Abstract
The Burkholderia cepacia complex currently comprises five genomic species, i.e., B. cepacia genomovar I, B. multivorans (formerly known as B. cepacia genomovar II), B. cepacia genomovar III, B. cepacia genomovar IV, and B. vietnamiensis (also known as B. cepacia genomovar V). In the absence of straightforward diagnostic tests for the identification of B. cepacia genomovars I, III, and IV, the last two genomic species were not formally classified as novel Burkholderia species (genomovar I contains the type strain and therefore retains the name B. cepacia). In the present study, we describe differential biochemical tests and a recA gene-based PCR assay for the routine identification of strains currently known as B. cepacia genomovar IV and propose formal classification of this organism as Burkholderia stabilis sp. nov. B. stabilis can indeed be differentiated from all other B. cepacia complex strains by the absence of beta-galactosidase activity, from strains of B. cepacia genomovars I and III and B. vietnamiensis by the inability to oxidize sucrose, and from B. multivorans by the lack of growth at 42°C. In addition, analysis with the recA gene-derived primers BCRG41 (5′-ACCGGCGAGCAGGCGCTT-3′) and BCRG42 (5′-ACGCCATCGGGCATGGCA-3′) specifically allows the detection of B. stabilis strains in a conventional PCR assay. Examination of a set of 21 B. stabilis strains by means of random amplified polymorphic DNA analysis and pulsed-field gel electrophoresis typing suggested that the genome of this organism is highly conserved, which is in sharp contrast to the generally accepted genomic diversity, variability, and plasticity among B. cepacia strains.
Accurate species-level identification of Burkholderia strains is often a tedious process. The discovery of a variety of novel species, new taxonomic insights, and the peculiar genomic characteristics of these organisms present diagnostic laboratories with a manifold of problems. Driving forces behind this rapid evolution are the biotechnological interest in biocontrol and bioremediation applications of Burkholderia-like organisms, their role as plant pathogens, and, not the least, their role as significant pathogens for particular patient groups such as cystic fibrosis patients (7). A study of the taxonomy of Burkholderia cepacia-like organisms revealed the complex nature of this organism (23). The name B. cepacia complex was proposed to comprise a cluster of five closely related species, originally referred to as B. cepacia genomovars I through V (23) (the term genomovar was introduced to denote phenotypically similar but genotypically distinct groups of strains [22]). Apart from genomovar I, which contains the type strain of B. cepacia, two of these genomovars have been formally named (Burkholderia multivorans and Burkholderia vietnamiensis for strains previously known as B. cepacia genomovars II and V, respectively); the others await assignment of a binomial species name pending the availability of distinguishing phenotypic identification criteria (23).
We recently reported on the application of a genomic fingerprinting technique, amplified fragment length polymorphism (AFLP) analysis, for the differentiation of members of the B. cepacia complex (5). In that study, we demonstrated that several strains classified as B. cepacia genomovar I by means of whole-cell protein electrophoresis (23) belonged, in fact, to B. cepacia genomovar IV. While examining additional putative B. cepacia strains, we repeatedly identified genomovar IV strains by AFLP analysis but not by whole-cell protein electrophoresis. In this report we describe effective procedures for the identification of B. cepacia genomovar IV strains and propose the name Burkholderia stabilis sp. nov. to accommodate B. cepacia genomovar IV strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Reference strains of B. cepacia genomovars I (13 strains) and III (3 strains) and of B. multivorans (3 strains), B. vietnamiensis (3 strains), and Burkholderia gladioli (3 strains), have been described in previous reports (5, 23). Three Burkholderia pseudomallei reference strains were obtained from the CCUG Culture Collection (Culture Collection of the Department of Clinical Bacteriology, University of Göteborg, Göteborg, Sweden). All 21 B. stabilis strains and their sources are listed in Table 1.
TABLE 1.
B. stabilis strains studied
| Strain | Other strain no.a | Source (country, yr of isolation) |
|---|---|---|
| LMG 6997 | CCUG 3461B | Ear (Sweden, 1974) |
| LMG 7000 | CCUG 13348 | Blood (Sweden, 1983) |
| LMG 13017 | Blood culture (Belgium, 1988) | |
| LMG 14086 | Respirator (United Kingdom, 1970) | |
| LMG 14291 | Cystic fibrosis patient (Belgium, 1993) | |
| LMG 14294T | NCTC 13011 | Cystic fibrosis patient (Belgium, 1993) |
| LMG 14295 | Cystic fibrosis patient (Belgium, 1993) | |
| LMG 14940 | Cystic fibrosis patient (Belgium, 1994) | |
| LMG 15949 | Cystic fibrosis patient (Belgium, 1993) | |
| LMG 15950 | Water bath, cystic fibrosis ward (Belgium, 1994) | |
| LMG 15951 | Spirometer (Belgium, 1994) | |
| LMG 18138 | E20 | Cystic fibrosis patient (Belgium, 1995) |
| R-3338 | M71-40 | Cystic fibrosis patient (Germany, 1997) |
| R-6617 | J687 | Human, non-cystic fibrosis patient (France) |
| R-6618 | J762 | Urine (United States) |
| LMG 18888 | HK 268a | Human blood (Belgium, 1995) |
| R-136 | J1750 | Cystic fibrosis patient (United States before 1989) |
| R-4059 | H107 | Cystic fibrosis patient (Germany, 1993) |
| R-732 | C6061 | Cystic fibrosis patient (Canada, 1994) |
| R-737 | CEP059 | Respiratory tract, non-cystic fibrosis patient (Canada) |
| R-741 | CEP194, J668 | Human, non-cystic fibrosis patient (Switzerland) |
CCUG, Culture Collection of the Department of Clinical Bacteriology, University of Göteborg, Göteborg, Sweden; LMG, Laboratorium Microbiologie Gent Culture Collection, Universiteit Gent, Ghent, Belgium; NCTC, National Collection of Type Cultures, London, United Kingdom.
Bacteriological purity was checked by plating and examining living cells by phase-contrast microscopy and Gram staining of cells. Strains were grown on nutrient agar (Oxoid CM3 agar) supplemented with 0.04% (wt/vol) KH2PO4 and 0.24% (wt/vol) Na2HPO4 · 12H2O (pH 6.8) and incubated aerobically at 28°C unless indicated otherwise.
AFLP fingerprinting.
DNA was isolated and purified as described by Pitcher et al. (15). AFLP analysis was performed as described by Coenye et al. (5). Preparation of template DNA for PCR analysis, preselective and selective PCR amplification with 6-carboxyfluorescein-labeled primers, separation of amplified fragments on an ABI Prism 377 DNA Sequencer, and data capture and analysis with GeneScan (version 2.1; Perkin-Elmer Applied Biosystems) and GelCompar (version 4.2; Applied Maths, Kortrijk, Belgium) software were performed as described previously (5).
PAGE of whole-cell proteins.
All reference strains and B. stabilis strains were included in the sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis. After an incubation period of 48 h, whole-cell protein extracts were prepared and SDS-PAGE was performed as described before (16). The densitometric analysis, normalization and interpolation of the protein profiles, and numerical analysis were performed with the GelCompar software package (version 4.2; Applied Maths). Similarity levels between the patterns were calculated by using the Pearson product moment correlation coefficient and are expressed as percent similarity for convenience. Data for the reference strains of B. cepacia genomovars I, II (B. multivorans), III, and V (B. vietnamiensis) and for B. gladioli were generated in a previous study (23).
Conventional biochemical tests.
The following tests were performed with all 21 B. stabilis strains listed in Table 1 as described previously (8). Briefly, pure cultures were stored at −70°C in Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) with 8% dimethyl sulfoxide. Frozen isolates were subcultured onto Columbia agar containing 5% sheep blood (PML Microbiologicals, Richmond, British Columbia, Canada) before testing. The primary identification system used was the API Rapid NE system (Biomerieux Vitek Inc., Hazelwood, Mo.) supplemented with glucose, maltose, lactose, xylose, sucrose, and adonitol oxidation-fermentation (OF) sugars (10) and an adaptation of Moeller lysine and ornithine decarboxylases (Difco) (the 2-ml test volume was overlaid with 0.5 ml of oil and a heavy, visible inoculum was used; a tube without amino acid was included as a negative control). The ability to grow on BCSA agar (8), indicating resistance to gentamicin and polymyxin, was tested. Incubation was at 35°C in ambient air; tubes with OF sugars were incubated for up to 7 days, and the tubes used for the other tests were incubated for 2 days. Any organism that was P-nitrophenyl-β-d-glucoside (PNPG) negative on the API strip was tested for the presence of beta-galactosidase (o-nitrophenyl-β-d-galactopyranoside [ONPG] test) (1). In addition, growth on tryptic soy agar (Becton Dickinson and Co., Cockeysville, Md.) at 35 and 42°C was observed for appearance and pigmentation.
In addition, a range of 68 conventional biochemical tests was performed by previously described methods (9) with eight B. stabilis strains (strains LMG 6997, LMG 7000, LMG 14086, LMG 14291, LMG 14294, LMG 14295, LMG 14940, and LMG 15949) as reported in a previous study (23). Except for the ONPG test, data for B. cepacia genomovars I and III, B. multivorans, and B. vietnamiensis were taken from a previous study (23). Unpublished data from D. Henry showed that strains from these reference species all exhibit beta-galactosidase activity.
PCR assay.
Assays for the identification of B. cepacia complex genomovars were based on the gene encoding RecA and are described in detail in a separate report (E. Mahenthiralingam, J. M. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme, submitted for publication). The nucleotide sequences of the entire recA genes from B. stabilis LMG 7000 and LMG 14291 were determined. These sequences were aligned by using the CLUSTAL W algorithm (21) with the sequences of recA genes from strains representative of B. cepacia genomovars I and III, B. multivorans, and B. vietnamiensis. This alignment facilitated the design of PCR primers BCRG41 (5′-ACCGGCGAGCAGGCGCTT-3′) and BCRG42 (5′-ACGCCATCGGGCATGGCA-3′) which were specific to the B. stabilis recA sequence and which were mismatched at the 3′ base with recA sequences from all remaining genomovars. Under standard PCR conditions (14) at an annealing temperature of 64°C, a product of 647 bp was specifically amplified from the B. stabilis strains. Strains of the other genomovars failed to produce a PCR product under these conditions (Mahenthiralingam et al., submitted).
Genetic typing.
Genetic typing of B. stabilis strains was performed both by random amplified polymorphic DNA (RAPD) analysis as described previously (13) and by pulsed-field gel electrophoresis (PFGE) fingerprinting. The PFGE method was adapted from that of Cheng and Lessie (3) and was performed as follows. After overnight growth in 5 ml of Luria-Bertani broth (19), the bacteria were harvested by centrifugation and were resuspended to an optical density at 620 nm of between 0.8 and 0.9 in SE buffer (75 mM NaCl, 25 mM EDTA [pH 7.4]). The suspension was warmed to 45°C for 5 min and was mixed with an equal volume of molten 2% low-melting-point agarose (Type 7; Sigma-Aldrich Canada, Oakville, Ontario, Canada) that was kept at the same temperature, and the mixture was poured into 70-μl disposable plug molds (Bio-Rad, Mississauga, Ontario, Canada). The plugs were briefly chilled to 4°C, and then three to five plugs were placed in 10 ml of PEN buffer (0.5 M EDTA [pH 9.6], 1% N-lauroyl sarcosine) containing 1 mg of pronase (Boehringer Mannheim, Laval, Quebec, Canada) per ml held within a 15-ml sterile tube. After 24 h of incubation with gentle rocking at 37°C, the plugs were washed with five volume changes (one per hour) of TE buffer (Tris-EDTA) (19). Slices (approximately 2 mm) were then cut from the plugs and were incubated overnight with 10 U of SpeI in a 150-μl digestion mixture at 37°C. The macrorestricted DNA was separated in 1.2% agarose gels made with 0.5× TBE buffer (Tris-borate-EDTA) (19) at 5 V/cm for 44 h, with pulse switch times ramped from 20 to 60 s according to the manufacturer's standard guidelines (CHEF-DR II apparatus; Bio-Rad). Bacteriophage lambda concatemers were included as size standards (Bio-Rad).
The normalization of the banding patterns that were obtained and the numerical analysis with the Pearson product moment correlation coefficient were performed with the GelCompar software package (version 4.2; Applied Maths).
16S rDNA sequence analysis.
Preparation of DNA, amplification of part of the rRNA gene (rDNA) operon comprising the nearly complete 16S DNA, and sequence analysis and assembly were performed as described before (4, 24).
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession number for the 16S rDNA sequence of strain LMG 14294 is AF148554. The nucleotide sequence accession numbers for the recA genes of B. stabilis strains LMG 7000 and LMG 14291 are AF143789 and AF143790, respectively.
RESULTS
AFLP fingerprinting.
The intergel reproducibility level was higher than 93%; the intragel correlation between patterns was higher than 95% (data not shown). Identification of strains was achieved by numerical analysis of the AFLP patterns of novel isolates with those of a well-characterized set of reference strains described by Coenye et al. (5) (data not shown).
PAGE of whole-cell proteins.
Duplicate protein extracts were prepared to check the reproducibility of the growth conditions and the preparation of the extracts. The correlation level between duplicate protein patterns was more than 93% (data not shown).
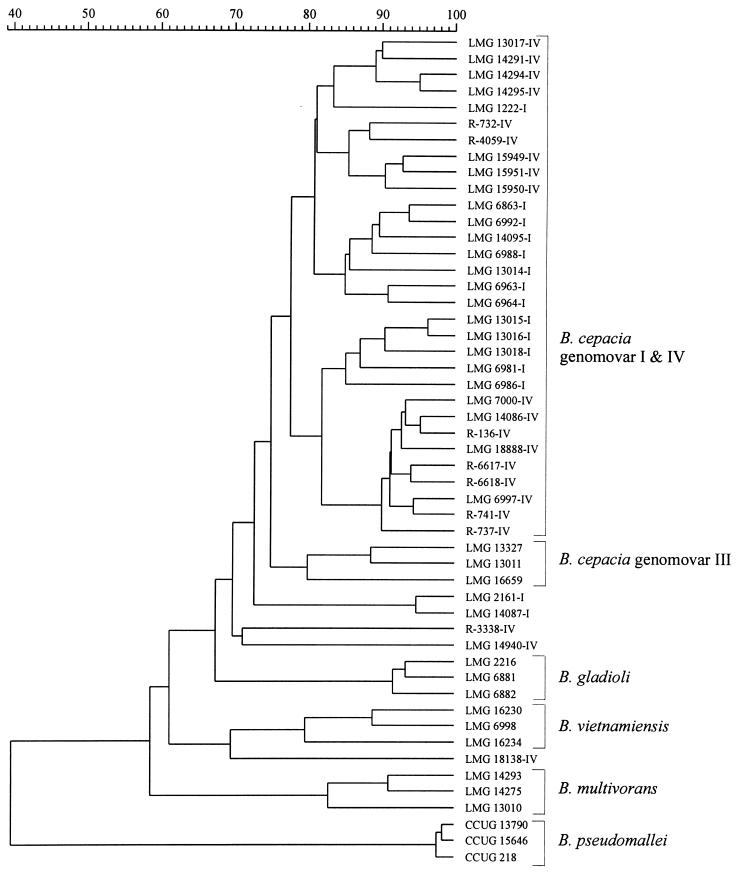
Figure 1 shows the result of the numerical analysis of the whole-cell protein patterns of reference strains of the various B. cepacia genomovars and of B. gladioli and B. pseudomallei. Strains of the last two species and of B. multivorans, B. vietnamiensis, and B. cepacia genomovar III each form well-delineated clusters. However, the majority of B. cepacia genomovar I and B. stabilis strains form a single heterogeneous cluster above a similarity level of 77.2%. In addition, some B. cepacia genomovar I strains (strains LMG 2161 and LMG 14087) and B. stabilis strains (strains LMG 14940, R-3338, and LMG 18138) occupy distinct positions in the dendrogram.
FIG. 1.
Dendrogram derived from the unweighted pair group average linkage of correlation coefficients between the protein patterns of all strains studied. Strain numbers with the suffix I or IV refer to B. cepacia genomovar I or B. stabilis strains, respectively.
Conventional biochemical tests.
The results of those tests that were performed with all 21 B. stabilis strains are as follows. All 21 B. stabilis strains were negative for nitrate reduction, ONPG, esculin hydrolysis, growth at 42°C, and sucrose oxidation. All 21 strains were positive for acidification of glucose, lactose, maltose, xylose, and adonitol, liquefaction of gelatin, and lysine and ornithine decarboxylation. All 21 strains demonstrated a weakly positive oxidase reaction. The API Rapid NE system gave profile numbers of 0046577 (for five strains that did not liquefy gelatin) or 0056577 (for the remaining strains that liquefied gelatin). Tube gelatin tests gave positive results for all except one (strain R-3338) of the strains that were negative with the API Rapid NE system. In the API identification database (API 20 NE, version 6), the identification result for strains with the profile 0056577 corresponded to a “good identification” score for B. cepacia, followed by Pseudomonas fluorescens as the second choice; the note “possibility of B. gladioli” was given, too. The same identification results were obtained for strains with profile 0046577, but with “low discrimination” instead of “good identification.” All strains grew on BCSA agar, indicating resistance to gentamicin and polymyxin. No production of pigments was observed.
The results of the 68 tests performed with the restricted set of eight strains are listed in the description of the B. stabilis strains given below.
PCR assay.
All of the B. stabilis strains tested reacted positively with the B. stabilis recA-specific primers. None of the strains representing other genomovars or Burkholderia species reacted positively. The PCR product of the correct size from strain LMG 7000 was confirmed to encode the recA gene by direct nucleotide sequence analysis (data not shown).
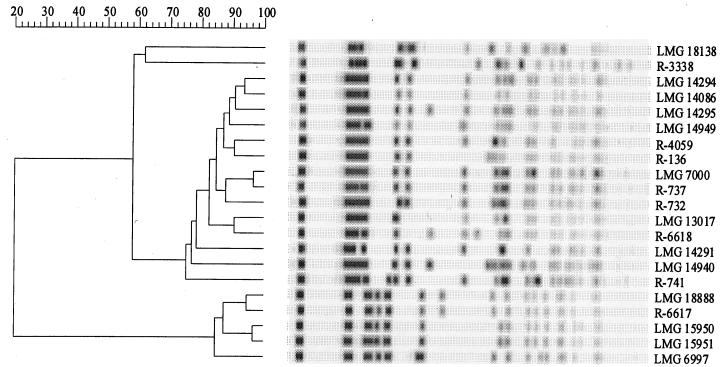
Genetic strain typing.
Reproducible and discriminatory genetic fingerprints were obtained for B. stabilis strains examined by RAPD analysis (data not shown) and PFGE. Figure 2 is a computer-based reproduction of the SpeI-PFGE profiles of all B. stabilis strains and the results of the corresponding numerical analysis of the banding patterns. Two main clusters with very similar banding patterns and two strains with unique profiles (strains LMG 18138 and R-3338) can be distinguished. RAPD fingerprinting identified the same strain clusters (data not shown).
FIG. 2.
Computer-generated SpeI-PFGE profiles of all of the B. stabilis strains and the corresponding numerical analysis of the banding patterns.
DISCUSSION
The B. cepacia complex currently comprises five genomic species originally referred to as B. cepacia genomovars I through V, respectively (23). One of these genomic species (genomovar V) was identified as B. vietnamiensis, a nitrogen-fixing bacterium originally isolated from the rice rhizosphere (6). Of the remaining four genomic species, only genomovar II could readily be differentiated from the others by the absence of sucrose utilization and variable lysine decarboxylase activity, which were typically present in other B. cepacia complex strains (23); this genomovar was named B. multivorans. In the absence of straightforward differential tests, the remaining genomic species were referred to as B. cepacia genomovars I, III, and IV (23).
Identification of B. stabilis strains.
Previous AFLP analyses and subsequent DNA-DNA hybridization experiments revealed that some of the strains used when searching for differential tests for the separation of B. cepacia genomovar I, III, and IV strains were misidentified, as some B. cepacia genomovar I strains were shown to be genomovar IV (5). Continued application of AFLP analysis as a first-line identification approach in a research laboratory identified a set of 21 strains as B. cepacia genomovar IV. Subsequent analyses of these strains by classical biochemical tests revealed that genomovar IV strains can be differentiated from other B. cepacia complex strains by the absence of the beta-galactosidase activity necessary for the breakdown of ONPG. In addition, the inability to oxidize sucrose separates them from strains of genomovars I and III and B. vietnamiensis but not from B. multivorans. The lack of growth at 42°C further separates B. cepacia genomovar IV strains from B. multivorans strains.
In addition, PCR technology offers alternative possibilities for the detection of organisms where species-level identification is particularly difficult. Bauernfeind et al. (2) recently described an assay that facilitated discrimination of B. multivorans and B. vietnamiensis strains from B. cepacia genomovar I, III, and IV strains and that was based on specific regions in the rRNA operon. This assay, however, did not allow differentiation of the last three genomovars. Mahenthiralingam et al. (submitted) described a different approach based on polymorphisms of the recA genes. Their B. cepacia genomovar IV-specific primers proved to be highly specific and sensitive and facilitated identification of all the B. cepacia genomovar IV strains examined in the present study.
The results of the present study corroborated the failure of one-dimensional whole-cell protein electrophoresis to distinguish between B. cepacia genomovar I and IV strains (Fig. 1). We previously reported on the failure of whole-cell fatty acid analysis to distinguish between members of the B. cepacia complex (23). Reevaluation of these same previously published data in light of the corrected classification of several B. cepacia genomovar IV strains corroborated our previous findings, as the different B. cepacia genomovars and B. gladioli remained virtually indistinguishable (Table 2).
TABLE 2.
Fatty acid compositions of the strains studieda
| Fatty acid | Fatty acid composition (%)b
|
|||||
|---|---|---|---|---|---|---|
| B. cepacia genomovar I (14 strains) | B. multivorans (14 strains) | B. cepacia genomovar III (48 strains) | B. stabilis (11 strains) | B. vietnamiensis (4 strains) | B. gladioli (4 strains) | |
| 14:0 | 3.7 ± 1.6 | 4.8 ± 0.6 | 4.3 ± 0.7 | 4.4 ± 0.5 | 3.8 ± 0.6 | 4.9 ± 0.2 |
| 16:1 ω7c | 4.9 ± 2.8 | 6.5 ± 2.5 | 3.3 ± 1.6 | 3.3 ± 1.2 | 9.8 ± 2.5 | 4.4 ± 2.3 |
| 16:0 | 26.8 ± 2.8 | 28.9 ± 3.5 | 26.5 ± 3.3 | 25.6 ± 4.3 | 19.5 ± 2.5 | 29.0 ± 1.1 |
| 17:0 cyclo | 17.9 ± 3.6 | 18.2 ± 4.9 | 22.3 ± 4.3 | 17.8 ± 3.3 | 14.0 ± 4.9 | 17.2 ± 1.9 |
| 16:1 2OH | Tr (11)c | 1.3 ± 0.8 (12) | 1.5 ± 1.0 (42) | Tr (8) | 1.3 ± 0.2 | 1.5 ± 0.2 |
| 16:0 2OH | 3.3 ± 0.9 | 2.8 ± 1.0 | 3.0 ± 0.9 | 4.5 ± 1.5 | 2.8 ± 0.4 | 3.2 ± 0.3 |
| 16:0 3OH | 6.6 ± 0.9 | 6.8 ± 0.5 | 6.1 ± 1.2 | 5.7 ± 0.9 | 6.4 ± 0.7 | 7.1 ± 1.0 |
| 18:1 | 12.4 ± 4.0 | 11.5 ± 4.8 | 9.4 ± 4.9 | 10.6 ± 1.6 | 19.7 ± 5.5 | 9.8 ± 3.9 |
| 18:0 | Tr (13) | Tr (5) | 2.3 ± 2.2 | Tr (8) | 5.2 ± 3.4 | Tr |
| 19:0 cyclo ω8c | 12.5 ± 4.7 | 9.7 ± 3.3 | 12.3 ± 4.3 | 15.3 ± 4.2 | 5.8 ± 2.5 | 10.0 ± 2.7 |
| 18:1 2OH | 3.4 ± 0.8 | 1.3 ± 0.7 (12) | 1.6 ± 1.4 | 2.4 ± 0.6 | 3.4 ± 0.5 | 4.1 ± 1.3 |
| 14:0 3OH | 6.2 ± 1.6 | 6.4 ± 1.1 | 6.1 ± 1.1 | 5.4 ± 0.8 | 6.3 ± 0.9 | 6.8 ± 0.9 |
Data were generated and taken from a previous study (23), but new average profiles were calculated in light of the corrected classification of several B. stabilis strains (see text).
Those fatty acids for which the average amount for all taxa was less than 1% are not given. Therefore, the percentages for each group do not total 100%. Tr, trace amount (less than 1%).
The numbers in parentheses refer to the numbers of strains containing this fatty acid.
By use of the procedures for the identification of B. cepacia genomovar IV listed above, all required criteria are available to propose an official binomial species name for this organism (22). We therefore propose the name B. stabilis to accommodate the former B. cepacia genomovar IV strains.
Population structure of B. stabilis.
A variety of typing studies have been performed with B. cepacia-like organisms (13, 17, 20, 25, 26). One of the general conclusions of such studies was that the extent of genetic diversity within this group of bacteria is extremely high. In the present study, we examined 21 B. stabilis strains by means of RAPD analysis and PFGE, both of which are highly discriminatory fingerprinting methods (3, 13). These strains were isolated over a period of nearly 30 years (1970 until 1997) from sputum samples of cystic fibrosis patients (10 strains), different infections in non-cystic fibrosis patients (8 strains), and a number of sources of the hospital environment (3 strains) (Table 1). The geographic origins of the strains were very diverse: Belgium (10 strains), Canada (2 strains), France (1 strain), Germany (2 strains), Sweden (2 strains), Switzerland (1 strain), the United Kingdom (1 strain), and the United States (2 strains) (Table 1). Three of these isolates (isolates LMG 14294, LMG 14295, and LMG 15949) were cultured from sputum specimens of one cystic fibrosis patient over a 6-month period, while strain LMG 14940 was obtained from another cystic fibrosis patient attending the same clinic. Furthermore, there is an epidemiological link between strains LMG 15950 and LMG 15951, as both were isolated in 1994 from equipment of different wards of a single Belgian hospital. There was, however, no apparent relationship between any of the other isolates. Yet, the genomic variability among these strains was remarkably restricted. Numerical analysis of the PFGE fingerprinting patterns of these strains revealed only two main clusters of strains (Fig. 2); two additional strains, LMG 18138 and R-3338, were characterized by unique PFGE fingerprints. Although there were obvious differences among some of the strains within each of the clusters, the overall profiles were remarkably conserved. This genomic stability is in sharp contrast to the reported genomic diversity among B. cepacia strains in general and to the generally accepted genomic plasticity and variability of these organisms (11, 12, 18, 26). These data indicate that findings that are valid for some B. cepacia-like strains should not be extrapolated to the entire B. cepacia complex or even to the related species. They also indicate that interpretation of the similarity of DNA profiles for epidemiological investigations should be done with extreme caution.
Description of B. stabilis sp. nov.
Burkholderia stabilis (sta′bi.lis. L. adj. stabilis, stable, permanent, referring to the relative genomic stability of this B. cepacia genomovar) cells are motile rods that are 1.0 to 2.0 μm long and 0.6 to 0.9 μm wide (the description is based on data obtained in the present study for all 21 strains and on data reported previously [23] for strains LMG 6997, LMG 7000, LMG 14086, LMG 14291, LMG 14294, LMG 14295, LMG 14940, and LMG 15949). Growth is observed at room temperature and at 37°C but not at 42 or 5°C. So far, no pigmented strains have been detected and no melanin-like pigment is produced on tyrosine agar. There is growth on MacConkey agar and Simmons citrate agar. There is oxidation in OF medium, an alkaline reaction on Christensen's citrate agar, and growth in the presence of cetrimide. There is no reduction of 0.4% selenite. Tolerance to KCN is strain dependent (two of eight strains tested showed weak growth; all others were negative). There is no fluorescence on King's B medium. Tyrosine, Tween 20, and Tween 80 are hydrolyzed. Catalase, oxidase, and lecithinase activities are present; urease and beta-galactosidase (ONPG test) activities are absent. Nitrate reduction is strain dependent when it is determined as described by Holmes et al. (9) (three of eight strains tested reduced nitrate; all others did not) but is uniformly negative with the API strip; nitrite is not reduced. There is no indole, hydrogen sulfide, or 3-ketolactose production, no hydrolysis of esculin or starch, and no DNase activity. Liquefaction of gelatin and hydrolysis of casein are strain dependent (16 of 21 strains tested liquefied gelatin by the API assay; all but 1 of the strains liquefied gelatin when liquefaction was determined by a tube test [8]; 2 of 8 strains tested liquefied gelatin when liquefaction was determined by a plate test [9]; 7 of 8 strains tested hydrolyzed casein). Arginine dihydrolase and arginine desimidase activities are absent. Lysine and ornithine decarboxylase activities are absent when the activities were tested for as described by Barrow and Feltham (1) but were present when the activities were tested for as described in the present study. There is no oxidation of gluconate and no production of phenylpyruvic acid, and all strains utilize malonate. Poly-beta-hydroxybutyrate is utilized and is present as inclusion granules. Acid but no gas is produced from glucose-peptone-water sugar. Acid is produced from 10% (wt/vol) glucose, from 10% (wt/vol) lactose, and from the following sugars in ammonium salt medium: glucose, adonitol, arabinose, cellobiose, dulcitol, glycerol, inositol, lactose, maltose, mannitol, sorbitol, trehalose, xylose, and fructose. Acid is not produced (or is only weakly produced) from raffinose, rhamnose, ethanol, or sucrose. Acid production from salicin is strain dependent (three of eight strains tested produced acid from salicin).
The DNA base ratio is 68 to 69 mol%. Major fatty acid components are 14:0 (about 4.4%), 14:0 3OH (about 5.4%), 16:1 ω7c (about 3.3%), 16:0 (about 25.6%), 17:0 cyclo (about 17.8%), 16:0 3OH (about 5.7%), 18:1 (about 10.6%), and 19:0 cyclo ω8c (about 15.3%).
B. stabilis strains have been isolated from the sputum of cystic fibrosis patients, from blood, ear, and respiratory tract infections in non-cystic fibrosis patients, and from the hospital environment. The full range of pathogenicity is incompletely understood. The type strain is LMG 14294, which was isolated in 1993 in Leuven, Belgium, from the sputum of a cystic fibrosis patient. Its DNA base ratio is 68 mol%, and its phenotypic characteristics are as described above for the species. The database accession number of its 16S rRNA gene sequence is AF148554.
The type strain and several additional strains are available from the Belgian Co-ordinated Collections of micro-organisms/Laboratorium Microbiologie Gent Culture Collection and National Collection of Type Cultures culture collections.
ACKNOWLEDGMENTS
P.V. and P.D.V. are indebted to the Fund for Scientific Research, Flanders, Belgium, for positions as postdoctoral fellow and research director, respectively. T.C. acknowledges the support received from the Vlaams Instituut voor Bevordering van Wetenschappelijk-Technologisch Onderzoek in de Industrie (Brussels, Belgium) in the form of a bursary for advanced study. We acknowledge the financial support by the Cystic Fibrosis Trust (Bromley, United Kingdom) (grant RS15). E.M., D.H., and D.P.S. acknowledge grant support from the Canadian Cystic Fibrosis Foundation.
We thank G. Probe, J. Bishof, and K. Vandemeulebroecke for technical assistance and A. Bauernfeind, E. Falsen, J. R. W. Govan, and S. Lauwers for providing us with Burkholderia strains.
REFERENCES
- 1.Barrow G I, Feltham R K A, editors. Cowan and Steel's manual for the identification of medical bacteria. Cambridge, United Kingdom: Cambridge University Press; 1993. [Google Scholar]
- 2.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV. J Clin Microbiol. 1999;37:1335–1339. doi: 10.1128/jcm.37.5.1335-1339.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng H-P, Lessie T. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye T, Falsen E, Vancanneyt M, Hoste B, Govan J R W, Kersters K, Vandamme P. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples; description of Ralstonia gilardii sp. nov. Int J Syst Bacteriol. 1999;49:405–413. doi: 10.1099/00207713-49-2-405. [DOI] [PubMed] [Google Scholar]
- 5.Coenye T, Schouls L M, Govan J R W, Kersters K, Vandamme P. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int J Syst Bacteriol. 1999;49:1657–1666. doi: 10.1099/00207713-49-4-1657. [DOI] [PubMed] [Google Scholar]
- 6.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 7.Govan J R W, Hughes J, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 8.Henry D A, Campbell M E, LiPuma J J, Speert D P. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes B, Lapage S P, Malnick H. Strains of Pseudomonas putrefaciens from clinical material. J Clin Pathol. 1975;28:149–155. doi: 10.1136/jcp.28.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugh R, Leifson E. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram-negative bacteria. J Bacteriol. 1953;66:24–26. doi: 10.1128/jb.66.1.24-26.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson W M, Tyler S D, Rozee K R. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J Clin Microbiol. 1994;32:924–930. doi: 10.1128/jcm.32.4.924-930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessie T G, Hendrickson W, Manning B D, Devereux R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 13.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by random amplified polymorphic DNA (RAPD) fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic strains of Burkholderia cepacia recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 16.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprinting. In: Goodfellow M, O'Donnell A G, editors. Modern microbial methods. Chemical methods in bacterial systematics. Chichester, United Kingdom: J. Wiley & Sons; 1994. pp. 493–521. [Google Scholar]
- 17.Rabkin C S, Jarvis W R, Anderson R L, Govan J R W, Klinger J, Lipuma J J, Martone W J, Monteil H, Richard C, Shigeta S, Sosa A, Stull T, Swenson J, Woods D. Pseudomonas cepacia typing systems: collaborative study to assess their potential in epidemiologic investigations. Rev Infect Dis. 1989;11:600–607. doi: 10.1093/clinids/11.4.600. [DOI] [PubMed] [Google Scholar]
- 18.Rodley P D, Römling U, Tümmler B. A physical map of the Burkholderia cepacia type strain. Mol Microbiol. 1995;17:57–67. doi: 10.1111/j.1365-2958.1995.mmi_17010057.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Segonds C, Bingen E, Couetdic G, Mathy S, Brahimi N, Marty N, Plesiat P, MichelBriand Y, Chabanon G. Genotypic analysis of Burkholderia cepacia isolates from 13 French cystic fibrosis centers. J Clin Microbiol. 1997;35:2055–2060. doi: 10.1128/jcm.35.8.2055-2060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ursing J B, Rossello-Mora R A, Garcia-Valdes E, Lalucat J. Taxonomic note: a pragmatic approach to the nomenclature of phenotypically similar genomic groups. Int J Syst Bacteriol. 1995;45:604. [Google Scholar]
- 23.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 24.Vandamme P, Goris J, Coenye T, Hoste B, Janssens D, Kersters K, De Vos P, Falsen E. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol. 1999;49:663–669. doi: 10.1099/00207713-49-2-663. [DOI] [PubMed] [Google Scholar]
- 25.Wise M G, Shimkets L J, McArthur J V. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–198. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yohalem D S, Lorbeer J W. Multilocus isoenzyme diversity among strains of Pseudomonas cepacia isolated from decayed onions, soils, and clinical sources. Syst Appl Microbiol. 1994;17:116–124. doi: 10.1007/BF00871753. [DOI] [PubMed] [Google Scholar]