Abstract
Lipopolysaccharide (LPS) is an essential glycolipid that covers the surface of gram-negative bacteria. The transport of LPS involves a dedicated seven-protein transporter system called the lipopolysaccharide transport system (Lpt) machinery that physically spans the entire cell envelope. The LptB2FG complex is an ABC transporter that hydrolyzes ATP to extract LPS from the inner membrane for transport to the outer membrane. Here, we extracted LptB2FG directly from the inner membrane with its original lipid environment using styrene-maleic acid polymers. We found that styrene-maleic acid polymers–LptB2FG in nanodiscs display not only ATPase activity but also a previously uncharacterized adenylate kinase (AK) activity, as it catalyzed phosphotransfer between two ADP molecules to generate ATP and AMP. The ATPase and AK activities of LptB2FG were both stimulated by the interaction on the periplasmic side with the periplasmic LPS transport proteins LptC and LptA and inhibited by the presence of the LptC transmembrane helix. We determined that the isolated ATPase module (LptB) had weak AK activity in the absence of transmembrane proteins LptF and LptG, and one mutation in LptB that weakens its affinity for ADP led to AK activity similar to that of fully assembled complex. Thus, we conclude that LptB2FG is capable of producing ATP from ADP, depending on the assembly of the Lpt bridge, and that this AK activity might be important to ensure efficient LPS transport in the fully assembled Lpt system.
Keywords: cell wall, lipopolysaccharide (LPS), ABC transporter, adenylate kinase, NMR
Abbreviations: AK, adenylate kinase; IM, inner membrane; LPS, lipopolysaccharide; Lpt, lipopolysaccharide transporter system; LptAm, monomeric version of LptA; OM, outer membrane; SMA, styrene-maleic acid; STD, saturation transfer difference; TBS, Tris-buffered saline; TM, transmembrane
Gram-negative bacteria possess a double membrane system delimiting an aqueous space, the periplasm. Although the cytoplasmic inner membrane (IM) is a canonical phospholipid bilayer, the outer membrane (OM) is asymmetric with its outer layer mostly composed of the lipopolysaccharide (LPS) glycolipid (1). These molecules are surface exposed, contribute to the high impermeability of the OM, and play a role in immune response, pathogenesis, and drug resistance (2). Maintenance of the OM asymmetry is essential for bacterial viability, and a continuous flow of LPS molecules needs to cross the periplasm and reach the external layer to keep up with the cell growth. A system of seven essential proteins, the lipopolysaccharide transport system (Lpt), is dedicated to trafficking LPS across the cell envelope. These proteins (LptA-G) are found in all cellular compartments (IM, periplasm, and OM), and in the periplasm, they associate through jellyroll domains (present in LptF, LptC, LptA, and LptD) to form a continuous periplasmic bridge connecting the IM and OM (3, 4).
LptB2FG IM complex is an ABC transporter in which LptB2 represents the nucleotide-binding domain component that binds and hydrolyzes ATP to generate mechanical force to transport LPS. Upon ATP binding, LptB dimerizes in a closed structure, and ATP hydrolysis relaxes the dimer into an open conformation. The conformational changes are transmitted to LptFG through coupling helices and allow LPS extraction (5). LptB2FG associates with LptC via two distinct interactions: (i) LptF/LptG interact with LptC transmembrane (TM) domain, which contributes to the formation of the cavity that accommodates LPS into the complex and regulates LptB2FG ATPase activity (5) and (ii) LptF and LptC jellyroll domains associate in the periplasm. LptA jellyroll then bridges LptB2FGC to the LptD/E OM complex that ultimately assembles the LPS to the OM outer leaflet (3).
The superfamily of ABC transporter proteins comprises more than 500 members and supports traffic of metabolites and molecules through ATP hydrolysis (6). Several prokaryotic ABC transporters (MsbA, TmrAB, and LmrA) couple another reaction in their ATPase domain, adenylate kinase (AK) (7). AK catalyzes a phosphotransfer reaction between two ADP molecules producing ATP and AMP with no energy consumption (8). MsbA in particular translocates LPS across the IM before its transport by LptB2FG (9, 10). The additional active site for the AK has been suggested to be located proximal to the ATP binding site because the reaction requires both ADP molecules close by in space (11).
LptB2FG was extracted directly from the Escherichia coli IM with styrene-maleic acid (SMA) polymers without the use of detergents. 1H-NMR on nucleotides showed that the SMA–LptB2FG complex has not only ATPase activity but also AK activity, and both activities are regulated by the assembly of LptB2FG with LptC and LptA partners. Point mutations were introduced in the LptB2FG complex and in the isolated LptB to address their effect on ATPase and AK activity by NMR and locate the AK site.
Results
LptB2FG extracted from the IM by SMA polymers displays ATPase and AK activity
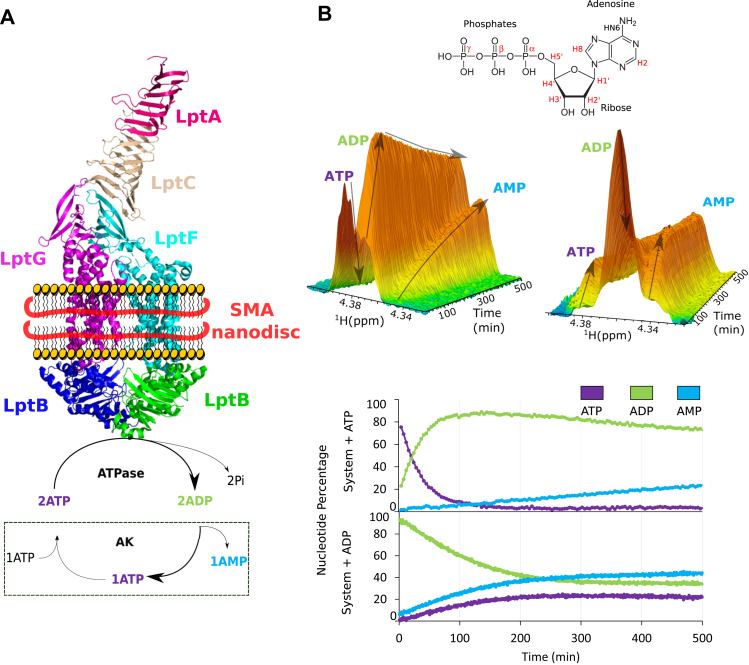
LptB2FG complexes from several organisms have been expressed and purified in detergent micelles (12, 13, 14, 15), reconstituted nanodiscs (5), or liposomes (16). These protocols all involve solubilization and purification of the LptB2FG complex in detergent micelles. The use of detergent-free protocols in the preparation of membrane protein, to maintain their original lipid environments, becomes increasingly important to ensure functionality and stability. In that aspect, the use of SMA polymers and its derivatives, which allow the direct extraction of membrane proteins from purified membranes, provides a versatile tool to study membrane proteins as close as possible to their natural environment (17, 18). The LptB2FG complex extraction directly from the IM of E. coli was tested using two SMA polymers with 2:1 and 2.3:1 SMA ratio. SMA–LptB2FG was successfully purified in nanodiscs of about 10-nm diameter only with SMA 2:1 polymer and with the expected 2:1:1 LptB–LptF–LptG stoichiometry (Fig. S1). The LptB2FG ATP hydrolysis was checked in presence of LptC and LptA, its periplasmic partners, using NMR to probe the reaction (Fig. 1A). The monomeric version of LptA (LptAm) and the soluble version of LptC (ΔTM-LptC), which are able to sustain activity and cell viability (4, 19, 20), were added to SMA–LptB2FG, and ATP hydrolysis was followed by real-time 1H-NMR (Fig. 1B). NMR is a spectroscopic technique that provides resonance frequency of active nuclei, in our case protons, depending on their chemical environments (21). It can discriminate between ATP and ADP as several 1H shift frequencies upon phosphate loss, here, hydrogen H4′ on the ribose (Fig. 1B). SMA–LptB2FG shows ATP hydrolysis, with fast disappearance of peak characteristics of ATP and concomitant appearance of ADP-specific peaks in the NMR spectrum. The rate of ATP hydrolysis is 6.5 mol of ATP consumed/min/mol of LptB. After the initial rapid accumulation of ADP due to ATPase activity, ADP level decreases at the same rate as the appearance of a new set of NMR peaks (8.60 ppm in the H8 region and 4.39 ppm in H4′ region). This suggests the conversion of ADP into another species over time and its 1H-NMR chemical shifts suggest that it could correspond to AMP (22). 31P NMR spectrum was collected at the end of the reaction and showed the characteristic peak of AMP at 3 ppm (Fig. S2). The conversion of ADP into AMP is compatible with an AK reaction 2ADP ATP + AMP, which has already been observed for several ABC transporters (7, 11). ATP was replaced by ADP as a substrate, and the reaction was followed in identical conditions in real time (Fig. 1B). ADP decrease is seen immediately, together with the appearance of AMP and ATP. The generation of ATP is consistent with AK activity and excludes the hydrolysis of ADP into AMP. The ATP levels observed are lower than those of AMP, as AK and ATPase activities occur simultaneously, and newly generated ATP is partly hydrolyzed back to ADP. The initial rates of AK activity are much slower than ATPase with 0.2 and 0.7 mol of AMP produced/min/mol of LptB with ATP or ADP as starting substrates (Fig. 1B).
Figure 1.
LptB2FG in SMA nanodiscs shows ATPase and AK activity.A, representation of the LptB2FGCA (with ΔTM-LptC and LptAm) system in SMA nanodiscs with its updated enzymatic cycle. B, real-time kinetics of LptB2FGCA observed by 1H-NMR with either ATP (left 16 scans/trace) or ADP (right four scans/trace) as the substrate over time with evolution of the characteristic H4′ peaks of ATP, ADP, and AMP. Nucleotide quantification along the experiment is represented (bottom). Lpt, lipopolysaccharide transporter system; SMA, styrene-maleic acid.
ATPase and AK activities are regulated by the assembly of the Lpt system
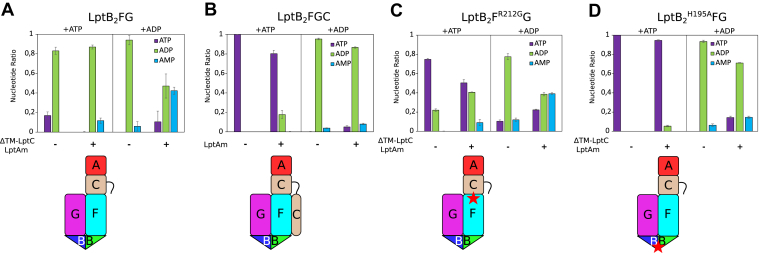
LptB2FG ATPase activity is repressed by the TM domain of LptC (5, 16); we thus examined the ATPase and AK activities of SMA–LptB2FG with/without ΔTM-LptC/LptAm and in complex with the full-length LptC with/without LptAm (Fig. 2, A and B). For the following experiments, nucleotides were quantified by NMR at the end of incubation (Fig. S3). SMA–LptB2FG displays less ATPase and AK activity in the absence of ΔTM-LptC and LptAm. Proper assembly of the periplasmic part of LptC and its complex with LptA (20) is thus important in stimulating both ATPase and AK activity. SMA–LptB2FGC containing full-length LptC has no ATPase activity, but the repression is relieved in the presence of LptAm (Fig. 2B). Similarly, SMA–LptB2FGC has little AK activity (Fig. 2B), which is increased by the addition of LptAm. AK activity and ATPase activities are thus both stimulated by the assembly of the LptB2FGCA complex without the TM LptC segment. In intact LptB2FGC, ATPase and AK are activated by assembly with LptA.
Figure 2.
Assembly with LptC and LptA influence ATPase and AK activity of LptB2FG.Top panel, assembly on the periplasmic or membrane level with LptC and LptA influences ATPase and AK activities of LptB2FG. SMA–LptB2FG is incubated with ATP or ADP, and nucleotide levels (ATP, ADP, and AMP) quantified by 1H-NMR from two or three independent experiments and the SD are shown. A, SMA–LptB2FG without/with ΔTM-LptC/LptAm. B, SMA–LptB2FGC complex without/with LptAm. C and D, SMA–LptB2FR212GG and SMA–LptB2H195AFG without/with ΔTM-LptC/LptAm. A schematic representation of the complexes used is shown at the bottom panel, with stars marking the location of mutations. Lpt, lipopolysaccharide transporter system; SMA, styrene-maleic acid.
Variants in periplasmic LptF domain and in cytoplasmic LptB differently affect AK activity
A number of mutations in LptB2FG were studied for their effect on the complex activity, either biochemically or for their in vivo functionality (23). Two SMA–LptB2FG complexes carrying already characterized mutations were purified, one on the LptF periplasmic side and one in the ATPase domain (LptB). The LptFR212G variant can overcome the absence of the essential LptC protein and allows cell viability (24). R212 is located in the LptF periplasmic domain at the interface with LptC and is in the path of the LPS flow (15). SMA–LptB2FR212GG has impaired ATPase activity when compared with WT complexes (Fig. 2C), and ΔTM-LptC and LptAm stimulate ATPase and AK activities (Fig. S3). With ADP as the substrate, SMA–LptB2FR212GG with and without ΔTM-LptC and LptAm show similar production of AMP as WT complexes (Fig. 2C). R212G mutation, although impairing the ATPase, does not affect the AK, suggesting different regulation mechanisms of the two activities.
LptB H195 is involved in γ phosphate binding of ATP (Fig. 3A), and the LptBH195A variant has decreased ATPase activity in the isolated LptB and is deleterious for cell growth (23). SMA–LptB2H195AFG has almost knocked out ATPase activity, even in the presence of ΔTM-LptC and LptAm. AK activity is still activated by ΔTM-LptC/Am but significantly decreased compared with the WT complex (Figs. 2D and S3). Alterations in the ATPase binding site of LptB affect the ATPase activity and AK activity, but the latter to a lesser degree.
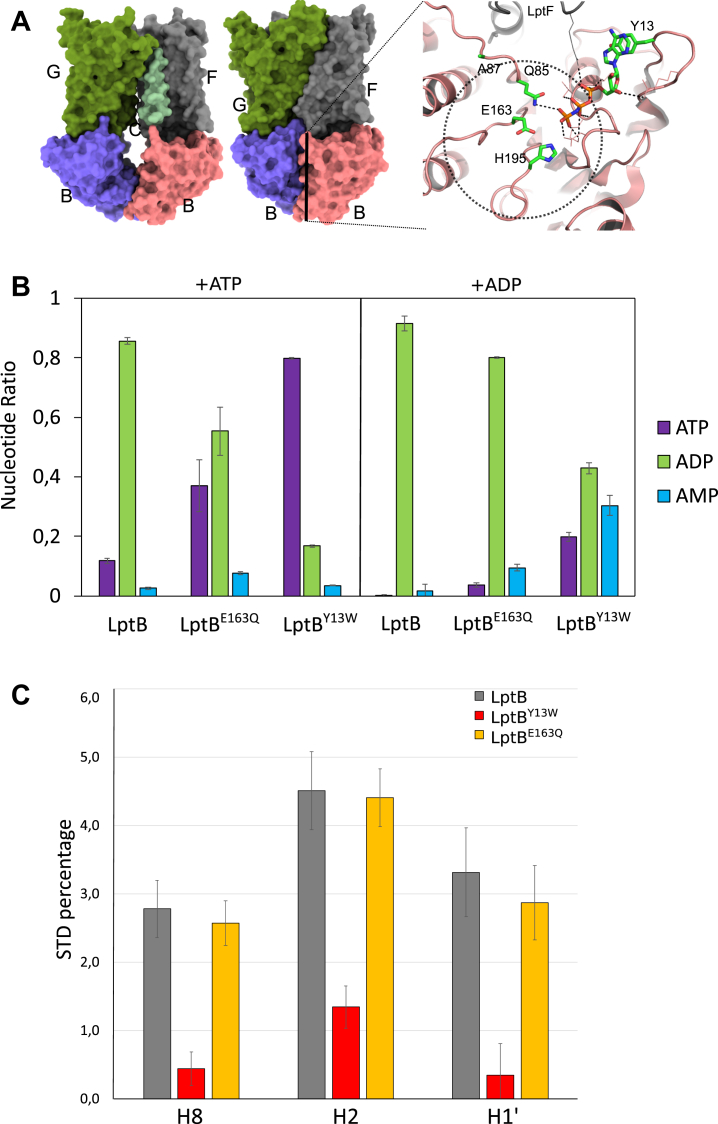
Figure 3.
Mutations in LptB stimulate adenylate kinase activity.A, structures of LptB2FGC in open (apo-form; PDB ID: 6S8N) and closed conformation (with bound ATP analog; PDB ID: 6S8G). The jellyroll domains were not resolved in these structures and the LptC transmembrane helix in the open conformation. The right panel shows the ATPase site with putative location of the second ADP in the AK reaction (dashed line area). The residues mutated in this study are shown. B, ATPase/AK of LptB2 and LptB2E163Q and LptB2Y13W mutants, with ATP or ADP as the substrate. The nucleotide levels (ATP, ADP, and AMP, in color code) were detected by the 1H-NMR experiment in two independent experiments, and the SD is shown. C, saturation transfer difference (STD) experiments of LptB2 and LptB2E163Q and LptB2Y13W on ADP-βS. The three resonances shown are from H8, H2, and H1′ on the adenosine of ADP-βS. The error bars represent two times the SD of the NMR experiments’ noise. AK, adenylate kinase; Lpt, lipopolysaccharide transport system.
Mutations in the A and B loops of LptB stimulate AK
The location of the AK site is shared with the ATPase site (25, 26). One ADP molecule occupies the ATP-binding site, whereas the other must be presented “in line” with the first one for the phosphotransfer to occur. Current models for AK suggest the second ADP substrate molecule to be located around the Q-loop (11) (Fig. 3A). The location of the second ADP molecule was probed in LptB using mutagenesis on the LptB-isolated protein. In addition to the H195A mutation in the H motif (or switch region), the other mutations were inserted in canonical ABC motifs, specifically at the A loop (Y13W), B loop (E163Q and 163A), and Q loop (A87Q) (Fig. 3A).
Purified soluble LptB WT and variant proteins were incubated with either ATP or ADP, and the nucleotides were quantified by 1H-NMR at the endpoint of the experiment (Fig. S4). LptB, similarly to the SMA–LptB2FG complex (without ΔTM-LptC and LptAm), displays ATPase activity and little AK activity (Fig. S5). All mutations introduced in LptB decrease ATPase activity, and E163A is catalytically fully inactive, with no ATPase or AK activity. Two LptB variants show increased AK activity, E163Q and more significantly Y13W, the latter accumulating AMP at levels similar to SMA–LptB2FG/ΔTM-LptC/LptAm (Fig. 3B). To assess how the binding of nucleotides relates to the activity of LptB, saturation transfer difference (STD) experiments were recorded on WT and LptB variants. STD is an NMR technique that allows observation of the free ligand in transient binding to a protein (27). The protein is selectively saturated, and part of the saturation is transferred to the ligand during binding, and a decrease of intensity is observed on the free ligand. STD on ADP by WT LptB shows saturation on protons H2, H8, and H1′ of ADP, showing significant interaction with LptB. The LptBE163Q variant shows no difference of STD with respect to the WT. STD measurement of the Y13W variant with ADP is not possible because AK activity is too fast relative to the experimental time (Fig. S6). ADP-βS, which is not a substrate of LptB, showed STD similar as ADP with the WT and LptBE163Q variant (Fig. S6). LptBY13W, on the other hand, showed a highly reduced STD (Figs. 3C, S6 and S7). A reduction in STD can be ascribed to a reduced affinity of LptBY13W for ADP-βS, with a reduced residence time of the ligand, or on the contrary to a reduction in koff leading to less saturation of the free ligand. A Y-W mutation in this position has already been found to reduce ATPase activity and ATP binding in other ABC transporters (28, 29). We thus conclude that the LptBY13W variant increase in AK activity can be assigned to a reduction in the affinity for ADP, likely in the canonical ATPase-binding site considering the position of Y13 (Fig. 3A). Apart from the E163A mutant, largely impaired in ATPase and AK activities, we could find no mutation that selectively affects AK activity, suggesting that a defined binding site for the second ADP molecule does not exist in LptB.
Discussion
Understanding bacterial mechanisms that coordinate envelope assembly is critical to harness ways to challenge persistent and clinically relevant infections. Here, LptB2FG and LptB2FGC complexes were extracted directly from the E. coli IM in their natural lipid context, without the need for detergent extraction. The LptB2FG complex ATPase activity is stimulated by interaction on the periplasmic side of LptB2FG with LptC and LptA and fully repressed when full-length LptC is coextracted with LptB2FG. The SMA–LptB2FG LptB subunit is also an AK. This activity is marginal until LptB2FG interacts with the jellyroll domains of LptC and LptA, thus forming an almost complete Lpt bridge (missing the LptD/E OM complex). Establishment of the bridge must be transmitted by conformational changes through the lipid bilayer to the cytoplasmic LptB and activate both ATPase and AK activities. The LptB2FGC complex that contains full-length LptC with its N-terminal TM domain has no ATPase activity and little AK activity until it interacts with LptA that increases both activities. The level of activity of LptB2FGC is much lower because of the presence of the TM LptC domain. This domain interacts with the LPS into the LptB2FG complex and is more dynamic in LptB2FG bound to ATP analogs (5). This could suggest a role of TM-LptC in the synchronization of LPS entry and ATP hydrolysis.
The Lpt system possesses several mechanisms that regulate its activity to ensure efficient LPS transport when the Lpt system is assembled. A mutant discovered in the LptF periplasmic jellyroll domain that can complement ΔlptC cells is particularly interesting. SMA–LptB2FR212GG ATPase activity is highly reduced compared with the WT complex, whereas AK activity is identical. This suggests that the regulation of the ATPase and AK activities by the assembly of the Lpt system is somewhat different. Nevertheless, the regulation mechanisms are currently not characterized at the molecular level.
AK reaction involves two ADP molecules, one located in the ATPase canonical site and one located nearby. Our mutagenesis study could not pinpoint the location of a binding site for the second ADP. LptBE163Q showed increased AK activity, and H195A in the full complex showed reduced AK activity. Although those residues could be important for the AK activity, they are all involved in the ATPase reaction, by binding ATP γ-phosphate, the magnesium ion, or coordinating water molecules (16). Our data suggest that, on the contrary to pure AK (30), there is no well-defined binding site for the AK activity in LptB, apart from the canonical ATP binding site. The high AK activity of the LptBY13W variant cannot be easily explained. The consequence of this mutation is to strongly reduce STD, likely because of the decreased affinity/residence time of the ligand in the canonical ATPase site. This could in turn accelerate the turnover of the reaction, but we cannot exclude that this variant in isolated LptB somehow mimics the conformation of LptB2FG in presence of the jellyroll domains of LptC and LptA.
The exact role of the AK activity in the LptB2FG–ABC transporter remains unknown. In one ABC transporter, the cystic fibrosis receptor (CFTR), the AK activity was shown to be important for the transport (31). Our characterization of the AK activity was performed at a high substrate concentration (5 mM), about ten times higher than the concentration of ADP in bacteria. The AK reaction was thus tested in physiologic concentrations with a mix of ATP, ADP, and AMP (Fig. S8). In these conditions, AK activity is similar as with 5 mM ADP as the substrate (Fig. 2). Similarly, LptB2FG has AK activity starting with only 500 μM of ADP, the concentration found in cells (Fig. S8). Although this reaction is possible in the concentrations of nucleotides found in vivo, its contribution to the LPS transport has not been tested yet. Nevertheless, the capacity of the AK reaction to generate ATP, which can be used to transport LPS, at a negligible energy cost must be considered in future studies, and specifically in conditions of bacterial stress.
Experimental procedures
Used plasmids
The plasmids used for soluble LptB, SMA–LptB2FG, ΔTM-LptC, and LptAm purifications were previously described: pET22-42-LptB-His8 (23) and pCDF-Duet1-LptB2FG (32), expressing LptB-His6/LptF/LptG (32), pQESH LptCΔ[1–23] (ΔTM-LptC) (19), and pET21b-LptAΔ160 (LptAm) (20). LptB point mutations were performed by GENEWIZ using pET22-42-LptB-His8 as the template. lptFR212G mutation in pCDF-Duet1-LptB2FG was introduced by site-directed mutagenesis using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) and AP613 (5′-ACCAGGGAACGGGCTTCGAAGGC-3′)–AP614 (5′-TGAGAGTGACGACCTGGGAG-3′) oligonucleotides. The plasmids pBAD/HisA-LptC-His (encoding LptC with a C-terminal His8 tag) and pCDF-Duet1-utLptB2FG (encoding untagged LptB, LptF, and LptG) were used for the purification of SMA–LptB2FGC. pBAD/HisA-LptC-His is a derivative of pBAD/HisA-LptC (32) obtained by replacing the lptC gene (cloned into NcoI-HindIII sites) with the PCR-amplified lptC-His8 gene using pET23/42-LptC-His (4) as the template and AP656 (5′-catgccatggGTAAAGCCAGACGTTGGGTT-3′)–AP657 (5′- cccaagcttTTAGTGGTGGTGGTGGTG-3′) oligonucleotides. pCDF-Duet1-utLptB2FG is a derivative of pCDF-Duet1-LptB2FG obtained by replacing lptB-His6 gene (cloned into NcoI-EcoRI sites) with the PCR-amplified lptB gene using E. coli MG1655 genomic DNA as the template and AP599 (5′-catgccatggCAACATTAACTGCAAAG-3′) and AP655 (5′-gagaggaattcTCAGAGTCTGAAGTCTTCCC-3′) oligonucleotides.
Strains used and protein expression
The LptB expression was carried out in E. coli BL21 (DE3) strain (Novagen) and LptB2FG in E. coli C43 (DE3) strain (Novagen). The coexpression of LptB2FG and LptC was carried out in E. coli KRX cell, as described (32). LptAm and ΔTM-LptC were expressed and purified in E. coli BL21 (DE3), as described (20). For LptB and LptB2FG, bacterial cells were grown in LB, supplemented with the correct antibiotic (ampicillin 100 μg ml−1 and spectinomycin 50 μg ml−1) at 37 °C, until optical density of 600 nm (OD600) around 0.7. For both sets of proteins, induction was performed with IPTG: for LptB2 proteins, with 0.1 mM at 20 °C for 16 h, whereas for LptB2FG proteins, with 0.5 mM at 37 °C for 3 h. The cells were harvested by centrifugation at 6000g for 20 min at 4 °C and frozen at −20 °C until purification. The coexpression of LptB2FG with LptC and cell harvesting was carried out as before, except induction that was carried out with 0.02% (w/v) L-rhamnose and 0.02% (w/v) L-arabinose.
Purification of LptB2 proteins
The cells were mixed with buffer A containing 20 mM Tris HCl, 150 mM NaCl, 20% (v/v) glycerol, pH 8.0, 0.5 mM Tris(2-carboxyethyl)phosphine, and cOmplete EDTA-free protease inhibitor cocktail (Sigma) and lysed by sonication. Soluble fraction was separated by centrifugation at 10,000g for 20 min at 4 °C and loaded after addition of 10 mM imidazole into Ni-NTA Agarose (QIAGEN). Resin was washed with buffer A with 20 mM imidazole and eluted with buffer A with 300 mM imidazole. LptB was then purified on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) in Tris-buffered saline (TBS) (50 mM Tris HCl and 150 mM NaCl, pH 8.0) supplemented with 0.5 mM Tris(2-carboxyethyl)phosphine. LptB was concentrated with a 10-kDa cut-off Amicon Ultra Centrifugal Filter (Merck), and the sample concentration was determined by running a 15% SDS-PAGE with known concentration samples of bovine serum albumin. The yields of purified proteins for the various LptB proteins were 30, 24, 15, 5, 3, and 1 mg/l of the culture for WT, H195A, E163A, E163Q, A87Q, and Y13W, respectively.
Purification of LptB2FG and LptB2FGC
The cells in the lysis buffer (50 mM Tris HCl, 300 mM NaCl, 1 mM MgCl2, pH 8.0, and cOmplete EDTA-free [Sigma]) were lysed on a microfluidizer at 15,000 psi. The cell debris are removed by centrifugation at 10,000g for 20 min at 4 °C, and the membranes are collected by centrifugation at 100,000g for 1 h at 4 °C. The membranes are resuspended in 50 mM Tris HCl, 250 mM NaCl, pH 8.0, with 0.5% SMA 25010 (Xiran SL25010 Polyscope) for 17 h at room temperature (RT). Soluble SMALP particles were obtained by ultracentrifugation at 100,000g for 30 min at 4 °C and loaded into a HisTrap 1-ml column equilibrated in 20 mM Tris HCl, 150 mM NaCl, and 30 mM imidazole, pH 8.0. Elution was carried out in a gradient with the previous buffer supplemented with 170 mM imidazole. Fractions containing the proteins were dialyzed against TBS (20 mM Tris and 150 mM NaCl, pH 8.0) at RT and concentrated with a 100-kDa cut-off Amicon Ultra Centrifugal Filter (Merck). The sample concentration was determined by running a 15% SDS-PAGE with known concentrations of bovine serum albumin.
Electron microscopy
SMA–LptB2FG at 59 μg/ml was prepared by the negative-stain mica-carbon flotation technique. The samples were absorbed to the clean side of a carbon film on mica, stained with 2% Na4O40SiW12 in distilled water, and then transferred to a 400-mesh copper grid. The images were taken under low-dose conditions (<10 e−/Å2) with defocus values between 1.2 and 2.5 μm on a Tecnai 12 LaB6 electron microscope at 120-kV accelerating voltage using CCD Camera Gatan Orius 1000.
NMR spectroscopy
The experiments were recorded on Bruker 600-, 700-, 850-, and 950-MHz spectrometers equipped with triple 1H, 13C, 15N resonance cryoprobes, at 25 °C for 1H and 31P and at 37 °C for real-time kinetic analysis, in TBS with 10% D2O. All 1D 1H experiments were based on the Bruker zgesgp experiment with water suppression using sculpting with gradients and 15-s interscan delay. The data were processed using TopSpin 3.5 and Ccpnmr Analysis 2.4.2. ATPase and AK activities were checked by supplying ATP or ADP as the substrate. For LptB2FG/C, 5 μM of complex was incubated with 5 mM of nucleotide and 1 mM MgCl2 in TBS. When necessary, LptCΔ[1–23]/and LptAΔ160 were added at 10 μM. These activity assays were incubated at 37 °C for 7 h, flash-frozen, and transferred to 3-mm NMR tubes to be analyzed. For LptB, 2 μM of protein was incubated with 5 mM of nucleotide/2.5 mM MgCl2 in TBS at 25 °C for 17 h, flash-frozen, and transferred to 3-mm NMR tubes. The STD experiments were recorded on a 700-MHz cryoprobe at 25 °C, with Bruker stddiffesgp.3 pulse sequence alternating on and off resonance at 0.5 ppm and −40 ppm, with a 60-ms spinlock to suppress protein signals. 500 μM of the nucleotide (ADP or ADPβS) is mixed with 12.5 μM LptB (WT or mutant) with 50 μM MgCl2 in TBS with 1.7% glycerol and 10% D2O.
Data availability
All data are contained within this article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank A. Vallet and A. Favier for support with NMR and B. Bersch with SMA. This work used the EM facilities at the Grenoble Instruct-ERIC Center (ISBG; UAR 3518 CNRS CEA-UGA-EMBL) with support from the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-0005-02) and GRAL, a project of the University Grenoble Alpes Graduate School CBH-EUR-GS (ANR-17-EURE-0003) within the Grenoble Partnership for Structural Biology. The IBS Electron Microscope facility is supported by the Auvergne Rhône-Alpes Region, the Fonds Feder, the Fondation pour la Recherche Médicale, and GIS-IBiSA. This project was funded by the Train2Target project from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie Grant agreement #721484.
Author contributions
T. B. and C. L. conceptualization; T. B., K. G.-B., I. A., E. C. C. M., P. S., A. P., and C. L. resources; T. B., J.-P. S., and C. L. formal analysis; T. B. and C. L. investigation; T. B. and C. L. writing–original draft; T. B., P. S., A. P., J.-P. S., and C. L. writing–review and editing; I. A., J.-P. S., and C. L. supervision; A. P. funding acquisition; A. P. and C. L. project administration; J.-P. S. methodology; C. L. software; C. L. validation; C. L. visualization.
Funding and additional information
A. P. and J.-P. S. are supported by the European Commission via the International Training Network Train2Target (721484).
Edited by Chris Whitfield
Supporting information
References
- 1.Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani B., Ruiz N. Function and biogenesis of lipopolysaccharides. EcoSal Plus. 2018 doi: 10.1128/ecosalplus.esp-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperandeo P., Martorana A.M., Polissi A. The lipopolysaccharide transport (Lpt) machinery: A nonconventional transporter for lipopolysaccharide assembly at the outer membrane of gram-negative bacteria. J. Biol. Chem. 2017;292:17981–17990. doi: 10.1074/jbc.R117.802512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villa R., Martorana A.M., Okuda S., Gourlay L.J., Nardini M., Sperandeo P., Dehò G., Bolognesi M., Kahne D., Polissia A. The Escherichia coli lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 2013;195:1100–1108. doi: 10.1128/JB.02057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y., Orlando B.J., Liao M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature. 2019;567:486–490. doi: 10.1038/s41586-019-1025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015;7:14. doi: 10.12703/P7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaur H., Lakatos-Karoly A., Vogel R., Nöll A., Tampé R., Glaubitz C. Coupled ATPase-adenylate kinase activity in ABC transporters. Nat. Commun. 2016;7:13864. doi: 10.1038/ncomms13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzeja P., Terzic A. Adenylate kinase and AMP signaling networks: Metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009;10:1729–1772. doi: 10.3390/ijms10041729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doerrler W.T., Raetz C.R.H. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 2002;277:36697–36705. doi: 10.1074/jbc.M205857200. [DOI] [PubMed] [Google Scholar]
- 10.Voss B.J., Stephen Trent M. LPS transport: Flipping out over MsbA. Curr. Biol. 2018;28:R30–R33. doi: 10.1016/j.cub.2017.10.067. [DOI] [PubMed] [Google Scholar]
- 11.Kaur H., Abreu B., Akhmetzyanov D., Lakatos-Karoly A., Soares C.M., Prisner T., Glaubitz C. Unexplored nucleotide binding modes for the ABC exporter MsbA. J. Am. Chem. Soc. 2018;140:14112–14125. doi: 10.1021/jacs.8b06739. [DOI] [PubMed] [Google Scholar]
- 12.Tang X., Chang S., Luo Q., Zhang Z., Qiao W., Xu C., Zhang C., Niu Y., Yang W., Wang T., Zhang Z., Zhu X., Wei X., Dong C., Zhang X., et al. Cryo-EM structures of lipopolysaccharide transporter LptB 2 FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism. Nat. Commun. 2019;10:4175. doi: 10.1038/s41467-019-11977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong H., Zhang Z., Tang X., Paterson N.G., Dong C. Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat. Commun. 2017;8:222. doi: 10.1038/s41467-017-00273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Q., Yang X., Yu S., Shi H., Wang K., Xiao L., Zhu G., Sun C., Li T., Li D., Zhang X., Zhou M., Huang Y. Structural basis for lipopolysaccharide extraction by ABC transporter LptB 2 FG. Nat. Struct. Mol. Biol. 2017;24:469–474. doi: 10.1038/nsmb.3399. [DOI] [PubMed] [Google Scholar]
- 15.Owens T.W., Taylor R.J., Pahil K.S., Bertani B.R., Ruiz N., Kruse A.C., Kahne D. Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature. 2019;567:550–553. doi: 10.1038/s41586-019-1039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman D.J., Xie R., Taylor R.J., George A.H., Okuda S., Foster P.J., Needleman D.J., Kahne D. Lipopolysaccharide is transported to the cell surface by a membrane-to membrane protein bridge. Science. 2018;359:798–801. doi: 10.1126/science.aar1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dörr J.M., Scheidelaar S., Koorengevel M.C., Dominguez J.J., Schäfer M., van Walree C.A., Killian J.A. The styrene–maleic acid copolymer: A versatile tool in membrane research. Eur. Biophys. J. 2016;45:3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dörr J.M., Koorengevel M.C., Schäfer M., Prokofyev A.V., Scheidelaar S., van der Cruijsen E.A.W., Dafforn T.R., Baldus M., Killian J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K + channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sperandeo P., Villa R., Martorana A.M., Šamalikova M., Grandori R., Dehò G., Polissi A. New insights into the lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex. J. Bacteriol. 2011;193:1042–1053. doi: 10.1128/JB.01037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laguri C., Sperandeo P., Pounot K., Ayala I., Silipo A., Bougault C.M., Molinaro A., Polissi A., Simorre J.-P. Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci. Rep. 2017;7:9715. doi: 10.1038/s41598-017-10136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marion D. An introduction to biological NMR spectroscopy. Mol. Cell. Proteomics. 2013;12:3006–3025. doi: 10.1074/mcp.O113.030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lian Y., Jiang H., Feng J., Wang X., Hou X., Deng P. Direct and simultaneous quantification of ATP, ADP and AMP by 1H and 31P nuclear magnetic resonance spectroscopy. Talanta. 2016;150:485–492. doi: 10.1016/j.talanta.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 23.Sherman D.J., Lazarus M.B., Murphy L., Liu C., Walker S., Ruiz N., Kahne D. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4982–4987. doi: 10.1073/pnas.1323516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedet M., Falchi F.A., Puccio S., Benedettodi C., Peano C., Polissi A., Dehò G. The lack of the essential LptC protein in the trans-envelope lipopolysaccharide transport machine is circumvented by suppressor mutations in LptF, an inner membrane component of the escherichia coli transporter. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abele U., Schulz G.E. High-resolution structures of adenylate kinase from yeast ligated with inhibitor Ap5A, showing the pathway of phosphoryl transfer. Protein Sci. 1995;4:1262–1271. doi: 10.1002/pro.5560040702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson U., Wolf-Watz M. Overlap between folding and functional energy landscapes for adenylate kinase conformational change. Nat. Commun. 2010;1:1–8. doi: 10.1038/ncomms1106. [DOI] [PubMed] [Google Scholar]
- 27.Mayer M., Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 28.Ambudkar S.V., Kim I.W., Xia D., Sauna Z.E. The A-loop, a novel conserved aromatic acid subdomain upstream of the Walker A motif in ABC transporters, is critical for ATP binding. FEBS Lett. 2006;580:1049–1055. doi: 10.1016/j.febslet.2005.12.051. [DOI] [PubMed] [Google Scholar]
- 29.Carrier I., Urbatsch I.L., Senior A.E., Gros P. Mutational analysis of conserved aromatic residues in the A-loop of the ABC transporter ABCB1A (mouse Mdr3) FEBS Lett. 2007;581:301–308. doi: 10.1016/j.febslet.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 30.Randak C.O., Welsh M.J. Adenylate kinase activity in ABC transporters. J. Biol. Chem. 2005;280:34385–34388. doi: 10.1074/jbc.R500009200. [DOI] [PubMed] [Google Scholar]
- 31.Dong Q., Ernst S.E., Ostedgaard L.S., Shah V.S., ver Heul A.R., Welsh M.J., Randak C.O. Mutating the conserved Q-loop glutamine 1291 selectively disrupts adenylate kinase-dependent channel gating of the ATP-binding cassette (ABC) adenylate kinase cystic fibrosis transmembrane conductance regulator (CFTR) and reduces channel function in prima. J. Biol. Chem. 2015;290:14140–14153. doi: 10.1074/jbc.M114.611616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narita S.-I., Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583:2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this article.