Abstract
Introduction:
There have been no specific guidelines regarding which genes should be tested in the clinical setting for PD or parkinsonism. We evaluated the types of clinical genetic testing offered for Parkinson’s disease (PD) as the first step of our gene curation.
Methods:
The National Institutes of Health (NIH) Genetic Testing Registry (GTR) was queried on 12/7/2020 to identify current commercial PD genetic test offerings by clinical laboratories, internationally.
Results:
We identified 502 unique clinical genetic tests for PD, from 28 Clinical Laboratory Improvement Amendments (CLIA)-approved clinical laboratories. These included 11 diagnostic PD panels. The panels were notable for their differences in size, ranging from 5 to 62 genes. Five genes for variant query were included in all panels (SNCA, PRKN, PINK-1, PARK7 (DJ1), and LRRK2). Notably, the addition of the VPS35 and GBA genes was variable. Panel size differences stemmed from inclusion of genes linked to atypical parkinsonism and dystonia disorders, and genes in which the link to PD causation is controversial.
Conclusion:
There is an urgent need for expert opinion regarding which genes should be included in a commercial laboratory multi-gene panel for PD.
Keywords: Parkinson’s disease, genetic testing, clinical laboratories, multi-gene panels
Introduction
The role that genetics plays in Parkinson’s disease (PD) etiology is increasingly acknowledged. In the last few decades, we have learned that pathogenic variants in certain genes such as LRRK2, GBA, and PRKN can be important contributing factors. For most forms of PD, other genetic factors, environmental agents and aging also play a role [1]. At least one major pathogenic variant in a PD-associated gene is identified in approximately 10% of patients with PD, depending on the testing used and population studied [1,2]. Parkinson’s disease, when inherited, can be autosomal dominant, related to variants in the genes SNCA, LRRK2, VPS35, or autosomal recessive caused by variants in PRKN, PINK1, or PARK7 (DJ1) [3] (Figure 1). Variants in the GBA gene are believed to be major risk factors for Parkinson’s disease, when present in heterozygous or homozygous state [4]. Other genes linked to monogenic atypical forms of parkinsonism have also been described [5, 6]. Recently, over 90 genetic variants that appear to be associated with an increased risk of PD have been identified in genome-wide association (GWAS) studies [7]. Individually, these variants are common in the general population and are not associated with a definable PD risk, and they are not typically included in commercially-available testing panels.
Figure 1.

Monogenic Causes of Typical Parkinson’s Disease
*Some researchers classify variants in this gene as major risk factors.
Genetic testing options have rapidly increased for movement disorders including PD, driven by new technology such as next generation sequencing (NGS), the interest in precision medicine, demand, and lower costs. This information obtained from testing can be integrated into clinical care allowing for better disease prognostication, management and qualification for clinical trials [8]. On a personal level, information from genetic testing can be useful for people with PD allowing for understanding about their disease etiology, improved risk assessment for family members, and life planning.
As of 2018, it was estimated that there were 75,000 genetic tests (single gene, multigene panel, whole exome, and other complex genetic testing products) marketed by Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories in the United States [9]. Commercial panels and gene tests for a disorder are not necessarily created according to clinical validity and utility. Furthermore, test selection does not always mirror reimbursement or insurance coverage for a condition. Recognizing the wide variation in gene panels, the American College of Medical Genetics (ACMG) recently published a technical guideline for clinical laboratories regarding the design of gene panels [10].
Multi-gene panel tests are often selected by clinicians, because of their perceived efficiency, completeness, and cost-effectiveness. However, there are no guidelines to direct the physician in test selection for PD; some groups have provided guidance and algorithms based on family history of PD, clinical features, and ethnicity [11,12]. The limited availability of genotype-phenotype correlations can make it difficult for the health care provider (HCP) to choose the right test. Ultimately, it can be very difficult to navigate the genetic testing menus for PD that features clinical and genetic heterogeneity [13], due to the lack of uniform testing and inclusion of genes linked to monogenic parkinsonism and other forms of parkinsonism.
The objective of this study is to describe the current landscape of PD genetic testing offered by clinical laboratories and to compare test offerings, as we begin gene curation for PD.
Methods
This past year, the ClinGen (Clinical Genome Resource) Parkinson’s Disease Gene Curation Expert Panel (GCEP) (https://clinicalgenome.org/affiliation/40079/) formed to create a consensus about the gene-disease validity of specific PD genes. The findings from this panel aim to guide key stakeholders, including patients, payers, clinicians, scientists, and clinical laboratory directors. The panel is composed of international, multidisciplinary experts: molecular geneticists, clinicians with genetic research focus, and PD-specific genetic counselors. In order to begin a preliminary gene list for curation, one of the first steps taken by the group was to briefly ascertain and compare current commercial PD genetic testing offerings. We used the Genetic Test Registry (GTR, www.ncbi.nlm.nih.gov/gtr), a database of orderable genetic tests supported by the National Institutes of Health (NIH), to identify current commercial PD genetic test offerings in the United States and internationally [14]. (This database, one of several available test information resources on the web, is a commonly used, self-reported, genetic testing resource for genetic counselors and physicians that is open access.) The search string “Parkinson” was used with the following filters: “Clinical testing/Confirmation of Mutations Identified Previously” and “CLIA Certified” for laboratories in the United States. Returns were reviewed and categorized by test type and laboratory. Testing offered was further confirmed by the company websites. Laboratories currently offering multi-gene panels that were labeled as “Parkinson’s disease” were chosen for further descriptive analysis. Laboratories that did not clearly offer a multi-gene, diagnostic panel for PD were excluded. In addition, companies whose PD test offerings were not located on their website were excluded.
We reviewed the selected panels, comparing the selection of genes across panels, observing where there was agreement or not. We also noted the disease category representing a gene that was included on a panel, such as if they were linked to differential diagnoses of PD (e.g., Wilson’s disease or dystonia) or to atypical parkinsonism (e.g., Kufor-Rakeb disease).
Results
A final GTR query to inform curation was performed on 12/7/2020 and returned the following: 502 unique clinical genetic tests were offered from 28 Clinical Laboratory Improvement Amendments (CLIA)-approved labs in the United States and internationally. PD gene test offerings were not located for 13 companies upon additional searching of their websites. Four companies did not have obvious diagnostic PD multi-gene panels listed on their websites and were excluded. We observed many types of panels for PD (up to 22 different test choices) offered by individual companies, to evaluate by specific genes, overall clinical features (atypical/parkinsonism); unique clinical features (e.g., dystonia or dementia); age of onset, and inheritance.
From this original test list, 11 company test offerings were chosen for further analysis. All offered a general, diagnostic, multi-gene panel for Parkinson’s disease that included 3 or more genes. General diagnostic PD panels were notable for their differences, especially including size, from small (5 genes) to very large (62 genes). There were 71 unique genes queried by the laboratories in total (Table 1). All panels offered by companies in the analysis included 5 genes consistently linked to major PD risk in multiple studies (SNCA, PRKN, PINK1, PARK7, and LRRK2). Beyond this consensus, panels varied in inclusion of other genes, most notably GBA and VPS35 (Figure 2); and some companies offered an enzyme assay for GBA as well. All PD panels except one included genes linked with juvenile or atypical parkinsonism, genes linked with diseases in the differential diagnosis of PD (i.e. Wilson’s disease or dystonia), and less well-established genes according to published literature: DNAJC13, TMEM230, GIGYF2, HTRA2, RIC3, EIF4G1, UCHL1, and CHCHD2 [15]. Of the analyzed companies, seven originated from the US and four from Europe; there was a trend for European companies to offer larger gene panels.
Table 1.
Genes on Selected PD Panels*
| GENE | LAB (TOTAL # OF GENES ON PARKINSON’S DISEASE PANEL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Asper Biogene (40) | Athena (5) | Blueprint Genetics (62a) | CeGaT (30) | Centogene (36) | Fulgent (26) | GeneDx (29) | Invitae (16) | Knight Diagnostic (19) | Prevention Genetics (24) |
U of W NCG Lab (45) | |
| ADH1C | ● | ||||||||||
| AFG3L2 | ● | ||||||||||
| ATP1A3 | ● | ● | ● | ● | ● | ● | ● | ||||
| ATP13A2 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ||
| ATP6AP2 | ● | ● | ● | ● | |||||||
| ATP7B | ● | ● | |||||||||
| ATXN2 | ● | ||||||||||
| C10ORF2 | ● | ||||||||||
| C19ORF12 | ● | ● | |||||||||
| C9ORF12 | ● | ||||||||||
| CHCHD2 | ● | ● | ● | ||||||||
| COASY | ● | ||||||||||
| COMT | ● | ||||||||||
| CP | ● | ||||||||||
| CSF1R | ● | ● | ● | ||||||||
| CYP27A1 | ● | ● | |||||||||
| DCTN1 | ● | ● | ● | ● | ● | ● | ● | ||||
| DJ1/PARK7 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| DNAJC5 | ● | ||||||||||
| DNAJC6 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ||
| DNAJC13 | ● | ● | |||||||||
| EIF4G1 | ● | ● | ● | ● | |||||||
| FBX07 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |
| FTL | ● | ● | ● | ● | |||||||
| FUS | ● | ||||||||||
| GBA | ● | ●b | ● | ● | ● | ● | ● | ● | ● | ||
| GCH1 | ● | ● | ● | ● | ● | ● | ● | ● | |||
| GIGYF2 | ● | ● | ● | ||||||||
| GNAL | ● | ||||||||||
| GRN | ● | ● | |||||||||
| HTRA2 | ● | ● | ● | ● | ● | ||||||
| LMNB1 | ● | ||||||||||
| LRRK2 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| MAPT | ● | ● | ● | ● | ● | ● | ● | ||||
| NOTCH3 | ● | ||||||||||
| OPA1 | ● | ||||||||||
| PANK2 | ● | ● | |||||||||
| PDGFB | ● | ● | |||||||||
| PDGFRB | ● | ● | |||||||||
| PINK1 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| PLA2G6 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ||
| PODXL | ● | ||||||||||
| POLG | ● | ● | ● | ● | |||||||
| PRKN | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| PRKRA | ● | ● | ● | ● | ● | ● | ● | ||||
| PRNP | ● | ||||||||||
| PSEN1 | ● | ||||||||||
| PTRHD1 | ● | ||||||||||
| RAB29 | ● | ||||||||||
| RAB39B | ● | ● | ● | ● | |||||||
| SLC6A3 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |
| SLC16A2 | ● | ||||||||||
| SLC20A2 | ● | ● | ● | ● | |||||||
| SLC30A10 | ● | ● | ● | ||||||||
| SLC39A14 | ● | ● | |||||||||
| SMPD1 | ● | ||||||||||
| SNCA | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| SNCB | ● | ● | |||||||||
| SPG11 | ● | ● | |||||||||
| SPR | ● | ● | ● | ● | ● | ● | ● | ||||
| SYNJ1 | ● | ● | ● | ● | ● | ● | ● | ||||
| TAF1 | ● | ● | ● | ● | ● | ||||||
| TBP | ● | ||||||||||
| TH | ● | ● | ● | ● | ● | ● | ● | ● | |||
| TMEM230 | ● | ● | ● | ||||||||
| UCHL1 | ● | ● | ● | ● | ● | ||||||
| VPS13A | ● | ● | ● | ||||||||
| VPS13C | ● | ● | ● | ● | ● | ||||||
| VPS35 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |
| WDR45 | ● | ● | |||||||||
| XPR1 | ● | ● | |||||||||
Based on data pulled from the GTR on 12/7/2020 and should not be used to base clinical test decisions as this information quickly changes over time
includes 37 mitochondrial genes
available by request with reflex testing
Figure 2.
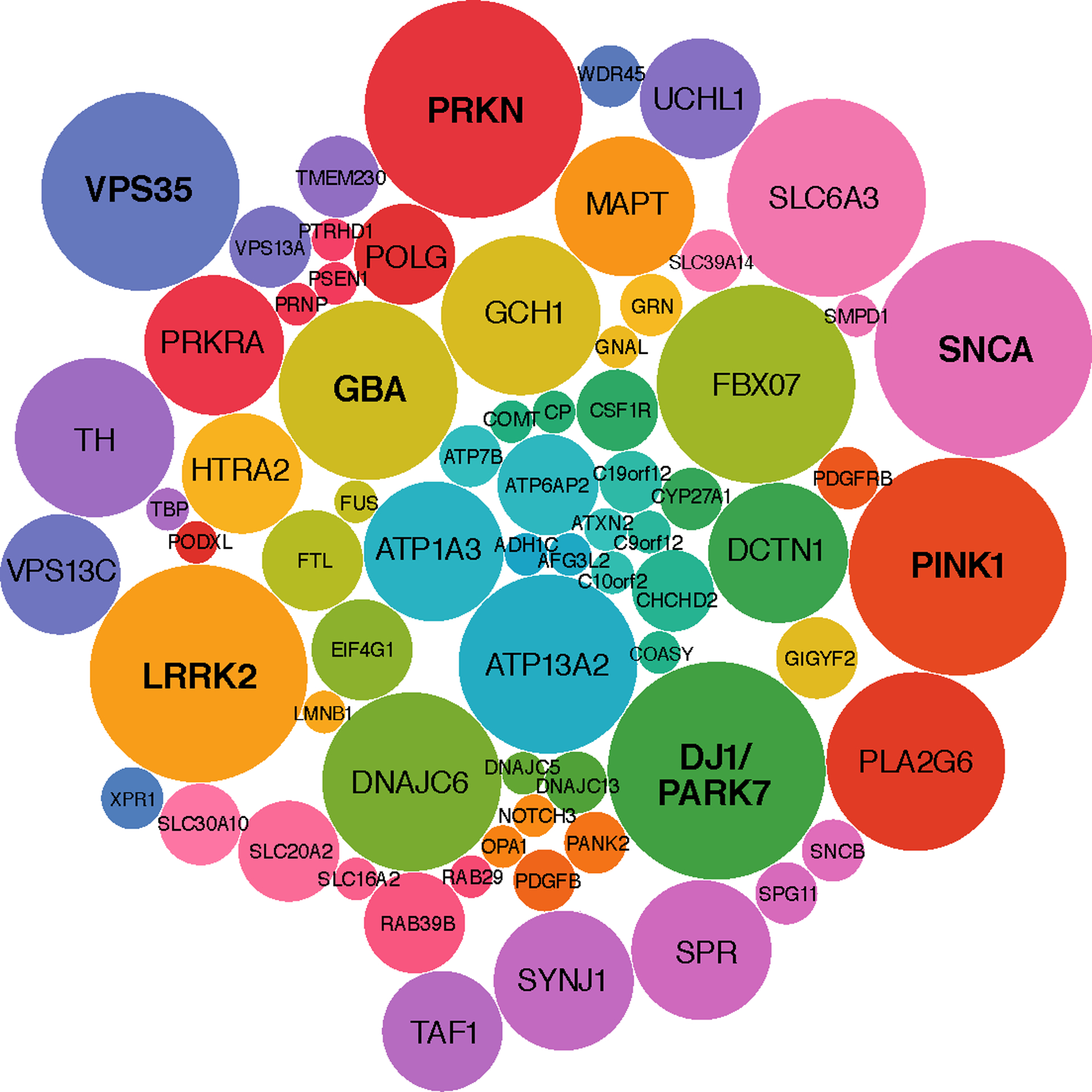
Genes Offered on PD Panels
Bubble plot of genes offered on general diagnostic panels of the 11 companies analyzed. Bubble size corresponds to the frequency a particular gene appears across panels. The genes SNCA, PRKN, PINK1, PARK7 and LRRK2 were most common and offered across all panels. GBA, a gene carrying significant risk for PD, was not consistently offered and only appeared on 8 panels, whereas other genes FBOX07 (atypical parkinsonism) and SLC6A3 (infantile parkinsonism-dystonia) appear more frequently.
None of the analyzed panels were designated for a particular geographic or ancestral population. Although among the 502 unique tests originally returned, at least one population-based panel was identified, specifically for Ashkenazi Jewish ancestry as part of broader disease screening. Among the 11 panels, additional variability was observed in the testing methodology listed by the laboratory on GTR; testing in some cases was referred to as targeted sequencing and in others, the testing/sequencing was more complete. Although all laboratory panels included some type of sequencing, three laboratories did not explicitly offer additional analysis of copy number variation for genes like PRKN or SNCA, known to have deletions and duplications.
Discussion
We have documented marked differences in diagnostic, multi-gene panels for PD offered by laboratories in the United States and internationally. In addition, we observed a remarkable array of PD gene panel options, even by the same company. Genes considered in the literature to be established as linked with monogenic PD or a major risk factor such as GBA, were not always included on a panel. Monogenic, atypical forms of parkinsonism were represented on the panels to a variable degree with the majority of labs including ATP13A2, DNAJC6, FBX07, SYNJ1, and DCTN1 (Table 1).
We believe that this diverse and unstandardized state of PD testing has consequences for test ordering, usefulness of the genetic information, and interpretation. Recent work has found that movement disorder specialists are not widely offering genetic testing to their patients with PD. A key reason cited by neurologists for this reduced utilization was the confusion surrounding genetic testing [16]. Further, the impact of negative results on test utility and interpretation for genetic counseling are markedly different if five versus more than 60 genes are tested. For instance, if only five major genes are tested – and then incompletely – the scope of testing may be too narrow to rule out a major genetic cause. The implications of testing that is too narrow for PD are very clear. Currently, precision medicine clinical trials for GBA mutation carriers are widely available for pathogenic variants carriers. However, some panels do not include GBA, (Table 1), or may not perform full sequencing of the gene. There also can be a difference as to which variants are reported out by laboratories.
Variants that are of uncertain significant (VUS) present a unique challenge. They do not fit into a benign or pathogenic category based on available data and, what is a particular difficulty, is that they may be reclassified later. Multigene and especially larger panels will have a potentially high rate of VUS—with commercial multigene panels approaching 10% for some neurologic disorders [17]. As a result, the large number of VUS likely to be discovered/revealed, combined with lack of consistency in reporting and classification, has the potential to further complicate test interpretation and counseling of patients.
To overcome these issues surrounding genetic testing, we suggest that physicians should have panel options for PD that are based on overall clinical features (typical versus atypical) with all genes included that are likely associated with monogenic PD. Additional options to include on the panel would be genes whose variants are associated with atypical parkinsonism and others associated with similar presentation, recognizing that there will be a cost/benefit analysis required for these larger panels. The technology used for PD panels will also be important to clarify, including the type of sequencing employed. In addition, laboratories should make clear to the physician if deletion/duplication testing will be performed since the majority of variants in the PRKN and SNCA gene involve copy number variation [2,12,18]. Another consideration for panel design and interpretation of results is the awareness about ethnic and population differences in PD, as we begin to better understand genetic diversity among populations and generate more supporting data through large research initiatives. There are indications that there are different genetic risks and clinical expressions of PD among population groups [19].
Future work by the ClinGen PD GCEP will aim to create consensus on casual genes for PD and those of uncertainty. The preliminary information obtained from the commonly used GTR will inform the process of creating a PD gene list for curation. In addition, the GCEP will review evidence from the Movement Disorder Society Genetic mutation database (MDSGene) as well as from published literature to further refine the gene list. We have ongoing curation of the following seven genes defined as Tier 1 for PD (not necessarily in this order): SNCA, PRKN, PINK1, PARK7, LRRK2, and VPS35. Ideally, this consensus will guide key stakeholders with the ultimate aim to improve the use of genetic information in the care of people with PD and improve patient outcomes.
Conclusion
We have identified marked heterogeneity in commercial gene tests offered for PD, specifically for multigene panels. This may create obstacles to test ordering and unnecessarily complicate genetic testing, interpretation, and counseling by HCPs. Our findings highlight the urgent need for expert opinion on which genes and variants commercial laboratory services should consider for general PD panels and other PD-related panels.
Acknowledgements:
Clinical Genome Resource (ClinGen) Parkinson’s Disease Gene Curation Expert Panel Authors:
Alexis Brice, MD1, Amasi Kumeh2, Andrew B. West, PhD3, Andrew Singleton, PhD4, Anna Naito, PhD2, Birgitt Schüle, MD5, Brian Fiske, PhD6, Carolin Gabbert, MA7, Christine Klein, MD7, Connie Marras, MD, PhD8, Cornelis Blauwendraat, PhD4, Courtney Thaxton, PhD9, Dario Alessi, PhD10, David Craig, PhD11, Edward A. Fon, MD12, Emily Forbes, DO13, Enza Maria Valente, MD, PhD14, Esther Sammler, MD, PhD10, Gill Chao, MMSc, LCGC15, Giulietta Riboldi, MD16, Houda Zghal Elloumi, PhD, FACMG17, Ignacio Mata, PhD18, James C. Beck, PhD2, Jamie C. Fong, MS19, Jean-Christophe Corvol, MD, PhD, HDR20, Jeanine Schulze, MS21, Jennifer Verbrugge, MS21, Joshua Shulman, MD, PhD19, Judith Peterschmitt22, Karen Marder, MPH, MD 23, Katja Lohmann, PhD7, Kelly Nudelman, PhD21, Lara Lange, MD7, Lola Cook, MS21, Mark R Cookson, PhD4, Martha Nance, MD24, Matthew Farrer, PhD25, Melina Grigorian, PhD15, Michael A. Schwarzschild, MD, PhD26, Niccolo Mencacci, MD, PhD27, Owen Ross, PhD28, Pramod Mistry, PhD29, Priscila Hodges, MS21, Rachel Blake15, Rachel Saunders-Pullman, MD, MPH30, Roy N. Alcalay, MD, MS23, S. Pablo Sardi, PharmD, PhD15, Sali Farhan, PhD12, Samuel Strom, PhD15, Shalini Padmanabhan, PhD6, Shruthi Mohan, PhD31, Simonne Longerich, PhD32, Susanne Schneider, MD, PhD33, Suzanne Lesage, PhD34,Tanya Bardakjian, MS, LCGC35,Tatiana Foroud, PhD21, Thomas Courtin, MD1, Thomas Tropea, DO, MPH35, Yunlong Liu, PhD21, Ziv Gan-Or, MD, PhD12
Affiliations:
1 Institut du Cerveau et de la Moelle Epinière (ICM), Paris, France
2 Parkinson’s Foundation, NY, NY
3 Duke University, Durham, NC
4 National Institute on Aging, Bethesda, MD
5 Stanford University, Stanford, CA
6 The Michael J. Fox Foundation for Parkinson’s Research, NY, NY
7 Institute of Neurogenetics, University of Luebeck, Luebeck, Germany
8 University of Toronto, Toronto, Ontario, Canada
9 ClinGen Clinical Genome Resource, National Institutes of Health/University of North Carolina, Chapel Hill, NC
10 University of Dundee, Dundee, Scotland, United Kingdom
11 University of Southern California, Los Angeles, CA
12 Montreal Neurological Institute, McGill University, Montreal, Quebec, Canada
13 UC Health Neurosciences Center - Anschutz Medical Campus, Colorado, CO
14 IRCCS Fondazione Mondino, Italy
15 Fulgent Genetics, Temple City, CA
16 NYU Langone Health, NY, NY
17 GeneDx, Gaithersburg, MD
18 Cleveland Clinic, Cleveland, OH
19 Baylor College of Medicine, Houston, TX
20 Sorbonne University, Paris, France
21 Indiana University School of Medicine, Indianapolis, IN
22Sanofi, Paris, France
23Columbia University Irving Medical Center, NY, NY
24 Struthers Parkinson’s Center, MN
25 University of Florida, FL
26 Massachusetts General Hospital/Harvard Medical School, Boston, MA
27 Northwestern University, Chicago, IL
28 Mayo Clinic, Rochester, Minneapolis, MN
29 Yale University, New Haven, MA
30 Mount Sinai Beth Israel, NY, NY
31 University of North Carolina, Chapel Hill, NC
32 Biogen, Cambridge, MA
33 University of Munich, Munich, Germany
34 University of Pennsylvania, Philadelphia, PA
Movement Society Disorder (MDS) Task Force on Recommendations for Clinical Genetic Testing in Parkinson’s Disease Authors:
Ali S. Shalash, MD, PhD1, Anne Hall, JD2, Avner Thaler, MD, PhD3, Carolyn M. Sue, MBBS, PhD4, Christine Klein, MD5, Deborah Mascalzoni, PhD6, Deborah Raymond, MS, CGC7, Emilia Mabel Gatto, MD8, Gian D. Pal, MD, MS9, Inke König, PhD10, Ivana Novakovic, PhD11, Karen Marder, MD, MPH12, Marcelo Merello, MD, PhD13, Mehri Salari, MD14, Niccolo Emanuele Mencacci, MD15, Nobutaka Hattori, MD, PhD16, Oksana Suchowersky, MD17, Rachel Saunders-Pullman, MD, MPH7, Roy N. Alcalay, MD, MS12, Soraya Bardien, PhD18, Sun Ju Chung, MD, PhD19, Tatiana Foroud, PhD20, Tatyana Simuni, MD15, Timothy Lynch, MB, BSc21, Vincenzo Bonifati, MD, PhD22
Affiliations:
1 Department of Neurology, Ain Shams University, Cairo, Egypt
2 Parkinson’s Foundation, NY, NY
3 Tel Aviv Sourasky Medical Center, Tel Aviv, Israel
4 University of Sydney, Sydney, Australia
5 Institute of Neurogenetics, University of Luebeck, Luebeck, Germany
6 EURAC Research, Bolzano, Italy
7 Mount Sinai Beth Israel, NY, NY
8 Sanatorio Trinidad Mitre, Buenos Aires, Argentina
9 Rush University Medical Center, Chicago, IL
10 University of Leubeck, Luebeck, Germany
11 University of Belgrade School of Medicine, Belgrade, Serbia
12 Columbia University Irving Medical Center, NY, NY
13 FLENI, Buenos Aires, Argentina
14 Shahid Beheshti University of Medical Science, Tehran, Iran
15 Northwestern University, Chicago, IL
16 Juntendo University School of Medicine, Tokyo, Japan
17 University of Alberta, Edmonton, Alberta, Canada
18 University of Stellenbosch, Cape Town, South Africa
19 Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
20 Indiana University School of Medicine, Indianapolis, IN
21 The Dublin Neurological Institute, Dublin, Ireland
22 Erasmus University Medical Center, Rotterdam, Netherlands
Financial disclosures:
Full financial disclosure for the previous 12 months: RNA’s research is supported by the NIH, the DoD, the Parkinson’s Foundation and the Michael J. Fox Foundation. He has received consultation fees from Genzyme/Sanofi, Restorbio and Roche. LC, JS, JV have no financial disclosures to report. JB and AN are employed by the Parkinson’s Foundation. KM receives research support from the NIH (NS100600, UL1TR001873. U24NS107168, RM1HG007257), Parkinson Disease Foundation, Michael J Fox Foundation and Lewy Body Disease Association. RSP receives funding from NIH NS-107016 and the Bigglesworth Family Foundation, and is the Bachmann Strauss Chair. CK serves as a medical advisor to Centogene for curation of genetic testing reports in the fields of movement disorders and dementia, but excluding Parkinson’s disease.
Funding sources for the study:
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Declaration of competing interests: None
References
- [1].Tysnes O-B, Storstein A, Epidemiology of Parkinson’s disease, J Neural Transm (Vienna). 124 (2017) 901–905. 10.1007/s00702-017-1686-y. [DOI] [PubMed] [Google Scholar]
- [2].Skrahina V, Gaber H, Vollstedt E-J, Förster TM, Usnich T, Curado F, Brüggemann N, Paul J, Bogdanovic X, Zülbahar S, Olmedillas M, Skobalj S, Ameziane N, Bauer P, Csoti I, Koleva-Alazeh N, Grittner U, Westenberger A, Kasten M, Beetz C, Klein C, Rolfs A, ROPAD Study Group, The Rostock International Parkinson’s Disease (ROPAD) Study: Protocol and Initial Findings, Mov Disord. 36 (2021) 1005–1010. 10.1002/mds.28416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kim CY, Alcalay RN, Genetic Forms of Parkinson’s Disease, Semin Neurol. 37 (2017) 135–146. 10.1055/s-0037-1601567. [DOI] [PubMed] [Google Scholar]
- [4].Reed X, Bandrés-Ciga S, Blauwendraat C, Cookson MR, The role of monogenic genes in idiopathic Parkinson’s disease, Neurobiol Dis. 124 (2019) 230–239. 10.1016/j.nbd.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weissbach A, Wittke C, Kasten M, Klein C, Chapter Seven - ‘Atypical’ Parkinson’s disease – genetic, in: Stamelou M, Höglinger GU (Eds.), International Review of Neurobiology, Academic Press, 2019: pp. 207–235. 10.1016/bs.irn.2019.10.011. [DOI] [PubMed] [Google Scholar]
- [6].Wittke C, Petkovic S, Dobricic V, Schaake S, Respondek G, Weissbach A, Madoev H, Trinh J, Vollstedt E-J, Kuhnke N, Lohmann K, Mahlow MD, Marras C, König IR, Stamelou M, Bonifati V, Lill CM, Kasten M, Huppertz H-J, Höglinger G, Klein C, Genotype–Phenotype Relations for the Atypical Parkinsonism Genes: MDSGene Systematic Review, Movement Disorders. March 19 2021. 10.1002/mds.28517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. , Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies, The Lancet Neurology. 18 (2019) 1091–1102. 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schneider SA, Hizli B, Alcalay RN, Emerging Targeted Therapeutics for Genetic Subtypes of Parkinsonism, Neurotherapeutics. 17 (2020) 1378–1392. 10.1007/s13311-020-00920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Phillips KA, Deverka PA, Hooker GW, Douglas MP, Genetic Test Availability And Spending: Where Are We Now? Where Are We Going?, Health Aff (Millwood). 37 (2018) 710–716. 10.1377/hlthaff.2017.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bean LJH, Funke B, Carlston CM, Gannon JL, Kantarci S, Krock BL, Zhang S, Bayrak-Toydemir P, ACMG Laboratory Quality Assurance Committee, Diagnostic gene sequencing panels: from design to report-a technical standard of the American College of Medical Genetics and Genomics (ACMG), Genet Med. 22 (2020) 453–461. 10.1038/s41436-019-0666-z. [DOI] [PubMed] [Google Scholar]
- [11].Harbo HF, Finsterer J, Baets J, Van Broeckhoven C, Di Donato S, Fontaine B, De Jonghe P, Lossos A, Lynch T, Mariotti C, Schöls L, Spinazzola A, Szolnoki Z, Tabrizi SJ, Tallaksen C, Zeviani M, Burgunder J-M, Gasser T, EFNS, EFNS guidelines on the molecular diagnosis of neurogenetic disorders: general issues, Huntington’s disease, Parkinson’s disease and dystonias, Eur J Neurol. 16 (2009) 777–785. 10.1111/j.1468-1331.2009.02646.x. [DOI] [PubMed] [Google Scholar]
- [12].Cook L, Schulze J, Kopil C, Hastings T, Naito A, Wojcieszek J, Payne K, Alcalay RN, Klein C, Saunders-Pullman R, Simuni T, Foroud T, Genetic testing for Parkinson disease, Neurology: Clinical Practice. (2020) 10.1212/CPJ.0000000000000831. 10.1212/CPJ.0000000000000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].von Coelln R, Shulman LM, Clinical subtypes and genetic heterogeneity: of lumping and splitting in Parkinson disease, Curr Opin Neurol. 29 (2016) 727–734. 10.1097/WCO.0000000000000384. [DOI] [PubMed] [Google Scholar]
- [14].Rubinstein WS, Maglott DR, Lee JM, Kattman BL, Malheiro AJ, Ovetsky M, Hem V, Gorelenkov V, Song G, Wallin C, Husain N, Chitipiralla S, Katz KS, Hoffman D, Jang W, Johnson M, Karmanov F, Ukrainchik A, Denisenko M, Fomous C, Hudson K, Ostell JM, The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency, Nucleic Acids Res. 41 (2013) D925–935. 10.1093/nar/gks1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lunati A, Lesage S, Brice A, The genetic landscape of Parkinson’s disease, Rev Neurol (Paris). 174 (2018) 628–643. 10.1016/j.neurol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- [16].Alcalay RN, Kehoe C, Shorr E, Battista R, Hall A, Simuni T, Marder K, Wills A-M, Naito A, Beck JC, Schwarzschild MA, Nance M, Genetic testing for Parkinson disease: current practice, knowledge, and attitudes among US and Canadian movement disorders specialists, Genet. Med (2019). 10.1038/s41436-019-0684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Roggenbuck J, Quick A, Kolb SJ, Genetic testing and genetic counseling for amyotrophic lateral sclerosis: an update for clinicians, Genetics in Medicine. 19 (2017) 267–274. 10.1038/gim.2016.107. [DOI] [PubMed] [Google Scholar]
- [18].Pankratz N, Kissell DK, Pauciulo MW, Halter CA, Rudolph A, Pfeiffer RF, Marder KS, Foroud T, Nichols WC, Parkinson Study Group-PROGENI Investigators, Parkin dosage mutations have greater pathogenicity in familial PD than simple sequence mutations, Neurology. 73 (2009) 279–286. 10.1212/WNL.0b013e3181af7a33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Global Parkinson’s Genetics Program, GP2: The Global Parkinson’s Genetics Program, Mov Disord. 36 (2021) 842–851. 10.1002/mds.28494. [DOI] [PMC free article] [PubMed] [Google Scholar]


