Abstract
Background:
Efforts to explain the burden of cardiovascular disease (CVD) often focus on genetic factors or social determinants of health. There is little evidence on the comparative predictive value of each, which could guide clinical and public health investments in measuring genetic versus social information. We compared the variance in CVD-related outcomes explained by genetic versus socioeconomic predictors.
Methods:
Data were drawn from the Health and Retirement Study (N=8,720). We examined self-reported diabetes, heart disease, depression, smoking, and body mass index, and objectively measured total and high-density lipoprotein cholesterol. For each outcome, we compared the variance explained by demographic characteristics, socioeconomic position (SEP), and genetic characteristics including a polygenic score for each outcome and principal components (PCs) for genetic ancestry. We used R-squared values derived from race-stratified multivariable linear regressions to evaluate the variance explained.
Results:
The variance explained by models including all predictors ranged from 3.7% to 14.3%. Demographic characteristics explained more than half this variance for most outcomes. SEP explained comparable or greater variance relative to the combination of the polygenic score and PCs for most conditions among both white and Black participants. The combination of SEP, polygenic score, and PCs performed substantially better, suggesting that each set of characteristics may independently contribute to prediction of CVD related outcomes.
Conclusions:
Focusing on genetic inputs into personalized medicine predictive models, without considering measures of social context that have clear predictive value, needlessly ignores relevant information that is more feasible and affordable to collect on patients in clinical settings.
Keywords: socioeconomic position, polygenic score, cardiovascular disease
INTRODUCTION
Cardiovascular disease (CVD) is the leading cause of death in the United States.1 Conceptual frameworks guiding efforts to reduce the burden of CVD are often motivated around either a precision medicine approach primarily focused on genetics or a social determinants of health perspective that discounts the potential relevance of genetic factors. Precision medicine typically focuses on precisely tailoring treatment based on an individual’s genetic and other biomedical characteristics.2 In contrast, social determinants frameworks emphasize the importance of addressing social factors, such as components of socioeconomic position (SEP), to reduce CVD burden and disparities at the population level.3 Although these need not be competing frameworks, in practice precision medicine is rarely used to motivate strategies leveraging social determinants of health, and social determinants of health initiatives rarely engage with biomarker or genetic metrics. To date, there is little evidence on whether more relevant predictive information is likely to be derived from genetic data or social factors, because few studies have included both types of data as explanatory variables in the same models.
Given the decrease in price, a whole genome assay is currently feasible as a routine part of clinical care and a precision medicine framework would suggest the information derived from such genetic data could be used to identify risk strata for clinical care.4 Genetic information is already being used to guide some treatment decisions.5,6 At the same time, some have proposed routinely incorporating information on social determinants of health into clinical records.7 Social determinants data may be useful to identify high-risk groups and guide clinical decisions based on the social context faced by the patient, although this is not widespread practice in the U.S. For example, closely related work has evaluated adding social characteristics (like SEP) to clinical risk prediction algorithms,8–10 and this is now common practice in some international settings (e.g., the ASSIGN score in Scotland) but not in the U.S.11
Integrating any type of new information into clinical care entails costs related to collecting, storing, and creating user-friendly access to the data. Given this, if we want to maximize our ability to anticipate high risk of CVD, what information is in fact most important to collect? Is there substantial added value of genetic tests in explaining the variation in CVD outcomes, over and above standard questions on demographics? Similarly, is there added value in assessing social determinants? Would the two types of data in combination substantially outperform either in isolation? There are increasing calls in population health research for a more robust engagement between genetic research and social epidemiology to address this gap in our understanding.12
In this study, we help to answer these questions by leveraging longitudinal data on a large sample of U.S. older adults. We examine the variance in CVD and related risk factors that is explained by demographic, genetic, and socioeconomic predictors. We do not attempt to identify causal determinants of CVD and related risk factors in our analysis or develop a clinical prediction model; instead, we focus here on the relative importance of prediction in the context of a precision health approach to disease prevention.
METHODS
Data
Our sample was drawn from the U.S. Health and Retirement Study (HRS), a longitudinal cohort study that has been conducted biennially since 1992 among a nationally representative sample of men and women over 50 years of age and their spouses (N = 37,495). Additional details on the survey design have been described previously.13 We used data through the 2014 survey wave, the latest available at the time data analysis began. Neighborhood-level socioeconomic characteristics at the census tract level were drawn from the 2000 Decennial Census,14 linked to HRS based on census tract of residence in 2000.
We restricted the sample to individuals who participated in genetic testing and for whom valid data were available on the polygenic scores of interest (N = 12,367, details on genetic testing below). We also restricted the sample to individuals for whom data were available on neighborhood of residence (N = 9,909). Finally, we restricted the sample to self-reported non-Hispanic white participants (N = 7,522, which HRS labels as those with “European ancestry” for the purposes of their genetic data) and self-reported Black participants (N = 1,198, which HRS labels as those with “African ancestry”). HRS has released polygenic scores—derived from prior genome-wide association studies—only for these two populations (see details below), and there are few individuals of other racial/ethnic subgroups in HRS. Because the majority of genome-wide association studies are done among European-ancestry populations, polygenic scores constructed from these data may not necessarily have the same predictive capacity for populations of non-European ancestry.15 In addition, Black and white participants may be of mixed genetic ancestry despite how they are categorized in HRS.
Variables
Outcomes
In each survey wave, HRS asks respondents whether they have ever been diagnosed with a list of specific conditions. We selected self-reported and objectively measured CVD outcomes and risk factors for which prevention or treatment is available in a clinical setting, and for which information on demographic and other characteristics may influence clinical guidelines. We also required that a relevant polygenic score for these outcomes was available in HRS. For self-reported outcomes, we defined someone as having that condition if they ever reported that they had the condition in any survey wave. These included self-reported measures of diabetes, heart disease, smoking, and body mass index (BMI) based on self-reported height and weight. We also included depression risk, as depression and poor mental health have been repeatedly documented as risk factors for CVD.16,17 We determined depression risk based on whether an individual ever scored 3 or more on the shortened 8-item Center for Epidemiologic Studies Depression scale used by HRS.18 Finally, we included objectively measured serum markers of total cholesterol and high-density lipoprotein (HDL, “good”) cholesterol that were collected in the 2006–2012 survey waves for a subset of study participants. For individuals who provided a blood sample in more than one survey wave, we took the mean of the observed values.
Demographic Characteristics
Demographic characteristics included a range of measures that have been associated with CVD: gender, foreign-born status, birth year, and census region of residence (Northeast, Midwest, South, and West). Census region was drawn from the first wave in which the subject participated in HRS, to reduce confounding by health status. Birth year was included as a cubic spline to account for possible non-linear relationships with the outcomes. Because age and birth year are highly correlated in our data due to the nature of the HRS sampling, we did not include both variables in the models.
Socioeconomic Position
To measure SEP as comprehensively as possible, we included variables that captured multiple socioeconomic dimensions at both the individual and neighborhood levels. The individual-level variables included educational attainment (less than high school, high school, some college, college or more), assets, income, longest held occupation (manager/professional versus other), and an index of childhood SEP. Household assets and income were drawn from the first wave in which the subject participated, to reduce confounding by health status (i.e., worsened CVD in early survey waves might lead to reduced income and assets in later waves), and the variable was transformed with a natural logarithm for our analysis due to variable skew. The childhood SEP index was constructed and validated in HRS data in prior work, and included measures of childhood social capital, financial capital, and human capital.19
Neighborhood socioeconomic status was captured using 16 census tract characteristics, drawn from the 2000 Decennial Census14 and linked based on census tract of residence in 2000 (see eAppendix). To reduce the dimensionality of these data, we conducted principal components analysis, similar to the construction of composite measures of neighborhood-level disadvantage used in prior work.20,21 Based on graphical examination of the elbow (i.e., kink) in the resulting scree plot,22 we selected the first five principal components (PCs) to include in subsequent regressions.
Genetic Characteristics
Genetic data were collected from HRS participants during the 2006–2012 waves using a mouthwash technique. Genotyping was conducted by the NIH Center for Inherited Disease Research using the Illumina Human Omni-2.5 Quad beadchip, which includes roughly 2.4 million single-nucleotide polymorphisms (SNPs). Additional details are available from HRS.23
Using genome-wide association study data, HRS constructed genome-wide polygenic scores for a range of phenotypes, aggregating thousands to millions of SNPs across the genome and weighting them by effect sizes derived from genome-wide association study data. HRS defined weights by the odds ratio or beta estimate from the genome-wide association study meta-analysis files corresponding to the phenotype of interest, and additional details are available in HRS documentation.23 Using genome-wide polygenic scores is preferred over including only SNPs deemed significant in prior genome-wide association studies, as the former are thought to explain more variance in the phenotype of interest.24 Each polygenic score therefore estimates an individual’s genetic risk of developing the disease of interest. HRS provides separate polygenic scores for self-reported white participants (which it labels as those with “European ancestry”) and self-reported Black participants (which it labels as those with “African ancestry”). For each participant, we used the polygenic score corresponding with their self-reported race, although it should be noted that racial identity is a social construct, and that systematic biological/health differences between Black and white adults reflect the embodiment of social inequalities and systemic racism targeting Black people in the U.S. context.
Using these SNPs, HRS also constructed race-specific PCs (i.e., separately for white and Black participants) to represent genetic ancestry.23 We included the first 10 components in regression models to account for population stratification.25
Data Analysis
We first tabulated characteristics of the sample. We then constructed a series of models, to test how different combinations of covariates altered the variance explained for each outcome. Model 1 regressed each health outcome of interest (e.g., diabetes) on the demographic characteristics above. Model 2 included demographic characteristics and measures of SEP. Model 3 included demographic characteristics and PCs for genetic ancestry. Model 4 included demographic characteristics and the disease-specific polygenic score (e.g., for diabetes). Model 5 included demographic characteristics and both PCs and the polygenic score. Finally, Model 6 included all predictors: demographic characteristics, SEP, PCs, and the polygenic score.
We assessed variance explained using adjusted R-squared, defined as the variance in the outcome explained by predictions from the estimated model divided by the total variance in the outcome, or fraction of the variance explained by the model. This metric has been used to quantify prediction in prior related work.26,27 Since R-squared (or an appropriate analog) is not available with an equivalent interpretation for logistic regressions,28,29 ordinary least squares linear regressions were used for both continuous and binary outcomes, to allow comparability of the measure of variance explained across all outcomes. Robust standard errors were clustered at the household level to account for correlated observations among spouses.
We also examined the additional variance explained by SEP, PCs, and the polygenic score, individually and in combination, over and above the variance explained by demographic characteristics alone. To do so, we subtracted the R-squared for Model 1 from the R-squared for Models 2–6, as Model 1 is nested within (i.e., has a subset of the covariates from) Models 2–6. We approximated confidence intervals for the R-squared values using the Fisher’s z-transformation and approximate standard error for a correlation coefficient.30
Finally, we conducted formal evaluations of model performance for binary outcomes by calculating sensitivity, specificity, accuracy, and Brier scores for each set of models, (see eAppendix). For logistic regression models with binary outcomes, we also computed likelihood ratio tests, comparing each model with the base model that included only demographic predictors.
Ethics Approval
Approval for this study was provided by the institutional review board of the first author’s institution (15–18340).
RESULTS
Sample Characteristics
More than half the sample was female, with a mean birth year of 1935 (SD 9.0) among white participants and 1937 (SD 8.8) for Black participants (Table 1). About half of white participants and two-thirds of Black participants had a high school education or less. Mean annual household income was $56,914 (SD $57,299) for white participants and $35,133 (SD $35,855) for Black participants. Nearly a third of white participants and 16% of Black participants were managers or professionals in their longest held occupations.
Table 1.
Sample Characteristics
| Mean (SD) or % | ||
|---|---|---|
| White Participants N = 7,522 | Black Participants N = 1,198 | |
| Demographic characteristics | ||
| Female (%) | 59 | 64 |
| Foreign born (%) | 4 | 6 |
| Birth year (mean (SD)) | 1935 (9.0) | 1937 (8.8) |
| Census region of residence (%) | ||
| Northeast | 17 | 18 |
| Midwest | 29 | 20 |
| South | 36 | 56 |
| West | 17 | 6 |
| Socioeconomic characteristics | ||
| Educational attainment (%) | ||
| Less than high school | 14 | 40 |
| High school | 40 | 32 |
| Some college | 23 | 17 |
| College or more | 23 | 12 |
| Assets (USD, mean (SD)) | 317,893 (643,613) | 79,604 (165,014) |
| Income (USD, mean (SD)) | 56,914 (57,299) | 35,133 (35,855) |
| Occupation: manager/professional (%) | 29 | 16 |
| Childhood SEP index (mean (SD)) | 0.22 (0.87) | −0.25 (0.86) |
| Health characteristics | ||
| Diabetes (%) | 25 | 41 |
| Heart disease (%) | 41 | 34 |
| Depression (%) | 38 | 53 |
| Ever smoker (%) | 58 | 59 |
| Body mass index (kg/m2, mean (SD)) | 27 (4.9) | 29 (5.6) |
| Total cholesterol (mg/dL, mean (SD)) | 196 (38) | 195 (37) |
| High-density lipoprotein cholesterol (mg/dL, mean (SD)) | 54 (15) | 55 (16) |
Note: Assets, income, occupational status, and census region of residence reported here are from the first wave in which the subject participated, to reduce confounding by health status. SEP: socioeconomic position
Nearly a quarter of white participants and 41% of Black participants self-reported diabetes, 41% of white participants and 34% of Black participants reported heart disease, and 38% of white participants and 53% of Black participants met criteria for high risk of depression. Over half were ever-smokers. Mean BMI was 27 (SD 4.9) among white participants and 29 (SD 5.6) among Black participants, mean total cholesterol was 196 (SD 38) among white participants and 195 (SD 37) among Black participants, and mean HDL was 54 (SD 15) among white participants and 55 (SD 16) among Black participants.
Variance in CVD Explained by Demographic Characteristics
R-squared values for regressions models incorporating only demographic characteristics (i.e., gender, foreign-born status, birth year, and census region of residence) ranged from 1% (for diabetes) to 8% (for total cholesterol) among white participants (blue diamonds in Figure 1A), and from 2% (for heart disease) to 8% (for BMI) among Black participants (blue diamonds in Figure 1B). The total variance explained when including all categories of predictors was below 15% for all outcomes, ranging from 4% for diabetes to 14% for BMI among white participants (Figure 1A), and from 6% for HDL to 11% for BMI among Black participants (Figure 1B). For heart disease, smoking, and total and HDL cholesterol, demographic factors accounted for at least half of the variance that could be explained by all predictors combined among white participants. Among Black participants, demographic factors accounted for at least half of the total variance explained by all predictors for smoking, BMI, and total and HDL cholesterol.
Figure 1. Percent of variance explained by demographic, socioeconomic, and genetic covariates.
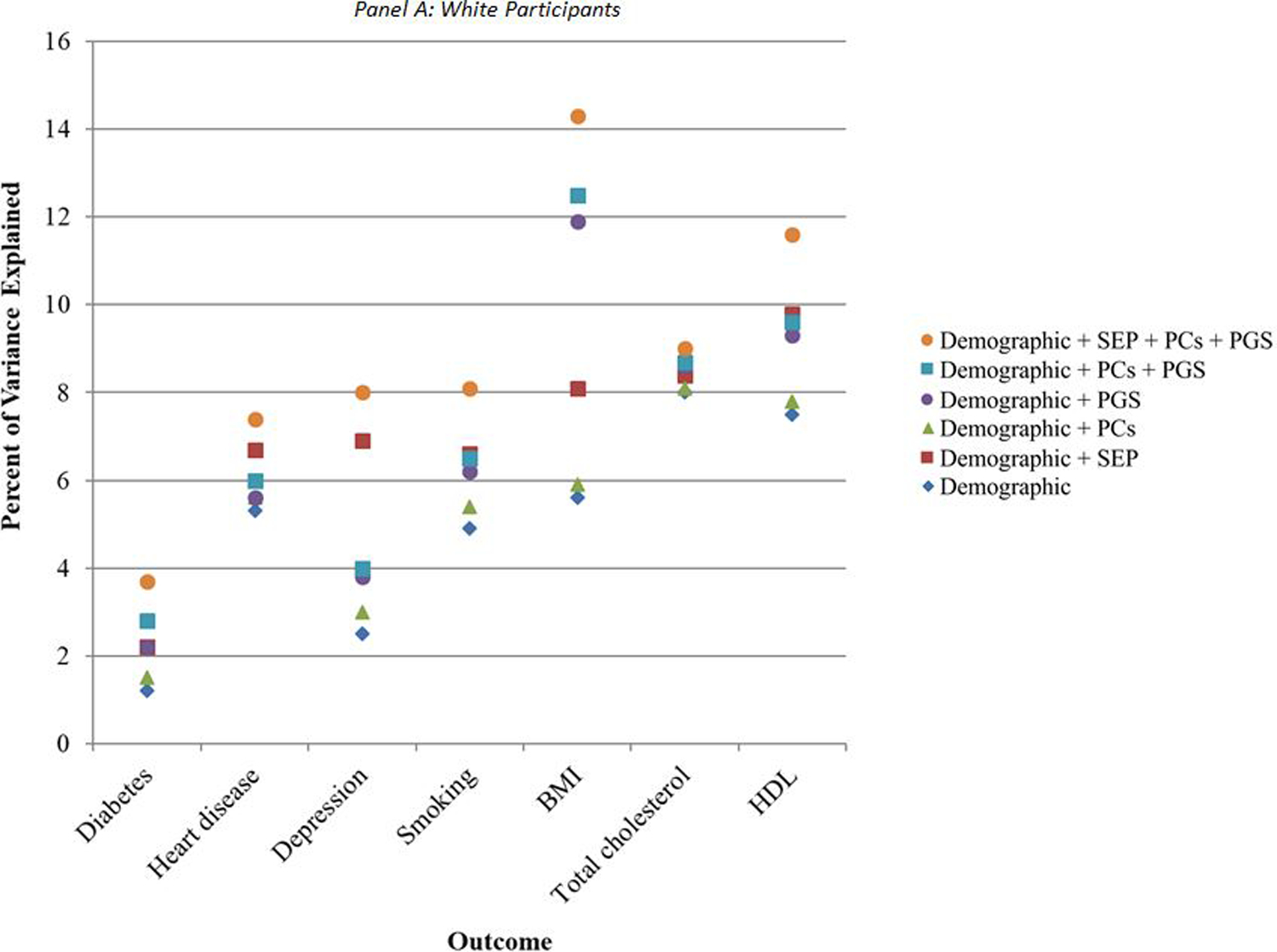
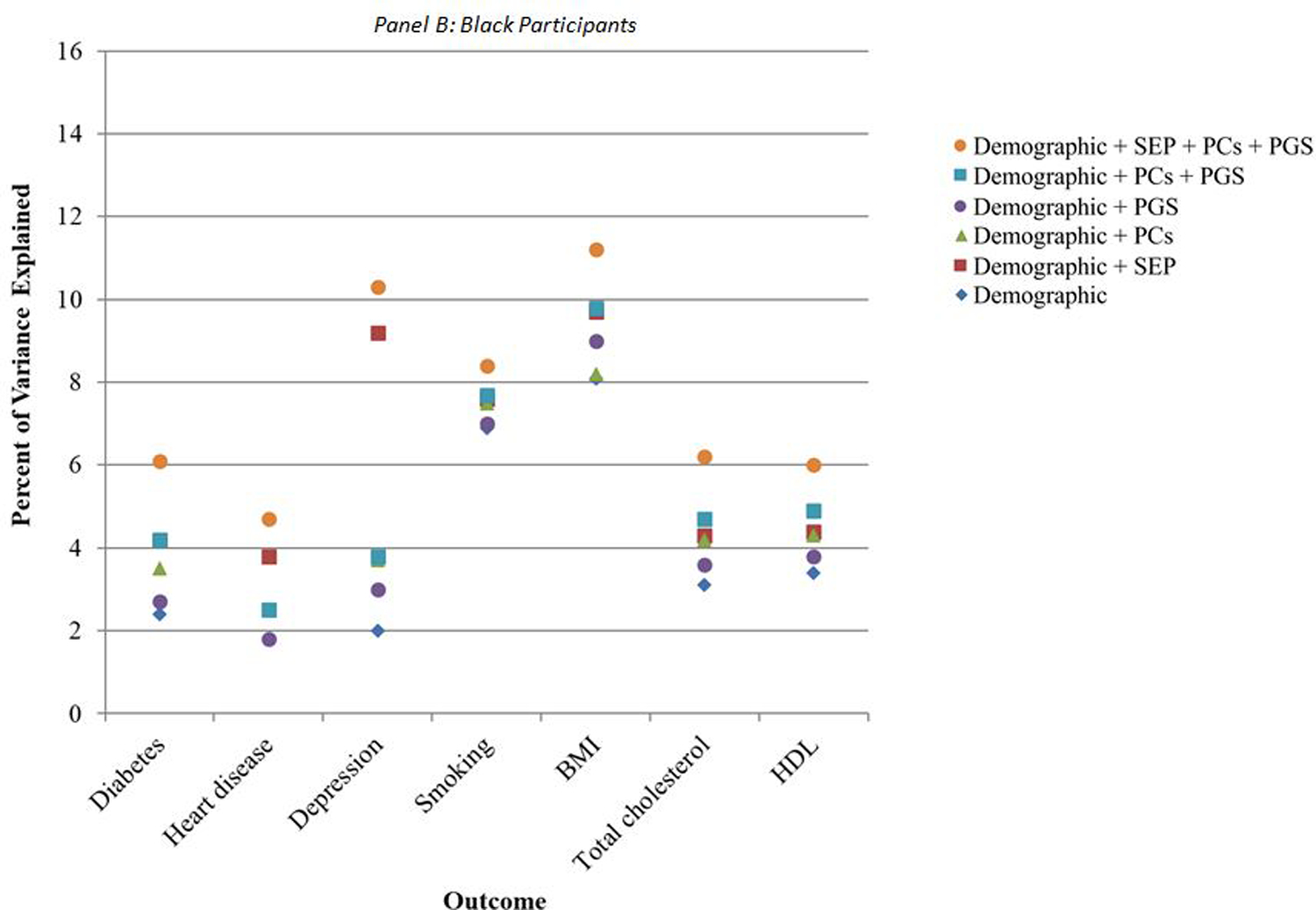
Note: N = 7,522 white participants and 1,198 Black participants. Percent of variance explained is the R-squared value from a multivariate linear regression of the given outcome on the given combination of covariates. BMI: body mass index; HDL: high-density lipoprotein cholesterol; PCs: principal components for genetic ancestry; PGS: polygenic score specific to relevant health condition; SEP: socioeconomic position.
Additional Variance in CVD Explained by Socioeconomic and Genetic Characteristics
The additional variance explained by the addition of SEP, genetic PCs, and polygenic scores over and above the model including only demographic predictors differed across outcomes and for white and Black participants (Figure 2, eTables 1–2). Among white participants, PCs for genetic ancestry consistently contributed the least amount of explained variance (green triangles in Figure 2A), such that estimates for R-squared were not different from models including only demographic characteristics (eTable 1). This was followed by polygenic scores for all conditions except BMI and total cholesterol (purple circles). Among Black participants, the polygenic score contributed the least amount of explained variation for all conditions except for BMI (purple circles in Figure 2B), such that estimates for R-squared were not significantly different from models including only demographic characteristics (eTable 2). This was followed by PCs for genetic ancestry (green triangles).
Figure 2.
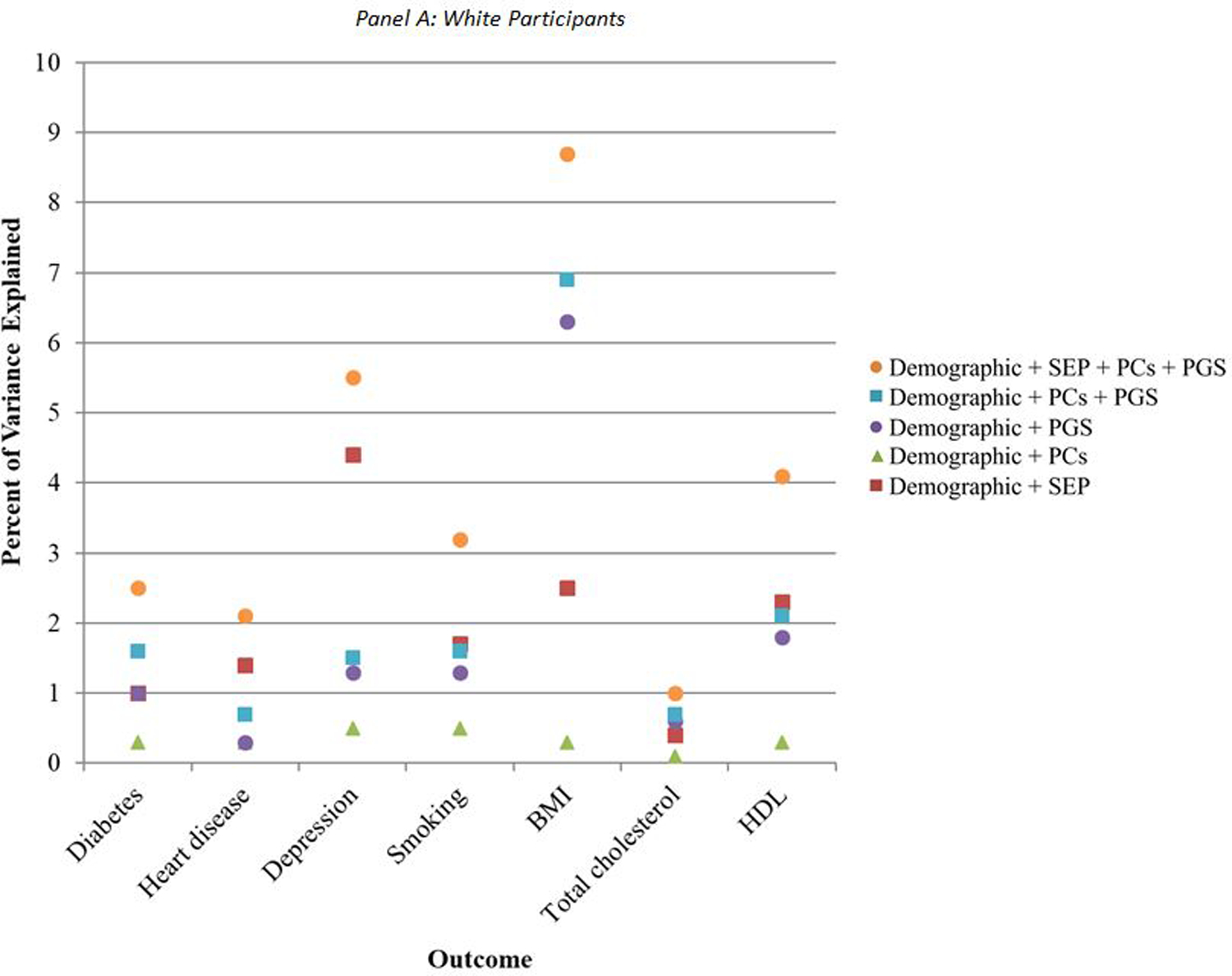
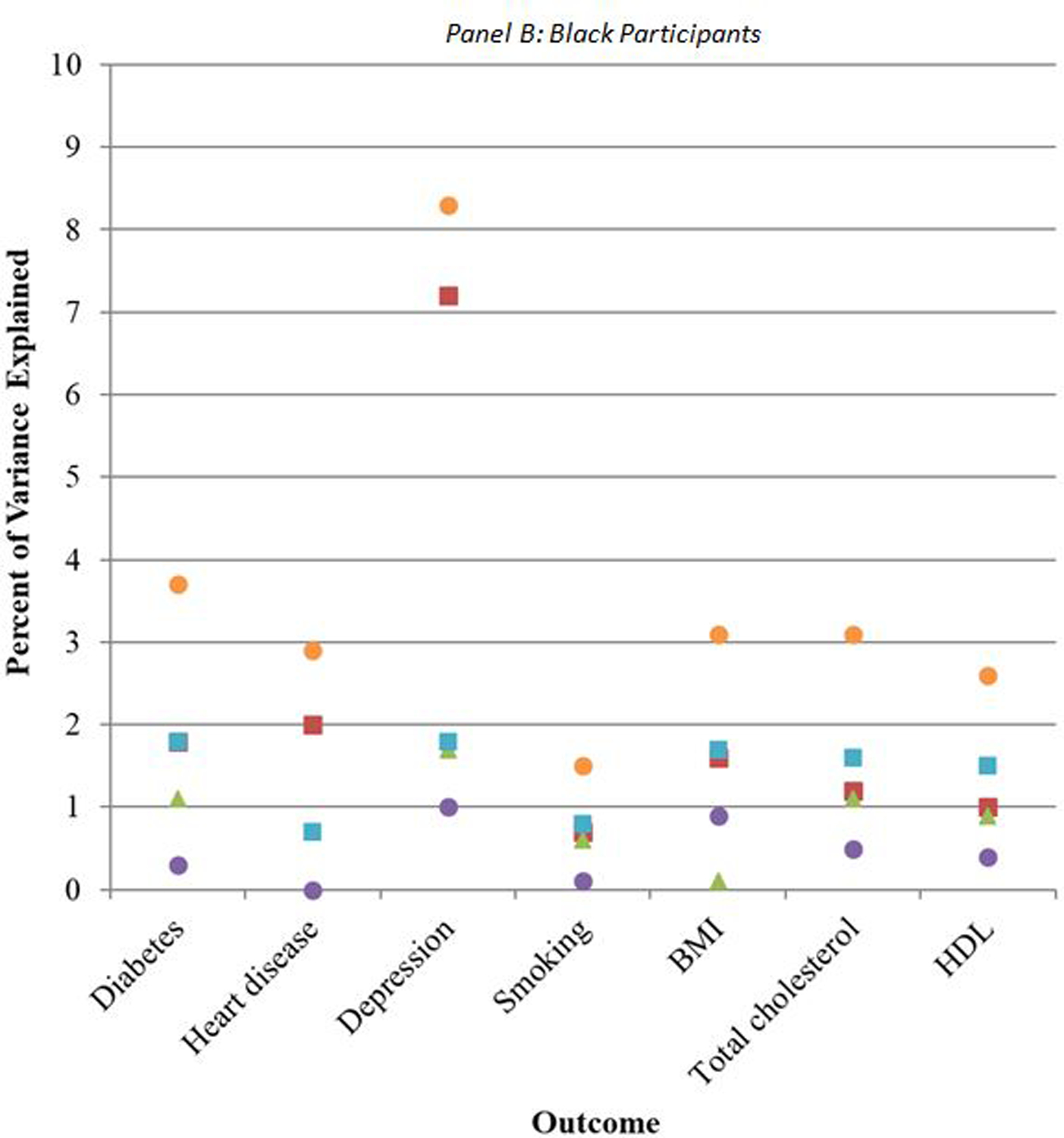
Additional percent of variance explained by socioeconomic and genetic covariates, relative to demographic covariates alone
When including both sets of genetic predictors—genetic PCs and polygenic scores (blue squares in Figures 2A, 2B)—the additional variance explained increased relative to the variance explained by demographic characteristics alone, and was comparable to the variance explained by SEP (eTables 1–2). For depression, however, SEP characteristics explained a higher percentage of the variance than the combination of PCs and polygenic scores for both white (7%, 95%CI: 6, 8 for SEP versus 4%, 95%CI: 3, 5 for PCs and polygenic scores) and Black participants (9%, 95%CI: 6, 13 for SEP versus 4%, 95%CI: 2, 6 for PCs and polygenic scores) (eTables 1–2). For BMI among white participants, PCs and polygenic scores together explained a higher percentage of the variance (13%, 95%CI: 11, 14) than SEP characteristics (8%, 95%CI: 7, 9).
For nearly all outcomes, among both white and Black participants, there was substantially more variance explained when all three of SEP, PCs, and polygenic scores were included in the models (orange circles in Figures 2A, 2B; eTables 1–2). This suggests that SEP is not collinear with the genetic factors, and that they each may contribute to prediction of CVD and related risk factors, although the magnitude of this additional prediction varied by risk factor.
Finally, likelihood ratio tests for binary outcomes demonstrated that adding predictors to the base model increased the goodness-of-fit of the model for the White sample for all outcomes (i.e., diabetes, heart disease, depression, and smoking). For the Black sample, only models that included SEP improved the goodness-of-fit for diabetes, heart, disease, and smoking, while all predictors increased the goodness-of-fit for depression (eTable 3).
DISCUSSION
This study is among the first to compare the variance explained by demographic, socioeconomic, and genetic characteristics across a range of CVD-related outcomes and risk factors, with the goal of informing investments in measuring genetic and social information for predictive modeling in clinical care settings. Despite the excitement and substantial investment in genetic testing and personalized medicine, this study demonstrated that in many cases SEP explained a greater or comparable amount of the variance in CVD and related risk factors relative to polygenic scores, and it was particularly important in explaining the variation in depression risk.
The predictive capacity of polygenic scores was lower among Black participants, even though we used scores that were specific to this population. This is likely due to the fact that the genome-wide association studies used to create these scores have been conducted predominantly in white populations and thus the samples for creating the scores specific to Black participants are less well powered.31 This leads to worse calibration of polygenic score-based models for Black people and could contribute to disparities in appropriate treatment if incorporated into routine clinical decisions. More generally, HRS only provides PC and polygenic score data stratified by race, which is itself a sociopolitical construct. Survey documentation from HRS indicates that this is “to control for confounding from population stratification, or to account for any ancestry differences in genetic structures within populations that could bias estimates,”23 although this does not account for the fact that biological differences within racial categories may be more important to consider than those between individuals of different races.32–34 For example, PCs may be associated with skin color, state of residence, and related experiences of racism for Black participants, which is known to affect health. Part or all of the effect attributed to genetic ancestry in Black participants could be therefore related to social factors. Practically speaking, this precluded our ability to carry out an analysis in the overall sample. Efforts are underway to diversify the samples included in genome-wide association studies, and future work should replicate this study when more genome-wide association study data from a larger number of Black participants become available. Racial inequalities in cardiovascular health are stark in the U.S., and by stratifying our models on race, we obscure one of the largest demographic predictors. This does not modify interpretations of what could be offered by adding genetic or SEP information over and above the demographic-only models. Given the national priority of addressing racial disparities in health, we consider the stratified models most relevant for prioritizing new sources of information. For Black participants, the addition of SEP matched or outperformed the addition of genetic information for all outcomes except total and HDL cholesterol.
A handful of prior studies have conducted analyses similar to those in this study. One prior study determined that a polygenic score for obesity improved prediction of BMI over and above demographic and socioeconomic characteristics. It also found that the predictive value of the polygenic score was not as great as that of SEP and concluded that it would likely have limited clinical utility.26 Of note, this prior study’s measure of SEP consisted solely of a categorical variable for educational attainment. As in our study, it also found that the polygenic score had limited utility in Black participants. Another study of schizophrenia compared the variance explained by a polygenic score, SEP, and an individual’s family history, finding them to be roughly comparable, although the polygenic score included only highly significant SNPs, and the measure of SEP was limited to information on each individual’s parents’ socioeconomic status in the year prior to his/her birth.27 This study was conducted in Denmark and racial heterogeneity was not examined.
In general, in our population sample of community-residing adults, the total variance explained for each outcome was still small, with R-squared values of less than 15% for all conditions even when including all of the covariates that represented a broad array of measures of demographics, SEP, and genetic characteristics. This modest percentage reflects the role of measurement error of constructs included, and other unmeasured factors, both genetic and socioeconomic. For example, whole genome sequencing and examination of rare variants has shown promise for prediction,35 as have prior generation measurements of SEP.36 However, the possibility remains that the majority of unexplained variation may be due to randomness.37 For most outcomes, more than half of the variation that could be explained with measured covariates was explained by demographic characteristics alone (i.e. gender, age, place of birth, and census region of residence) which likely represents the tremendous importance of age as a predictor and likely causal determinant of CVD and related risk factors.
This study has several limitations. First, it was restricted to a sample of white and Black participants due to lack of availability of PCs and the small sample size for other racial/ethnic groups in HRS. Similarly, due to the use of HRS, there may be selection bias due to the inclusion of only older adults and possibly higher-SEP individuals that are likely to participate in surveys and genetic testing,38 and the size of the sample precluded the ability to conduct rigorous tests of overfitting. However, there are very few data sets other than HRS—particularly in the U.S.—that include this level of detail on demographic, socioeconomic, as well as genetic characteristics; future studies can replicate these analyses when data become available on younger, lower-SEP populations, and larger samples. Additionally, it is important to note that these models are not focused on determining the causal determinants of CVD and related risk factors, although predictive modeling is often a first step in understanding causal relationships. Thus, while social factors may be a direct target of intervention in clinical settings and in sectors outside of healthcare to reduce disease burden and disparities,39 this work does not necessarily speak to the possible effectiveness of such strategies. It is also unclear how the current results would generalize to health conditions other than CVD and its related risk factors, as we demonstrated heterogeneity in the variance explained by demographics, SEP, and genetic characteristics even among the handful of related outcomes included in this study. Of note, most outcomes were self-reported and likely correspond to prevalent cases. The use of prevalent outcomes will mix prediction of incidence and survival, and misclassified outcomes are likely to reduce the ability of the predictors to be useful. Additionally, for some of the health outcomes examined, more recent polygenic scores may have been developed, but these were not available for analysis in HRS. Finally, we did not evaluate interactions between the various categories of predictors, which might improve predictive capacity.
In conclusion, this is among the first studies to compare the variance explained by demographic, socioeconomic, and genetic characteristics for CVD and related risk factors. Social factors explained a large amount of variance in CVD-related outcomes, independent of genetic factors. However, polygenic scores also typically added to predictive precision. To focus only on incorporating genetic information into personalized medicine models, without considering social context, is needlessly ignoring relevant information that may be more feasible and affordable to gather in many cases. These findings may help to inform conversations about investments in measuring genetic versus social information for predictive modeling so that clinics and public health practitioners can more effectively identify risk at both the individual and the population level.
Supplementary Material
Sources of funding:
This work was supported by grant K08-HL132106 from the National Institutes of Health to Dr. Hamad, and by grant Talent Attraction Program 2018-T1/BMD-11226 from the Community of Madrid, Spain, to Dr. Walter.
Footnotes
Conflicts of interest: None declared.
Data sharing statement:
The data used for our study can be requested by submitting an application for restricted data from the Health and Retirement Study at the University of Michigan (https://hrs.isr.umich.edu).
REFERENCES
- 1.Heron M Deaths: Leading Causes for 2016. National Vital Statistics Reports 2018;67(6). [PubMed] [Google Scholar]
- 2.Ashley EA. The Precision Medicine Initiative: A New National Effort. JAMA 2015;313(21):2119–2120. [DOI] [PubMed] [Google Scholar]
- 3.Fiscella K, Tancredi D. Socioeconomic Status and Coronary Heart Disease Risk Prediction. JAMA 2008;300(22):2666–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, Rowley R, Williams MS, Roden D, Ginsburg GS, Bult C, Chisholm RL, Deverka PA, McLeod HL, Mensah GA. Opportunities, resources, and techniques for implementing genomics in clinical care. The Lancet 2019;394(10197):511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sibbing D, Aradi D, Alexopoulos D, ten Berg J, Bhatt DL, Bonello L, Collet J-P, Cuisset T, Franchi F, Gross L, Gurbel P, Jeong Y-H, Mehran R, Moliterno DJ, Neumann F-J, Pereira NL, Price MJ, Sabatine MS, So DYF, Stone GW, Storey RF, Tantry U, Trenk D, Valgimigli M, Waksman R, Angiolillo DJ. Updated Expert Consensus Statement on Platelet Function and Genetic Testing for Guiding P2Y12 Receptor Inhibitor Treatment in Percutaneous Coronary Intervention. JACC: Cardiovascular Interventions 2019;12(16):1521–1537. [DOI] [PubMed] [Google Scholar]
- 6.Peters N, Opherk C, Bergmann T, Castro M, Herzog J, Dichgans M. Spectrum of Mutations in Biopsy-Proven CADASIL: Implications for Diagnostic Strategies. Archives of Neurology 2005;62(7):1091–1094. [DOI] [PubMed] [Google Scholar]
- 7.Institute of Medicine. Capturing social and behavioral domains and measures in electronic health records: Phase 2. Washington, D.C.: The National Academies Press, 2014. [PubMed] [Google Scholar]
- 8.Collins GS, Altman DG. An independent external validation and evaluation of QRISK cardiovascular risk prediction: a prospective open cohort study. BMJ 2009;339:b2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiscella K, Tancredi D, Franks P. Adding socioeconomic status to Framingham scoring to reduce disparities in coronary risk assessment. Am Heart J 2009;157(6):988–94. [DOI] [PubMed] [Google Scholar]
- 10.Irvin JA, Kondrich AA, Ko M, Rajpurkar P, Haghgoo B, Landon BE, Phillips RL, Petterson S, Ng AY, Basu S. Incorporating machine learning and social determinants of health indicators into prospective risk adjustment for health plan payments. BMC public health 2020;20:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodward M, Brindle P, Tunstall-Pedoe H. Adding social deprivation and family history to cardiovascular risk assessment: the ASSIGN score from the Scottish Heart Health Extended Cohort (SHHEC). Heart 2007;93(2):172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belsky DW, Moffitt TE, Caspi A. Genetics in Population Health Science: Strategies and Opportunities. American Journal of Public Health 2013;103(S1):S73–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juster FT, Suzman R. An overview of the Health and Retirement Study. Journal of Human Resources 1995;30:S7–S56. [Google Scholar]
- 14.U.S. Census Bureau. American FactFinder. https://factfinder.census.gov Accessed 12 December, 2017.
- 15.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. The American Journal of Human Genetics 2017;100(4):635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley SM, Rumsfeld JS. Depression and cardiovascular disease. Trends in Cardiovascular Medicine 2015;25(7):614–622. [DOI] [PubMed] [Google Scholar]
- 17.Joynt KE, Whellan DJ, O’Connor CM. Depression and cardiovascular disease: mechanisms of interaction. Biological Psychiatry 2003;54(3):248–261. [DOI] [PubMed] [Google Scholar]
- 18.Radloff LS. The CES-D scale: A self-report depression scale for research in the general population. Applied Psychological Measurement 1977;1(3):385–401. [Google Scholar]
- 19.Vable AM, Gilsanz P, Nguyen TT, Kawachi I, Glymour MM. Validation of a theoretically motivated approach to measuring childhood socioeconomic circumstances in the Health and Retirement Study. PLOS ONE 2017;12(10):e0185898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White JS, Hamad R, Li X, Basu S, Ohlsson H, Sundquist J, Sundquist K. Long-term effects of neighbourhood deprivation on diabetes risk: quasi-experimental evidence from a refugee dispersal policy in Sweden. The Lancet Diabetes & Endocrinology 2016;4(6):517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messer LC, Laraia BA, Kaufman JS, Eyster J, Holzman C, Culhane J, Elo I, Burke JG, O’campo P. The development of a standardized neighborhood deprivation index. Journal of Urban Health 2006;83(6):1041–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabrigar LR, Wegener DT, MacCallum RC, Strahan EJ. Evaluating the use of exploratory factor analysis in psychological research. Psychological methods 1999;4(3):272. [Google Scholar]
- 23.Ware E, Schmitz L, Gard A, Faul J. HRS Polygenic Scores - Release 3. Ann Arbor, Michigan: University of MIchigan, 2018. [Google Scholar]
- 24.Ware EB, Schmitz LL, Faul J, Gard A, Mitchell C, Smith JA, Zhao W, Weir D, Kardia SL. Heterogeneity in polygenic scores for common human traits. bioRxiv 2017:106062. [Google Scholar]
- 25.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 26.Belsky DW, Moffitt TE, Sugden K, Williams B, Houts R, McCarthy J, Caspi A. Development and evaluation of a genetic risk score for obesity. Biodemography and social biology 2013;59(1):85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agerbo E, Sullivan PF, Vilhjálmsson BJ, Pedersen CB, Mors O, Børglum AD, Hougaard DM, Hollegaard MV, Meier S, Mattheisen M, Ripke S, Wray NR, Mortensen PB. Polygenic Risk Score, Parental Socioeconomic Status, Family History of Psychiatric Disorders, and the Risk for Schizophrenia: A Danish Population-Based Study and Meta-analysisVariables Associated With Schizophrenia RiskVariables Associated With Schizophrenia Risk. JAMA Psychiatry 2015;72(7):635–641. [DOI] [PubMed] [Google Scholar]
- 28.Menard S Coefficients of determination for multiple logistic regression analysis. The American Statistician 2000;54(1):17–24. [Google Scholar]
- 29.Mittlböck M, Schemper M. Explained variation for logistic regression. Statistics in medicine 1996;15(19):1987–1997. [DOI] [PubMed] [Google Scholar]
- 30.Cohen J, Cohen P, West SG, Aiken LS. Applied multiple regression/correlation analysis for the behavioral sciences Routledge, 2013. [Google Scholar]
- 31.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature News 2016;538(7624):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feldman MW, Lewontin RC. Race, ancestry, and medicine. In: Koenig BA, Lee SS-J, Richardson SS, eds. Revisiting race in a genomic age, 2008;89–101. [Google Scholar]
- 33.Lee C “Race” and “ethnicity” in biomedical research: How do scientists construct and explain differences in health? Social Science & Medicine 2009;68(6):1183–1190. [DOI] [PubMed] [Google Scholar]
- 34.Lewontin RC. The apportionment of human diversity. In: Dobzhansky T, Hecht MK, Steere WC, eds. Evolutionary biology. New York City, New York: Springer, 1972;381–398. [Google Scholar]
- 35.Ashley EA. Towards precision medicine. Nature Reviews Genetics 2016;17(9):507. [DOI] [PubMed] [Google Scholar]
- 36.Mare RD. A multigenerational view of inequality. Demography 2011;48(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith GD. Epidemiology, epigenetics and the ‘Gloomy Prospect’: embracing randomness in population health research and practice. International Journal of Epidemiology 2011;40(3):537–562. [DOI] [PubMed] [Google Scholar]
- 38.Domingue BW, Belsky DW, Harrati A, Conley D, Weir DR, Boardman JD. Mortality selection in a genetic sample and implications for association studies. International Journal of Epidemiology 2017;46(4):1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottlieb L, Sandel M, Adler NE. Collecting and applying data on social determinants of health in health care settings. JAMA internal medicine 2013;173(11):1017–1020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for our study can be requested by submitting an application for restricted data from the Health and Retirement Study at the University of Michigan (https://hrs.isr.umich.edu).


