Abstract
The regulation of electrical membrane potential is a fundamental property of living cells. This biophysical parameter determines nutrient uptake, intracellular potassium and turgor, uptake of toxic cations, and stress responses. In fungi and plants, an important determinant of membrane potential is the electrogenic proton-pumping ATPase, but the systems that modulate its activity remain largely unknown. We have characterized two genes from Saccharomyces cerevisiae, PTK2 and HRK1 (YOR267c), that encode protein kinases implicated in activation of the yeast plasma membrane H+-ATPase (Pma1) in response to glucose metabolism. These kinases mediate, directly or indirectly, an increase in affinity of Pma1 for ATP, which probably involves Ser-899 phosphorylation. Ptk2 has the strongest effect on Pma1, and ptk2 mutants exhibit a pleiotropic phenotype of tolerance to toxic cations, including sodium, lithium, manganese, tetramethylammonium, hygromycin B, and norspermidine. A plausible interpretation is that ptk2 mutants have a decreased membrane potential and that diverse cation transporters are voltage dependent. Accordingly, ptk2 mutants exhibited reduced uptake of lithium and methylammonium. Ptk2 and Hrk1 belong to a subgroup of yeast protein kinases dedicated to the regulation of plasma membrane transporters, which include Npr1 (regulator of Gap1 and Tat2 amino acid transporters) and Hal4 and Hal5 (regulators of Trk1 and Trk2 potassium transporters).
The plasma membrane H+-ATPase plays a crucial role in the physiology of fungi and plants (51, 52), and the activity of this electrogenic proton pump must be finely regulated to match the requirements for nutrient uptake, osmotic balance, ion homeostasis, and stress tolerance. Accordingly, H+-ATPase activities correlate with growth rates (42) and stress responses (4, 32, 39).
One mechanism of plasma membrane H+-ATPase regulation occurs at the transcriptional level. In the yeast Saccharomyces cerevisiae, the transcription factors Rap1 and Gcr1 mediate the increase in H+-ATPase expression triggered by glucose metabolism. On the other hand, the transcription factor Gln3 determines increased expression of the enzyme under conditions of ammonium starvation. In addition to these nutritional responses, the H+-ATPase gene (PMA1) is also regulated by the Mcm1 transcription factor, which connects the proton pump with regulatory pathways for cell growth cycle and cell wall integrity (40).
The H+-ATPase activity is reversibly regulated by several nutritional, environmental, and stress factors. The mechanism of this posttranslational regulation is similar in yeasts and plants: the activators overcome negative regulation by an inhibitory domain located at the carboxyl terminus of the enzyme (40, 43). In S. cerevisiae, glucose metabolism activates the ATPase (49) and increases its proton-pumping efficiency (59), as required by the high growth rate induced by this preferred carbon source (15). Several conserved residues at the carboxyl terminus play an important role in the glucose activation of yeast H+-ATPase, and they include the potential phosphorylation sites Ser-899 and Thr-912 (12, 44).
Very little is known about the machinery mediating activation of fungal and plant H+-ATPases. Preliminary evidence points to phosphorylation of the inhibitory domain as part of the activating mechanism (40). In yeast cells, the results of site-directed mutagenesis suggest that phosphorylation of Ser-899 activates the H+-ATPase (12), but the nature of the protein kinase(s) participating in this activation is not clear. Glucose metabolism in yeast is known to activate protein kinase A (61), but this kinase does not seem to participate in the glucose activation of the H+-ATPase (30, 37).
Genetic analysis of the machinery which activates yeast H+-ATPase has been complicated by the fact that mutations in multiple genes affecting the level of expression of the enzyme (see above) mimic the effect of mutations in the activation system (16). In addition, the presence in centromeric genomic libraries of several allele-nonspecific suppressors which increase the expression level of the H+-ATPase further complicates genetic analysis (N. de la Fuente, A. Goossens, R. Serrano, and F. Portillo, unpublished results). Biochemical analysis of H+-ATPase phosphorylation in isolated membranes has demonstrated the participation of casein kinase I. The activity of this essential and multifunctional kinase is inhibitory for the H+-ATPase, and the existence of another, yet uncharacterized, “upregulating” kinase activated by glucose was postulated (13). Actually, a correlation between glucose-induced phosphorylation and activation of yeast H+-ATPase has been demonstrated (6).
In the present work, a genetic screen based on lithium tolerance and a systematic analysis of mutant phenotypes of yeast protein kinases have converged on the identification of a protein kinase, Ptk2, which mediates the Ser-899-dependent part of the glucose activation of yeast H+-ATPase. This kinase belongs to a subfamily of protein kinases dedicated to the regulation of plasma membrane transporters.
MATERIALS AND METHODS
Yeast strains, culture conditions, and analysis of salt tolerance.
The S. cerevisiae strains used for this work, with the exception of those described in the next section, are listed in Table 1. Strains were derived by transformation (24), and crosses and standard methods for yeast culture and manipulation were used (18, 54). In order to test for tolerance to toxic salts, the different strains were grown to saturation (48 h) in liquid synthetic minimal medium (SD) containing 2% glucose, 0.7% yeast nitrogen base without amino acids (Difco, Detroit, Mich.), 50 mM succinic acid adjusted to pH 5.5 with Tris base, and adenine (30 μg/ml), histidine (30 μg/ml), leucine (100 μg/ml), tryptophan (80 μg/ml), and uracil (30 μg/ml) as required by the different strains. Cultures were diluted 10-, 100-, and 1,000-fold, and volumes of about 3 μl were dropped with a stainless steel replicator (Sigma, St. Louis, Mo.) on plates containing 2% Bacto Agar (Difco) and either SD or rich medium with the toxic cations. Rich medium (YPD) contained 1% yeast extract (Difco), 2% Bacto Peptone (Difco), and 2% glucose. MnCl2, norspermidine, and hygromycin B were added to the autoclaved medium before it was poured. All the other salts and sorbitol were added before autoclaving. In the case of SD plates with acetic acid, no succinic acid was included, but pH was also adjusted to pH 5.5 with Tris.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Referencea |
|---|---|---|
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 60 |
| W303-1B | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 60 |
| SKY697 | W303-1A ena1-4::HIS3 | 33 |
| AG86 | W303-1B ena1-4::HIS3 | |
| AG149 | W303-1B ena1-4::HIS3 ptk2::TRP1 | |
| AG151 | W303-1A ena1-4::HIS3 sen1-1 | |
| AG205 | W303-1B ena1-4::HIS3 ptk2::TRP1 [pRS496] | |
| AG211 | W303-1B ena1-4::HIS3 ptk2::TRP1 pma1::URA3-GAL1 promoter-PMA1 | |
| AG224 | W303-1B ena1-4::HIS3 ptk2::TRP1 pma1::URA3-GAL1 promoter-PMA1 [pRS496] | |
| FPY1506 | W303-1B[YEp352] | |
| FPY1507 | W303-1B[YEp352::YOR267c] | |
| FPY1508 | W303-1B[YEp352::PTK2] | |
| FPY1456 | W303-1B yor267c::KanMX[YEp352] | |
| FPY1457 | W303-1B yor267c::KanMX[YEp352::YOR267c] | |
| FPY1458 | W303-1B yor267c::KanMX[YEp352::PTK2] | |
| FPY1459 | W303-1B ptk2::KanMX[YEp352] | |
| FPY1460 | W303-1B ptk2::KanMX[YEp352::YOR267c] | |
| FPY1461 | W303-1B ptk2::KanMX[YEp352::PTK2] | |
| FPY398 | W303-1B pma1-Ser899Asp::URA3 | |
| FPY402 | W303-1B pma1-Ser899Ala::URA3 | |
| FPY1434 | W303-1B ptk2::KanMX | |
| FPY1500 | W303-1B ptk2::KanMX pma1-Ser899Ala::URA3 | |
| FPY1502 | W303-1B ptk2::KanMX pma1-Ser899Asp::URA3 |
Unless otherwise indicated, the strains are from this study.
Isolation and genetic analysis of sen mutants.
Spontaneous mutations which suppressed the lithium sensitivity of strain SKY697 (ena1-4::HIS3 derivative of W303-1A) were isolated as colonies growing after 7 days in SD plates supplemented with methionine (100 μg/ml) and 50 mM LiCl. The salt tolerance mutations were designated sen for suppression of ena1-4 and appeared with a frequency of about 10−5. Such a high frequency may reflect the large number of genes related to salt homeostasis in yeast (10). Of 250 independent mutants, 6 were selected as the most resistant in plates with 75 mM LiCl. After crossing with strain AG86 (ena1-4 but opposite mating type to SKY697), sporulation, and tetrad dissection, all six mutations proved to be monogenic recessive and defined two complementation groups: sen1 with one allele (sen1-1) and sen2 with five alleles (sen2-1 to sen2-5).
Sequencing of the sen1-1 mutation.
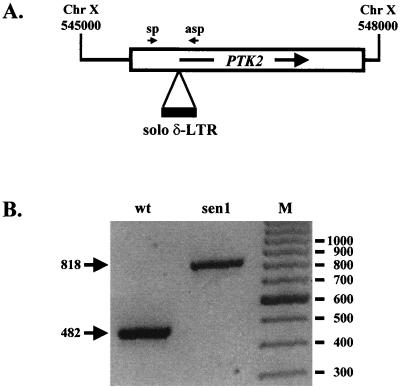
A complementation test crossing the sen1-1 mutant (strain AG151) with the ptk2 mutant (strain AG149) demonstrated that these mutations are allelic. A genomic PCR with primers sp (5′-ATTCACCTTCGTCTTCTGCTG) and asp (5′-AAGGGGAACATCCGTATCTT), corresponding to positions +202 and +683 from the start codon of the PTK2 open reading frame, respectively (see Fig. 3), amplified a fragment in the sen1-1 mutant of 818 bp, with an insertion of 336 bp compared to the wild type. This fragment was sequenced using the primers described above with a 310 ABI Prism Genetic Analyzer and cycle sequencing (Applied Biosystems, Foster City, Calif.).
FIG. 3.
sen1-1 mutation is a solo δ-LTR insertion at the PTK2 locus. (A) Scheme of the region of chromosome X between positions 545000 and 548000, containing the PTK2 locus and the site of insertion of a solo δ-LTR. The ORF spans positions 545475 to 547931 on the w-strand, and the 336-bp insertion is at position 545942. sp and asp correspond to the primers used for the PCR in panel B. (B) Products of the PCR performed with chromosomal DNA from strains SKY697 (wt) and AG151 (sen1-1 mutant) with the primers indicated in panel A. Lane M, size markers; their sizes (in base pairs) and that of the amplified bands are indicated at the right and at the left of the figure, respectively.
Yeast strains with disruptions of the protein kinase genes of the NPR1 subfamily.
Yeast strains derived from W303-1A (60) with disruptions of the SAT4/HAL4 (YCR008w) and HAL5 (YJL165c) genes have already been described (SKY655, hal4::LEU2; SKY656, hal5::HIS3) (33). KanMX disruptions of the other NPR1-related genes (22) were obtained from EUROSCARF (European Saccharomyces cerevisiae Archive for Functional Analysis; http://www.rz.uni -frankfurt.de/FB/fb16/mikro/euroscarf/). The disruptions in PTK2/STK2 (YJR059w), YDL214c, YDL025c, and YOR267c were made in the FY1679 background (MATa ura3-52 trp1-Δ63 leu2-Δ1 his3-Δ200) while those of KKQ8 (YKL168c), PTK1 (YKL198c), and NPR1 (YNL183c) were made in the BY4741 background (MATα his3-Δ1 leu2-Δ0 ura3-Δ0 lys2-Δ0). The in vivo and in vitro plasma membrane H+-ATPase activities of the different mutants were referred to those of the corresponding wild-type strain.
Cloning and disruption of PTK2 and YOR267c.
Plasmid YEp352::PTK2 was made by subcloning a 3.4-kb BamHI fragment from plasmid pYCGYJR056w (EUROSCARF) in the BamHI site of plasmid YEp352 (multicopy and URA3 marker) (20). Plasmid YEp352::YOR267c was made by subcloning a 3.3-kb HindIII fragment from plasmid pYCGYOR267c (EUROSCARF) in the HindIII site of YEp352.
Disruption of YOR267c with the KanMX marker was made by transformation of yeast strains with a 2.3-kb NotI fragment from plasmid pYORCYOR267c (EUROSCARF). Gene disruption was tested by genomic PCR with primers YOR267-A1 (5′-CAAGACAGTTCCCAACCGCTTAA; 394 bp upstream of start codon) and YOR267-A4 (AATTCAGAATTGGTAGCTACGA; 329 bp downstream of stop codon). The primers for the KanMX gene were K2 (5′-CGATAGATTGTCGCACCTG) and K3 (5′-CCATCCTATGGAACTGCCTC).
Disruption of PTK2 with the KanMX marker was made by transformation of yeast strains with a 3-kb NotI fragment from plasmid pYORCYJR059w (EUROSCARF). Alternatively, a disruption cassette of PTK2 with the TRP1 marker gene was a generous gift of R. Poulin (Quebec, Canada). It consisted of the 3.4-kb BamHI fragment (26) subcloned in pBluescript (Stratagene, La Jolla, Calif.) and with the HpaI-NdeI fragment of 2.6 kb containing all of the PTK2 open reading frame (ORF) but the first 80 bp replaced by a 3-kb fragment containing the TRP1 gene. Gene disruption was tested by genomic PCR with primers CSHT1 (5′-CATACCCGCGTCCTATAGGC; 360 bp upstream of start codon) and CSKY2 (5′-GGTGGTCCCGCCTGATCCGA; 277 bp downstream of stop codon).
Expression of constitutively active Pma1.
The chromosomal copy of PMA1 was placed under control of the galactose-dependent GAL1 promoter by integrative transformation with the URA3 plasmid pRS-61 as described (7). The constitutively active pma1-245 mutant ATPase, lacking the last 18 amino acids of the enzyme, was expressed from plasmid pRS-496 (centromeric and with LEU2 marker) (43).
Replacement of pma1-Ser899Asp and pma1-Ser899Ala mutant alleles with chromosomal wild-type PMA1.
The pma1-Ser899Asp and pma1-Ser899Ala mutant alleles (12) were subcloned in plasmid pFP36 by exchange of a 2.2-kb XbaI fragment as described (29). This plasmid contains a 5-kb HindIII fragment including the PMA1 gene and a downstream URA3 marker. The pma1::URA3 fragments containing the mutations were used for yeast transformation.
Assays for the Pma1 ATPase.
Proton efflux from the cells was measured at pH 4.0 after starvation and readdition of glucose (48). This assay has been demonstrated to correlate with the in vivo activity of the Pma1 ATPase (41, 42, 48). Yeast total membrane fraction and purified plasma membranes were isolated with or without treatment of the cells with glucose, and specific ATP hydrolysis corresponding to Pma1 activity was assayed (with 2 mM ATP unless otherwise indicated) (49, 50). Protein concentration was measured with the Bio-Rad (Hercules, Calif.) protein assay reagent and bovine globulin as the standard. The amount of Pma1 in membranes was quantified by immunoassay with specific antibody as described (11).
Uptake of lithium and methylammonium. (i) Lithium uptake.
Cells growing in YPD medium were supplemented with 40 mM LiCl and incubated for 90 min; the steady-state intracellular lithium concentration was measured by atomic absorption spectrometry after centrifugation, washing, and extraction of the cells as described (33).
(ii) Methylammonium uptake.
Cells were grown in SD medium, washed, and incubated for 90 min in 50 mM succinate-Tris buffer (pH 5.5) with 2% glucose and 0.25 mM [14C]methylammonium; the steady-state intracellular [14C]methylammonium concentration was measured by liquid scintillation counting after filtration and washing as described (33).
RESULTS
Mutation suppressing the lithium sensitivity of ena1-4 disruptants.
The ENA1 gene, encoding a cation extrusion pump, is a major determinant of salt tolerance in S. cerevisiae (19). Accordingly, most screenings for either gain or loss of salt tolerance result in mutations of either this gene or the multiple components of its complex regulatory system (53). In order to identify novel determinants of salt tolerance and ion homeostasis, we have performed a screening based on the isolation of mutations suppressing the lithium sensitivity of ena1-4 disruptants in medium supplemented with methionine. Deletion of the ENA1 gene obviated the complex regulatory system of this important determinant of salt tolerance (53), and methionine supplementation precluded mutations of the salt-sensitive methionine biosynthetic pathway (17). The use of lithium as the toxic cation instead of sodium offered the advantage that this cation inhibits growth at concentrations low enough to pose no osmotic problems. We considered that this approach could lead to the regulatory system of the plasma membrane H+-ATPase because of the observation that mutations which decrease the activity of the enzyme result in tolerance to toxic cations (38, 56, 57, 62).
We have characterized two complementation groups of spontaneous recessive mutations resulting in lithium tolerance. Mutations at the SEN2 locus (suppressor of Ena) are the most abundant, with frequencies of about 10−6. They confer specific lithium tolerance and have no effect on the sensitivity of yeast cells to other toxic cations, such as sodium, hygromycin B, manganese, and tetramethylammonium. Until now it has not been possible to clone the responsible gene by complementation.
One of the lithium tolerance mutations corresponded to the SEN1 locus and exhibited pleiotropic phenotypes related to cation homeostasis. As indicated in Fig. 1, the sen1-1 mutant is tolerant to concentrations of lithium, sodium, manganese, hygromycin B, and tetramethylammonium which are highly toxic to wild-type cells. On the other hand, the mutation causes sensitivity to acetic acid and has no effect on sorbitol stress.
FIG. 1.
sen1-1 and ptk2::TRP1 mutations confer tolerance to different toxic cations and sensitivity to acetic acid. Strain SKY697 (wt) and its derivatives AG149 (ptk2) and AG151 (sen1) were grown in liquid SD medium to saturation, and serial dilutions were dropped on SD plates with either NaCl (0.2 M), LiCl (40 mM), KCl (1.2 M), sorbitol (1.8 M), acetic acid (0.12 M), or tetramethylammonium chloride (TMA, 1 M) or on YPD plates with MnCl2 (2 mM) or hygromycin B (HygB, 125 μg/ml), as indicated. Growth was recorded after 2 days in the absence of toxic cations and after 3 to 4 days in its presence.
Reduced cation accumulation and ATPase activity in the sen1-1 mutant.
Toxic cation tolerance may be due to reduced cellular uptake and, as indicated in Table 2, steady-state uptake of lithium and methylammonium is reduced by the sen1-1 mutation. Therefore, a reasonable hypothesis to explain the phenotype of sen1-1 mutants (Fig. 1) is that the uptake of diverse toxic cations is reduced. The cellular uptake of cations is driven by the plasma membrane electrical potential (negative inside) (28, 52, 57), and as these toxic cations are unlikely to use a common transporter, the results could be explained by a decrease in this biophysical parameter as the primary effect of the sen1-1 mutation.
TABLE 2.
Effect of the sen1-1 mutation on the uptake of lithium and methylammoniuma
| Strain | Avg cation uptake (nmol/mg of cells) ± SD
|
|
|---|---|---|
| Lithium | Methylammonium | |
| SKY6697 (ena1-4 SEN1) | 27.2 ± 1.3 | 26.2 ± 0.2 |
| AG151 (ena1-4 sen1-1) | 17.3 ± 0.8 | 22.3 ± 0.4 |
Results are the average (± standard deviation) of three determinations, and the uptake experiments were repeated twice with similar (± 10%) results. The external concentrations of lithium and methylammonium were 40 and 0.25 mM, respectively. Uptake times were 90 min, corresponding to the steady-state level of intracellular cation (33).
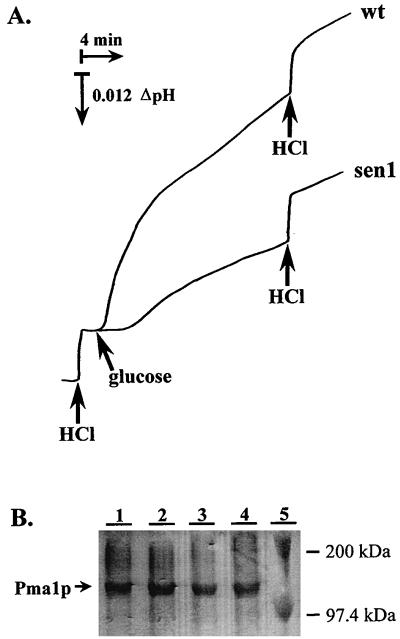
A simple model for the regulation of membrane potential in yeast is that it is determined by the concerted activities of the electrogenic proton-pumping H+-ATPase (the generator of electrical potential) and of the Trk1,2 potassium uptake system, a major consumer of electrical potential (28, 33). The high rates of proton efflux balanced by potassium uptake which can be measured in yeast cells suggest that these could be the most active electrogenic systems (48, 52). As indicated in Fig. 2A, the sen1-1 mutant exhibited decreased proton-pumping activity in vivo (26% of wild-type value). Western blot and quantitative scanning analysis, on the other hand, indicated normal levels of the enzyme (Fig. 2B). Assay of ATP hydrolysis in isolated membranes also indicated decreased plasma membrane H+-ATPase activity in the sen1-1 mutant (70% of wild-type value; data not shown). Therefore, the hypothesis was advanced that the sen1-1 mutation, by reducing the activity of the plasma membrane H+-ATPase, decreases the electrical potential and the uptake of toxic cations. The sensitivity to acetic acid of this mutant is in agreement with previous work demonstrating that the level of ATPase activity is a positive determinant of tolerance to acid stress (42, 57). In addition, the sen1-1 mutant exhibited no growth phenotype in medium with limiting potassium concentrations (data not shown), at variance with mutations in the Trk1,2 transport system (14, 33).
FIG. 2.
Effect of the sen1-1 mutation on the yeast Pma1 proton pump. Cells of strains SKY697 (wt) and AG151 (sen1 mutant) were grown in YPD, and the in vivo activity (A) and amount (B) of the Pma1 proton-pumping ATPase were determined as described in the text. (A) pH changes of yeast suspensions induced by glucose. Cells (50 mg) were suspended in 10 ml of assay buffer, and pH changes were recorded after addition of 200 μmol of glucose. Calibration with 400 nmol of HCl was made as indicated. (B) Immunological quantification of Pma1 in yeast. Total membrane protein (25 μg) from strains SKY697 (wild type [wt] for PTK2, lanes 1 and 2) and AG151 (sen1-1, lanes 3 and 4) was immunodetected with anti-Pma1 antibody after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotting. Lane 5 contains molecular size standards as indicated.
sen1-1 mutation is a “solo δ-LTR” insertion in the PTK2 gene.
As no complementation of the sen1 mutation could be achieved with the PMA1 H+-ATPase gene, we hypothesized that it was the regulatory system of the enzyme which was affected. Recently, a protein kinase subfamily has been identified in yeast, the Npr1 subgroup (22), which seems to be dedicated to the regulation of plasma membrane transporters. Members of this subfamily regulate the Gap1 amino acid permease (Npr1) (55, 58), polyamine uptake mediated by an uncharacterized transporter (Ptk1,2) (25, 26, 36), and the Trk1,2 potassium transporter (Hal4,5) (33). It was tempting to speculate that a kinase of this subfamily could regulate the plasma membrane H+-ATPase and be responsible for the sen1-1 mutation.
The Npr1 subfamily of yeast protein kinases has nine members, but disruption of only two of these genes significantly affected proton pumping in vivo. They corresponded to YJR059w and YOR267c, whose disruptions resulted in rates of proton pumping that were 27 and 50% of the wild-type value, respectively. ATP hydrolysis by purified plasma membranes was also reduced in the disruptions of YJR059w and YOR267c to 65 and 75% of the wild-type value, respectively, but not in the mutations of the other seven genes of the subfamily. YJR059w corresponds to PTK2/STK2, the protein kinase gene previously described as being required for polyamine transport (26, 36). A phenotype for YOR267c has not previously been described.
In the process of checking the gene disruption of YJR059w, we made the fortuitous observation that the sen1-1 strain had an insertion at its PTK2 locus (Fig. 3). Cloning and sequencing demonstrated that this insertion corresponded to a “solo δ-LTR” derived from the yeast Ty retrotransposon (3) interrupting the PTK2 ORF at position 467. Accordingly, disruption of the PTK2 gene resulted in the same pleiotropic growth phenotypes and H+-ATPase alterations as the original sen1-1 mutation (Fig. 1). Crossing the sen1-1 and ptk2 strains demonstrated that these recessive mutations were allelic, and these results indicate that sen1-1 is a null allele of PTK2. The sen1-1 and ptk2 mutations could be complemented with the 3.4-kb BamHI genomic fragment (26, 36) containing the PTK2 gene in the 2μm multicopy plasmid. However, complementation by this fragment in a single-copy, centromeric plasmid was only partial, as previously reported (36). This can be explained if regulatory regions of the PTK2 gene extend into 5′- and/or 3′-flanking genes (CDC8 and CBF1, respectively).
The ptk2 mutation affects the glucose-induced change in Km of the plasma membrane H+-ATPase. The glucose activation of the H+-ATPase can be separated into two kinetic effects assigned to two independent regulatory sites within the carboxyl terminus: a glucose-induced decrease in the Km for ATP probably depends on phosphorylation of Ser-899, while a glucose-induced increase in the Vmax of the enzyme requires Arg-909 and Thr-912 by a mechanism more complex than simple phosphorylation (10, 40, 44). As indicated in Table 3, the ptk2 mutation blocks the affinity (Km) change in the H+-ATPase induced by glucose, with no effect on the increase in Vmax. Therefore, we next investigated the effect of mutations at the carboxyl terminus of the ATPase on the phenotype of the ptk2 mutant.
TABLE 3.
Effect of glucose and of the ptk2 and yor267c mutations on the kinetic properties of the plasma membrane H+-ATPase
| Genotypea |
Km (mM)b
|
Vmax (μmol/min per mg of protein)b
|
||
|---|---|---|---|---|
| GS | GF | GS | GF | |
| PTK2 YOR267c | 4.0 | 1.2 | 0.15 | 1.5 |
| ptk2 YOR267c | 4.0 | 4.0 | 0.13 | 1.9 |
| PTK2 yor267c | 4.0 | 2.5 | 0.14 | 1.7 |
Strains W303-1B (PTK2 YOR267c), FPY1434 (ptk2 YOR267c), and FPY1506 (PTK2 yor267c) are described in Table 1.
ATPase activity of purified plasma membranes was assayed at pH 6.5 with a range of Mg and ATP concentrations from 0.8 to 6 mM and an excess Mg over ATP of 5 mM (10). The apparent Km and Vmax were extrapolated from double-reciprocal plots fitted using a standard least-squares method (linear regression coefficients, r = 0.96 to 0.98). Similar values (within 10%) were obtained with two different plasma membrane preparations isolated independently. GS, glucose-starved cells; GF, glucose-fermenting cells. Cells were grown, collected, and treated or not with glucose before homogenization, and plasma membranes were isolated as described in Materials and Methods.
Suppression of ptk2 by mutations at the carboxyl terminus of Pma1.
In order to investigate if Ptk2 regulates the H+-ATPase by acting on its C-terminal regulatory domain, we tested if deletion of this inhibitory domain, which results in a constitutively active H+-ATPase (43), suppressed the phenotypes of the ptk2 mutation. As indicated in Fig. 4 (column 3), ectopic expression of a truncated Pma1 ATPase in a single-copy, centromeric plasmid partially suppressed the tolerance to sodium and lithium caused by the ptk2 mutation. The Pma1 ATPase is known to be oligomeric, probably hexameric (1, 31, 52), and the partial nature of the suppression could be explained if the simultaneous expression of wild-type and truncated copies resulted in mixed ATPase oligomers with only partial deregulation. Because of the essential nature of the PMA1 gene, exclusive expression of the truncated ATPase required the use of a yeast strain in which the wild-type chromosomal PMA1 gene is under galactose control and which, in glucose medium, expresses only the truncated ATPase (43). Complete suppression of ptk2 phenotypes could be obtained with this strain (Fig. 4, column 4). The tolerance to hygromycin B and tetramethylammonium of the ptk2 mutant was also suppressed by deletion of the regulatory domain of Pma1 (not shown).
FIG. 4.
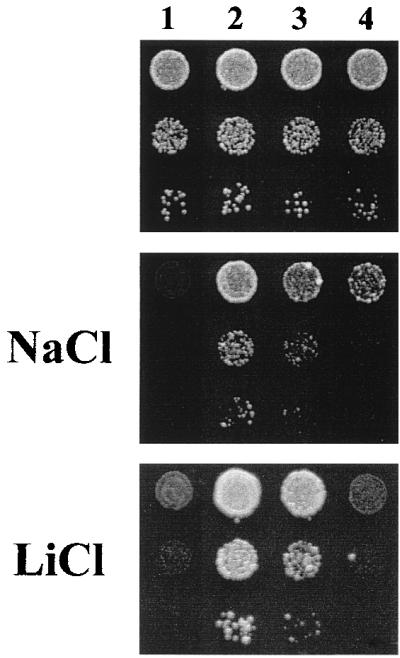
Deletion of the inhibitory domain of Pma1 suppresses the growth phenotypes of the ptk2 mutation. Strains SKY697 (wild type for PTK2, column 1), AG149 (ptk2, column 2), AG205 (ptk2 with plasmid pRS496 expressing truncated Pma1, column 3) and AG224 (ptk2 pma1::URA3-GAL1 promoter-PMA1 with chromosomal ATPase under galactose control and plasmid pRS496 expressing truncated Pma1, column 4) were grown in liquid SD medium to saturation, and serial dilutions were dropped on SD plates with either NaCl (0.2 M) or LiCl (40 mM) as indicated. Growth was recorded after 2 or 4 days in the absence or presence of toxic cations, respectively.
Phosphorylation of Ser-899 at the C terminus has been proposed to result in Pma1 activation because the Ser899Ala mutation reduced glucose activation and the Ser899Asp mutation mimicked the glucose-activated state (12). As indicated in Fig. 5, the Ser899Ala mutation of the H+-ATPase produced a phenotype of resistance to sodium, hygromycin B (Fig. 5), and lithium (not shown) similar to that of the ptk2 mutation. On the other hand, the Ser899Asp-mutated H+-ATPase suppressed the phenotypes caused by the ptk2 mutation. These results are consistent with the idea that Ptk2 acts on the C terminus of the ATPase through phosphorylation of Ser-899.
FIG. 5.
Effect of mutations of Pma1 Ser-899 on tolerance to hygromycin B and NaCl of Ptk2+ (upper panels) and Ptk2− (lower panels) yeast cells. Wild-type yeast strain W303-1B (PMA1 PTK2) and derivatives with pma1-Ser899 and ptk2 mutations as indicated (strains FPY398, FPY402, FPY1434, FPY1500, and FPY1502) were grown in liquid medium to saturation and diluted 20-fold, and 3 μl was dropped on YPD plates with no addition or containing hygromycin B (50 μg/ml) or NaCl (1.2 M), as indicated. Growth was recorded after 4 days.
Protein kinase YOR267c affects the sensitivity of yeast cells to hygromycin B.
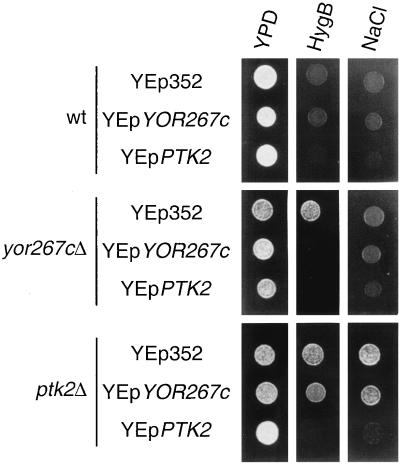
As indicated above, the protein kinase YOR267c also affected Pma1 activity, although to a lesser extent than Ptk2. As indicated in Table 3, in the ptk2 mutant, glucose does not at all improve the affinity of the ATPase for ATP, while in the case of the yor267c mutant, glucose still increases this affinity somewhat, although to a lesser extent than in the control strain. Also, the growth phenotypes of mutants with gain and loss of function of YOR267c were weaker than those of PTK2 mutants. Disruption of YOR267c increased tolerance to hygromycin B but not to sodium (Fig. 6), lithium, or norspermidine (not shown). Disruption of PTK2 increased the tolerance of yeast cells to all these toxic cations. Also, overexpression of YOR267c only partially reduced the tolerance to hygromycin B of ptk2 mutants, without affecting tolerance to NaCl (Fig. 6), lithium, or norspermidine (not shown). Overexpression of PTK2 in either wild-type yeast, YOR267c mutants, or ptk2 mutants decreases the tolerance of yeast cells to all these toxic cations (Fig. 6 and data not shown).
FIG. 6.
Effect of gain and loss of function of YOR267c and PTK2 on tolerance of yeast cells to hygromycin B and NaCl. Wild-type yeast strain (wt, FPY1506) and derivatives with disruptions (Δ) of either yor267c (strain FPY1456) or ptk2 (FPY1459) were transformed with either empty plasmid (YEp352) or multicopy plasmids with YOR267c and PTK2 as indicated. Experimental conditions were as described in the legend to Fig. 5.
DISCUSSION
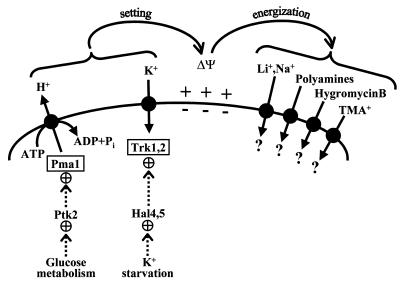
The participation of the protein kinase Ptk2 in the regulation of ion transport in yeast represents the merging of two apparently independent lines of research, polyamine transport and salt tolerance. Ptk2 was initially identified as a yeast protein kinase required for polyamine transport (26, 36), and we now report that Ptk2 is required for the sensitivity of yeast cells to several toxic cations, including lithium, sodium, hygromycin B, manganese, tetramethylammonium, and polyamines. Although previous work suggested a direct regulation of the polyamine transporter by Ptk2 (26, 36), we interpret the effects of this kinase on cation transport as probably being indirect. Ptk2 is required for the full activation of the yeast plasma membrane proton pump (Pma1) by glucose and therefore for the glucose-energized uptake of different cations such as lithium, methylammonium, and polyamines. A plausible interpretation is that, by activating Pma1, Ptk2 increases the electrical membrane potential of the yeast plasma membrane, negative inside, and that this biophysical parameter determines the uptake of toxic cations mediated by different transport systems (Fig. 7). As it is unlikely that lithium, tetramethylammonium, hygromycin B, and polyamines use the same transport system, the energization of different cation transport systems by the electrical membrane potential could explain the pleiotropic nature of ptk2 mutations. The nature of the polyamine transporter in yeast is not known (23). Monovalent cations may enter yeast cells by a low-affinity transporter which has only been identified at the electrophysiological level (45) and which may correspond to the plant LCT1 gene (46).
FIG. 7.
Model for the role of Pma1 and Trk1,2 on yeast salt tolerance by modulation of the electrical membrane potential (ΔΨ), which determines the uptake of toxic cations by different voltage-sensitive transporters.
As indicated in the model in Fig. 7, the major determinants of plasma membrane potential in yeast are the Pma1 proton pump (the electrical generator) and the Trk1,2 high-affinity K+ transport system, a major consumer of electrical potential because of the high rates of K+ uptake in K+-starved cells (52). Our hypothesis is that the relative activity of these two systems sets the steady-state value of the electrical potential and in so doing modulates the activity of secondary active transport systems such as those involved in nutrient and toxic cation uptake. In addition, the Pma1 and Trk1,2 systems modulate important biophysical parameters, such as internal and external pH, cell turgor, and intracellular K+ concentrations. Therefore, the regulation of major determinants of ion homeostasis such as Pma1 and Trk1,2 is crucial for cell metabolism and growth and for the sensitivity to toxic cations. Although the absolute values of the yeast plasma membrane potential cannot be measured by electrophysiological methods (28), a series of indirect measurements indicate that this biophysical parameter correlates with the uptake of cations such as hygromycin B, methylammonium, and tetraphenylphosphonium (28, 33, 38, 57).
Mutations of the Pma1 ATPase which decrease the electrogenic activity of the enzyme cause the same pleiotropic growth phenotypes of tolerance to toxic cations as the ptk2 mutations described in the present work (38, 56, 62). It would be interesting to determine the causal relationship between the different mutations affecting the activity of Pma1 and the different growth phenotypes exhibited by cells. As a tentative interpretation, the electrogenic activity of the proton pump, modified by either mutation or altered phosphorylation, would set different levels of the electrical membrane potential (Fig. 7). Cation transporters would then exhibit a differential response according to their specific voltage sensitivities. The different phenotypes of YOR267c and Ptk2 protein kinase mutants, as well as the different phenotypes exhibited by pma1 mutants (31), may be explained by their different levels of proton-pumping activity. Additional complications include the fact that decreased activity of the plasma membrane ATPase somehow enhances vacuolar compartmentation of toxic cations (34).
Npr1, the founding member of a subfamily of yeast protein kinases, regulates the activity and stability of the amino acid permeases Gap1 and Tat2 in response to ammonium availability, probably by direct phosphorylation of these membrane proteins (47). We have previously described two redundant protein kinases of the Npr1 subfamily (Hal4 and Hal5) which regulate Trk1,2, the major high-affinity K+ transport system of yeast, in response to K+ starvation (33). Now we report that another member of the Npr1 group, Ptk2, regulates the yeast proton pump Pma1 in response to glucose metabolism. In addition, two other protein kinases of the Npr1 group, Ptk1 and YOR267c, seem to modulate Pma1. Ptk1 is highly homologous to Ptk2, and its gene was discovered as a multicopy suppressor of the polyamine uptake defect of ptk2 mutants (25). Disruption of ptk1 causes no growth or Pma1 activity phenotype (F. Portillo, unpublished results). It could correspond to a protein kinase similar in function to but less active than Ptk2. Although YOR267c is much more related to Npr1 than to Ptk2, it also regulates Pma1 activity. However, the growth phenotypes of gain and loss of function for this kinase are restricted to hygromycin B, and we propose to rename YOR267c HRK1, for hygromycin resistance kinase. According to the model developed above, the unknown transporter for hygromycin B may be more sensitive than other cation transporters to the small changes in membrane potential determined by the YOR267c mutations. Finally, Npr1 itself seems to regulate ion homeostasis, because although its disruption has no significant effect on Pma1 activity (F. Portillo, unpublished results), it reduces polyamine transport (27) and slightly increases sodium tolerance (S. Kron, personal communication), as if this kinase also contributed to Pma1 activity.
The emerging picture is that yeast cells contain a subfamily of protein kinases which are dedicated to the regulation of plasma membrane transporters and which may exhibit some promiscuity. Ptk2 and Hal4,5 are the major regulators of Pma1 and Trk1,2, respectively, while Npr1 modulates the amino acid transporters Gap1 and Tat2, although it may also act on Pma1, together with less active kinases such as Ptk1 and Hrk1. The specificity of these protein kinases is unknown, and it remains to be demonstrated at the biochemical level that they act directly on the Pma1 protein and other transporters, because the observed effects could be indirect. The existence of kinase cascades dedicated to ion homeostasis is plausible, and the signalling pathways should have sensors of basic cellular parameters such as energy metabolism, membrane potential, pH, K+ concentration, and turgor. These mechanisms, and the nature of the protein phosphatases counteracting the activating kinases, are currently under investigation.
The glucose-induced affinity change of the ATPase requires, in addition to the protein kinase Ptk2, the ubiquitin-protein ligase Rsp5 (8), suggesting that the turnover of some protein(s) is also important for the regulation of the ATPase. The glucose-induced increase in the Vmax of the ATPase depends on Arg-909 and Thr-912, which define a potential phosphorylation site for calmodulin-dependent protein kinase II. However, mutational analysis has indicated that the mechanism of Vmax regulation is more complex than simple phosphorylation (12) and involves the membrane protein YOR137c (9) and the small heat shock protein Hsp30 (4). Other pieces of the puzzle of ion homeostasis in yeast include the calcium-regulated protein phosphatase calcineurin, which regulates both Trk1,2 and the ATPase (62), and, as demonstrated in fission yeast, some type I protein phosphatases and mitogen-activated protein kinases (2).
Ptk2 (present work) and Hal4,5 (33) represent the first protein kinases which modulate the major electrogenic transporters of yeast cells (Fig. 7). Future studies should help to convert this glimpse of the mechanisms of ion homeostasis into a detailed picture of the complex network of signaling pathways which, since early times of evolution, were probably needed to adjust the basic biophysical parameters of cells in response to growth and environmental changes (21, 53). As the modulation of membrane potential is crucial for the uptake of toxic cations (Fig. 7), it may be part of the cellular responses to salt stress. The recent identification of a conserved proteolipid which seems to depolarize the plasma membrane (35) and which is induced in plants by salt stress (5) supports this hypothesis.
ACKNOWLEDGMENTS
This work was supported by grants from the Spanish DGICYT (Madrid) (PB97-0054 and PB98-0565-C04-1). A.G. was a fellow of the Marie Curie Research Training Grants (European Commission, Brussels, Belgium). N.D.L.F. was a fellow of the Ministerio de Educación y Ciencia (Madrid, Spain), and J.J.F. was a fellow of the Conselleria de Educació i Ciencia (Valencia, Spain).
We thank Richard Poulin (Quebec, Canada) for the ptk2::TRP1 disruption cassette, André Goffeau (Louvain-la-Neuve, Belgium) for a copy of his manuscript on PMP3 before publication (35), and Lynne Yenush for critical reading of the manuscript.
The first two authors contributed equally to this work.
REFERENCES
- 1.Auer M, Scarborough G A, Kühlbrandt W. Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature. 1998;392:840–843. doi: 10.1038/33967. [DOI] [PubMed] [Google Scholar]
- 2.Balcells L, Martin R, Ruiz M C, Gomez N, Ramos J, Ariño J. The Pzh1 protein phosphatase and the Spm1 protein kinase are involved in the regulation of the plasma membrane H+-ATPase in fission yeast. FEBS Lett. 1998;435:241–244. doi: 10.1016/s0014-5793(98)01082-5. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, Sandmeyer S B. Yeast transposable elements. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 193–261. [Google Scholar]
- 4.Braley R, Piper P W. The C-terminus of yeast plasma membrane H+-ATPase is essential for the regulation of this enzyme by heat shock protein Hsp30, but not for stress activation. FEBS Lett. 1997;418:123–126. doi: 10.1016/s0014-5793(97)01359-8. [DOI] [PubMed] [Google Scholar]
- 5.Capel J, Jarillo J, Salinas J, Martinez-Zapater L. Two homologous low-temperature genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiol. 1997;115:569–576. doi: 10.1104/pp.115.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang A, Slayman C W. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cid A, Perona R, Serrano R. Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr Genet. 1987;12:105–110. doi: 10.1007/BF00434664. [DOI] [PubMed] [Google Scholar]
- 8.de la Fuente N, Maldonado A M, Portillo F. Glucose activation of the yeast plasma membrane H+-ATPase requires the ubiquitin-proteasome proteolytic pathway. FEBS Lett. 1997;411:308–312. doi: 10.1016/s0014-5793(97)00721-7. [DOI] [PubMed] [Google Scholar]
- 9.de la Fuente N, Maldonado A M, Portillo F. Yeast gene YOR137c is involved in the activation of the yeast plasma membrane H+-ATPase by glucose. FEBS Lett. 1997;420:17–19. doi: 10.1016/s0014-5793(97)01478-6. [DOI] [PubMed] [Google Scholar]
- 10.Entian K-D, et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 11.Eraso P, Cid A, Serrano R. Tight control of the amount of yeast plasma membrane ATPase during changes in growth conditions and gene dosage. FEBS Lett. 1987;224:193–197. doi: 10.1016/0014-5793(87)80446-5. [DOI] [PubMed] [Google Scholar]
- 12.Eraso P, Portillo F. Molecular mechanism of regulation of yeast plasma membrane H+-ATPase by glucose: interaction between domains and identification of new regulatory sites. J Biol Chem. 1994;269:10393–10399. [PubMed] [Google Scholar]
- 13.Estrada E, Agostinis P, Vandenheede J R, Goris J, Merlevede W, Francois J, Goffeau A, Ghislain M. Phosphorylation of yeast plasma membrane H+-ATPase by casein kinase I. J Biol Chem. 1996;271:32064–32072. doi: 10.1074/jbc.271.50.32064. [DOI] [PubMed] [Google Scholar]
- 14.Gaber R F. Molecular genetics of yeast ion transport. Int Rev Cytol. 1992;137A:299–353. doi: 10.1016/s0074-7696(08)62679-0. [DOI] [PubMed] [Google Scholar]
- 15.Gancedo C, Serrano R. Energy-yielding metabolism. In: Rose A H, Harrison J S, editors. The yeasts. 2nd ed. 3: metabolism and physiology of yeast. London, England: Academic Press; 1989. pp. 205–259. [Google Scholar]
- 16.Garcia-Arranz M, Maldonado A M, Mazón M J, Portillo F. Transcriptional control of yeast plasma membrane H+-ATPase by glucose: cloning and characterization of new genes involved in this regulation. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- 17.Gläser H-U, Thomas D, Gaxiola R, Montrichard F, Surdin-Kerjan Y, Serrano R. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 1993;12:3105–3110. doi: 10.1002/j.1460-2075.1993.tb05979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 19.Haro R, Garciadeblas B, Rodriguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 20.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman J F, editor. The cellular functions of membrane transport. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1964. [Google Scholar]
- 22.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:14–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 23.Igarashi K, Kashiwagi K. Polyamine transport in bacteria and yeast. Biochem J. 1999;344:633–642. [PMC free article] [PubMed] [Google Scholar]
- 24.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakinuma Y, Maruyama T, Nozaki T, Wada Y, Ohsumi Y, Igarashi K. Cloning of the gene encoding a putative serine/threonine protein kinase which enhances spermine uptake in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;216:985–992. doi: 10.1006/bbrc.1995.2717. [DOI] [PubMed] [Google Scholar]
- 26.Kaouass M, Audette M, Ramotar D, Verma S, de Montigny D, Gamache I, Torossian K, Poulin R. The STK2 gene, which encodes a putative Ser/Thr protein kinase, is required for high-affinity spermidine transport in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2994–3004. doi: 10.1128/mcb.17.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaouass M, Gamache I, Ramotar D, Audette M, Poulin R. The spermidine transport system is regulated by ligand inactivation, endocytosis, and by the Npr1 Ser/Thr protein kinase in Saccharomyces cerevisiae. J Biol Chem. 1998;273:2109–2117. doi: 10.1074/jbc.273.4.2109. [DOI] [PubMed] [Google Scholar]
- 28.Madrid F, Gomez M J, Ramos J, Rodriguez-Navarro A. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 29.Maldonado A M, de la Fuente N, Portillo F. Characterization of an allele-nonspecific intragenic suppressor in the yeast plasma membrane H+-ATPase gene (PMA1) Genetics. 1998;150:11–19. doi: 10.1093/genetics/150.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazon M J, Behrens M M, Portillo F, Piñón R. cAMP- and RAS-independent nutritional regulation of plasma-membrane H+-ATPase activity in Saccharomyces cerevisiae. J Gen Microbiol. 1989;135:1453–1460. doi: 10.1099/00221287-135-6-1453. [DOI] [PubMed] [Google Scholar]
- 31.McCusker J H, Perlin D S, Haber J E. Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskvina E, Imre E-M, Ruis H. Stress factors acting at the level of the plasma membrane induce transcription via the stress response element (STRE) of the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;32:1263–1272. doi: 10.1046/j.1365-2958.1999.01438.x. [DOI] [PubMed] [Google Scholar]
- 33.Mulet J M, Leube M P, Kron S J, Rios G, Fink G R, Serrano R. A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol Cell Biol. 1999;19:3328–3337. doi: 10.1128/mcb.19.5.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nass R, Cunningham K W, Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 35.Navarre C, Goffeau A. Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 2000;19:2515–2524. doi: 10.1093/emboj/19.11.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozaki T, Nishimura K, Michael A J, Maruyama T, Kakinuma Y, Igarashi K. A second gene encoding a putative serine/threonine protein kinase which enhances spermine uptake in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1996;228:452–458. doi: 10.1006/bbrc.1996.1681. [DOI] [PubMed] [Google Scholar]
- 37.Passos J B, Vanhalewyn M, Brandao R L, Castro I M, Nicoli J R, Thevelein J M. Glucose-induced activation of plasma membrane H+-ATPase in mutants of the yeast Saccharomyces cerevisiae affected in cAMP metabolism, cAMP-dependent protein phosphorylation and the initiation of glycolysis. Biochim Biophys Acta. 1992;1136:57–67. doi: 10.1016/0167-4889(92)90085-p. [DOI] [PubMed] [Google Scholar]
- 38.Perlin D S, Brown C L, Haber J E. Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J Biol Chem. 1988;263:18118–18122. [PubMed] [Google Scholar]
- 39.Piper P W. Molecular events associated with acquisition of heat tolerance by the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 1993;11:339–356. doi: 10.1111/j.1574-6976.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 40.Portillo F. Regulation of plasma membrane H+-ATPase in fungi and plants. Biochim Biophys Acta. 2000;1469:31–42. doi: 10.1016/s0304-4157(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 41.Portillo F, Serrano R. Dissection of functional domains of the yeast proton-pumping ATPase by directed mutagenesis. EMBO J. 1988;7:1793–1798. doi: 10.1002/j.1460-2075.1988.tb03010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Portillo F, Serrano R. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur J Biochem. 1989;186:501–507. doi: 10.1111/j.1432-1033.1989.tb15235.x. [DOI] [PubMed] [Google Scholar]
- 43.Portillo F, de Larrinoa I F, Serrano R. Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl-terminus. FEBS Lett. 1989;247:381–385. doi: 10.1016/0014-5793(89)81375-4. [DOI] [PubMed] [Google Scholar]
- 44.Portillo F, Eraso P, Serrano R. Analysis of the regulatory domain of yeast plasma membrane H+-ATPase by directed mutagenesis and intragenic suppression. FEBS Lett. 1991;287:71–74. doi: 10.1016/0014-5793(91)80018-x. [DOI] [PubMed] [Google Scholar]
- 45.Roberts S K, Fischer M, Dixon G K, Sanders D. Divalent cation block of inward currents and low-affinity K+ uptake in Saccharomyces cerevisiae. J Bacteriol. 1999;181:291–297. doi: 10.1128/jb.181.1.291-297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schachtman D P, Kumar R, Schroeder J I, Marsh E L. Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc Natl Acad Sci USA. 1997;94:11079–11084. doi: 10.1073/pnas.94.20.11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serrano R. Effects of ATPase inhibitors on the proton pump of respiratory-deficient yeast. Eur J Biochem. 1980;105:419–424. doi: 10.1111/j.1432-1033.1980.tb04516.x. [DOI] [PubMed] [Google Scholar]
- 49.Serrano R. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 1983;156:11–14. doi: 10.1016/0014-5793(83)80237-3. [DOI] [PubMed] [Google Scholar]
- 50.Serrano R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- 51.Serrano R. Structure and function of plasma membrane ATPase. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:61–94. [Google Scholar]
- 52.Serrano R. Transport across yeast vacuolar and plasma membranes. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 523–585. [Google Scholar]
- 53.Serrano R. Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int Rev Cytol. 1996;165:1–52. doi: 10.1016/s0074-7696(08)62219-6. [DOI] [PubMed] [Google Scholar]
- 54.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 55.Stanbrough M, Magasanik B. Transcriptional and posttranscriptional regulation of the general amino acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulaszewski S, Balzi E, Goffeau A. Genetic and molecular mapping of the pma1 mutation conferring vanadate resistance to the plasma membrane ATPase from Saccharomyces cerevisiae. Mol Gen Genet. 1987;207:38–46. doi: 10.1007/BF00331488. [DOI] [PubMed] [Google Scholar]
- 57.Vallejo C, Serrano R. Physiology of mutants with reduced expression of plasma membrane H+-ATPase. Yeast. 1989;5:307–319. doi: 10.1002/yea.320050411. [DOI] [PubMed] [Google Scholar]
- 58.Vandenbol M, Jauniaux J-C, Grenson M. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encodes a protein kinase homologue. Mol Gen Genet. 1990;222:393–399. doi: 10.1007/BF00633845. [DOI] [PubMed] [Google Scholar]
- 59.Venema K, Palmgren M G. Metabolic modulation of transport coupling ratio in yeast plasma membrane H+-ATPase. J Biol Chem. 1995;270:19659–19667. doi: 10.1074/jbc.270.33.19659. [DOI] [PubMed] [Google Scholar]
- 60.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyperrecombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 61.Winde J H, Thevelein J M, Winderickx J. From feast to famine: adaptation to nutrient depletion in yeast. In: Hohmann S, Mager W H, editors. Yeast stress responses. Heidelberg, Germany: Springer; 1997. pp. 7–52. [Google Scholar]
- 62.Withee J L, Sen R, Cyert M S. Ion tolerance of Saccharomyces cerevisiae lacking the Ca2+/CaM-dependent phosphatase (calcineurin) is improved by mutations in URE2 or PMA1. Genetics. 1998;149:865–878. doi: 10.1093/genetics/149.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]