Abstract
Round cell sarcomas with EWSR1-PATZ1 fusion are rare polyphenotypic sarcomas that typically show both neural and myogenic differentiation on immunohistochemistry. The histology features lobular admixture of cellular fascicles of relatively monotonous spindle cells and small blue round cells separated by fibrotic stroma. The clinical behavior of EWSR1-PATZ1 sarcoma is uncertain currently with mixed outcomes reported even in cases with metastases. We herein report an additional case of EWSR1-PATZ1 fusion-related round cell sarcoma in the face of a 5-year-old boy with unusual histologic features of pale zones, rosette/gland-like structures and expression of epithelial markers. Fluorescent in-situ hybridization study (FISH) using EWSR1 breakapart probes was negative and molecular study with RNA sequencing was required to confirm the diagnosis. These findings highlight the diagnostic challenge and potential pitfall of FISH study in EWSR1-PATZ1 sarcoma. Further studies are required to increase the understanding of their behavior, morphologic spectrum and molecular features that will help devise new treatment strategies to these rare tumours.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12105-021-01285-w.
Keywords: Sarcoma, Round cell sarcoma, Spindle cell, Polyphenotypic, Epithelioid, Ewing-like sarcoma, RNAseq
Introduction
Undifferentiated round cell sarcomas comprise heterogenous groups of tumours unified by relatively monotonous small round cells that are undifferentiated or show limited differentiation. These include the distinctive sarcomas with CIC fusions, and those with BCOR gene fusion or internal tandem duplication [1–3]. Other sarcomas in this category include those with EWSR1 fused to non-ETS family partners, such as EWSR1-NFATC2 and EWSR1-PATZ1 fusions. Despite initial controversy, increasing evidence suggests that these tumours are separate entities from Ewing sarcoma. Most EWSR1-NFATC2 sarcomas are typified by nested to corded arrangement of small round cells within hyaline to myxoid stroma, with and without spindle cells, and a significant proportion shows positivity to cytokeratin. Characteristically, amplification of the EWSR1-NFATC2 gene is often present and they are resistant to conventional neoadjuvant chemotherapy that are used in Ewing sarcoma [4–9]. On the other hand, EWSR1-PATZ1 sarcomas feature lobular admixture of cellular fascicles of relatively monotonous spindle cells and small blue round cells separated by fibrotic stroma. They are polyphenotypic and typically show both neural and myogenic differentiation on immunohistochemistry [10–14]. The clinical behavior of EWSR1-PATZ1 sarcoma is uncertain, with mixed outcomes reported even in cases with metastases (Table 1). We herein report an additional case of EWSR1-PATZ1 fusion-related round cell sarcoma in the face of a 5-year-old boy with unusual histologic features of pale nodules, rosette/ gland-like structures and expression of epithelial markers. Fluorescent in-situ hybridization study (FISH) using EWSR1 breakapart probes was negative and molecular study with RNA sequencing was required to confirm the diagnosis. These findings highlight the diagnostic challenge and potential pitfall of FISH study in EWSR1-PATZ1 sarcoma.
Table 1.
Clinical features of EWSR1-PATZ1 fusion sarcoma (with treatment and outcome)
| Case | Age/Sex | Location/ Presentation | Treatment | Outcome | References |
|---|---|---|---|---|---|
| 1 | 5/M | Facial mass with distant metastases | Marginal resection with clear margin and adjuvant chemotherapy | Alive with metastases at 6 months | Index case |
| 2 | 36/F | Muscle under scapula with distant metastases | Biopsy without resection. Chemotherapy with Mesna, ifosfamide, VAC/IE, gemcitabine, docetaxel, pazopanib | Mixed initial response with later progression and succumbed at 30 months | [11] |
| 3 | 53/M | Upper arm/ axillary mass with distant metastases | Biopsy without resection. Chemotherapy with carboplatin, paclitaxel and palliative radiotherapy | Initial 20% reduction in size, followed by progression and succumbed | [11] |
| 4 | 81/F | Posterior neck mass between C2 and C3 spinous processes | Marginal resection with clear margin | Alive without disease at 19 months | [11] |
| 5 | 31/F | Retroperitoneal mass | Resection followed by adjuvant chemotherapy with vincristine, doxorubicin, cyclophosphamide | Progressed at 5 months after surgery with metastases and succumbed | [12] |
| 6 | 53/F | Pelvic/ Iliac fossa mass | Resection followed by adjuvant chemotherapy with Mesna, ifosfamide, doxorubicin | Alive without disease at 3 months after surgery | [12] |
| 7 | 52/F | Neck mass attached to cervical rootlet C4 | Resection followed by adjuvant radiation and chemotherapy with doxorubicin and ifosfamide | Unknown outcome | [13] |
| 8 | 44/M | Abdominal wall | Marginal resection with clear margin | Alive without disease at 19 months after surgery | [14] |
| 9 | 49/M | Abdominal wall | Surgery | Unknown outcome | [14] |
| 10 | 81/F | Posterior neck (between C2/C3) | Marginal resection with clear margin | Alive without disease at 19 months after surgery | [14] |
| 11 | 36/M | Abdominal wall | Complete excision with clear margin | Unknown outcome | [14] |
| 12 | 10/F | Chest wall with parietal pleural nodules and implants on visceral pleura and pericardium | Neoadjuvant chemotherapy with Ewing sarcoma protocol, rib resection and surgical debulking with unknown margin status | Alive without disease at 60 months after surgery | [14] |
| 13 | 74/F | Lung (left upper lobe) with pleural and mediastinal invasion | Biopsy only | Unknown outcome | [14] |
| 14 | 46/F | Abdominal wall and diaphragm, multiple lung metastases developed later | Radiation therapy (adjuvant) and complete excision of primary tumour; Wedge excision and lobectomy for lung metastasis; Neoadjuvant chemotherapy for metastases with adriamycin, ifosfamide, mesna | Alive with stable disease at 60 months | [14] |
| 15 | 34/F | Trapezius muscle, lung metastases | Neoadjuvant chemotherapy with cyclophosphamide, vincristine, doxorubicin, later added etoposide, ifosfamide; Excision with residual disease. Adjuvant radiation and chemotherapy with ifosfamide, etoposide, vincristine, cyclophosphamide; gemcitabine, docetaxel added at 4 months due to progression | Progression at 4 months, unknown response to the most recent chemotherapy | [14] |
| 16 | 66/M | Abdominal wall, lung metastases | Biopsy only | Unknown outcome | [14] |
Materials and Methods
Immunohistochemistry
Immunohistochemistry was performed on 4um-thick paraffin sections after deparaffinization. Histologic sections were stained with antibodies according to standard protocols. Multi-tissue roll was stained in parallel with test tissue for both negative and positive controls. Clones, manufacturers and dilutions of antibodies used were given in Supplementary Table 1.
Fluorescence In-Situ Hybridization
FISH was performed on the formalin-fixed paraffin-embedded tissue section as previously described [15, 16]. The FISH probes used in this study were listed in the Supplementary Table 2. The FISH signals were captured by ISIS software (Metasystems) on Carl Zeiss’s Axio Imager Z2. FISH signals from 100 non-overlapping nuclei were scored. For break-apart FISH, nuclei were scored as positive for locus break-apart when one green and one orange signal were separated from each other by at least 2-signal diameter. The specimen was classified as positive for rearrangement when at least 15% of the nuclei showing positive break-apart signal [15]. For CDKN2A deletion FISH, the slides were scored as previously described [16].
Next Generation Sequencing (RNAseq and Somatic Targetable Panel)
The fusion transcripts were analyzed by TruSight Pan Cancer RNA Panel (Illumina). Briefly, RNA was extracted from the formalin-fixed paraffin-embedded sections using RNeasy FFPE kit (Qiagen). The RNA-seq libraries were prepared using 100 ng RNA with the TruSight Pan Cancer RNA Panel (Illumina). The library was sequenced with 76 base-pair paired-end reads on an Illumina MiSeq for 3 million reads pairs. The sequencing reads were mapped to reference human genome hg19 using the STAR aligner and the fusion variants were called STAR Fusion caller [17]. The fusion variants were visualized on integrative genomic viewer (IGV).
In addition, the specimen was analyzed by CUHK somatic cancer panel for single nucleotide variants and small indels of 85 genes as previously described [18]. Briefly, the DNA was extracted and sonicated to 200 bp by Covaris FFPE DNA kit (Covaris). Three hundred nanogram of DNA was subjected to library preparation using KAPA HyperPrep kit (Roche) and enriched with a custom-designed solution based hybrid capture panel (Roche). The baits targeted 700 kb of the human genome including the coding exons of 85 genes and introns of 13 genes. The gene list is shown in Supplementary Table 3. The libraries were sequenced on the Illumina MiSeq instrument at 75 bp pair-end to a goal coverage of 500X. Sequencing reads were aligned with default parameters using BWA-mem v.0.7.10 to the hg19 reference genome and then duplicate reads were removed using Picard v1.97. Local realignment around indels was performed using the Genome Analysis Tool Kit software. Five variant calling programs were used to call single nucleotide variants and indels: MuTect2, Varscan2, SAMtools, Platypus, and Freebayes. Variants were functionally annotated using ANNOVAR.
Results
Clinical Findings
A 5-year-old boy was found by his mother to have a right facial mass near the jaw region 3 years ago. Previous investigations at another hospital had claimed to show a cystic lesion on ultrasound measuring 1.9 cm in greatest dimension. Aspiration of cystic fluid yielded no malignant cells and was treated with polidocanol injection. The mass increased in size rapidly in 2 months’ time. Repeat ultrasound at 1 year showed mixed solid and cystic lesion measuring 3.3 × 2.4 cm, and was aspirated again with no malignant cell revealed. Repeat injection of bleomycin into the mass was performed. At 3 years after first presentation, he was presented to us for a large right facial mass, which was marginally excised with limited local lymph nodes removal.
Pathologic Findings
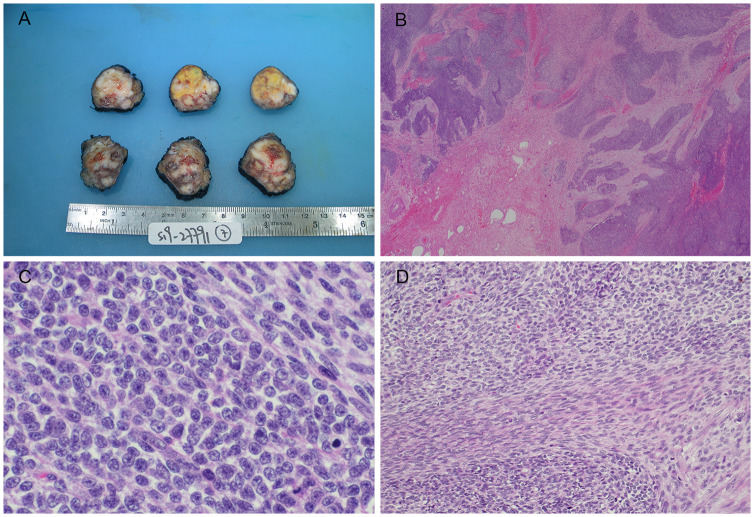
The surgically resected tumour was oval in shape and measured 4.5 × 3.5 × 3 cm and weighed 23.6 g. Cut surfaces revealed a tan-coloured to whitish, solid nodular tumour with patchy hemorrhage and necrosis (Fig. 1a). It was partly surrounded by skeletal muscle. A total of 21 lymph nodes from right neck dissection, superficial and facial artery regions showed no evidence of metastasis. Resection margins were clear.
Fig. 1.
Pathological features of tumour. (a) Gross examination of tumour showing a tan-coloured to whitish solid nodular appearance with patchy hemorrhages and necrosis. (b) Microscopy revealed a lobular architecture with variably cellular fibrous stroma mimicking desmoplastic small round cell tumour. (c) Tumour featuring admixture of primitive small round cells and spindle cells, (d) some of the latter were associated with intercellular collagen
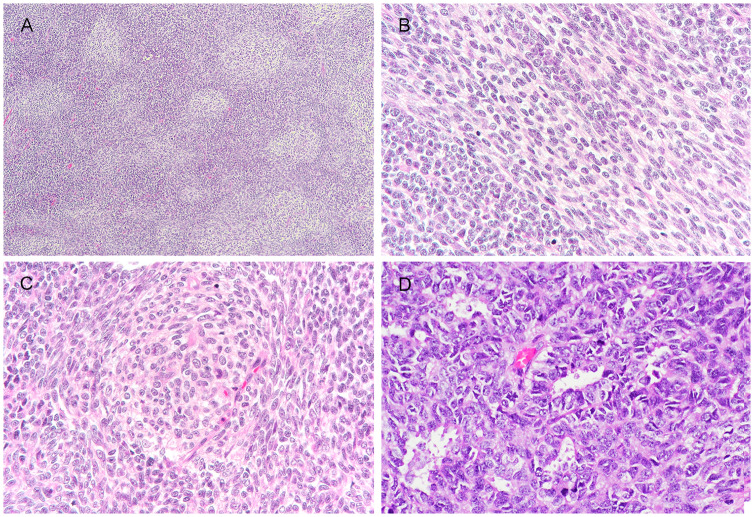
Microscopic examination revealed highly irregular tumour lobules widely infiltrating the skeletal muscle (Fig. 1b). The lobules were separated by hyaline to fibrocellular septa and comprised a diffuse mixture of small blue round cells and spindle cells. The small round cells showed relatively monomorphic roundish hyperchromatic nuclei, indistinct nucleoli and very scanty cytoplasm (Fig. 1c). The spindle cells were arranged in tight narrow fascicles and had modest amount of pale eosinophilic cytoplasm. They were intimately admixed with the round cell component and associated with intercellular collagen at areas (Fig. 1d). In focal areas, there were pale zones featuring pale-staining spindle cell fascicles that continued and sometimes whirled around small pale nodules of cells with increased pale to clear cytoplasm (Fig. 2a–c). Focal rosette or gland-like structures with well-defined lumen were also seen (Fig. 2d). The latter structures merged imperceptibly with the adjacent small round cells. Mitosis was brisk, counting up to 33 per 10 high power fields. Patchy hemorrhage and necrosis were noted. There was no morphologic evidence of skeletal muscle, cartilage, bone or squamous differentiation (Figs. 3 and 4).
Fig. 2.
Representative images of unusual histologic features. (a) Focal “biphasic” appearance imparted by alternating pale-staining zones and small blue primitive round cell areas; The pale staining zones comprised both (b) fascicles of spindle cells with increased amount of pale staining cytoplasm and (c) small pale nodules, with focal whirling by pale spindle cells at periphery; (d) Focal rosette/ gland-like structures
Fig. 3.
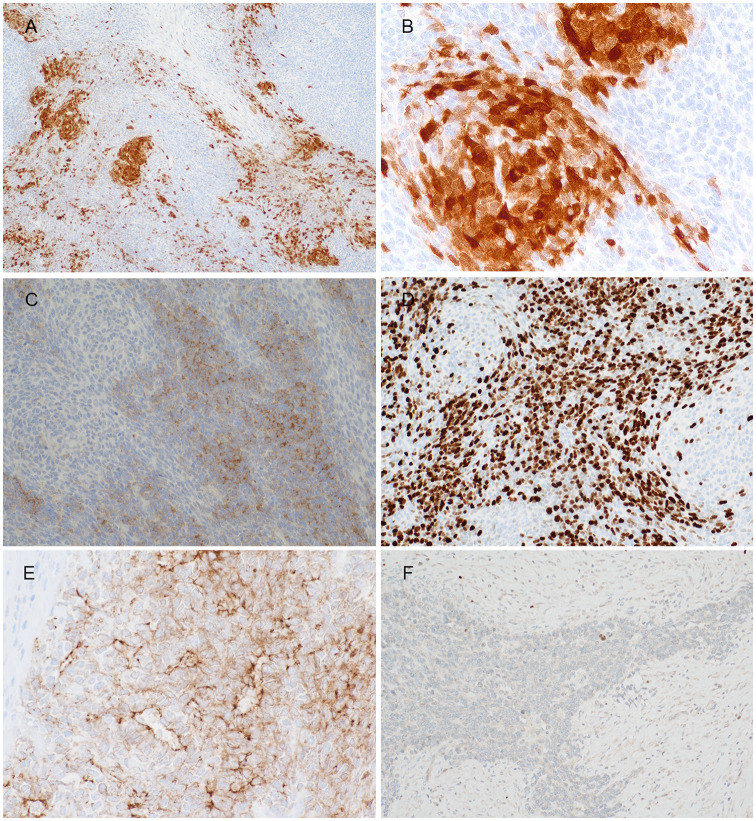
Representative images of immunohistochemistry results. S100 showed (a) patchy positivity as well as (b) focally highlighted the pale nodules. The primitive small round cells were positive to (c) synaptophysin with (d) high Ki67 index, whilst the opposite staining pattern was seen in the pale zone. (e) Claudin-4 highlighted the focal rosette/ gland-like structures, while (f) myogenin showed rare cells at the periphery of tumour lobules, probably representing entrapped skeletal muscle cells
Fig. 4.
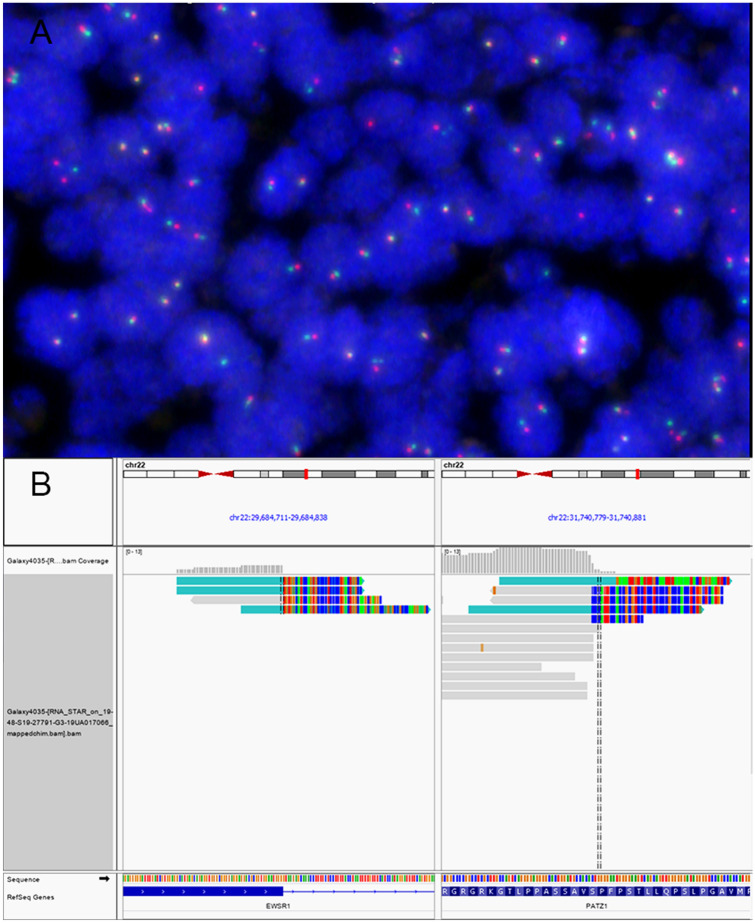
Molecular genetics study results: (a) EWSR1 break-apart FISH failed to detect the rearrangement of EWSR1-PATZ1. Due to the proximity of EWSR1 and PATZ1genes, the break-apart of the EWSR1 locus was not observed in the majority of tumor cells. (b) The EWSR1-PATZ1 fusion was visualized on IGV. The soft-clipped reads supporting the junction between the end of exon 9 of EWSR1 gene (NM_013986) and exon 1 of PATZ1 gene (NM_014323) was shown. The fusion product was predicted to be translated in-frame
Immunohistochemistry revealed diffuse but weak positivity to CD99 (cytoplasmic and membranous), with patchy strong positivity to S100 (Fig. 2a). The latter focally highlighted the small pale nodules and rarer adjacent spindle cells (Fig. 2b). Patchy positivity to GFAP, multifocal positivity to NeuN and SOX10 (not shown) were also present in tumour. Intriguingly, there was synaptophysin expression in the small primitive round cells, with reciprocal negativity in the pale nodules or fascicles (Fig. 2c). The Ki67 index was very high in the small round cells, whilst low in the pale zones (Fig. 2d). In addition, the tumour showed multifocal positivity to cytokeratins, e.g. AE1/3 and CK7 (not shown). Claudin-4 staining showed patchy positivity, and at the same time highlighting the rosette/ gland-like structures (Fig. 2e). Myogenic markers including desmin, MyoD1 and myogenin (Fig. 2f), however, only stained focal rare cells near the interface between the fibrous septa and tumour lobules. These probably represent nuclei of entrapped skeletal muscle with degeneration. Details of more immunohistochemistry results were shown in supplementary Table 1.
Molecular Genetics Findings
Fluorescence in-situ hybridization using break-apart probe revealed no evidence of translocation of EWSR1, FUS, CIC, BCOR, BRAF genes. CDKN2A deletion was not identified. Subsequent RNA sequencing using next generation sequencing designed for fusion gene detection subsequently identified in-frame chimeric transcript between exon 9 of EWSR1 and exon 1 of PATZ1 genes. Customed NGS cancer somatic mutation panel showed no targetable gene mutation.
Discussion
Round cell sarcomas with EWSR1-PATZ1 fusion are typically described as polyphenotypic, expressing both neural and myogenic markers on immunohistochemistry [11–14]. Although initial reports on these tumours described typical mixture of primitive small round and spindle cells, the morphologic spectrum is much wider and may comprise pure population of bland small round to ovoid cells with or without collagen matrix [14]. This reported case is unusual in that it lacks definite myogenic marker expression whilst it expresses epithelial markers including cytokeratins and claudin-4. The latter also highlighted focal rosette/ gland-like structures present in the tumour. Although multifocal positivity to cytokeratins is non-specific and can be seen in Ewing sarcoma and other round cell sarcomas including those with EWSR1-PATZ1 fusion [14, 19, 20], claudin-4 has been shown to be a relatively good epithelial marker and able to highlight epithelial structures in the sarcoma settings [21]. Morphologically similar rosette/gland-like structures have also been reported in other undifferentiated sarcoma such as sarcoma with NUTM1 gene fusion [22] and therefore are not unique to this case. In addition, the presence of pale zones comprising pale spindle fascicles continued as nodules amidst sheets of primitive small round cells produced a biphasic morphology on low power. Although of uncertain histogenesis, the pale nodules/fascicles were less mitotically active and negative to synaptophysin, in contrast with the surrounding primitive round cell component. Whether these pale spindle fascicles/ nodules could represent focal tendency towards a more differentiated phenotype is contentious, albeit round cell sarcomas with EWSR1-PATZ1 fusion can sometimes show a lower-grade morphology with scanty mitosis and increased collagen matrix [14]. Given the unusual morphologic and immunophenotypic features of this case, a number of differential diagnoses has been considered. The tumour mimics Ewing sacoma/ peripheral primitive neuroectodermal tumour (PNET) due to its morphology of monotonous small round cells and a remarkable degree of neural differentiation including expression of S100, SOX10, synaptophysin and GFAP. Although EWSR1-PATZ1 fusion sarcomas are usually distinguished from PNET/ Ewing sarcomas by the additional presence of spindle cells and variable expression of different myogenic markers on immunohistochemistry, the absence of the latter in our case has mandated molecular detection of the fusion transcript to confirm the diagnosis. The presence of well demarcated lobules of small round cells separated by fibrocellular septa, together with apparent mixed epithelial and neural differentiation on immunohistochemistry, also raises the differential diagnosis of desmoplastic small round cell tumour. However, desmoplastic small round cell tumour is typically negative to synaptophysin, and shows different EWSR1-WT1 gene fusion [23]. The presence of hypercellular fascicles of spindle cells and small round cells with cytokeratin expression of this case also raises the possibility of synovial sarcoma. The latter can be distinguished by its characteristic SS18-SSX fusion gene and strong positivity to TLE1 [24], which was negative in our case. Similarly admixed small round and spindle cells can be seen in BCOR-related sarcoma, although they typically show rich capillary network, pale nuclear chromatin and increased amount of eosinophilic cytoplasm to vacuolated appearance. Fusion or internal tandem duplication of BCOR gene, and strong BCOR protein expression would be seen in these cases [3, 25]. Although rhabdomyosarcomas with spindle cell morphology is possible by morphology, most cases of rhabdomyosarcomas do not show EWSR1 gene rearrangement. The exception to this are the recently described rhabdomyosarcomas comprising both round epithelioid and spindle cells that are associated with the fusion transcript of EWSR1-TFCP2 and strong expression of ALK protein [26–28]. These tumours could be distinguished by its mostly osseous origin, compared to the typical soft tissue site of EWSR1-PATZ1 sarcomas. The diagnosis of this case is particularly challenging, as FISH study showed no definite evidence of EWSR1 gene translocation using breakapart probes. This is not unexpected as the partner genes involved in the fusion reside on the same arm of chromosome 22 and are separated by 2 Mb only, of which the separated signals cannot be reliably detected due to the limitation on resolution of FISH study. Whilst the results of FISH for EWSR1 breakapart were positive in one study, another series showed that all four tested cases were below the cut-off threshold for positive results [12, 14]. This reveals the potential pitfall of utilizing breakapart FISH probes for diagnosing this group of round cell sarcoma. Indeed, FISH results using breakapart probes could be falsely negative in sarcomas with BCOR-CCNB3 fusion for similar reason [29], and in some CIC-fusion sarcoma [30] or rarely classical Ewing sarcoma due to cryptic insertion or translocation [31, 32]. Although a proportion of sarcomas with EWSR1-PATZ1 fusion have been reported to harbour MDM2 amplification or CDKN2A deletion [11, 14], these were not found in our case. Next generation sequencing using customized somatic DNA panel revealed no targetable gene mutation.
At present, the clinical behavior of EWSR1-PATZ1 sarcoma is not certain due to the small number of cases. Deadly outcome has been reported in patients with metastases, although a recent larger series reported patients who were alive without disease after radical resection of both the primary tumours and metastases (Table 1). Our patient is notable for his relatively long history of tumour for 3 years before presentation to us. In corroboration to a previous report, a history of slow growing tumour can be seen in some cases [11]. Most cases are also resistant to chemotherapy, both in the neoadjuvant or adjuvant therapy settings [11–14]. The presence of MDM2 amplification and CDKN2A deletion found by these studies may suggest the potential utility of target therapy such as CDK4/6 inhibitor, although these alterations were not present in all cases. A previous study on six cases of EWSR1-PATZ1 sarcomas demonstrated that these tumours were separate from classical Ewing sarcomas by gene expression signature with high expression of a specific G protein coupled receptor transcript, GPR12 [10]. Interestingly, GPR12 has been shown to bind to sphingosine-1-phosphate as its predicted ligand [33] and potentially its activity could be modulated by phytocannabinoid cannabidiol [34]. Further study on this sphingosine-1-phosphate/ G protein coupled receptor axis may open up new treatment strategy for this group of sarcoma.
Conclusion
We report on a case of round cell sarcoma with EWSR1-PATZ1 fusion in the face of a five-year-old boy. In addition to the more typical small primitive round and spindle cells, this tumour also showed pale nodules continuous with pale-staining spindle fascicles, rosette/ gland-like structures with expression of claudin-4, but without definite myogenic marker expression. These findings are unusual for EWSR1-PATZ1 sarcoma and could raise diagnostic confusion with other sarcomas. FISH study using breakapart probes may not detect the EWSR1-PATZ1 fusion event and hence presents potential diagnostic pitfall. Literature review shows that surgical resection remains the mainstay of treatment, whilst there is general lack of tumour response to chemotherapy. Studies with larger number of cases are needed to improve on the understanding and exploring alternative treatment opportunities for these rare tumours.
Supplementary information
Below is the link to the electronic supplementary material.
Funding
The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Compliance wthe Ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical Approval
All procedures were performed as part of diagnostic workup and for treatment purpose. All procedures performed involving human participant were in accordance with the ethical standards of the institutional review board.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Antonescu CR, Owosho AA, Zhang L, et al. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: a Clinicopathologic and molecular study of 115 cases. Am J Surg Pathol. 2017;41(7):941–949. doi: 10.1097/PAS.0000000000000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kao YC, Owosho AA, Sung YS, et al. BCOR-CCNB3 fusion positive sarcomas: a Clinicopathologic and molecular analysis of 36 cases with comparison to morphologic Spectrum and clinical behavior of other round cell sarcomas. Am J Surg Pathol. 2018;42(5):604–615. doi: 10.1097/PAS.0000000000000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao YC, Sung YS, Zhang L, Jungbluth AA, et al. BCOR overexpression is a highly sensitive marker in round cell sarcomas with BCOR genetic abnormalities. Am J Surg Pathol. 2016;40(12):1670–1678. doi: 10.1097/PAS.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GY, Thomas DG, Davis JL, et al. EWSR1-NFATC2 translocation-associated sarcoma Clinicopathologic findings in a rare aggressive primary bone or soft tissue tumor. Am J Surg Pathol. 2019;43(8):1112–1122. doi: 10.1097/PAS.0000000000001260. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Perez JA, Nielsen GP, Antonescu C, et al. EWSR1/FUS-NFATc2 rearranged round cell sarcoma: clinicopathological series of 4 cases and literature review. Hum Pathol. 2019;90:45–53. doi: 10.1016/j.humpath.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bode-Lesniewska B, Fritz C, Exner GU, et al. EWSR1-NFATC2 and FUS-NFATC2 gene fusion-associated mesenchymal tumors: Clinicopathologic correlation and literature review. Sarcoma. 2019;2019:9386390. doi: 10.1155/2019/9386390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perret R, Escuriol J, Velasco V, et al. NFATc2-rearranged sarcomas: clinicopathologic, molecular, and cytogenetic study of 7 cases with evidence of AGGRECAN as a novel diagnostic marker. Mod Pathol. 2020; 10.1038/s41379-020-0542-z. [DOI] [PubMed]
- 8.Yoshida KI, Machado I, Motoi T et al. NKX3–1 Is a Useful Immunohistochemical Marker of EWSR1-NFATC2 Sarcoma and Mesenchymal Chondrosarcoma. [DOI] [PMC free article] [PubMed]
- 9.Yau DTW, Chan JKC, Bao S, et al. Bone sarcoma with EWSR1-NFATC2 fusion: sarcoma with varied morphology and amplification of fusion gene distinct from Ewing sarcoma. Int J Surg Pathol. 2019;27(5):561–567. doi: 10.1177/1066896919827093. [DOI] [PubMed] [Google Scholar]
- 10.Watson S, Perrin V, Guillemot D, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245(1):29–40. doi: 10.1002/path.5053. [DOI] [PubMed] [Google Scholar]
- 11.Bridge JA, Sumegi J, Druta M, et al. Clinical, pathological, and genomic features of EWSR1-PATZ1 fusion sarcoma. Mod Pathol. 2019;32(11):1593–1604. doi: 10.1038/s41379-019-0301-1. [DOI] [PubMed] [Google Scholar]
- 12.Chougule A, Taylor MS, Nardi V, et al. Spindle and round cell sarcoma with EWSR1-PATZ1 gene fusion: a sarcoma with Polyphenotypic differentiation. Am J Surg Pathol. 2019;43(2):220–228. doi: 10.1097/PAS.0000000000001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park KW, Cai Y, Benjamin T, et al. Round cell sarcoma with EWSR1-PATZ1 gene fusion in the neck: case report and review of the literature. Laryngoscope. 2020; 10.1002/lary.28554. [DOI] [PubMed]
- 14.Michal M, Rubin BP, Agaimy A, et al. EWSR1-PATZ1-rearranged sarcoma: a report of nine cases of spindle and round cell neoplasms with predilection for thoracoabdominal soft tissues and frequent expression of neural and skeletal muscle markers. Mod Pathol. 2020; 10.1038/s41379-020-00684-8. Epub ahead of print. PMID: 33012788 [DOI] [PubMed]
- 15.Yeung SF, Tong JHM, Law PPW, et al. Profiling of oncogenic driver events in lung adenocarcinoma revealed MET mutation as independent prognostic factor. J Thorac Oncol. 2015;10(9):1292–1300. doi: 10.1097/JTO.0000000000000620. [DOI] [PubMed] [Google Scholar]
- 16.Wu D, Hiroshima K, Matsumoto S, et al. Diagnostic usefulness of p16/CDKN2A FISH in distinguishing between sarcomatoid mesothelioma and fibrous pleuritis. Am J Clin Pathol. 2013;139(1):39–46. doi: 10.1309/AJCPT94JVWIHBKRD. [DOI] [PubMed] [Google Scholar]
- 17.Haas BJ, Dobin A, Stransky N, et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20(1):213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan AW, Chau SL, Tong JH, et al. The landscape of actionable molecular alterations in Immunomarker-defined large-cell carcinoma of the lung. J Thorac Oncol. 2019;14(7):1213–1222. doi: 10.1016/j.jtho.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Folpe AL, Goldblum JR, Rubin BP, et al. Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. Am J Surg Pathol. 2005;29(8):1025–1033. doi: 10.1097/01.pas.0000167056.13614.62. [DOI] [PubMed] [Google Scholar]
- 20.Machado I, Cruz J, Lavernia J, et al. Superficial EWSR1-negative undifferentiated small round cell sarcoma with CIC/DUX4 gene fusion: a new variant of Ewing-like tumors with locoregional lymph node metastasis. Virchows Arch. 2013;463(6):837–842. doi: 10.1007/s00428-013-1499-9. [DOI] [PubMed] [Google Scholar]
- 21.Schaefer IM, Agaimy A, Fletcher CD, et al. Claudin-4 expression distinguishes SWI/SNF complex-deficient undifferentiated carcinomas from sarcomas. Mod Pathol. 2017;30(4):539–548. doi: 10.1038/modpathol.2016.230. [DOI] [PubMed] [Google Scholar]
- 22.Dickson BC. Sung, Rosenblum et al. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 2018;42(5):636–645. doi: 10.1097/PAS.0000000000001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher CD, Bridge JA, Hogendoorn PCW, Mertens F, et al. Synovial sarcoma. In: WHO classification of Tumours of soft tissue and bone. 4th ed. Geneva: World Health Organization; 2013. [Google Scholar]
- 24.Terry J, Saito T, Subramanian S, et al. TLE1 as a diagnostic immunohistochemical marker for synovial sarcoma emerging from gene expression profiling studies. Am J Surg Pathol. 2007;31(2):240–246. doi: 10.1097/01.pas.0000213330.71745.39. [DOI] [PubMed] [Google Scholar]
- 25.Pierron G, Tirode F, Lucchesi C, et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44(4):461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- 26.Agaram NP, Zhang L, Sung YS, et al. Expanding the Spectrum of intraosseous rhabdomyosarcoma: correlation between 2 distinct gene fusions and phenotype. Am J Surg Pathol. 2019;43(5):695–702. doi: 10.1097/PAS.0000000000001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dashti NK, Wehrs RN, Thomas BC, et al. Spindle cell rhabdomyosarcoma of bone with FUS-TFCP2 fusion: confirmation of a very recently described rhabdomyosarcoma subtype. Histopathology. 2018;73(3):514–520. doi: 10.1111/his.13649. [DOI] [PubMed] [Google Scholar]
- 28.Wong DD, van Vliet C, Gaman A, et al. Rhabdomyosarcoma with FUS re-arrangement: additional case in support of a novel subtype. Pathology. 2019;51(1):116–120. doi: 10.1016/j.pathol.2018.09.056. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida A, Arai Y, Hama N, et al. Expanding the clinicopathologic and molecular spectrum of BCOR-associated sarcomas in adults. Histopathology. 2020;76(4):509–520. doi: 10.1111/his.14023. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida A, Arai Y, Kobayashi E, et al. CIC break-apart fluorescence in-situ hybridization misses a subset of CIC-DUX4 sarcomas: a clinicopathological and molecular study. Histopathology. 2017;71(3):461–469. doi: 10.1111/his.13252. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko Y, Kobayashi H, Handa M, et al. EWS-ERG fusion transcript produced by chromosomal insertion in a Ewing sarcoma. Genes Chromosomes Cancer. 1997;18(3):228–231. doi: 10.1002/(SICI)1098-2264(199703)18:3<228::AID-GCC9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 32.Newby R, Rowe D, Paterson L, et al. Cryptic EWSR1-FLI1 fusions in Ewing sarcoma: potential pitfalls in the diagnostic use of fluorescence in situ hybridization probes. Cancer Genet Cytogenet. 2010;200(1):60–64. doi: 10.1016/j.cancergencyto.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Foster SR, Hauser AS, Vedel L, et al. Discovery of human signaling systems: Pairing peptides to G protein-coupled receptors. Cell. 2019;179(4):895–908. doi: 10.1016/j.cell.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laun AS, Shrader SH, Brown KJ, et al. GPR3, GPR6, and GPR12 as novel molecular targets: their biological functions and interaction with cannabidiol. Acta Pharmacol Sin. 2019;40(3):300–308. doi: 10.1038/s41401-018-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.