrifugal ultrafiltration (Millipore). The concentration was determined by A280 using NanoDrop ONE (Thermo Scientific). The final purified protein samples were concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C until further use.
DNA substrates
All DNA substrates used for crystallization and ITC experiments were purchased from Sangon Biotech (Shanghai). Each sample of single-stranded DNA oligonucleotides was dissolved in an annealing buffer containing 10 mM Tris-HCl pH 7.5, 50 mM NaCl, heated at 100 °C for 1 min, and then cooled immediately on ice. To form the double-stranded DNA oligonucleotides (dsDNA), two single strands were mixed at the same molar concentration and were heated at 95 °C for 30 s and then cooled to room temperature naturally. All dsDNA oligonucleotides used for crystallization and biochemical experiments in this study are summarized in Table S1.
Crystallization and data collection
The final concentrations of the native and SeMet-labeled Aca1 proteins for crystallization trials were 2 mg/ml in a buffer containing 20 mM Tris-HCl pH 7.5, 500 mM NaCl. To form the Aca1 and DNA complex, the Aca1 protein and 19-bp 5′-overhang IR2 DNA (5′-TATAGGCACAATGTGCCTAAT-3′) were mixed and incubated at a molar ratio of 1:1.2. The final concentration of the Aca1–IR2 complex for crystallization was approximately 5 mg/ml (estimated by protein only) in a buffer containing 20 mM Tris-HCl pH 7.5, 200 mM NaCl. The final concentrations of the native and SeMet-labeled Aca2 proteins for crystallization trials were 10 mg/ml in a buffer containing 20 mM Tris-HCl pH 7.5, 500 mM NaCl. To form the Aca2 and DNA complex, the Aca2 protein and 26-bp 5′-overhang ZF40-IR1 DNA (5′-TTGCTTGTTCGCGATTGCGAACATATA-3′) were mixed and incubated at a molar ratio of 1:1.2. The final concentration of the Aca2–26-bp DNA complex for crystallization was approximately 5 mg/ml (estimated by protein only) in a buffer containing 20 mM Tris-HCl pH 7.5, 200 mM NaCl. All crystals were obtained using the sitting-drop vapor-diffusion method at 20 °C.
-
(1)
The crystals of apo-Aca1 were crystallized in 0.2 M di-potassium hydrogen phosphate; 20% (w/v) PEG 3350.
-
(2)
The crystals of apo-SeMet-labeled Aca1 were crystallized in 0.2 M sodium formate; 20% (w/v) PEG 3350.
-
(3)
The crystals of the Aca1–IR2 complex were crystallized in 0.2 M di-ammonium tartrate; 20% (w/v) PEG 3350.
-
(4)
The crystals of apo-Aca2 were crystallized in 0.2 M magnesium chloride hexahydrate; 0.1 M TRIS hydrochloride pH 8.5; 20% (w/v) PEG 8000.
-
(5)
The crystals of apo-SeMet-labeled Aca2 were crystallized in 0.1 M HEPES sodium pH 7.5; 2% (w/v) PEG400; 2.0 M ammonium sulfate.
-
(6)
The crystals of the Aca2–IR1 complex were crystallized in 0.1 M TRIS hydrochloride pH 8.0; 1.6 M lithium sulfate.
All crystals were soaked in cryoprotectants made from mother liquors supplemented with 15 to 20% glycerol before being flash frozen in liquid nitrogen. Diffraction data for all crystals were collected at a wavelength of 0.979 Å at beamline 19U1 (BL19U1) at Shanghai Synchrotron Radiation Facility, China. X-ray datasets were subsequently processed with the programs HKL3000 or iMosflm in the CCP4 suite.
Structure determination and refinement
The Autosol program in the PHENIX suite was used to calculate the phase of apo-Aca1 by the SAD method. Automatic protein model building was performed with PHENIX (37). The structure of the Aca1–IR2 complex was determined by molecular replacement with PHASER MR (38) in the CCP4 suite (39) using the apo-structure as a search model. The final models were iteratively rebuilt, refined, and validated with COOT (40), REFMAC5 (41), and PHENIX. The phase of apo-Aca2 was determined by HKL2MAP from the SAD dataset (42). The Aca2 model was built, refined, and validated with COOT (40), REFMAC5 (41), and PHENIX. The structure of Aca2–26-bp DNA was solved by PHASER with apo-Aca2 as the search model. A summary of crystallographic statistics is shown in Table 1. All structure figures were prepared with PyMOL (The PyMOL Molecular Graphics System, Schrödinger).
ITC experiments
All the samples of wild-type Aca1 and mutants, as well as the dsDNA substrates used in the ITC assays, were diluted to proper concentrations, then buffer-exchanged to 50 mM Tris-HCl pH 7.5, 150 mM NaCl by centrifugal ultrafiltration. The ITC experiments were performed at 25 °C on a MicroCal PEAQ-ITC (Malvern Panalytical Ltd) using 19 injections of 2.0 μl. Then, 250 μl of 30 μM solution of protein (the protein concentration was calculated as dimer) was loaded in the cell, and 50 μl of 245 μM solution of dsDNA substrate was loaded into the syringe. All samples of wild-type Aca2 and mutants, as well as dsDNA substrates used in ITC assays, were diluted to proper concentrations and then were buffer-exchanged to 50 mM Tris-HCl pH 7.5, 200 mM NaCl by centrifugal ultrafiltration. The ITC experiments were performed at 25 °C on a MicroCal PEAQ-ITC (Malvern Panalytical Ltd) using 19 injections of 2.0 μl. Then, 250 μl of 10 μM solution of dsDNA substrate was loaded into the cell, and 50 μl of 150 μM solution of Aca2 protein (the protein concentration was calculated as dimer) was loaded into the syringe. As the concentrations of the Aca proteins might be intrinsically not determined accurately, or the recombinant proteins might be not 100% active, we fixed the N = 1 during data fitting to get better estimation of Kd values (43). Then the measured heat changes of the binding reactions were processed using a standard “one set of sites” model implemented in the Origin package.
Size-exclusion chromatography analysis
The oligomeric statuses of wild-type Aca1 and double-mutant T64D/F67D and wild-type Aca2 together with the triple mutant K4A/E5A/Y108A were analyzed by a HiLoad 10/300 Superdex 75 Increase column (GE Healthcare) at a flow rate of 0.7 ml/min with a buffer containing 20 mM Tris-HCl pH7.5, 1 M NaCl. A molecular weight standard (Gel Filtration Standard Mix from Bio-Rad) was analyzed under the same conditions. The oligomeric statuses of all mentioned protein samples were estimated based on the standard.
In vivo reporter assays
The sequence encoding wild-type or mutant Aca1 protein was cloned into the pRSFDuet1 vector (kanamycin) under control of the T7 promoter. The egfp gene was cloned into a modified pUCP20 vector (ampicillin) containing the promoter region of acrIF-aca1 to control the expression of egfp. The pUCP20-acr-egfp and aca1-pRSFDuet1 plasmids were cotransformed into the E. coli BL21 (DE3)-RIL strain and incubated on ampicillin–kanamycin double-resistant plates. A single clone was selected and transferred into a culture tube with 5 ml LB medium supplied with 50 mg/l kanamycin and 100 mg/l ampicillin, and then cultured for 12 h at 37 °C. Next, 3 ml sample was transferred to 100 ml LB medium under the same culture condition supplying 0.5 mM IPTG and continue culturing at 37 °C. After 20 h, all sample cells were collected and cooled on ice. Then, 1 ml of each sample is used to dilute to the same density OD600 = 0.5, and 100 μl of these dilutions was further selected to load into a 96-well transparent plate to test the expression level of eGFP. The eGFP fluorescence intensity was determined with a BioTek Microplate Reader; the excitation wavelength/emission wavelength was 480/520. The values of fluorescence intensity were processed and plotted with Prism7. Each experiment has three independent repeats. The in vivo reporter assay of Aca2 was done following the same procedure as that for Aca1.
Data availability
The structures were deposited into the Protein Data Bank (PDB) with accession codes 7VJN for native Aca1, 7FA3 for SeMet-Aca1, 7VJM for the Aca1–IR2 complex, 7VJO for native-Aca2, 7VJP for SeMet-Aca2, and 7VJQ for the Aca2–26-bp-DNA complex.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the staff from BL19U1 and BL18U1 beamlines at the Shanghai Synchrotron Radiation Facility for assistance during data collection.
Author contributions
Y. L. and H. H. conceptualization; Y. L., L. Z., M. G., and L. C. data curation; B. W. and H. H. formal analysis; H. H. funding acquisition; Y. L., L. Z., M. G., and L. C. investigation; Y. L., B. W., and H. H. writing and revision of the manuscript.
Funding and additional information
This work was supported by the National Key R&D Program of China (2018YFC1004500); the Shenzhen Government ‘Peacock Plan’ (Y01226136 to H. H.); Thousand Young Talents Program (to H. H.); Chinese National Natural Science Foundation of China (31900435 to B. W.); and Guangdong Science and Technology Department (2020B1212060018 and 2020B1212030004 to B. W.).
Edited by Wolfgang Peti
Contributor Information
Baixing Wu, Email: wubx28@mail.sysu.edu.cn.
Hongda Huang, Email: huanghd@sustech.edu.cn.
Supporting information
References
- 1.Hampton H.G., Watson B.N.J., Fineran P.C. The arms race between bacteria and their phage foes. Nature. 2020;577:327–336. doi: 10.1038/s41586-019-1894-8. [DOI] [PubMed] [Google Scholar]
- 2.Rostol J.T., Marraffini L. (Ph)ighting phages: How bacteria resist their parasites. Cell Host Microbe. 2019;25:184–194. doi: 10.1016/j.chom.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., Moineau S., Mojica F.J.M., Scott D., Shah S.A., Siksnys V., et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 7.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 9.Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J. CRISPR-Cas: Adapting to change. Science. 2017;356 doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 10.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K., de Vos W.M., Dame R.T., de Vries R., Brouns S.J., van der Oost J. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W., Samai P., Marraffini L.A. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell. 2016;164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sashital D.G., Wiedenheft B., Doudna J.A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson A.R., Lu W.T., Stanley S.Y., Wang J., Mejdani M., Trost C.N., Hicks B.T., Lee J., Sontheimer E.J. Anti-CRISPRs: Protein inhibitors of CRISPR-Cas systems. Annu. Rev. Biochem. 2020;89:309–332. doi: 10.1146/annurev-biochem-011420-111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley S.Y., Maxwell K.L. Phage-encoded anti-CRISPR defenses. Annu. Rev. Genet. 2018;52:445–464. doi: 10.1146/annurev-genet-120417-031321. [DOI] [PubMed] [Google Scholar]
- 19.Hwang S., Maxwell K.L. Meet the anti-CRISPRs: Widespread protein inhibitors of CRISPR-Cas systems. CRISPR J. 2019;2:23–30. doi: 10.1089/crispr.2018.0052. [DOI] [PubMed] [Google Scholar]
- 20.Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawluk A., Bondy-Denomy J., Cheung V.H., Maxwell K.L., Davidson A.R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio. 2014;5 doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawluk A., Staals R.H., Taylor C., Watson B.N., Saha S., Fineran P.C., Maxwell K.L., Davidson A.R. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 2016;1:16085. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- 23.Marino N.D., Zhang J.Y., Borges A.L., Sousa A.A., Leon L.M., Rauch B.J., Walton R.T., Berry J.D., Joung J.K., Kleinstiver B.P., Bondy-Denomy J. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science. 2018;362:240–242. doi: 10.1126/science.aau5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L., Davidson A.R. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016;167:1829–1838.e9. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birkholz N., Fagerlund R.D., Smith L.M., Jackson S.A., Fineran P.C. The autoregulator Aca2 mediates anti-CRISPR repression. Nucleic Acids Res. 2019;47:9658–9665. doi: 10.1093/nar/gkz721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley S.Y., Borges A.L., Chen K.H., Swaney D.L., Krogan N.J., Bondy-Denomy J., Davidson A.R. Anti-CRISPR-associated proteins are crucial repressors of anti-CRISPR transcription. Cell. 2019;178:1452–1464.e13. doi: 10.1016/j.cell.2019.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osuna B.A., Karambelkar S., Mahendra C., Sarbach A., Johnson M.C., Kilcher S., Bondy-Denomy J. Critical anti-CRISPR locus repression by a Bi-functional Cas9 inhibitor. Cell Host Microbe. 2020;28:23–30.e5. doi: 10.1016/j.chom.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watters K.E., Shivram H., Fellmann C., Lew R.J., McMahon B., Doudna J.A. Potent CRISPR-Cas9 inhibitors from Staphylococcus genomes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6531–6539. doi: 10.1073/pnas.1917668117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Csörgő B., Leon L.M., Chau-Ly I.J., Vasquez-Rifo A., Berry J.D., Mahendra C., Crawford E.D., Lewis J.D., Bondy-Denomy J. A compact cascade-Cas3 system for targeted genome engineering. Nat. Methods. 2020;17:1183–1190. doi: 10.1038/s41592-020-00980-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L., Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadzi S., Garcia-Pino A., Haesaerts S., Jurenas D., Gerdes K., Lah J., Loris R. Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a beta-strand sliding mechanism. Nucleic Acids Res. 2017;45:4972–4983. doi: 10.1093/nar/gkx138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown B.L., Grigoriu S., Kim Y., Arruda J.M., Davenport A., Wood T.K., Peti W., Page R. Three dimensional structure of the MqsR:MqsA complex: A novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid–base interactions: A three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Martin J., Rojo F., de Lorenzo V. Promoters responsive to DNA bending: A common theme in prokaryotic gene expression. Microbiol. Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin Y., Yang B., Entwistle S. Bioinformatics identification of anti-CRISPR loci by using homology, guilt-by-association, and CRISPR self-targeting spacer approaches. mSystems. 2019;4 doi: 10.1128/mSystems.00455-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., et al. Phenix: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pape T., Schneider T.R. HKL2MAP: A graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. [Google Scholar]
- 43.Kantonen S.A., Henriksen N.M., Gilson M.K. Evaluation and minimization of uncertainty in ITC binding measurements: Heat error, concentration error, saturation, and stoichiometry. Biochim. Biophys. Acta Gen. Subj. 2017;1861:485–498. doi: 10.1016/j.bbagen.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures were deposited into the Protein Data Bank (PDB) with accession codes 7VJN for native Aca1, 7FA3 for SeMet-Aca1, 7VJM for the Aca1–IR2 complex, 7VJO for native-Aca2, 7VJP for SeMet-Aca2, and 7VJQ for the Aca2–26-bp-DNA complex.