Abstract
The involvement of genetic factors in the pathogenesis of KC has long been recognized but the identification of variants affecting the underlying protein functions has been challenging. In this study, we selected 34 candidate genes for KC based on previous whole-exome sequencing (WES) and the literature, and resequenced them in 745 KC patients and 810 ethnically matched controls from Belgium, France and Italy. Data analysis was performed using the single variant association test as well as gene-based mutation burden and variance components tests. In our study, we detected enrichment of genetic variation across multiple gene-based tests for the genes COL2A1, COL5A1, TNXB, and ZNF469. The top hit in the single variant association test was obtained for a common variant in the COL12A1 gene. These associations were consistently found across independent subpopulations. Interestingly, COL5A1, TNXB, ZNF469 and COL12A1 are all known Ehlers–Danlos Syndrome (EDS) genes. Though the co-occurrence of KC and EDS has been reported previously, this study is the first to demonstrate a consistent role of genetic variants in EDS genes in the etiology of KC. In conclusion, our data show a shared genetic etiology between KC and EDS, and clearly confirm the currently disputed role of ZNF469 in disease susceptibility for KC.
Subject terms: Disease genetics, Genetics research, Disease genetics
Introduction
Keratoconus (KC) is a non-inflammatory corneal ectasia, characterized by the conical shape of the cornea due to thinning of this structure. The average reported prevalence of KC in Caucasians is about 1 in 2000 [1] but a recent study in a large Dutch population reports a larger prevalence of 1 in 375 [2]. KC is usually diagnosed during adolescence and appears to stabilize in the 3rd or 4th decade of life [1]. The treatment options depend on the severity of the disease, which shows variable expression. For the mildest forms of KC, contact lenses and spectacles are sufficient to correct visual acuity. When these treatment options are insufficient, sclera lenses and, in the most advanced cases, corneal transplantations are necessary [1].
There are several indications for a role of genetic variation in KC. First of all, higher concordance for KC severity has been reported in monozygotic twins [3]. Additionally, the prevalence in family members of KC patients is 15–67 times higher compared to the general population, with a broad sense heritability of up to 60% [4]. Positive family history has been reported in 18% of patients in the large KC population included in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study [5]. Finally, 16 loci for keratoconus were identified through linkage analysis in large pedigrees with segregation of the disease in multiple generations, but the responsible genes have never been identified. In most of these families, the disease seems to display an autosomal dominant pattern of inheritance with reduced penetrance and/or variable expression [6], although other types of inheritance have also been described [4].
In a large group of patients, KC seems to be a multifactorial genetic disease [7]. Several association studies in either KC or related syndromes involving corneal thinning point to an involvement of variants in collagen genes, in particular COL5A1, and the zinc-finger gene ZNF469. COL5A1 (Collagen type V alpha 1 chain) has also been associated with central corneal thickness through genome-wide association studies (GWAS) [8]. Variants in this gene cause Ehlers–Danlos syndrome (EDS) [9] and corneal thinning has been described in patients with COL5A1 haploinsufficiency [10]. Some of the significant Single Nucleotide Polymorphisms (SNPs) in this gene, previously identified in GWAS, have been associated with KC [11], but this was not confirmed across all investigated populations [12–14]. The ZNF469 gene was selected as a candidate for KC since bi-allelic loss-of-function mutations cause Brittle Cornea Syndrome (BCS), wherein extreme corneal thinning is a key symptom [15]. Furthermore, genetic variation near this gene has been associated with Central Corneal Thickness (CCT) with the lowest p value obtained for SNP rs9938149 (p value of 2.4 × 10−49 in the largest meta-analysis [8]). However, the exact role of genetic variation in both ZNF469 and COL5A1 in the pathogenesis of KC remains to be elucidated. Therefore, we analyzed these genes and 32 other candidate genes (selected based on literature and prior unpublished whole-exome sequencing (WES) experiments (Table 1 and Table S1) in a large population of patients and ethnically matched phenotypically unscreened controls.
Table 1.
Genes analyzed in the study.
| Gene | Full name | Gene | Full name |
|---|---|---|---|
| COL12A1 | Collagen type XII alpha 1 chain | miR184 | MicroRNA 184 |
| COL2A1 | Collagen type II alpha 1 chain | MMP9 | Matrix metallopeptidase 9 |
| COL5A1 | Collagen type V alpha 1 chain | NLRP1 | NLR family pyrin domain containing 1 |
| COL5A3 | Collagen type V alpha 3 chain | P3H4 | Prolyl 3-hydroxylase family member 4 (non-enzymatic) |
| DCAF11 | DDB1 and CUL4 associated factor 11 | PARP1 | Poly(ADP-ribose) polymerase 1 |
| DOCK1 | Dedicator of cytokinesis 1 | PARP2 | Poly(ADP-ribose) polymerase 2 |
| DOCK9 | Dedicator of cytokinesis 9 | PRPH2 | Peripherin 2 |
| EPPK1 | Epiplakin 1 | RDH13 | Retinol dehydrogenase 13 |
| FASLG | Fas ligand | SLC4A11 | Solute carrier family 4 member 11 |
| FNDC3B | Fibronectin type III domain containing 3B | SOD1 | Superoxide dismutase 1 |
| FOXO1 | Forkhead box O1 | TF | Transferrin |
| HLA-G | Major histocompatibility complex, class I, G | TGFBI | Transforming growth factor beta induced |
| IL1RN | Interleukin 1 receptor antagonist | TNXB | TENASCIN XB |
| LAMC3 | Laminin subunit gamma 3 | VSX1 | Visual system homeobox 1 |
| LCA5 | Lebercilin | WNT10A | Wnt family member 10A |
| LOX | Lysyl oxidase | ZEB1 | Zinc finger E-box binding homeobox 1 |
| LOXHD1 | Lipoxygenase homology domains 1 | ZNF469 | Zinc finger protein 469 |
Methods
Study population
Patients were recruited from three different European countries. The Belgian patients were collected at the Ophthalmology Department of the Antwerp University Hospital (UZA, Edegem, Belgium) and the Belgian control individuals were recruited from a collection available at the Center of Medical Genetics Antwerp (CMG, Antwerp, Belgium). The French patients and controls were recruited at the Centre Hospitalier Universitaire de Toulouse (Toulouse, France). In both centers, the diagnosis was based on slit-lamp biomicroscopy and tomographic evaluation of the cornea using the Pentacam (Oculus, Wetzlar, Germany) Scheimpflug camera. Italian patients and controls were collected at the Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Hospital Casa Sollievo della Sofferenza (San Giovanni Rotondo, Italy). The diagnosis was based on slit-lamp biomicroscopy and videokeratographic evaluation by using the OPD-Scan ARK-10000 (NIDEK, Tokyo, Japan) and ALLEGRO Oculyzer (WAVELIGHT AG, Erlangen, Germany). Within the case group, there was no selection regarding uni- or bilaterality, age at onset, or familiality of the trait, as this information was only sporadically available. We made sure, however, that no related subjects were included. Controls were not screened for the absence of keratoconus, but since the prevalence of KC in the general population is low, this is unlikely to have an effect on the results [16]. In total, 787 patients and 856 ethnically matched controls were included in this study. The numbers are shown in Table S2. Informed consents were obtained for all participants and all procedures were approved by the local ethical committees, in accordance with the ethics of the World Medical Association Declaration of Helsinki. DNA was isolated from fresh or frozen peripheral blood using standard techniques.
Candidate gene selection
Candidate genes were selected based on either a reported contribution to the pathogenesis of KC in the literature and/or from the results of WES analysis (unpublished results). In brief, in this unpublished study, we carried out a WES on 22 families in which KC segregates. WES was performed on one to five individuals per family starting from a long list of genes for which a link to KC was found in the literature, and that were known to be expressed in the cornea. The variants in this long list of genes were filtered using the criteria listed in detail in Table S1, to obtain a short list of candidate genes harboring one or more rare variants potentially affecting protein function, and segregating – at least partially – with the KC phenotype. An overview of the genes selected to be analyzed in this study can be found in Table 1.
Target enrichment and sequencing
Candidate genes were sequenced using molecular inversion probes (MIPs). MIP design was performed using the MIPgen pipeline [17]. The coding regions and exon-intron boundaries were included in the design, and for SOD1 probes targeting intron 2 were also designed, since a deletion in this intron has been reported in KC [18]. All isoforms available through NCBI RefSeq were taken into account during the design of the MIPs. An overview of all isoforms studied is provided in Supplementary Report 1. To provide uniform reporting of variant positions, we consistently selected the first isoform per gene, mentioned in Supplementary Report 1.
The MIP sample preparation was performed using the protocol described elsewhere [19, 20]. The final library was diluted to 1.7 pM and sequenced on the NextSeq 500 (Illumina, CA, USA) using custom sequencing and index primers in three 2 × 76 bp, dual indexed runs using a 150 cycles High-Output Illumina kit (Illumina, CA, USA). Afterward, the data from all three runs was merged.
Data analysis
VCF-files were generated using an in-house bioinformatics pipeline. Briefly, alignment of the fastq reads to the human genome was performed using BWA (v0.7.4). Afterwards, overlapping fragments of each read pair were trimmed and multi-sample variant calling was performed by Unified Genotyper from GATK (v3.5.0). We used hg19 as reference genome throughout this study and Supplementary Table S3 contains an overview of the exact reference sequences.
Quality parameters
Quality parameters for next-generation sequencing (NGS) were optimized based upon Sanger sequencing in 34 different samples, across different regions of the area targeted by the MIPs. Cutoffs for average base coverage, genotype calling based upon allelic fraction, sequencing depth, and quality by depth were optimized in order to exclude as many false positives from the analysis as possible. Full details on the optimization and Sanger-based validation are given in the Supplementary Methods and Supplementary Fig. S1.
The quality of the sequencing results in terms of coverage was expressed in 3 ways, by calculating (i) the median coverage within each transcript, across all positions and all individuals, (ii) the average coverage of each position (across all individuals within one transcript), and subsequently calculated the percentage of positions with an average coverage of at least 50x. (iii) the average coverage for each individual (across all positions) within each MIP. MIPs where more than 20% of the individuals have coverage of less than 50x, were omitted from all further analyses. A graphical overview of the performance of each MIP is given in Supplementary Report 2. A list of the MIPs not reaching the aforementioned quality criteria, is provided in Supplementary Report 3.
Statistical analysis
The association between the individual variants and the affection status (KC case or control) was carried out using Fisher’s exact test, as implemented in the software vTools. Gene-based tests included the Combined and Multivariate Collapsing test (CMC) [21], kernel-based adaptive cluster (KBAC) test [22], and Variable Thresholds methods tests (VT) [23] and SNP-set (Sequence) Kernel Association Test Method (SKAT) [24] and cAlpha test [25], as implemented in the software package vTools [26]. Gene-based tests used the following subsets of variants as input: (1) nonsense and frameshift variants, (2) nonsynonymous and in frame variants, (3) synonymous variants, (4) intronic variants, (5) 5’ UTR variants, (6) 3’ UTR variants, (7) noncoding RNA exonic variants, (8) noncoding RNA upstream variants, and (9) noncoding RNA downstream variants (the latter for miR184). All tests listed were carried out twice, using either the variants with a MAF < 1% in controls, or using the variants with a MAF < 0.1% in controls.
The significance level of the results, accounting for multiple hypothesis testing, was carried out using False Discovery Rate (FDR) analysis. Through a QQ-plot and a histogram, the distribution of the observed p values was compared to the uniform distribution (U(0,1)), which is the expected null distribution if none of the genes is associated with the phenotype. Q values, describing the expected false discovery rate in case a given p value is declared significant, were calculated as described by Storey and Tibshirani [27], via the R-package q value. In line with the recommendations of the developers of the q value technique, we are deliberately not using strict cutoffs for significance, but rather use the q value as a metric of how likely it is that an association signal represents a genuine association signal.
For variants from the single SNP association test located in the exon-intron boundaries, predictions of the effect of the variant on splicing were performed using Alamut Visual v.2.8. rev. 1.
Results
Recruitment
In a collaborative effort between 3 recruiting centers, a total of 787 independent keratoconus patients and 856 ethnically matched controls were collected. Based upon the results from a previous whole-exome study on small families segregating KC (unpublished results), combined with a literature search into genes with a reported contribution to KC, we composed a set of 34 candidate genes for targeted resequencing. An overview of these genes is given in Table 1, and a more detailed overview of the selection of these genes can be found in Table S1.
Sequencing
The yield and quality of the sequencing runs was: 74.38 Gbp and 94.53% ≥ Q30 (run 1), 69.88 Gbp and 95.65% ≥ Q30 (run 2) and 67.58 Gbp and 95.71% ≥ Q30 (run 3). After merging of the three runs, the average base coverage per sample for the target region was 350.66×. Eighty-eight samples had an average base coverage for the target region below 100× and these were excluded from the analysis. Supplementary Reports 1 and 2 give an overview of the coverage across all the transcripts from all candidate genes, including the median coverage of each transcript (across all positions and all individuals), and the percentage of positions within the transcript, that reaches an average coverage of at least 50× across all individuals. Supplementary Report 3 provides an overview of the MIPS where more than 20% of the individuals showed an average coverage (across all positions within that MIP) below 50×. These 439 MIPS, covering a total of 17.876 basepair positions, were omitted from the analysis.
Quality parameters for including samples and variants were fine-tuned by comparing the NGS results – subject to varying quality parameter cutoffs – to Sanger sequencing results, whereby this latter technique was considered to be the gold standard. Details on the optimization of the quality parameters and the metrics used is provided in Supplementary Methods. After quality control, 745 patients and 810 controls were retained that were included in the statistical analyses. A total of 2699 unique variants fulfilled the quality criteria.
To test the effects of genetic variants in the candidate genes on susceptibility for KC, we have carried out a separate analysis for the common variants (MAF > 1%) on the one hand, and the rare and very rare variants (MAF < 1% and MAF < 0.1%, respectively) on the other hand. Common variants are individually tested for association (single-variant association test), whereas for the rare and very rare variants the cumulative effect of the variants within a gene is tested (gene-based tests). To account for the multiple testing burden, we evaluated the significance level of the results using an FDR analysis, as described in the methods [27].
Single variant association tests
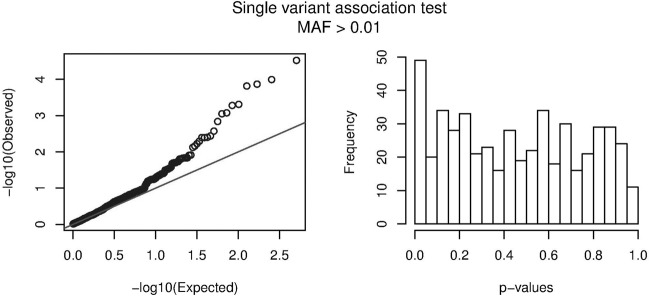
All variants having passed the quality control criteria and having a MAF in controls of at least 1%, were individually tested for association with the disease status. The QQ-plot and the histogram shown in Fig. 1, show a clear enrichment in low p values compared to the expected uniform distribution in case of no associations. The association results of the 15 most significant variants are shown in Table 2. A nominally significant p value below 0.05 was reached in 49 variants, 8 of which retained a q value below 0.05 upon FDR correction. The variant ranking 15th in the table with association results has a q value of 0.11. This means that 11% (i.e., one or two) of the 15 most significant variants, is expected to be a false positive, whereas the other 13 or 14 variants probably represent genuine association signals.
Fig. 1. Evaluation of significance for single variant association tests.
The observed distribution of the p-values for the single variant association tests (for variants with MAF > 1% in controls) is plotted in a Q-Q plot (left) and a histogram (right), showing an enrichment of low p-values. The black line in the Q-Q plot represents the expected null distribution in the absence of any association.
Table 2.
Results of the Fisher’s-exact tests on single variants with MAF > 0.01.
| Gene | Genomic position (hg19) | Nucleotide changea | Amino Acid change | Variant alleles in patients/Total number of patient alleles | Variant alleles in controls/Total number of control alleles | p value | q value | EDS gene |
|---|---|---|---|---|---|---|---|---|
| COL12A1 | chr6:75834971T>A | c.6479A>T | p.(Glu2160Val) | 132/1480 | 78/1550 | 3.01 × 10−5 | 0.0102 | Yes |
| COL5A1 | chr9:137687038A>G | c.2745+66G>A | NA | 911/1490 | 878/1620 | 0.0001 | 0.0129 | Yes |
| COL5A1 | chr9:137686906C>T | c.2701−22C>T | NA | 887/1450 | 869/1600 | 0.0001 | 0.0129 | Yes |
| TNXB | chr6:32025863C>T | c.7797G>A | p.(=) | 71/1046 | 131/1138 | 0.0002 | 0.0129 | Yes |
| COL5A1 | chr9:137688624A>G | c.2845−70A>G | NA | 758/1418 | 733/1558 | 0.0005 | 0.0296 | Yes |
| COL5A1 | chr9:137688657A>G | c.2845−37A>G | NA | 694/1414 | 664/1556 | 0.0005 | 0.0296 | Yes |
| COL5A1 | chr9:137690318A>T | c.2952+11A>T | NA | 770/1490 | 740/1620 | 0.0008 | 0.0378 | Yes |
| FNDC3B | chr3:172046861T>C | c.1374T>C | p.(=) | 733/1490 | 692/1602 | 0.0009 | 0.0378 | No |
| DOCK9 | chr13:99555188_99555189del | c.1176+37_1176+38del | NA | 48/550 | 24/586 | 0.0015 | 0.0548 | No |
| COL2A1 | chr12:48389023C>T | c.762+15G>A | NA | 487/1490 | 613/1620 | 0.0027 | 0.0907 | No |
| HLA-G | chr6:29797211C>T | c.636C>T | p.(=) | 82/1490 | 54/1620 | 0.0037 | 0.0977 | No |
| DOCK9 | chr13:99505632C>T | c.3946+30G>A | NA | 842/1490 | 831/1620 | 0.0040 | 0.0977 | No |
| RDH13 | chr19:55568084C>T | c.277G>A | p.(Ala93Thr) | 66/1490 | 111/1618 | 0.0040 | 0.0977 | No |
| RDH13 | chr19:55568085G>T | c.276C>A | p.(Asn92Lys) | 66/1490 | 111/1618 | 0.0040 | 0.0977 | No |
| COL2A1 | chr12: 48389643C>A | c.654+15C>A | NA | 267/1474 | 228/1596 | 0.0051 | 0.1146 | No |
Bold: variants for which effect on splicing was predicted using Alamut (Supplementary Fig. S2).
NA not applicable, EDS Ehlers–Danlos Syndrome.
aHg19 was used as reference genome. Transcripts: NM_004370.5 for COL12A1; NM_000093.4 for COL5A1; NM_019105.6 for TNXB; NM_022763.3 for FNDC3B; NM_001130048.1 for DOCK9; NM_001844.4 for COL2A1; NM_002127.4 for HLA-G; NM_001145971.1 for RDH13.
Most variants remaining significant upon FDR correction have an intermediate MAF. The most significant signal was obtained for a nonsynonymous common variant in COL12A1. Two of the significant common variants (c.2952+11A>T in COL5A1 and c.762+15G>A in COL2A1) are located in the exon-intron boundaries and are predicted to affect splicing (Fig. S2). Amongst these 15 most significant variants, the indel in DOCK9 is located in a polyT-stretch and is probably a sequencing artifact (rather than a true variant). This is supported by the fact that three deletions of different sizes are present in the data on this position. Remarkably, despite excellent coverage of intron 2 of SOD1 in our population of patients and controls (average coverage = 229x, Fig. S3), the previously reported SOD1 c.169+50_169+56del was not identified.
Variants with a MAF below 1% were omitted from the single variant association testing, as these variants offer limited statistical power to detect association. Only one SNP with a MAF below 1% would rank within the top 15 of the single SNP test: a synonymous c.573C>T variant in COL5A1, observed nine times in cases but never in controls (p = 0.0013).
Gene-based tests
Only common variants offer sufficient power for single-variant association testing. The effect of rare variation on the phenotype was tested gene-by-gene, using the cumulative effect of multiple rare variants within a gene. Since there is no information on neither the effect sizes of the variants (deleterious or beneficial) nor on the fraction of causal variants within the genes, it is recommended to carry out several statistical tests, to have robust power across a wide range of disease models [28]. As described in the methods, we have used three implementations of the variant burden tests and two variance components analyses. To avoid association results being driven by the more common variants that were already tested in the single SNP testing, we only used variants with a MAF below either 1% or 0.1% (in the controls) as input for the gene-based test. The remaining variants were split into 9 subsets by variant type and position, as described in the methods. Hence, we carried out the 5 association tests, starting from 9 subsets of variants, with 2 cutoffs for MAF, for each of the 34 genes.
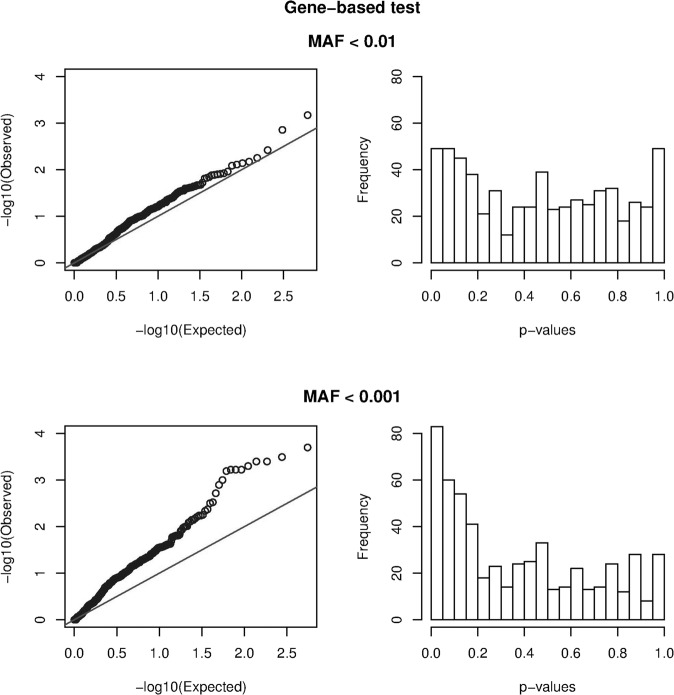
In Fig. 2, we plotted the distribution of the observed p values in a histogram and a QQ-plot. The graphs from the gene-based tests using variants with MAF < 1% (upper row) show no indication for an enrichment of low p values, as the observed distribution of p values closely resembles the null distribution. Calculation of the q values showed no variant with a q value below 0.05 (lowest q value = 0.41; Table S4 and S4bis). Remarkably, much more enrichment in low p values was observed for the gene-based test using the variants with MAF < 0.1% (bottom row), with a clear deviation from the expected null distribution. Calculation of the q values showed that, among the 83 nominally significant associations, 10 show a q value below 0.05. In Table 3, we show the 25 most significant hits from this analysis. Full results are given in Supplementary Table S4ter.
Fig. 2. Evaluation of significance in gene-based tests.
The observed distribution of the p-values for the gene-based tests is plotted in a Q-Q plot (left) and a histogram (right), to visualize the enrichment in low p-values. The upper and lower panel show the test based upon variants with MAF < 1%, and MAF < 0.1%, respectively. The black line in the Q-Q plot represents the expected null distribution in the absence of any association.
Table 3.
Gene-based tests (MAF < 0.1%) separately by variant category.
| Gene | p value | q value | Test | Variant category | EDS gene |
|---|---|---|---|---|---|
| COL2A1 | 2.00E−04 | 0.0318 | kbac | Intronic variants | Non-EDS gene |
| TNXB | 3.00E−04 | 0.0318 | CMC | Nonsynonymous variants and in-frame indels | EDS gene |
| TNXB | 4.00E−04 | 0.0318 | kbac | Nonsynonymous variants and in-frame indels | EDS gene |
| ZNF469 | 4.00E−04 | 0.0318 | kbac | Nonsynonymous variants and in-frame indels | EDS gene |
| TNXB | 5.00E−04 | 0.0318 | skat | Nonsynonymous variants and in-frame indels | EDS gene |
| ZNF469 | 6.00E−04 | 0.0318 | kbac | Synonymous variants | EDS gene |
| TNXB | 6.00E−04 | 0.0318 | vt | Nonsynonymous variants and in-frame indels | EDS gene |
| ZNF469 | 6.00E−04 | 0.0318 | vt | Nonsynonymous variants and in-frame indels | EDS gene |
| COL5A1 | 6.00E−04 | 0.0318 | skat | Synonymous variants | EDS gene |
| COL2A1 | 0.001 | 0.0446 | vt | Intronic variants | Non-EDS gene |
| COL2A1 | 0.0013 | 0.0516 | CMC | Intronic variants | Non-EDS gene |
| ZNF469 | 0.0019 | 0.0714 | CMC | Nonsynonymous variants and in-frame indels | EDS gene |
| ZNF469 | 0.003 | 0.1004 | vt | Synonymous variants | EDS gene |
| ZNF469 | 0.0032 | 0.1004 | CMC | Synonymous variants | EDS gene |
| PARP2 | 0.0043 | 0.1268 | skat | Nonsynonymous variants and in-frame indels | Non-EDS gene |
| MMP9 | 0.0046 | 0.1282 | kbac | Intronic variants | Non-EDS gene |
| TF | 0.0056 | 0.1365 | kbac | Synonymous variants | Non-EDS gene |
| TF | 0.0058 | 0.1365 | vt | Synonymous variants | Non-EDS gene |
| COL5A1 | 0.0058 | 0.1365 | skat | Intronic variants | EDS gene |
| COL12A1 | 0.0062 | 0.1382 | vt | Intronic variants | EDS gene |
| COL12A1 | 0.0067 | 0.1396 | skat | Synonymous variants | EDS gene |
| PARP2 | 0.0072 | 0.1396 | kbac | Nonsynonymous variants and in-frame indels | Non-EDS gene |
| COL5A1 | 0.0072 | 0.1396 | kbac | Synonymous variants | EDS gene |
| COL5A1 | 0.0078 | 0.1449 | kbac | Intronic variants | EDS gene |
| MMP9 | 0.0082 | 0.1462 | calpha | Intronic variants | Non-EDS gene |
Full results are shown in Supplementary Table S4ter.
Vt Variable Thresholds methods, kbac kernel-based adaptive cluster, CMC Combined and Multivariate collapsing, skat SNP-set (Sequence) Kernel Association Method, EDS Ehlers–Danlos Syndrome.
Discussion
In this study, we investigated the genetic architecture underlying keratoconus, using targeted resequencing and association tests on rare and common genetic variation in 34 candidate genes. Candidate genes were selected based upon previous associations in the literature and our own preliminary results in small families with KC. The results from the statistical analysis show a substantial consistency in the association signals across the single-SNP association testing on the common variants and the gene-based testing based upon the rare variants. Both types of tests show several association signals with low q values, from the TNXB, COL5A1, and COL2A1 genes (Supplementary Tables S5, S6 and S7). ZNF469 shows a consistent association through several gene-based tests, but no single SNP within this gene ranked within the top 15 of single SNP associations. The strongest association among the single-SNP tests was found for a nonsynonymous variant in the COL12A1 gene (p = 3.0E−5), while gene-based signals from this gene ranked within the top-25. Furthermore, the DOCK9 and RDH13 gene both show two single SNP with a q value below 0.05, but no gene-based signal.
Our choice of candidate genes was driven by either our own preliminary results on familial cases with KC or previously published association results and hypotheses about the genetic background of KC. For example, one particular set of candidate genes had some reported link with central corneal thickness. Keratoconus is one of the eye disorders that involves a reduction of the CCT, other syndromes that are associated with a reduction of CCT include Brittle Cornea syndrome, Ehlers–Danlos syndrome and primary open-angle glaucoma (POAG). Estimated heritabilities for CCT reach up to 95%, making it one of the most heritable human traits [29]. A systematic review by Swierkowska et al showed that genetic variation in genes associated with CCT could contribute to the corneal thinning in BCS, KC, and EDS, suggesting an underlying genetic connection between these three disorders [30]. A cross-ancestry genome-wide association study into CCT showed that the effect sizes of CCT-associated variants are strongly correlated to keratoconus susceptibility, with alleles associated with an increase in CCT, tend to lower the susceptibility to KC [31]. Many results from the current study lend further support to the hypothesis that corneal thinning is at least partly responsible for the KC phenotype, with several associations between the CCT genes and keratoconus.
The ZNF469 gene was included as a candidate gene in our study due to its reported involvement in BCS [15], which has been recently reclassified as a subtype of EDS [32], and the association with CCT [8]. In addition, we detected several rare variants in ZNF469 in our prior WES experiments (unpublished results), although they did not completely segregate with the phenotype in the families. Here we find a consistent association between KC susceptibility and ZNF469 variants across several gene-based tests, based upon the nonsynonymous, in frame and the synonymous variants (Supplementary Table S8). None of the single variants in ZNF469 shows a significant association with the KC phenotype, which seems to indicate that ZNF469 variants act through a cumulative effect of several very rare variants.
The gene-based association with ZNF469 is only found through variant burden tests, and not through variance component tests. Differences in significance between the gene-based tests are attributable to the difference in assumptions between the tests, and how these assumptions match with the actual underlying genetic architecture. The burden tests (CMC, VT, KBAC) make the implicit assumption that all variants are deleterious, whereas the variance component tests (SKAT, cALPHA) assume the effect sizes of the rare variants represent a spectrum, ranging from benign, over neutral, to deleterious. Which test offers the most power to detect an association, depends on the genetic architecture of the disease. Since this genetic architecture is unknown, we carried out several gene-based association tests with different assumptions, testing for association under a wide range of possible models [33].
The COL5A1 gene was previously associated with corneal thinning and with EDS. In the current study, we found several association signals in this gene, both through gene-based tests (based upon synonymous and intronic variants) and through single variant tests. Single variant association tests showed significant enrichment in cases of five variants in this gene, all of which are located in an intron (Supplemental Table S5). None of these variants have been previously described in KC. Despite being noncoding, a causative role in KC is not unlikely. Recent studies into complex disease show that variants associated with complex traits are often located in regulatory regions, with noncoding variant having a subtle effect on the transcription of the downstream gene. The dosage of this downstream gene then has an effect on disease susceptibility [34]. Whether this is the case for these variants remains to be elucidated.
The TNXB gene was included in our study because homozygous variants in this gene also cause EDS [35]. Haploinsufficiency of TNXB is associated with hypermobility of the joints [35], but corneal abnormalities have not (yet) been described for patients with this subtype of EDS, and no involvement with KC was ever reported. In our analyses, we identify a significant single variant association between a TNXB variant and KC. In addition, we find a significant enrichment of very-rare nonsynonymous variants, indicating that rare variants in TNXB are risk factors for KC (Tables S4 and S7). None of the identified variants are located in exons 32–44 of TNXB for which a highly similar pseudogene exists [36]. Based on these observations we hypothesize that loss-of-function variants lead to an EDS-phenotype while missense variants can be risk factors for KC. The underrepresentation of the synonymous c.7797G>A variant in KC patients versus controls (Single variant association test q value = 0.01297) provides an additional argument for a role of this gene in KC.
The top hit among the single variant association tests is the nonsynonymous c.6479A>T; p.(Glu2160Val) variant in the COL12A1 gene. This gene is expressed in the cornea and interacts with TNXB (in which we also detected enrichment) [37]. Homozygous loss-of-function variants and a heterozygous missense variant in this gene cause a syndrome showing an EDS and myopathy phenotype [38].
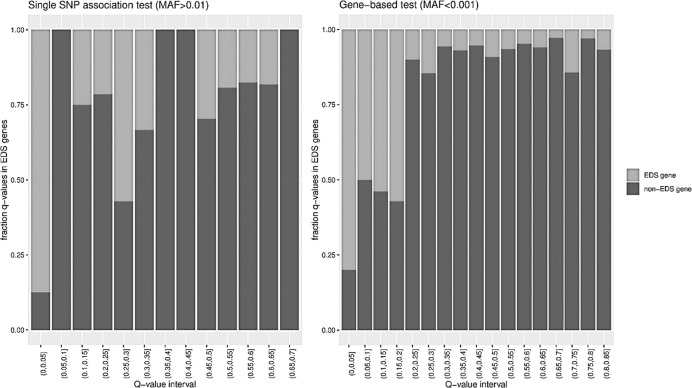
One of the most striking findings in our study, is the overrepresentation of known EDS genes, among the genes for which we find significant association signals. Figure 3 shows the fraction of EDS genes among all tested associations, binned according to the q value. Both for the single-gene tests and for the gene-based tests, the associations with a q value below 0.05, are vastly enriched in EDS genes. In general, it seems that the more severe variants on the protein level (such as frameshift and nonsense variants or glycine substitutions in the collagen helix) in these genes cause EDS, while less severe variants seem to contribute to susceptibility for KC, although we could not identify a clear genotype-phenotype correlation. Additionally, KC has been described as a symptom that can co-occur with EDS [39]. Joint hypermobility in KC has been reported [40] but this association has also been refuted [41]. We have no information on e.g., joint mobility or skin hyperextensibility of the included patients in this study to further investigate the hypothesis that KC and EDS might show a phenotypic continuum. A study examining the contribution of genetic variation in all described EDS genes in the pathogenesis of KC would undoubtedly provide novel insights.
Fig. 3. Overrepresentation of known Ehlers-Danlos Syndrome genes.
Bar chart of the fraction of EDS genes among all tested associations, binned according to the q value.
An additional gene in which we detected enrichment but that has not yet been described in KC is COL2A1. Heterozygous variants in this gene cause Stickler syndrome [42] and various skeletal dysplasias [43]. We observe significant enrichment of intronic variants in this gene in our patient population, but the pathogenic effect of these intronic variants is currently unclear (Supplementary Table S6).
Quite remarkably, our study does not confirm a role for SOD1, VSX1, and LOX in keratoconus, despite earlier reports attributing a role to these genes in the pathogenesis. This demonstrates that, if they are indeed involved in KC development, they only account for a minority of cases.
A weakness of this study is the scarcity of information on the severity of the KC phenotype in patients. For most patients, information on familial occurrence of the trait, comorbidity, age at onset, and unilaterality or bilaterality is not available. It is unlikely though that the scarcity of this information has influenced the conclusions of the current study. None of this information was used to select patients for inclusion, except for the exclusion of cases that were known to be related, and it is unlikely that this led to any recruitment biases. A study that includes more detailed information on the clinical picture would enable us to find more subtle genotype-phenotype correlations between the variants in the candidate genes and the clinical picture.
A second weakness of this study is that only a small number of candidate genes was tested. After finishing the analyses for this study, a genome-wide association analysis into KC was published by McComish et al. [44]. The 3 genes reaching genome-wide significance in this latter study (PNPLA2, MAML2, and CSNK1E), were not tested here and there is no obvious functional link between these genes and KC. The previously reported genes, involved in CCT, did not reach genome-wide significance. Although our study did not identify any possible monogenic variants causing KC, several significant associations in different genes were identified. One of the strengths of our study results is that these were consistent across the three subpopulations from different countries, which were collected completely independently by different research teams. Our most consistent finding is that variants in several EDS genes play a role in the pathogenesis of the multifactorial genetic form of KC, which seems to be in favor of a role for CCT in KC. Combined with the GWAS results from McComish et al. the conclusion seems that corneal thinning is one of the mechanisms through which KC arises, but certainly not the only one [44].
Supplementary information
Acknowledgments
Funding
This study was supported by funding from the BELSPO-IAP program (project IAP P7/43-BeMGI to GVC), the FRO and Braille Liga. HV was supported by a PhD grant from the IWT (grant no. 131526). GVC was supported by the FWO (grant no. 12D1717N). VS was supported by a Bourse Rétina France 2013 and Bourse Rétina France 2017. The supporting organizations had no role in the design or conduct of this research.
Data availability
The datasets generated and/or analyzed during the current study have been submitted to the European Genome-Phenome Archive, accession number EGAD00001006825.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Erik Fransen, Hanne Valgaeren
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00849-2.
References
- 1.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 2.Godefrooij DA, de Wit GA, Uiterwaal CS, Imhof SM, Wisse RP. Age-specific incidence and prevalence of keratoconus: a nationwide registration study. Am J Ophthalmol. 2017;175:169–72. doi: 10.1016/j.ajo.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Tuft SJ, Hassan H, George S, Frazer DG, Willoughby CE, Liskova P. Keratoconus in 18 pairs of twins. Acta Ophthalmol. 2012;90:e482–6. doi: 10.1111/j.1755-3768.2012.02448.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000;93:403–9. [PubMed] [Google Scholar]
- 5.Szczotka-Flynn L, Slaughter M, McMahon T, Barr J, Edrington T, Fink B, et al. Disease severity and family history in keratoconus. Br J Ophthalmol. 2008;92:1108–11. doi: 10.1136/bjo.2007.130294. [DOI] [PubMed] [Google Scholar]
- 6.Morrow GL, Stein RM, Racine JS, Siegel-Bartelt J. Computerized videokeratography of keratoconus kindreds. Can J Ophthalmol J canadien d’ophtalmologie. 1997;32:233–43. [PubMed] [Google Scholar]
- 7.Kriszt A, Losonczy G, Berta A, Vereb G, Takacs L. Segregation analysis suggests that keratoconus is a complex non-mendelian disease. Acta Ophthalmol. 2014;92:e562–8. doi: 10.1111/aos.12389. [DOI] [PubMed] [Google Scholar]
- 8.Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45:155–63. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toriello HV, Glover TW, Takahara K, Byers PH, Miller DE, Higgins JV, et al. A translocation interrupts the COL5A1 gene in a patient with Ehlers-Danlos syndrome and hypomelanosis of Ito. Nat Genet. 1996;13:361–5. doi: 10.1038/ng0796-361. [DOI] [PubMed] [Google Scholar]
- 10.Segev F, Heon E, Cole WG, Wenstrup RJ, Young F, Slomovic AR, et al. Structural abnormalities of the cornea and lid resulting from collagen V mutations. Investig Ophthalmol Vis Sci. 2006;47:565–73. doi: 10.1167/iovs.05-0771. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Bykhovskaya Y, Canedo AL, Haritunians T, Siscovick D, Aldave AJ, et al. Genetic association of COL5A1 variants in keratoconus patients suggests a complex connection between corneal thinning and keratoconus. Investig Ophthalmol Vis Sci. 2013;54:2696–704. doi: 10.1167/iovs.13-11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahebjada S, Schache M, Richardson AJ, Snibson G, MacGregor S, Daniell M, et al. Evaluating the association between keratoconus and the corneal thickness genes in an independent Australian population. Investig Ophthalmol Vis Sci. 2013;54:8224–8. doi: 10.1167/iovs.13-12982. [DOI] [PubMed] [Google Scholar]
- 13.Hao XD, Chen P, Chen ZL, Li SX, Wang Y. Evaluating the association between keratoconus and reported genetic loci in a Han Chinese population. Ophthalmic Genet. 2015;36:132–6. doi: 10.3109/13816810.2015.1005317. [DOI] [PubMed] [Google Scholar]
- 14.Liskova P, Dudakova L, Krepelova A, Klema J, Hysi PG. Replication of SNP associations with keratoconus in a Czech cohort. PloS One. 2017;12:e0172365. doi: 10.1371/journal.pone.0172365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohrbach M, Spencer HL, Porter LF, Burkitt-Wright EM, Burer C, Janecke A, et al. ZNF469 frequently mutated in the brittle cornea syndrome (BCS) is a single exon gene possibly regulating the expression of several extracellular matrix components. Mol Genet Metab. 2013;109:289–95. doi: 10.1016/j.ymgme.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moskvina V, Holmans P, Schmidt KM, Craddock N. Design of case-controls studies with unscreened controls. Ann Hum Genet. 2005;69:566–76. doi: 10.1111/j.1529-8817.2005.00175.x. [DOI] [PubMed] [Google Scholar]
- 17.Boyle EA, O’Roak BJ, Martin BK, Kumar A, Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udar N, Atilano SR, Brown DJ, Holguin B, Small K, Nesburn AB, et al. SOD1: a candidate gene for keratoconus. Investig Ophthalmol Vis Sci. 2006;47:3345–51. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 19.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta D, Gagliano Taliun SA, Weinstock JS, Zawistowski M, Sidore C, Fritsche LG, et al. Meta-MultiSKAT: Multiple phenotype meta-analysis for region-based association test. Genet Epidemiol. 2019;43:800–14. doi: 10.1002/gepi.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–8. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet. 2013;92:841–53. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, et al. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.San Lucas FA, Wang G, Scheet P, Peng B. Integrated annotation and analysis of genetic variants from next-generation sequencing studies with variant tools. Bioinformatics. 2012;28:421–2. doi: 10.1093/bioinformatics/btr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landers JA, Hewitt AW, Dimasi DP, Charlesworth JC, Straga T, Mills RA, et al. Heritability of central corneal thickness in nuclear families. Investig Ophthalmol Vis Sci. 2009;50:4087–90. doi: 10.1167/iovs.08-3271. [DOI] [PubMed] [Google Scholar]
- 30.Swierkowska J, Gajecka M. Genetic factors influencing the reduction of central corneal thickness in disorders affecting the eye. Ophthalmic Genet. 2017;38:501–10. doi: 10.1080/13816810.2017.1313993. [DOI] [PubMed] [Google Scholar]
- 31.Iglesias AI, Mishra A, Vitart V, Bykhovskaya Y, Hohn R, Springelkamp H, et al. Cross-ancestry genome-wide association analysis of corneal thickness strengthens link between complex and Mendelian eye diseases. Nat Commun. 2018;9:1864. doi: 10.1038/s41467-018-03646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brady AF, Demirdas S, Fournel-Gigleux S, Ghali N, Giunta C, Kapferer-Seebacher I, et al. The Ehlers-Danlos syndromes, rare types. Am J Med Genet Part C Semin Med Genet. 2017;175:70–115. doi: 10.1002/ajmg.c.31550. [DOI] [PubMed] [Google Scholar]
- 33.Graham SE, Nielsen JB, Zawistowski M, Zhou W, Fritsche LG, Gabrielsen ME, et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat Commun. 2019;10:1847. doi: 10.1038/s41467-019-09861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51:76–87. doi: 10.1038/s41588-018-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zweers MC, Bristow J, Steijlen PM, Dean WB, Hamel BC, Otero M, et al. Haploinsufficiency of TNXB is associated with hypermobility type of Ehlers-Danlos syndrome. Am J Hum Genet. 2003;73:214–7. doi: 10.1086/376564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schalkwijk J, Zweers MC, Steijlen PM, Dean WB, Taylor G, van Vlijmen IM, et al. A recessive form of the Ehlers-Danlos syndrome caused by tenascin-X deficiency. N Engl J Med. 2001;345:1167–75. doi: 10.1056/NEJMoa002939. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–9. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou Y, Zwolanek D, Izu Y, Gandhy S, Schreiber G, Brockmann K, et al. Recessive and dominant mutations in COL12A1 cause a novel EDS/myopathy overlap syndrome in humans and mice. Hum Mol Genet. 2014;23:2339–52. doi: 10.1093/hmg/ddt627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuming BS, Joffe L. Ehlers-Danlos syndrome associated with keratoconus. A case report. South Afr Med J. 1977;52:403–5. [PubMed] [Google Scholar]
- 40.Woodward EG, Morris MT. Joint hypermobility in keratoconus. Ophthalmic Physiol Opt. 1990;10:360–2. doi: 10.1111/j.1475-1313.1990.tb00882.x. [DOI] [PubMed] [Google Scholar]
- 41.Street DA, Vinokur ET, Waring GO, 3rd, Pollak SJ, Clements SD, Perkins JV. Lack of association between keratoconus, mitral valve prolapse, and joint hypermobility. Ophthalmology. 1991;98:170–6. doi: 10.1016/s0161-6420(91)32320-0. [DOI] [PubMed] [Google Scholar]
- 42.Knowlton RG, Weaver EJ, Struyk AF, Knobloch WH, King RA, Norris K, et al. Genetic linkage analysis of hereditary arthro-ophthalmopathy (Stickler syndrome) and the type II procollagen gene. Am J Hum Genet. 1989;45:681–8. [PMC free article] [PubMed] [Google Scholar]
- 43.Walter K, Tansek M, Tobias ES, Ikegawa S, Coucke P, Hyland J, et al. COL2A1-related skeletal dysplasias with predominant metaphyseal involvement. Am J Med Genet Part A. 2007;143A:161–7. doi: 10.1002/ajmg.a.31516. [DOI] [PubMed] [Google Scholar]
- 44.McComish BJ, Sahebjada S, Bykhovskaya Y, Willoughby CE, Richardson AJ, Tenen A, et al. Association of genetic variation with keratoconus. JAMA Ophthalmol. 2020;138:174–81. doi: 10.1001/jamaophthalmol.2019.5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study have been submitted to the European Genome-Phenome Archive, accession number EGAD00001006825.