Abstract
Oropharyngeal squamous cell carcinoma (SCC) is increasing in incidence and, in Western countries, strongly associated with transcriptionally-active high-risk human papillomavirus (HPV). Within HPV-positive tumors, there is wide morphologic diversity with numerous histologic subtypes of SCC. There are also variable degrees of keratinization, anaplasia, stromal fibrosis, and maturing squamous differentiation. Unlike in the uterine cervix, where associations between HPV types and lineages/sublineages within types have been investigated with some clear correlations identified, little to no data exists for oropharyngeal SCC. In this study, for a large cohort of oropharyngeal SCC patients, we performed RTPCR for high-risk HPV. For the HPV positive patients, we sequenced the DNA of the entire HPV16 genome and determined lineages and sublineages, correlating HPV status, genotype, and HPV16 lineages/sublineages with SCC subtype and various histologic features. Of the 259 patients, 224 (86.5%) were high-risk HPV positive, of which 210/224 (93.8%) were HPV type 16 and 6/224 (2.7%) HPV type 33. Of the four HPV16 lineages, A was the most frequent (192/214 or 89.8%) and of the HPV16 A sublineages, A1 was the most frequent (112/210 or 53.3%). Patients with HPV negative tumors were more often keratinizing vs other types (23/35 or 65.7%) and thus more likely to have more maturing squamous differentiation and stromal desmoplasia. There was no significant correlation between HPV type (16 versus other), between HPV16 lineage (A versus others), or HPV16 A sublineages (A1 or A2 versus others) and morphologic type of SCC nor the various morphologic features of anaplasia/multinucleation, degree of keratinization, nor amount of stromal desmoplasia. In summary, in our cohort, there was no correlation between the type of HPV, the HPV 16 lineage or sublineage, and any of the histologic features or morphologic SCC subtypes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12105-021-01318-4.
Keywords: Human papillomavirus, Squamous cell carcinoma, Oropharynx, Morphology, Type
Introduction
In Western countries such as the United States, the majority of patients with oropharyngeal squamous cell carcinoma (SCC) are positive for transcriptionally-active high-risk human papillomavirus (HPV) [1]. This confers a marked survival benefit, likely related to response to treatment [2–4].
While most OPSCCs are associated with HPV genotype 16, there are lineages and even sublineages of HPV16 defined by differences in the HPV genome which may be variably associated with OPSCC. Interestingly, recent studies of cervical cancer have suggested that even small genetic differences within the HPV16 genome can have major impacts on the risk of developing invasive carcinoma. HPV16 can be classified into 4 distinct main lineages (A [European-Asian], B [African-1], C [African-2] and D [North American, Asian-American]), and 16 finer sublineages within these lineages (A1-4, B1-4, C1-4, D1-4), based on differences (1-10%) in nucleotide sequences across the viral genome [5, 6]. In a study of 3200 uterine cervical specimens, Mirabello et al reported that HPV16 sublineages A4, D2, and D3 were more carcinogenic than the A1/A2 variants and that risk increased further when genetic ancestry of host and virus matched [7]. In particular, these three sublineages were associated with strongly increased risks of specifically glandular lesions, with the D2 sublineage associated with > 100-fold increased risk of adenocarcinoma. These findings suggest that genetic variation within the HPV genome may be a major driver of the carcinogenic process and histologic outcome.
In the uterine cervix, data also shows that certain morphologic types of carcinoma are associated with specific HPV genotypes. For example, adenocarcinomas have been consistently found to be more commonly associated with HPV type 18 than SCC [8–10. Little is known about how the type of high-risk HPV and the genetic variation within HPV16 itself correlate with morphology in the oropharyngeal SCC. In this study, we performed reverse transcription PCR (RTPCR) for high-risk HPV E6/E7 mRNA detection and high-risk HPV typing along with next generation sequencing of the high-risk HPV16 genome in a large cohort of well-characterized oropharyngeal SCC patients and correlated the results with morphologic subtypes and specific histomorphologic features [11].
Materials and Methods
Patients and Samples
After approval by the Vanderbilt University Medical Center Human Research Protection Program, incident, previously untreated patients with oropharyngeal SCC from Vanderbilt University Medical Center were identified though the Vanderbilt Research Derivative (RD), an IRB-approved, identified, searchable database of more than 3.5 million electronic health records (EHRs) from patients seen at VUMC [12]. The RD contains clinical data collected as part of routine patient care but reorganized to be easily searchable and usable for research purposes. The RD also links with the Vanderbilt Cancer Registry (VCR), which collects detailed clinical information from all reportable neoplasms diagnosed and/or treated at VUMC. Patients diagnosed between June 1, 2000 and July 9, 2018 were identified using the following International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) codes: C01.9, C02.4, C05.1, C05.2, C05.8, C09.0, C09.1, C09.8, C09.9, C10.0, C10.2, C10.3, C10.8, C10.9, C14.0 and C14.2. Clinical information prior to the year 2000 was not reliably captured within the VUMC EHR system; thus, cases diagnosed prior to 2000 were excluded. Each patient’s EHR was manually reviewed. Patients for whom an OPC diagnosis could not be confirmed, those with a prior history of cancer (other than non-melanoma skin cancer), multifocal carcinomas at presentation, patients who were immunocompromised, and those without a tumor specimen collected prior to treatment were excluded.
Morphologic Review
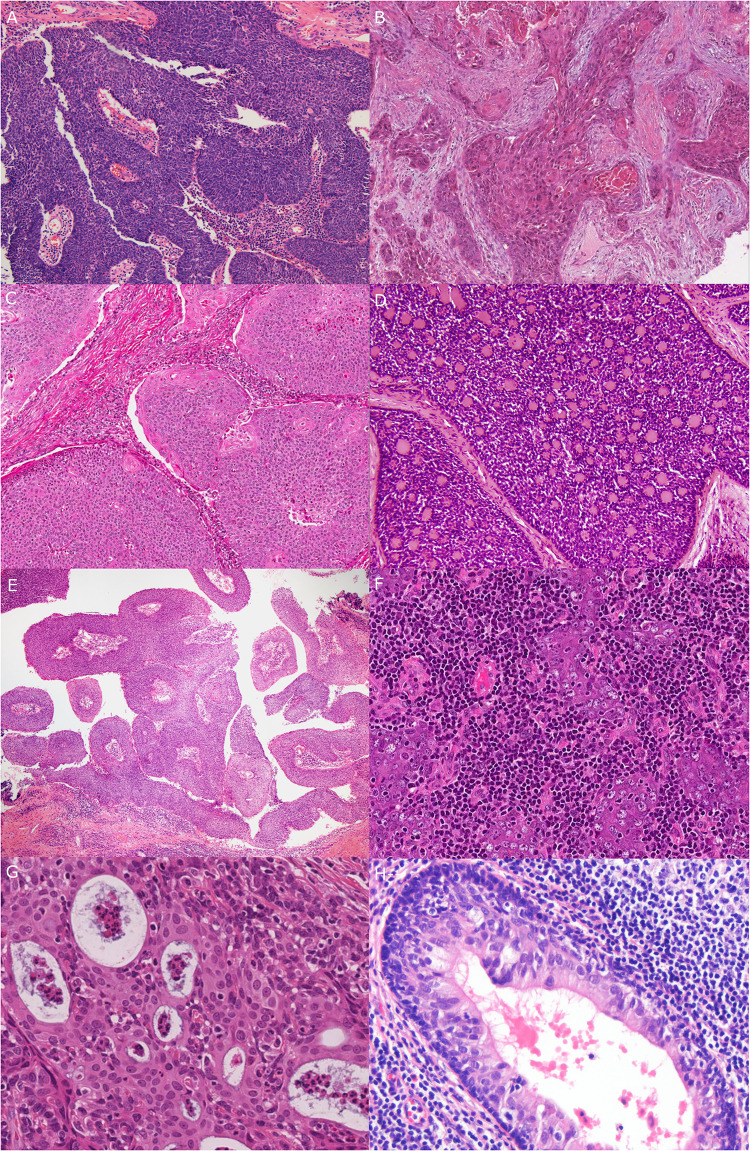
For patients that met inclusion criteria, representative tumor slides were reviewed by one or both of the study pathologists (JSL and MM) to confirm the diagnosis of SCC, to evaluate for SCC type or subtype according to WHO definitions (JSL) [13], and to characterize additional morphologic features (JSL). Typing of “conventional” oropharyngeal SCC was performed as previously described [14, 15] (Fig. 1). Briefly, keratinizing-type SCC consists entirely of maturing squamous epithelium with no areas with NK SCC or “basal” morphology. The cells have polygonal shapes with abundant, eosinophilic (keratinizing) cytoplasm, distinct cell borders, and intercellular bridges. The nests are usually angulated and irregular, and there is frequently marked stromal desmoplasia. The cytoplasm of maturing squamous cells is filled with keratin intermediate filaments imparting a dense appearance. Nonkeratinizing SCC consists of sheets, nests or trabeculae of oval and frequently spindled, hyperchromatic cells with indistinct cell borders and lacking prominent nucleoli. They have very little or only modest amounts of eosinophilic cytoplasm. Brisk mitotic activity is usually present. There is typically no (or minimal) stromal reaction to the invading tumor. Portions of the tumor can show squamous maturation, characterized by polygonal cells with mature, eosinophilic cytoplasm, distinct cell borders, intercellular bridges, and keratin pearls, but these mature areas must constitute less than 10% of the total surface area. Nonkeratinizing SCC with maturation is an intermediate group and consists of definitive areas with nonkeratinizing SCC morphology but that also has maturing squamous differentiation comprising greater that 10% of tumor surface area. It also frequently shows "reverse maturation", where the basal appearing cells are central in the nests, but the cells at the periphery show maturing squamous maturation, a pattern opposite that seen in conventional keratinizing type SCC [14–17]. Other rare histologic subtypes such as basaloid, undifferentiated, papillary, and adenosquamous carcinoma were diagnosed based on their published features [18–22] (Fig. 1).
Fig. 1.
All major types and subtypes of squamous cell carcinoma occurring in the oropharynx. a Nonkeratinizing (10X magnification). b Keratinizing (6X magnification). c Nonkeratinizing with maturation (10X magnification). d Basaloid (20X magnification). e Papillary (4X magnification). f Lymphoepithelial (10X magnification). g Adenosquamous (20X magnification). h Ciliated adenosquamous (20X magnification)
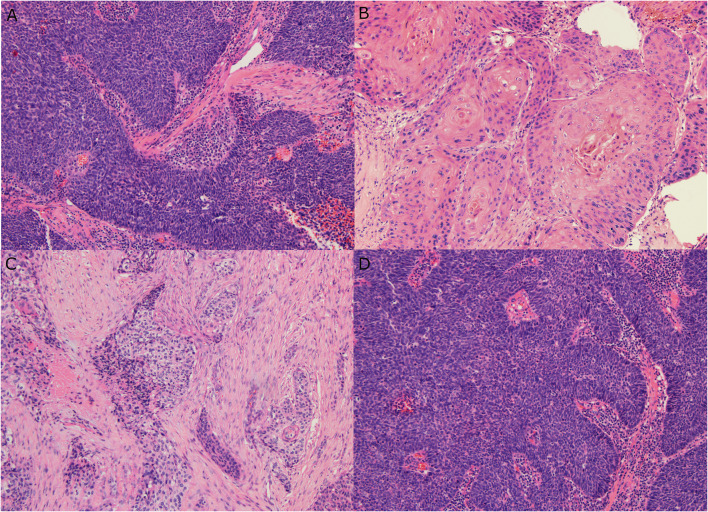
The presence of anaplasia or multinucleation was defined as previously [23]. Specifically, nuclear anaplasia was defined as any 400X magnification field (area = 0.2 mm2) with three or more nuclei with diameters equal to or wider than four lymphocyte nuclei (~25 microns). Tumor cell multinucleation was defined as any 400X magnification field (area = 0.2 mm2) with three or more tumor cells clearly having multiple nuclei [23]. In addition, a multinucleation index was generated by identifying the “hot spot” for multinucleated tumor cells where they were most prevalent and then counting 10 consecutive high power fields from there, generating an index from 0 to the maximum number of multinucleated cells in the 10 high power fields. In addition, the fractional surface area of tumor with maturing squamous differentiation as well as the fractional surface area of tumor consisting of desmoplastic, fibrocellular stroma (excluding standard normal stroma that may have been within the invasive tumor focus) were estimated in increments of 5% generating a result between 0 and the maximum fraction of either feature (Fig. 2).
Fig. 2.
Histologic features such as fraction of maturing squamous differentiation and stromal desmoplasia. a Nonkeratinizing SCC with no maturing squamous differentiation (10X magnification). b Keratinizing type SCC with 100% maturing squamous differentiation (20X magnification). c Keratinizing type SCC with extensive stromal desmoplasia (10X magnification). d Nonkeratinizing SCC with no stromal desmoplasia (10X magnification)
Specimen Collection, Processing, DNA and RNA Isolation
Formalin fixed paraffin embedded (FFPE) tissue blocks were retrieved from surgical pathology archives and cut into 10 um sections on glass slides. Tumor rich areas were circled on a representative H&E slide with a dotting pen by one of the head and neck pathologists (JSL or MM) for macro-dissection. Then, total RNA was extracted from the identified tumor regions with the miRNeasy FFPE Kit (Qiagen Inc, Valencia, Calif) according to the manufacturer’s protocol. Total DNA was extracted from the same identified tumor regions using a QIAamp DNA FFPE Tissue Kit (Qiagen). In this way, we were able to focus on the profiling of the tumors with minimal contamination from adjacent normal tissues [24].
HPV Genome Sequencing
Extracted tumor DNA was sent to the Cancer Genomics Research Laboratory (Frederick, Maryland) for sequencing. A custom Thermo Fisher Ion Torrent AmpliSeq HPV16 panel approach was used to amplify the HPV16 genome as previously described [25]. This next-generation sequencing (NGS) assay uses Thermo Fisher Life Sciences’ Ion Torrent Proton in combination with a custom HPV16 Ion Ampliseq panel of 48 multiplexed primers designed to cover the entire viral genome for all HPV16 variant lineages (Thermo Fisher Scientific, Waltham, MA, Ampliseq HPV16 Custom White Glove assay, panel #WG00038_HPV16_2). A custom annotation database was used to annotate identified nucleotide variants to HPV gene/region. HPV16 was classified according to the 4 major evolutionary branches (A, B, C, D) and 10 sublineages (A1, A2, A3, A4, B1, C1, D1, D2, D3, D4) using a maximum likelihood (ML) tree topology [26]. In the event that multiple HPV16 variants were detected, the predominant variant was assigned based on presence of at least 60% of the sequence reads. Any specimens with poor read depth or incomplete coverage across the genome was excluded.
Expression Profiling of HPV and Functional-Related Human Genes Using RT-PCR
HPV RT-qPCR assays were used to profile the expression of HPV E6 and E7 in OPSCC with total RNA extracted from the tumor blocks as previously published [24] (Supplemental Table 1). All oligo primers in the assays were purchased from Sigma-Aldrich. Reverse transcription (RT) reaction was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was then performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and 500 nM HPV type-specific primers. HPV assays (profiling E6 and E7 transcripts separately from each of the 13 HPV types) were individually performed on a 384-well PCR plate. The PCR running protocol was 95 ℃ for 10 min, followed by 36 cycles of amplification (95 ℃ for 10 s, 58 ℃ for 15 s and 60 ℃ for 15 s).
Statistical Analysis
The significance of the associations between HPV status (positive versus negative), HPV16 lineage, and HPV16 sublineage, and morphologic attributes were assessed by Pearson contingency table Chi-squared statistics. These attributes included histologic subtypes, mitotic index, stromal desmoplasia, maturing squamous differentiation, and the presence of anaplasia or multinucleation. These statistics were also used to assess how HPV positivity was associated with these morphologic attributes.
Results
Overall results for the entire cohort and for the HPV-positive and HPV-negative cohorts are presented in Table 1. As is typical of other United States OPSCC cohorts, the majority (86.5%) of patients were high-risk HPV-positive. As expected, HPV16 was the overwhelmingly most common followed by a handful of others, predominantly HPV33 and HPV35. Interestingly, we had no HPV18 positive tumors. Most of the HPV16 patients were lineage A, and most of these were the sublineage A1 or A2. The most common HPV16 non-A sublineage was D3.
Table 1.
All results for HPV type, subtype, and sublineage
| Frequency | % | |
|---|---|---|
| HPV type | ||
| 16 | 211 | 94.2 |
| Other | 12 | 5.4 |
| Combined (16 + other) | 1 | 0.5 |
| HPV Type (Others) | ||
| 16 | 212 | 94.6 |
| 33 | 5 | 2.2 |
| 35 | 5 | 2.2 |
| 26 | 2 | 0.9 |
| HPV 16 Lineage | ||
| A | 191 | 90.1 |
| B | 1 | 0.5 |
| C | 4 | 1.9 |
| D | 16 | 7.5 |
| HPV 16 Sublineage | ||
| A1 | 112 | 53.3 |
| A2 | 63 | 30.0 |
| A3 | 3 | 1.4 |
| A4 | 14 | 6.7 |
| C | 1 | 0.5 |
| D1 | 1 | 0.5 |
| D3 | 13 | 6.7 |
| D4 | 2 | 1.0 |
Most of the tumors were nonkeratinizing (52.1%), and this nonkeratinizing morphology was strongly associated with high-risk HPV-positivity (97.1%). A small fraction were keratinizing (or conventional) type SCC (12.4%) of which 28.1% were high-risk HPV-positive. The other most common morphology was the mixed or hybrid category, so-called “nonkeratinizing SCC with maturation” (23.2%), and 90.0% of these were high-risk HPV-positive. Thus, nonkeratinizing morphology, including both strictly nonkeratinizing and nonkeratinizing with maturation together, was strongly associated with the presence of transcriptionally-active HPV (187/197, 94.9%; p < 0.001) (Table 2). Specific SCC subtypes constituted a minority of cases (30/259, 11.6%), and, as a group, were strongly associated with high-risk HPV (93.3%). Among the uncommon subtypes, all but one was associated with HPV16, and all but three of these were HPV16 A, sublineage A1 or A2, essentially matching the distribution of HPV16 lineages for the overall cohort.
Table 2.
Morphologic types and features by high risk HPV status (positive versus negative)
| HPV positive | HPV negative | Entire cohort (%) | p value | |
|---|---|---|---|---|
| All patients | 224 (86.5) | 35 (13.5) | 259 | |
| Histologic type | ||||
| Conventional | ||||
| Nonkeratinizing | 133 (59.4) | 4 (11.4) | 137 (52.9) | < 0.001* |
| Nonkeratinizing with maturation | 54 (22.3) | 6 (17.1) | 60 (23.2) | 0.26 |
| Keratinizing | 9 (4.0) | 23 (65.7) | 32 (12.4) | < 0.001* |
| Subtypes | ||||
| Adenosquamous (Including Ciliated) | 6 (2.7) | 0 | 6 (2.3) | 0.24** |
| Lymphoepithelial | 7 (3.1) | 1 (2.9) | 8 (3.1) | |
| Basaloid | 3 (1.3) | 0 | 3 (1.2) | |
| Papillary | 12 (5.4) | 1 (2.9) | 13 (5.0) | |
| Anaplasia present? | 53 (23.7) | 10 (28.6) | 63 (24.3) | 0.53 |
| Multinucleation present? | 87 (38.8) | 6 (17.1) | 93 (35.9) | 0.013 |
| Anaplasia and/or Multinucleation present? | 105 (46.9) | 13 (37.1) | 118 (45.6) | 0.28 |
| Multinucleation Index (MNI) (Average) | 5.0 | 2.7 | 4.7 | |
| MNI 1 or more | 123 (54.9) | 15 (42.9) | 112 (43.2) | 0.88 |
| MNI 3 or more | 93 (41.5) | 7 (20.0) | 100 (38.6) | 0.98 |
| MNI 11 or more | 34 (15.1) | 3 (8.6) | 37 (14.3) | 0.34 |
| Stromal desmoplasia (%) (Average) | 8.6% | 18.0% | 9.8% | |
| 1% or more | 137 (61.1) | 31 (88.6) | 168 (64.9) | < 0.001 |
| 11% or more | 54 (24.1) | 17 (48.6) | 71 (27.4) | 0.001 |
| 31% or more | 12 (5.4) | 5 (14.2) | 17 (6.6) | 0.035 |
| Maturing squamous differentiation (%) (Average) | 16.2% | 75.4% | 24.2% | |
| 1% or more | 165 (73.7) | 32 (91.4) | 197 (76.1) | 0.022 |
| 11% or more | 67 (29.9) | 30 (85.7) | 97 (37.5) | < 0.0001 |
| 31% or more | 36 (16.1) | 26 (74.3) | 62 (23.9) | < 0.0001 |
| 51% or more | 23 (10.3) | 26 (74.3) | 49 (18.9) | < 0.0001 |
*Value for nonkeratinizing vs nonkeratinizing with maturation vs keratinizing
**Value for all subtypes combined vs all conventional SCC types
HPV-positive carcinomas were strongly associated with certain histologic features, largely because these are the ones that help define (and differentiate) keratinizing and nonkeratinizing SCC. HPV-negative tumors were statistically significantly more likely to have stromal desmoplasia (p < 0.001), maturing squamous differentiation (p = 0.022), and multinucleation (binary yes/no), although the latter was not statistically significant (p = 0.88) (Table 2).
Among the HPV-positive patients, there was no association between HPV type and any of the histologic features (Table 3). We did find, albeit in very small numbers, that keratinizing SCC was more likely than the other subtypes to be non-HPV16 related. Further, although not statistically significantly different, we did find that all but one of the major SCC subtypes, considered collectively, were HPV16 positive. There was no association between the HPV16 lineages (A-D) nor with the sublineages, histologic features, or SCC subtypes, either (Tables 4 and 5).
Table 3.
Morphologic types and features by high risk HPV type
| HPV16 | Non HPV16 | p value | |
|---|---|---|---|
| All patients | 212 (94.6) | 12 (5.4) | |
| Histologic type | |||
| Conventional | |||
| Nonkeratinizing | 128 (60.4) | 6 (50.0) | 0.25* |
| Nonkeratinizing with maturation | 51 (24.1) | 3 (25.0) | 0.77* |
| Keratinizing | 7 (3.3) | 2 (16.7) | 0.036* |
| Subtypes | |||
| Adenosquamous (Including Ciliated) | 6 (2.8) | 0 | 0.90** |
| Lymphoepithelial | 7 (3.3) | 0 | |
| Basaloid | 3 (1.4) | 0 | |
| Papillary | 11 (5.2) | 1 (8.3) | |
| Anaplasia present? | 50 (23.6) | 3 (25.0) | 0.77 |
| Multinucleation present? | 83 (39.2) | 4 (33.3) | 0.73 |
| Anaplasia and/or Multinucleation present? | 99 (46.7) | 6 (50.0) | 0.45 |
| Multinucleation Index (MNI) (Average) | |||
| MNI 1 or more | 117 (55.2) | 6 (50.0) | 0.51 |
| MNI 3 or more | 88 (41.5) | 5 (41.7) | 0.39 |
| MNI 11 or more | 31 (14.6) | 3 (25.0) | 0.39 |
| Stromal desmoplasia (%) (Average) | |||
| 1% or more | 129 (60.8) | 7 (58.3) | 0.61 |
| 11% or more | 51 (24.1) | 2 (16.7) | 0.20 |
| 31% or more | 10 (4.7) | 2 (16.7) | 0.11 |
| Maturing squamous differentiation (%) (Average) | |||
| 1% or more | 157 (74.1) | 8 (66.7) | 0.22 |
| 11% or more | 62 (29.2) | 5 (41.7) | 0.30 |
| 31% or more | 33 (15.6) | 3 (25.0) | 0.43 |
| 51% or more | 20 (9.4) | 3 (25.0) | 0.10 |
*Value for nonkeratinizing vs nonkeratinizing with maturation vs keratinizing
**Value for all variants combined vs all conventional SCC types
Table 4.
Morphologic types and features by high risk HPV 16 lineage
| A | B | C | D | p value* | |
|---|---|---|---|---|---|
| All patients | 191 | 1 | 4 | 16 | |
| Histologic type | |||||
| Conventional | |||||
| Nonkeratinizing | 116 (60.7) | 0 | 1 (25.0) | 11 (64.7) | 0.57 |
| Nonkeratinizing with maturation | 44 (23.0) | 1 | 2 (50.0) | 3 (18.8) | 0.33 |
| Keratinizing | 7 (3.7) | 0 | 0 | 0 | 0.36 |
| Subtypes | |||||
| Adenosquamous (Including Ciliated) | 6 (3.1) | 0 | 0 | 0.89 | |
| Lymphoepithelial | 6 (3.1) | 0 | 1 (25.0) | 0 | |
| Basaloid | 2 (1.0) | 0 | 0 | 1 (5.9) | |
| Papillary | 10 (5.2) | 0 | 0 | 1 (5.9) | |
| Anaplasia present? | 48 (25.1) | 0 | 1 (25.0) | 1 (5.9) | 0.09 |
| Multinucleation present? | 76 (39.8) | 0 | 2 (50.0) | 5 (29.4) | 0.47 |
| Anaplasia and/or Multinucleation present? | 89 (46.6) | 0 | 3 (75.0) | 7 (41.2) | 0.92 |
| Multinucleation index (MNI) (Average) | |||||
| MNI 1 or more | 104 (54.5) | 0 | 4 (100.0) | 9 (52.9) | 0.76 |
| MNI 3 or more | 81 (42.4) | 0 | 2 (50.0) | 5 (29.4) | 0.30 |
| MNI 11 or more | 28 (14.7) | 0 | 2 (50.0) | 1 (5.9) | 0.87 |
| Stromal desmoplasia (%) (Average) | |||||
| 1% or more | 115 (60.2) | 1 | 3 75.0) | 10 (58.8) | 0.76 |
| 11% or more | 46 (24.1) | 1 | 1 (25.0) | 3 (17.6) | 0.89 |
| 31% or more | 9 (4.7) | 0 | 1 (25.0) | 0 | 0.97 |
| Maturing squamous differentiation (%) (Average) | |||||
| 1% or more | 141 (73.8) | 1 | 3 (75.0) | 12 (70.6) | 0.91 |
| 11% or more | 54 (28.3) | 1 | 2 (50.0) | 5 (29.4) | 0.43 |
| 31% or more | 28 (14.7) | 1 | 0 | 3 (17.6) | 0.32 |
| 51% or more | 17 (8.9) | 1 | 0 | 2 (11.8) | 0.47 |
*Value for HPV 16 subtype A vs other subtypes
Table 5.
Morphologic types and features by high risk HPV type 16 sublineage
| A1 | A2 | A3 | A4 | C | D3 | Any A | p value* | |
|---|---|---|---|---|---|---|---|---|
| All patients | 112 | 62 | 3 | 14 | 4 | 13 | 191 | |
| Histologic Type | ||||||||
| Conventional | ||||||||
| Nonkeratinizing | 62 (55.4) | 41 (66.1) | 2 (66.7) | 10 (71.4) | 1 (25.0) | 9 (69.2) | 115 (60.2) | 0.57 |
| Nonkeratinizing with maturation | 29 (25.9) | 11 (17.5) | 1 (33.3) | 4 (28.6) | 2 (50.0) | 3 (23.1) | 43 (22.5) | 0.38 |
| Keratinizing | 6 (5.4) | 1 (1.6) | 0 | 0 | 0 | 0 | 7 (3.7) | 1.0 |
| Subtypes | ||||||||
| Adenosquamous (Including Ciliated) | 6 (5.4) | 0 | 0 | 0 | 0 | 0 | 6 (3.1) | 1.0** |
| Lymphoepithelial | 3 (2.7) | 3 (4.8) | 0 | 0 | 1 (25.0) | 0 | 6 (3.1) | |
| Basaloid | 0 | 2 (3.2) | 0 | 0 | 0 | 1 (7.7) | 2 (1.0) | |
| Papillary | 6 (5.4) | 4 (6.3) | 0 | 0 | 0 | 0 | 10 (5.2) | |
| Anaplasia present? | 25 (22.3) | 19 (30.2) | 2 (66.7) | 2 (14.3) | 1 (25.0) | 1 (7.7) | 48 (25.1) | 0.25 |
| Multinucleation present? | 41 (36.6) | 27 (42.9) | 2 (66.7) | 6 (4.3) | 2 (50.0) | 5 (38.5) | 76 (39.8) | 1 |
| Anaplasia and/or Multinucleation present? | 49 (43.8) | 32 (50.8) | 2 (66.7) | 6 (4.3) | 3 (75.0) | 7 (53.8) | 89 (46.6) | 0.47 |
| Multinucleation Index (MNI) (Average) | 4.5% | 5.2% | 13.7% | 7% | 9.8% | 2.6% | 5% | |
| MNI 1 or more | 58 (51.8) | 36 (57.1) | 2 (66.7) | 8 (57.1) | 4 | 9 (69.2) | 104 (54.5) | 0.21 |
| MNI 3 or more | 43 (38.4) | 29 (46.0) | 2 (66.7) | 7 (0.5) | 2 (50.0) | 5 (38.5) | 81 (42.4) | 1 |
| MNI 11 or more | 12 (10.7) | 10 (15.9) | 2 (66.7) | 4 (28.6) | 2 (50.0) | 1 (7.7) | 28 (14.7) | 0.73 |
| Stromal desmoplasia (%) (Average) | 8.5% | 8.7% | 8.3% | 5.8% | 12.5% | 6.8% | 8.4% | |
| 1% or more | 60 (53.6) | 44 (69.8) | 2 (66.7) | 10 (71.4) | 3 (75.0) | 9 (69.2) | 116 (60.7) | 0.8 |
| 11% or more | 26 (23.2) | 18 (28.6) | 1 (33.3) | 2 (14.3) | 1 (25.0) | 3 (23.1) | 47 (24.6) | 1 |
| 31% or more | 8 (7.1) | 1 (1.6) | 0 | 0 | 1 (25.0) | 0 | 9 (4.7) | 0.6 |
| Maturing squamous differentiation (%) (Average) | 18.6% | 9.3% | 2% | 14.6% | 10% | 19.5% | 15% | |
| 1% or more | 83 (74.1) | 46 (73.0) | 2 (66.7) | 10 (71.4) | 3 (75.0) | 10 (76.9) | 141 (73.8) | 1 |
| 11% or more | 39 (34.8) | 12 (19.0) | 0 | 3 (21.4) | 2 (50.0) | 5 (38.5) | 54 (28.3) | 0.42 |
| 31% or more | 22 (19.6) | 3 (4.8) | 0 | 3 (21.4) | 0 | 4 (30.8) | 28 (14.7) | 0.49 |
| 51% or more | 13 (11.6) | 2 (3.2) | 0 | 2 (14.3) | 0 | 2 (15.4) | 17 (8.9) | 0.67 |
*Value for HPV 16 A sublineage As vs other sublineages
**Value for all variants combined vs all conventional SCC types
Discussion
As the latest anatomic subsite for SCC to be found to be strongly associated with transcriptionally-active high-risk HPV, oropharyngeal SCC has shown itself to be similar to cervical, anal, and vaginal SCC in many ways while different in key others. In cervical SCC, HPV16 constitutes approximately 60% of high-risk HPV, and HPV18 constitutes approximately 10%, followed by between 2 and 6% for many other high risk types, such as 31, 33, 35, etc. In oropharyngeal SCC, HPV16 accounts for 90 to 95% of high-risk HPV, HPV18 constitutes for 2% or less, and HPV33 for 3 to 5% [8–10], 27. Other high risk HPV types are rare in oropharyngeal SCC, constituting 2% or less each [24]. Anal SCC appears to have a distribution of high-risk HPV types more similar to oropharyngeal SCC [28]. In cervical SCC, HPV18 is modestly associated with adenocarcinoma morphology [9, 10], and there are many large studies examining HPV types and various features of patients and patient’s tumors [8–10]. There have been few studies of oropharyngeal SCC examining the association of HPV genotype or lineage with SCC type [27], degree of keratinization, or other morphologic features.
We found that patients with high-risk HPV related tumors had less keratinization, less stromal desmoplasia, and slightly less tumor cell multinucleation than those with HPV negative tumors, but found no other associations with HPV positivity, HPV type, HPV16 lineage, or amongst the various HPV16 A sublineages. Rare subtypes of oropharyngeal SCC, such as ciliated adenosquamous, basaloid, and lymphoepithelial were just too uncommon to make any clear assessments about associations with specific HPV type or HPV16 lineage/sublineage. We did find that all ciliated adenosquamous carcinomas, a peculiar subtype where there is SCC but also gland foci with well-formed ciliated tumor cells, were associated with high-risk HPV16 lineage A, sublineage A1. This may simply be a function of small numbers, however. Further study would be needed and in larger numbers to draw any reliable conclusions.
What does any of this potentially mean for patient care? It does provide further evidence that HPV16, and particularly HPV16 sublineages A1 and A2, are the major drivers of oropharyngeal SCC, at least in the United States. This data also further helps confirm that SCC histologic subtypes are uncommon and, despite looking quite different, don’t have a different HPV distribution than more conventional SCC types. HPV16 sublineages B, C, and D are just so uncommon that to be established as significant for patient care will take very large, very well designed studies in determine. However, since we know HPV16 lineages/sublineage vary based on human race/ethnicity [5], 25, the predominance of the European HPV16 A1/A2 sublineages here is not unexpected in this predominantly ‘white’ population. It is possible that a more racially diverse population with oropharyngeal SCC would allow a more thorough evaluation of the other HPV16 lineages/sublineages.
In summary, in our 259 patient oropharyngeal SCC cohort, using RTPCR for HPV status and type, DNA sequencing of the entire HPV16 genome, and detailed morphologic assessment including features of keratinization, anaplasia/multinucleation, and stromal desmoplasia, we found that HPV-positive tumors were statistically significantly more likely to be nonkeratinizing, have low maturing squamous differentiation, and have lower degrees of stromal desmoplasia. For HPV-positive tumors, we found no significant associations between HPV type, HPV16 lineage, or HPV16 sublineage with histologic features or SCC type. It appears that all high-risk HPV types and HPV16 lineages/sublineages can generate the variety of SCC morphologic types.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to acknowledge the Translational Pathology Shared Resource (TPSR) at Vanderbilt University Medical Center which is supported by NCI/NIH Cancer Center Support Grant 5P30 CA68485-19 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant 2 U24 DK059637-16. This work was also supported by funds from the NIH Grant R01DE026471 (Wang) and by funds from the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH (Mirabello).
Funding
Research was performed using the Translational Pathology Shared Resource (TPSR) at Vanderbilt University Medical Center which is supported by NCI/NIH Cancer Center Support Grant 5P30 CA68485-19 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant 2 U24 DK059637-16. This work was supported by funds from the NIH grant R01DE026471 (Wang), and also by funds from the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH (Mirabello).
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest as it relates to this research.
Ethical Approval
This study was performed with approval of the respective institutional review boards and complies with required ethical standards. Given the retrospective nature of the study, patients were never contacted, and we did not require informed consent.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Instit. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, Chaturvedi AK, Anderson WF, et al. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol: Off J Am Soc Clin Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Instit. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 5.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirabello L, Clarke MA, Nelson CW, et al. The Intersection of HPV epidemiology, genomics and mechanistic studies of HPV-mediated carcinogenesis. Viruses. 2018;10:80. doi: 10.3390/v10020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirabello L, Yeager M, Cullen M, et al. HPV16 sublineage associations with histology-specific cancer risk using hpv whole-genome sequences in 3200 women. J Natl Cancer Inst. 2016;108. [DOI] [PMC free article] [PubMed]
- 8.Coutlee F, Ratnam S, Ramanakumar AV, et al. Distribution of human papillomavirus genotypes in cervical intraepithelial neoplasia and invasive cervical cancer in Canada. J Med Virol. 2011;83:1034–1041. doi: 10.1002/jmv.22081. [DOI] [PubMed] [Google Scholar]
- 9.Clifford GM, Smith JS, Plummer M, et al. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88:63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Insinga RP, Liaw KL, Johnson LG, et al. A systematic review of the prevalence and attribution of human papillomavirus types among cervical, vaginal, and vulvar precancers and cancers in the United States. Cancer Epidemiol Biomarkers Prev. 2008;17:1611–1622. doi: 10.1158/1055-9965.EPI-07-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Sullivan B, Lydiatt WM, Haughey BH, et al. HPV-Mediated (p16+) Oropharyngeal Cancer. In: Amin MB, ed. AJCC Cancer Staging Manual. Switzerland: Springer Nature; 2016.
- 12.Danciu I, Cowan JD, Basford M, et al. Secondary use of clinical data: the vanderbilt approach. J Biomed Inform. 2014;52:28–35. doi: 10.1016/j.jbi.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO Classification of Head and Neck Tumours. In: El-Naggar A, Chan JKC, Grandis JR, et al., eds. Lyon, France: IARC Press; 2017.
- 14.Chernock RD, El-Mofty SK, Thorstad WL, Parvin CA, Lewis JS., Jr HPV-related nonkeratinizing squamous cell carcinoma of the oropharynx: utility of microscopic features in predicting patient outcome. Head Neck Pathol. 2009;3:186–194. doi: 10.1007/s12105-009-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gondim DD, Haynes W, Wang X, et al. Histologic typing in oropharyngeal squamous cell carcinoma: a 4-year prospective practice study with p16 and high-risk HPV mrna testing correlation. Am J Surg Pathol. 2016;40:1117–1124. doi: 10.1097/PAS.0000000000000650. [DOI] [PubMed] [Google Scholar]
- 16.Chernock RD. Morphologic features of conventional squamous cell carcinoma of the oropharynx: 'keratinizing' and 'nonkeratinizing' histologic types as the basis for a consistent classification system. Head Neck Pathol. 2012;6(Suppl 1):S41–47. doi: 10.1007/s12105-012-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis JS, Jr, Khan RA, Masand RP, et al. Recognition of nonkeratinizing morphology in oropharyngeal squamous cell carcinoma - a prospective cohort and interobserver variability study. Histopathology. 2012;60:427–436. doi: 10.1111/j.1365-2559.2011.04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardesa A, Zidar N, Alos L. Adenosquamous carcinoma. In: Barnes L, Eveson J, Reichart P, Sidransky D, editors. Pathology and Genetics Head and Neck Tumours. Lyon, France: IARC Press; 2005. pp. 130–131. [Google Scholar]
- 19.Cardesa A, Zidar N, et al. Spindle cell carcinoma. In: Barnes EL, Eveson JW, Reichart P, et al., editors. World Health Organization Pathology and Genetics - Head and Neck Tumours. Lyon, France: IARC Press; 2005. pp. 127–128. [Google Scholar]
- 20.Cardesa A, Zidar N, Ereno C, et al. Basaloid squamous cell carcinoma. In: Barnes EL, Eveson JW, Reichart P, et al., editors. World Health Organization Classification of Tumours - Pathology and Genetics of Head and Neck Tumours. Lyon, France: IARC Press; 2005. pp. 125–126. [Google Scholar]
- 21.Cardesa A, Zidar N, Nadal A, et al. Papillary squamous cell carcinoma. In: Barnes L, eveson JW, Reichart P, et al., eds. World Health Organization Classification of Tumours - Pathology and Genetics Head and Neck Tumours. Lyon, France: IARC Press. 2005;126.
- 22.Singhi AD, Stelow EB, Mills SE, et al. Lymphoepithelial-like carcinoma of the oropharynx: a morphologic variant of HPV-related head and neck carcinoma. Am J Surg Pathol. 2010;34:800–805. doi: 10.1097/PAS.0b013e3181d9ba21. [DOI] [PubMed] [Google Scholar]
- 23.Lewis JS, Jr, Scantlebury JB, Luo J, et al. Tumor cell anaplasia and multinucleation are predictors of disease recurrence in oropharyngeal squamous cell carcinoma, including among just the human papillomavirus-related cancers. Am J Surg Pathol. 2012;36:1036–1046. doi: 10.1097/PAS.0b013e3182583678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao G, Chernock RD, Gay HA, et al. A novel RT-PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int J Cancer J Int du Cancer. 2013;132:882–890. doi: 10.1002/ijc.27739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen M, Boland JF, Schiffman M, et al. Deep sequencing of HPV16 genomes: a new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res. 2015;1:3–11. doi: 10.1016/j.pvr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 27.Jordan RC, Lingen MW, Perez-Ordonez B, et al. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol. 2012;36:945–954. doi: 10.1097/PAS.0b013e318253a2d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baricevic I, He X, Chakrabarty B, et al. High-sensitivity human papilloma virus genotyping reveals near universal positivity in anal squamous cell carcinoma: different implications for vaccine prevention and prognosis. Eur J Cancer. 2015;51:776–785. doi: 10.1016/j.ejca.2015.01.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.