Abstract
The stage selector protein (SSP) is a heteromeric complex involved in preferential expression of the human γ-globin genes in fetal-erythroid cells. We have previously identified the ubiquitous transcription factor CP2 as a component of this complex. Using the protein dimerization domain of CP2 in a yeast two-hybrid screen, we have cloned a novel gene, NF-E4, encoding the tissue-restricted component of the SSP. NF-E4 and CP2 coimmunoprecipitate from extract derived from a fetal-erythroid cell line, and antiserum to NF-E4 ablates binding of the SSP to the γ promoter. NF-E4 is expressed in fetal liver, cord blood, and bone marrow and in the K562 and HEL cell lines, which constitutively express the fetal globin genes. Enforced expression of NF-E4 in K562 cells and primary erythroid progenitors induces endogenous fetal globin gene expression, suggesting a possible strategy for therapeutic intervention in the hemoglobinopathies.
The human β-globin cluster is the classic paradigm of a multigene locus. The globin genes (ɛ, Gγ, Aγ, δ, and β) are expressed at high levels throughout ontogeny in a stringently regulated developmental stage- and tissue-specific pattern. From conception until week 5 of gestation, the embryonic globin (ɛ) gene is expressed in the yolk sac, the major site of erythropoiesis. After this time, the first switch in globin subtype occurs, as the fetal globin (Gγ and Aγ) genes become the dominant transcripts in the erythropoietic cells of the fetal liver. This expression pattern persists until birth, when the switch from fetal to adult globin (β-globin) synthesis occurs, coincident with the bone marrow becoming the predominant erythropoietic organ (26, 43, 57).
Studies of the human β-globin locus in transgenic mice and in patients carrying the Hispanic and Dutch thalassemic mutations have revealed that the key regulatory sequences required for high-level globin expression reside 6 to 20 kb upstream of the ɛ gene (15, 31, 61, 64, 65). These sequences, characterized by the presence of five 5′ DNase I hypersensitivity sites (5′HS1 to -5), are known as the locus control region (LCR) (16, 20, 62). Studies by Wijgerde et al. suggest that the HSs act cooperatively as a holocomplex which focuses the vast enhancing potential of the LCR to a single globin gene at any given time point during ontogeny (67). In murine fetal liver cells transgenic for the β-globin locus, the LCR flip-flops back and forth between the γ and β genes at the time of the fetal-adult switch. As the cellular transcription factor milieu changes to favor adult globin expression, the stability of the γ gene-LCR interaction decreases and β-globin becomes the predominantly transcribed gene.
Competition between globin genes for a single regulatory sequence was first proposed as a mechanism of developmental regulation by Choi and Engel (8). In these studies, a stage selector element (SSE) in the chick β-gene (β) promoter was essential for the preferential interaction of that promoter with the locus enhancer during adult erythropoiesis. The activity of the promoter element was mediated through the binding of a stage-specific factor, known as NF-E4 (19, 69). Promoter sequences and stage-specific factors have also been shown to be critical for correct developmental regulation of the human and murine β-globin clusters. Mice carrying a transgene of the human locus lacking the LCR, or embryonic stem cells in which the native LCR has been removed by homologous recombination, still display appropriate temporal patterns of globin expression, albeit at reduced levels (13, 58). Conversely, deletion of the human γ promoter or its substitution with a non-developmental-stage-specific erythroid promoter in transgenic mice abolishes the correct temporal profile of both γ- and β-gene expression (3, 53).
Two regions of the γ promoter appear to be responsible for its competitive advantage in the fetal-erythroid environment. The first, the CACCC box, binds a recently described member of the Krüppel family, fetal Krüppel-like factor (FKLF) (5). Expression of this gene is detectable in fetal liver and to a lesser extent adult bone marrow, but its functional effects appear to predominantly involve the ɛ and γ genes. The second region in the γ promoter was defined in transfection studies in the K562 cell line, a model of fetal erythropoiesis (37). In these experiments, an 18-bp stage selector element immediately 5′ of the TATA box was sufficient for preferential transcription from the γ promoter when in competition with the β promoter for a single enhancer element (HS2) from the LCR (28). Analogous to the chicken cluster, the activity of this SSE was dependent on the binding of a stage-specific factor, the fetal- erythroid cell-specific stage selector protein (SSP) (29).
Several lines of evidence support the importance of the SSP-SSE interaction in the developmental regulation of globin gene expression. Evolutionary phylogenetic footprinting studies demonstrate absolute conservation of the SSP binding site in species with a distinct stage of fetal globin expression of the γ genes and loss in species where the γ genes are embryonic (21, 60). Multiple SSP binding sites have also been identified in phylogenetic footprints in the ɛ promoter, HS2 and HS3 (22). The formation of a new binding site for the SSP by the −202(C→G) HPFH mutation also lends credence to a role for this factor in γ-gene regulation (27). Biochemical purification of the SSP revealed that the ubiquitously expressed transcription factor CP2 (also known as LBP-1c/LSF) formed a major component of the SSP binding activity (29, 30, 35, 70). Antiserum to CP2 ablated the SSP-SSE complex in electrophoretic mobility shift assays (EMSAs). It also reacted with highly purified chicken NF-E4 in Western analysis, indicating that this developmental complex is conserved in evolution (69). However, CP2 alone was incapable of binding to the SSE, suggesting that the SSP consisted of a heteromeric complex between CP2 and an unknown factor that provided the tissue specificity and DNA binding activity of the complex. The presence of this factor (named NF-E4, after the putative chick homologue) was confirmed in EMSA and UV cross-linking experiments, and its molecular mass was estimated as 40 to 45 kDa (29).
Using the protein dimerization domain of CP2 as bait in a yeast two-hybrid library screen, we now report the cloning and characterization of human NF-E4, the tissue-restricted component of the SSP. We demonstrate that enforced expression of this factor in fetal-erythroid cells induces fetal and embryonic globin expression.
MATERIALS AND METHODS
Yeast two-hybrid screen.
The cDNA sequence encoding the COOH-terminal 240 amino acids (aa 260 to 502) of CP2 was inserted into the yeast expression vector pGBT9 (Clontech). The resultant plasmid encodes a hybrid protein containing the DNA binding domain of GAL4 (GAL4DBD) fused in frame to CP2 residues. The yeast reporter strain, HF7C, rendered competent by the lithium acetate method, was sequentially transformed with this vector and a plasmid cDNA library derived from K562 cells constructed in the yeast expression vector pACT2 (14). The cDNAs in this vector were fused with the GAL4 transactivation domain (GAL4AD). The yeast cells were plated on leucine-, tryptophan-, and histidine-deficient plates and incubated at 30°C for 4 days. Potential protein interactions were indicated by activation of the histidine reporter gene and growth on these plates and by activation of the second reporter gene, β-galactosidase, and a positive 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) assay. Library plasmids were rescued from yeast clones using the acid-washed glass beads procedure and electroporated into the competent Escherichia coli strain MC1061.
Sequencing reactions were performed using the Taq Dyedeoxy terminator cycle sequencing method (Applied Biosystems) and analyzed on an Applied Biosystems 373 automated sequencer.
Yeast one-hybrid assay.
A concatemer of four copies of the SSE was cloned into the EcoRI/SalI sites of the yeast vector, pLacZ. As a control, a four-copy concatemer of the direct repeat elements (DREs) from the proximal β promoter was also cloned into this vector. Small-scale transformations of each vector were performed into Saccharomyces cerevisiae YM4271, which is auxotrophic for histidine, uracil, leucine, and tryptophan. Prior to transfection, the vector was linearized with NotI to allow genomic integration into the ura3-52 site, which confers auxotrophy to uracil, allowing selection of transformants. A single yeast colony which displayed no basal reporter gene activity was chosen for subsequent experiments. This colony was expanded and transformed with pACT-CP2, pACT106, or pACT117. The transformants were selected on minimal medium lacking leucine and uracil, and colonies were lifted on filters and assayed for β-galactosidase activity.
Mammalian two-hybrid assay.
Mammalian expression vectors containing the dimerization domain of CP2 fused in frame to the GAL4DBD and NF-E4 fused in frame to the VP16AD were generated. These plasmids were cotransfected with pG5CAT, a reporter construct with five GAL4DBDs linked to the chloramphenicol acetyltransferase (CAT) gene, into 293 cells, using calcium phosphate precipitation. Vectors lacking either CP2 or NF-E4 were transfected as controls. After 48 h, cells were harvested and whole-cell lysate was prepared. CAT activity was measured by CAT enzyme-linked immunosorbent assay as instructed by the manufacturer (Boehringer Mannheim).
5′ RACE.
A marathon 5′ RACE (rapid amplification of 5′ cDNA ends) cDNA library was constructed from mRNA from K562 cells as instructed by the manufacturer (Clontech). Nested PCR was performed with the following vector- and gene-specific primers: gene specific 1 (5′-CCCTTGGCTCAGATGAAGCGATGGTAGT-3′), gene specific 2 (5′-TGGCCTGCAGGGCCCCAGTAGGT-3′), vector specific 1 (5′-CCATCCTAATACGACTCACTATAGGGC-3′), and vector specific 2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′). PCR conditions were as follows: 95°C for 1 min, 1 cycle; 94°C for 10 s and 68°C for 2 min, 30 cycles; and 68°C for 5 min, 1 cycle. Nested PCR was performed under identical conditions except that the cycle number was reduced to 20. PCR products were electrophoresed on 1% agarose, blotted onto nitrocellulose, and probed with internal gene-specific oligonucleotides. Final PCR products were cloned into the TOPO 2.1 vector as instructed by the manufacturer (Invitrogen) and sequenced.
Generation of MSCV-based supernatant and transduction of mammalian cell lines.
The NF-E4 coding region was cloned into the retroviral vector plasmid MSCV-HA at a unique XhoI or EcoRI site. This bicistronic vector contains (i) the amphotropic retrovirus murine stem cell virus (MSCV) 5′ long terminal repeat (LTR), (ii) a hemagglutinin (HA) epitope tag with the NF-E4 coding sequence in frame either 5′ or 3′ to the tag, (iii) the encephalomyocarditis internal ribosomal entry site (IRES), (iv) the green fluorescent protein (GFP) cDNA, and (v) the MSCV 3′ LTR (see Fig. 2C). The plasmid was cotransfected with an amphotropic packaging plasmid into 293T cells by calcium phosphate precipitation. After 48 h, the supernatant containing amphotropic particles was harvested, filtered, and added to K562 or MEL cells every 12 h for 3 days. The cells were allowed to recover for 72 h and then analyzed for GFP expression by flow cytometry. The highest-expressing 10% of cells were sterilely sorted, expanded, resorted, and subsequently expanded in oligoclonal pools. A biological titer of the supernatant on NIH 3T3 cells was equivalent to 106 CFU/ml.
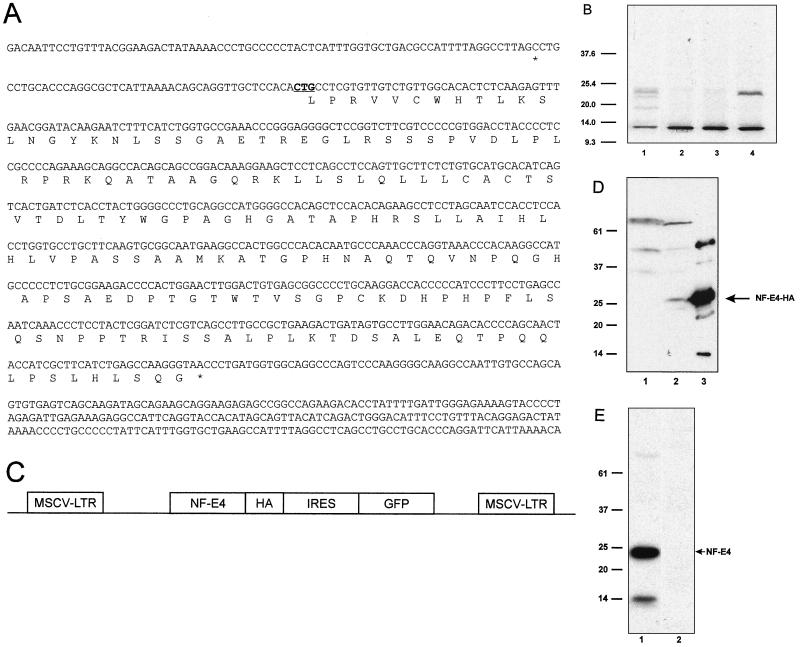
FIG. 2.
NF-E4 is a 22-kDa protein which may initiate at a CUG codon. (A) Nucleotide and amino acid sequences of NF-E4. The potential initiator CUG is underlined and in boldface. The 5′ and 3′ termination codons are marked with asterisks. (B) In vitro transcription/translation of NF-E4. The NF-E4 cDNA with its native Kozak sequence was cloned into pSP72, and [35S]methionine-labeled protein was produced (lane 1). We also generated radiolabeled protein from NF-E4 constructs in which the CTG was mutated to a GGG or TTG but retained the native Kozak sequence (lanes 2 and 3) or was mutated to an ATG with a consensus Kozak sequence (lane 4); 2 μl of each sample was resolved by SDS-PAGE on a 12% gel. The gel was dried and subjected to autoradiography. The migration of molecular size standards is marked in kilodaltons. (C) Diagrammatic representation of the MSCV-NF-E4-HA vector. The vector consists of the MSCV backbone containing the NF-E4 coding sequence tagged at the COOH terminus with HA, followed by an encephalomyocarditis virus IRES linked to the GFP cDNA. (D) Western analysis of K562 cell pools expressing NF-E4-HA. Whole-cell lysates from K562 cells transduced with MSCV-NF-E4-HA (lane 2) or MSCV alone (lane 1) were resolved on a 12% polyacrylamide gel, transferred to a PVDF membrane, probed with polyclonal anti-HA antiserum, and developed with ECL. The specific NF-E4-HA band is arrowed. Recombinant NF-E4-HA served as a control (lane 3). The migration of molecular mass standards is indicated in kilodaltons. (E) Western analysis of native K562 cells. Nuclear extract from K562 cells was resolved on a 12% polyacrylamide gel, transferred to a PVDF membrane, and probed with polyclonal anti-NF-E4 (lane 1) or preimmune (lane 2) serum. Signal was developed with ECL. The migration of molecular mass standards is indicated in kilodaltons.
Generation of RD18 producer cell lines.
Amphotropic supernatant generated in 293T cells (as described above) was used to transfect the FLYRD18 packaging cell line (10). Briefly, fresh filtered supernatant from 293T cells was added to RD18 cells plated at a density of 103 cells every 12 h for 3 days. Subsequently, the top 20% of GFP-positive RD18 cells were obtained by fluorescence-activated cell sorting (FACS) and cultured until confluent. Amphotropic supernatant harvested from these plates was used to transfect CD34+ progenitors. The expression of NF-E4 in the producer cell line was verified by immunoblotting with anti-HA antiserum (data not shown).
Isolation and retroviral transduction of human CD34+ cells.
Human cord blood was generously provided by the Bone Marrow Donor Institute Cord Blood Bank. CD34+ cells were isolated using a MiniMACS magnetic cell sorting system (Miltenyi Biotec Inc.). Cells were then cultured overnight with expansion medium, which contains 1% deionized bovine serum albumin (BSA; Stem Cell Technologies), insulin (5 μg/ml; Sigma), transferrin (100 μg/ml; BRL), low-density lipoprotein (10 μg/ml; Sigma), 10−4 M β-mercaptoethanol (BRL), recombinant human interleukin-3 (rhIL-3; 10 ng/ml; R&D), rhIL-6 (10 ng/ml; R&D), recombinant human stem cell factor (300 ng/ml; R&D), and Flt-3 (300 ng/ml; R&D). Non-tissue culture-treated 35-mm-diameter dishes were coated with RetroNectin CH286 solution (TaKaRa Biochemicals, Shiga, Japan) at the concentration of 20 μg/cm2 for 2 h at room temperature and then blocked with 2% BSA fraction V (Fisher Scientifics) for 30 min at room temperature (40). The coated dishes were preloaded with virus supernatant from RD18 producer lines (2 ml/well) for 30 min, after which the supernatant was removed. Another 1 ml of supernatant was added along with expanded CD34+ cells at <5 × 105 cells/dish; then 1 ml of expanded medium (in which the amount of each ingredient was doubled) was added and mixed well. Cells were cultured for 24 h and then harvested and carefully washed three times with a large volume of 1× phosphate-buffered saline. The cells were then cultured at 105 cells/ml in Iscove modified differentiation medium containing 30% fetal calf serum (HyClone), 1% deionized BSA (Stem Cell Technologies), recombinant human stem cell factor (100 ng/ml; R&D), rhIL-3 (0.1 pg/ml; R&D), human erythropoietin (10 U/ml; Amgen), insulin (10 μg/ml; Sigma), 10−4 M β-mercaptoethanol (BRL), 10−6 M hydrocortisone (Stem Cell Technologies), and penicillin-streptomycin and glutamine (1:1,000; BRL) as previously described (59). At day 12, GFP- and glycophorin A-positive cells were isolated by FACS, and RNA was prepared using RNAzol (Tel-Test Inc.).
Expression of GST fusion proteins and affinity chromatography.
CP2 and NF-E4 cDNAs were cloned in frame with the glutathione S-transferase (GST) coding sequence in the pGEX vectors (Pharmacia). The GST fusion proteins were expressed in E. coli BL21. Fusion proteins were purified on glutathione-Sepharose (Pharmacia), and their integrity was confirmed with Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For in vitro protein-protein interaction assays, 1 μg of GST or GST fusion protein was incubated for 1 h at 4°C with 10 μl of glutathione-Sepharose beads, which had been preblocked with 0.5% milk. After extensive washing, the beads were resuspended in 200 μl of binding buffer (10 mM Tris-HCl [pH 7.9], 500 mM KCl, 0.1 mM EDTA, 150 μg of BSA/ml, 0.1% Nonidet P-40, 10% glycerol) and incubated for 1 h at room temperature with [35S]methionine-labeled NF-E4. After extensive washing, retained proteins were eluted by boiling in SDS loading buffer and analyzed by SDS-PAGE and autoradiography.
Extract preparation, immunoprecipitation, and EMSA.
Nuclear extracts were prepared by the method of Dignam (11). Highly purified SSP was obtained by fractionating crude extract over heparin-Sepharose and DNA affinity columns as described previously (29).
For immunoprecipitation studies, nuclear extracts were initially precleared with normal rabbit serum (10 μg/ml) and then incubated with preimmune serum or antiserum to CP2 or NF-E4 overnight at 4°C. A 50% slurry of protein G-Sepharose was added and incubated at 4°C for 1 h. The mixture was then centrifuged at 3,000 × g for 1 min, and the pellet was washed in 50 mM Tris-HCl (pH 7.9) containing 150 mM NaCl prior to being resuspended in SDS loading buffer. Samples were subjected to SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and blotted with antiserum to NF-E4. Signal detection was achieved with the Amersham Pharmacia ECL (enhanced chemiluminescence) system according to the manufacturer's instructions. EMSAs were performed by incubating various amounts of nuclear extract with 105 cpm of [32P]dCTP-end-labeled double-stranded oligonucleotides encoding the SSE region of the γ promoter in a 20-μl reaction containing 500 ng of poly(dI-dC), 6 mM MgCl2, 16.5 mM KCl, and 100 μg of BSA. For antibody studies, 3 μl of preimmune serum or rabbit anti-mouse CP2 or NF-E4 antibody was preincubated for 10 min with the binding reaction prior to addition of the probe. After incubation on ice for 15 min and 25°C for 15 min, samples were electrophoresed on a 4% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer for 90 min at 10 V/cm.
RNase protection, reverse transcription-PCR (RT-PCR), and Northern analysis.
RNase protection analysis was performed using an Ambion RNase protection assay kit according to the manufacturer's instructions. Probes used in these studies were as described previously (40a). Probe input was 106 cpm/sample for γ- and β-globin probes and 0.25 × 106 cpm/sample for the 18S probe.
For RT-PCR, first-strand cDNA was prepared from 2 μg of mRNA from primary tissues using random hexamers. Each cDNA sample was appropriately diluted to give similar amplification of S14 RNA under the same PCR conditions. The primer sequences and PCR conditions were as follows: S14 sense (5′-GGCAGACCGAGATGAATCCTCA-3′) and antisense (5′-CAGGTCCAGGGGTCTTGGTCC-3′), 95°C for 1 min, followed by various cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min; NF-E4 sense (5′-ACCCGGGAGGGGCTCCGGTCTT-3′) and antisense (5′-CCCTTGGCTCAGATGAAGCGATGGTAGT-3′), 95°C for 1 min, followed by various cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. All PCR products were electrophoresed on 1.5% agarose gels, transferred to nitrocellulose, and analyzed by Southern blotting using 32P-radiolabeled internal oligonucleotides as probes. Membranes were then autoradiographed for 2 h at −70°C. Northern analysis of K562 pools was performed as described previously (54).
RESULTS
Isolation of CP2-interacting proteins from a K562 cell cDNA library.
Previous studies had demonstrated that the ubiquitous transcription factor CP2 formed a major component of the SSP. We and others have mapped the protein dimerization domain of CP2 to the 242 amino acid residues at its carboxy terminus (56, 63). Within this region, a 17-aa stretch (aa 292 to 309) is essential for protein-protein interactions (S. M. Jane and J. M. Cunningham, unpublished data). A cDNA sequence encoding the COOH-terminal 242 aa (aa 260 to 502) of CP2 was inserted into the yeast expression vector pGBT9. The resultant plasmid (GAL4CP2-260) encodes a hybrid protein containing the GAL4DBD fused to CP2 residues 260 to 502 (Fig. 1A). The yeast reporter strain HF7C was transformed with this vector and an expression library derived from K562 cell line cDNAs fused to the sequences encoding the GAL4AD. This library was chosen because K562, a human cell line, is a model of fetal erythropoiesis, constitutively expressing the ɛ and γ genes but not the adult β genes (51). In addition, abundant SSP binding activity is evident in nuclear extract from these cells, which was the source for the biochemical purification of the CP2 component of the SSP. From 5 × 106 clones screened, we isolated 100 clones that appeared to interact with the CP2 bait. From this collection we identified about 40 clones that encoded CP2, an expected result in view of the protein's ability to homodimerize. Another 50 clones represented previously identified false positives from the two-hybrid screen. Of the 10 remaining clones, 8 corresponded to known genes or ESTs (expressed sequence tags) whose tissue distribution suggested that they were unlikely to represent the tissue-restricted component of the SSP. Only two clones, c106 and c117, were novel, and hence their further evaluation was prioritized.
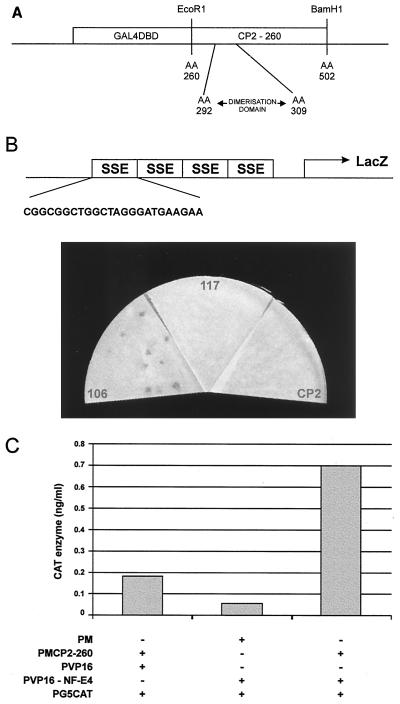
FIG. 1.
Isolation of CP2-interacting proteins. (A) Schematic of the bait construct used in the yeast two-hybrid screen of a K562 cDNA library. The coding sequence of CP2 from aa 260 to 502 was fused in frame with the GAL4DBD. The minimal dimerization domain (aa 292 to 309) is shown. (B) Yeast one-hybrid assay of CP2-interacting clones defined in the two-hybrid screen. Plasmids containing CP2 or c106 or c117 fused to the GAL4AD were transfected into a yeast reporter strain carrying four concatemerized SSE sites linked to a lacZ reporter gene. The sequence of a single SSE site is expanded. Transformants were selected on minimal medium plates lacking leucine and uracil, lifted onto filters, and assayed for β-galactosidase activity. Filters were photographed after 30 min, and equivalent transfection efficiencies were observed for all constructs. (C) Mammalian two-hybrid analysis of interaction between CP2 and NF-E4. 293 cells were transiently cotransfected with a reporter plasmid containing five GAL4DBD sites linked to the CAT gene (pG5CAT) and the indicated expression vectors. PM contains the GAL4DBD, PMCP2-260 contains the GAL4DBD fused to aa 260 to 502 of CP2, PVP16 has the VP16AD, and PVP16-NF-E4 has the NF-E4 sequence isolated from the two-hybrid clone fused to the VP16AD. Transfections were performed in triplicate, and cell lysates were assayed in triplicate for CAT activity.
As the DNA binding activity of the SSP appears to depend on the presence of the CP2 partner protein, we evaluated the ability of c106 and c117 to bind to the SSE in the yeast one-hybrid assay. Yeast strain YM4271, containing four concatemerized SSE sites linked to a lacZ reporter gene, was generated and transfected with the K562 cDNA library plasmids encoding the GAL4AD-c106, GAL4AD-c117, or GAL4AD-CP2 fusion protein. A strain containing four concatemerized DREs from the proximal β promoter linked to the same reporter was transfected as a control. After transfection, both strains were plated on medium lacking leucine and uracil, and resultant colonies were assayed for β-galactosidase activity. As seen in Fig. 1B, a positive result was observed with c106 but not with c117 or CP2 using the SSE binding site. No enzymatic activity was observed with the DRE binding sites with any of the three plasmids (data not shown). Based on these findings, we postulated that c106 was a strong candidate for the partner protein of CP2 in the SSP complex and hereafter refer to it as NF-E4.
To validate the interaction between CP2 and NF-E4 in a eukaryotic expression system, we used the mammalian two-hybrid assay. Mammalian expression vectors containing the dimerization domain of CP2 fused in frame to the GAL4DBD and NF-E4 fused in frame to the VP16AD were generated. These plasmids were cotransfected with pG5CAT, a reporter construct with five GAL4 DBDs linked to the CAT gene. Vectors lacking either CP2 or NF-E4 were transfected as controls. As seen in Fig. 1C, a marked induction of CAT activity was observed only in the presence of both proteins.
The NF-E4 gene encodes a 22-kDa protein which may initiate at a CUG codon.
To facilitate further studies of NF-E4, we used 5′ RACE from K562 cell cDNA to obtain a full-length clone. A 966-bp fragment was generated using nested gene- and vector-specific primers in multiple experiments. Comparison of the sequences derived from multiple clones with the databases using the BLAST algorithm revealed a high degree of homology with a sequence from a bacterial artificial chromosome containing a region of the human X chromosome. Sequence analysis revealed a long open reading frame contiguous with that defined in the original yeast two-hybrid GAL4AD-c106 fusion vector (Fig. 2A). Although one potential initiation codon (AUG) was observed beginning at nucleotide 421, several observations suggested that translation of full-length NF-E4 might not start at this AUG. First, the NF-E4 reading frame remains open for an additional 115 codons upstream of the first AUG before an in-frame termination codon is encountered. Second, the predicted size of the protein from the first AUG is markedly less than that suggested by our previous studies. Finally, a CUG codon preceded by a Kozak sequence and termination codon is present in the correct reading frame 100 codons upstream of the first AUG (32). Translation from this codon would generate a protein with a predicted molecular mass of approximately 22 kDa.
Many studies have documented translation of human mRNAs from initiation codons other than AUG, most often CUG (33, 38, 46). These include transcription factors such as TEF-1 and Krox-24 and proto-oncogene products such as c-myc, Int2, and others (1, 23, 34, 68). Comparison of the NF-E4 sequence with a human genomic clone isolated in our laboratory confirmed the presence of the single AUG in the midregion of the sequence and the upstream CUG and termination codon (data not shown). To determine whether effective translation was achieved from this CUG in vitro, we subcloned the NF-E4 cDNA into the in vitro transcription/translation vector pSP72 and generated [35S]methionine-labeled protein. We also generated NF-E4 clones in which the CTG was mutated to a GGG or TTG or to an ATG with a consensus Kozak sequence to determine whether these sequence changes resulted in altered efficiency in translation (32). As shown in Fig. 2B, the dominant protein species translated from the AUG containing NF-E4 vector (lane 4) had a molecular weight identical to that observed with the native CUG-initiated NF-E4 (lane 1). A marked reduction in efficiency of transcription and translation was observed with both the GGG- and TTG-containing constructs (lanes 2 and 3).
To determine whether effective translation was achieved from this CUG in vivo, we generated an MSCV-based vector containing the NF-E4 cDNA (24). This bicistronic vector contains the GFP cDNA linked by the encephalomyelitis virus IRES to the NF-E4 cDNA tagged at its COOH terminus with HA (Fig. 2C). K562 cells were transduced with this virus (MSCV-NF-E4-HA) or the parental virus carrying the GFP cDNA alone (MSCV); after 5 days, GFP-positive cells were selected by FACS. As shown in Fig. 2D, Western analysis with an anti-HA antibody demonstrated a band in extract from the MSCV-NF-E4-HA-transduced cells (lane 2) which comigrated with recombinant NF-E4-HA generated in bacteria (lane 3). No corresponding band was observed in K562 cells transduced with MSCV alone (lane 1).
Further support for CUG-initiated translation was obtained with the generation of polyclonal antiserum to the native NF-E4 protein. A dominant band of 22 kDa was observed in Western analysis of native K562 cells, consistent with initiation at the CUG codon (Fig. 2E). A second minor band was observed at approximately 14 kDa, which could reflect initiation at the downstream AUG.
NF-E4 interacts with CP2 in vitro and in vivo.
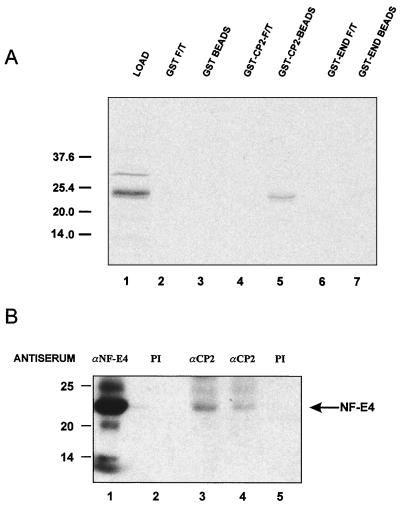
To confirm the interaction between CP2 and full-length NF-E4, we used GST chromatographic assays. GST, GST fused in frame with full-length endophilin (GST-END), or GST fused in frame with full-length CP2 (GST-CP2) was coupled to glutathione-Sepharose beads and incubated under stringent conditions with [35S]methionine-labeled in vitro-transcribed/translated NF-E4. Specific retention of NF-E4 was observed with the GST-CP2 beads but not control GST or GST-END beads (Fig. 3A). To confirm this interaction in an in vivo setting, coimmunoprecipitation studies were performed. Nuclear extract from K562 cells was immunoprecipitated with either anti-CP2 antiserum or preimmune serum and blotted with anti-NF-E4 antiserum (Fig. 3B). Immunoprecipitation and blotting with anti-NF-E4 antiserum served as the positive control (lane 1). A specific band of 22 kDa was observed after immunoprecipitation with 8 μl (lane 3) or 4 μl (lane 4) of anti-CP2 antiserum. No band was observed with preimmune serum derived from NF-E4- or CP2-inoculated rabbits (lanes 2 and 5). This finding indicates that CP2 and NF-E4 form a physiological complex in vivo.
FIG. 3.
CP2 and NF-E4 interact in vitro and in vivo. (A) Direct interaction between CP2 and NF-E4. GST-CP2, GST-END, or GST alone was expressed in E. coli, and 1 μg of protein bound to glutathione-Sepharose beads. The beads were then incubated with 2 μl of 35S-labeled in vitro-translated NF-E4 in binding buffer for 1 h at room temperature (see Materials and Methods). After extensive washing, the beads were resuspended in SDS loading buffer and subjected to SDS-PAGE (lanes 1, 3, and 5). The supernatants (flowthrough [F/T]) of the binding reactions were also analyzed (lanes 2, 4, and 6). Positions of migration of the NF-E4 load and the molecular weight standards are labeled in kilodaltons. (B) Coimmunoprecipitation of CP2 and NF-E4 from native K562 cells. Nuclear extract from K562 cells was immunoprecipitated with polyclonal antiserum to NF-E4 (lane 1) or CP2 (lanes 3 [8 μl] and 4 [4 μl]) or the corresponding preimmune (PI) serum (lane 2 or 5, respectively). The immunoprecipitates were fractionated by SDS-PAGE and blotted with antisera to NF-E4. Positions of migration of NF-E4 and the molecular mass standards (in kilodaltons) are labeled.
NF-E4 is a component of the SSP complex.
To confirm that NF-E4 contributed to the formation of the SSP, we examined the effect of anti-NF-E4 antiserum on the SSP-SSE interaction in an EMSA. As shown in Fig. 4A, addition of either anti-CP2 (lane 2) or anti-NF-E4 (lane 4) antiserum to crude K562 cell nuclear extract specifically ablated the formation of the SSP-SSE complex, leaving the Sp1-SSE complex unaltered. Addition of preimmune sera had no effect (lanes 1 and 3).
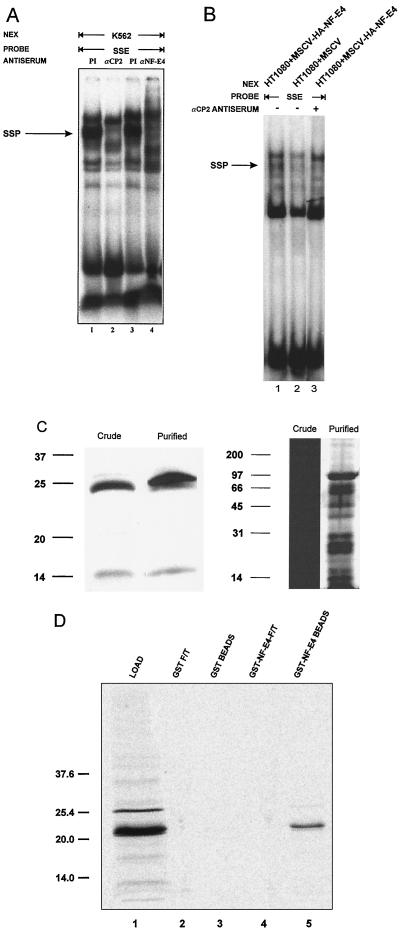
FIG. 4.
NF-E4 is a component of the SSP. (A) NF-E4 antiserum disrupts the SSP-SSE complex. Crude K562 cell nuclear extract (NEX) was studied in EMSA with the SSE probe in the presence of anti-CP2 antiserum (lane 2), anti-NF-E4 antiserum (lane 4), or the corresponding preimmune (PI) serum (lane 1 or 3, respectively). Migration of the SSP-SSE complex is arrowed. (B) Expression of NF-E4 in a null cell line generates a de novo SSP complex. Nuclear extract was prepared from the human sarcoma cell line HT1080 transduced with MSCV-HA-NF-E4 (lane 1) or MSCV (lane 2) and analyzed by EMSA using an SSE probe. The effect of anti-CP2 antisera on the EMSA from MSCV-HA-NF-E4 was examined in lane 3. Migration of the SSP complex is arrowed. (C) Western analysis and silver stain of NF-E4 in purified SSP. Crude K562 cell nuclear extract or purified SSP was resolved on a 12% polyacrylamide gel, transferred to a PVDF membrane, probed with polyclonal anti-NF-E4 antiserum and de-veloped with ECL (left); the samples were also electrophoresed, and the gel was silver stained (right). Molecular size standards are indicated in kilodaltons. (D) NF-E4 forms homodimeric complexes. GST–NF-E4 or GST alone was expressed in E. coli, and 1 μg of protein was bound to glutathione-Sepharose beads. The beads were then incubated with 2 μl of 35S-labeled in vitro-translated NF-E4 in binding buffer for 1 h at room temperature (see Materials and Methods). After extensive washing, the beads were resuspended in SDS loading buffer and subjected to SDS-PAGE (lanes 1 and 3). The supernatants (flowthrough [F/T]) of the binding reactions were also analyzed (lanes 2 and 4). Positions of migration of NF-E4 and the molecular mass standards (in kilodaltons) are labeled.
To determine whether de novo expression of NF-E4 in a CP2-expressing NF-E4 null cell line could result in the formation of the SSP complex, we transduced the human sarcoma cell line HT1080 with an MSCV-based retrovirus containing the NF-E4 cDNA tagged at the NH2 terminus with HA (MSCV-HA-NF-E4). This cell line contains abundant CP2 but no NF-E4 at the RNA or protein level (data not shown). HT1080 cells transduced with MSCV alone served as the control. GFP-positive cells were selected by FACS, and expression of NF-E4 was confirmed by Western analysis (data not shown). EMSA using an SSE probe revealed the presence of a new complex in nuclear extract derived from MSCV-HA-NF-E4-transduced cells which comigrated with native SSP (Fig. 4B, lane 1). No complex was observed in the line transduced with MSCV alone (lane 2). Addition of anti-CP2 antisera to this EMSA ablated the SSP-SSE complex (lane 3). Further evidence for the role of NF-E4 in formation of the SSP complex was derived from Western analysis of an SSP fraction purified by heparin-Sepharose and DNA affinity chromatography. As shown in the left panel of Fig. 4C, both the 22-kDa and the lower-molecular-mass NF-E4 species were detected in crude and purified samples.
One confounding result from our previous UV cross-linking studies was the prediction that the molecular mass of the CP2 partner protein in the SSP was 40 to 45 kDa (29). In view of this, we examined whether the 22-kDa NF-E4 protein could form homodimeric complexes in GST chromatography experiments. GST alone or fused in frame with full-length NF-E4 (GST–NF-E4) was coupled to glutathione-Sepharose beads and incubated under stringent conditions with [35S]methionine-labeled in vitro-transcribed-translated NF-E4. Specific retention of NF-E4 was observed with the GST–NF-E4 beads (lane 3) but not control GST beads (lane 1) (Fig. 4D). This finding coupled with our UV cross-linking data suggests that the SSP complex is composed of two NF-E4 molecules linked to a molecule of CP2.
NF-E4 demonstrates a highly restricted pattern of expression.
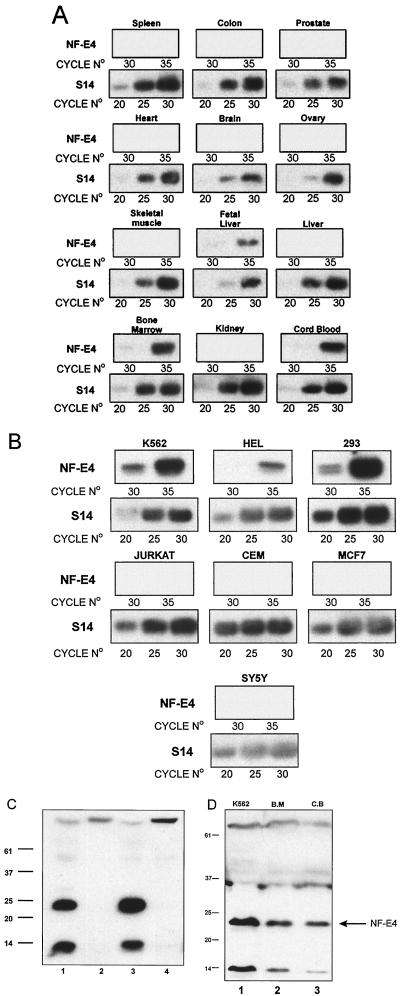
To determine the tissue distribution of NF-E4 expression, we initially performed Northern analysis on mRNA derived from K562 cells. Despite the fact that we had demonstrated the presence in these cells of NF-E4 mRNA (by RT-PCR) and protein (by Western blotting and EMSA), we were unable to detect a signal with a variety of NF-E4 cDNA probes (data not shown). Additional analysis of multitissue Northern blots also failed to detect a signal in a variety of tissues, as did RNase protection analysis on mRNA from tissues and cell lines (data not shown). We therefore proceeded to define the expression pattern of NF-E4 using RT-PCR. Based on the genomic sequence, we designed primers from exons 1 and 2 which are separated by an 1,800-bp intron. The identity of the correct-size PCR product was confirmed by Southern analysis using an internal primer as a probe. As shown in Fig. 5A, NF-E4 is expressed in fetal liver, cord blood, and bone marrow. No expression was observed from a variety of other organs, including colon, heart, spleen, kidney, liver, lymph node, and thymus (Fig. 5A and data not shown). In RT-PCR analysis of cell lines, expression was demonstrated with mRNA derived from the fetal and erythroid cell lines K562 and HEL and the embryonic kidney cell line 293T. No product was amplifiable from a variety of other lines, including Jurkat, CEM, MCF7, DU528, SY5Y, and COS (Fig. 5B and data not shown). To confirm the expression of NF-E4 at the protein level, nuclear extract from 293T, COS, K562, and HeLa cell lines was analyzed by Western blotting. As shown in Fig. 5C, the previously defined immunoreactive species were detected in K562 (lane 3) and 293T (lane 1) extracts. Additional Western analysis (Fig. 5D) of cord blood (lane 3)- and bone marrow (lane 2)-derived nuclear extract demonstrated a dominant band of 22 kDa which comigrated with that observed in control K562 extract (lane 1). In addition, the previously recognized smaller species was also detected in all lanes.
FIG. 5.
Expression of NF-E4 in primary tissues and established cell lines. First-strand cDNA transcribed from poly(A)+ RNA from multiple primary tissues or cell lines was used as a template to PCR amplify a product using primers specific for S14. Samples were then diluted and reamplified to give comparable band intensities for the same number of amplification cycles of S14 RNA within the linear range of the assay; this represents 20, 25, and 30 cycles. (A) Expression of NF-E4 in primary human tissues. Based on the S14 quantitation, comparable amounts of cDNA from multiple primary human tissues were PCR amplified using primers specific for NF-E4. These primers span a 1.8-kb intron and thus discriminate between mRNA and genomic DNA-derived signal. Cycle numbers were chosen to represent the linear range of amplification; this represents 30 and 35 cycles. (B) Expression of NF-E4 in cell lines. Based on the S14 quantitation, comparable amounts of cDNA from multiple established cell lines were PCR amplified using primers specific for NF-E4. RNA was prepared from erythroid lines (K562 and HEL), T-cell lines (CEM and Jurkat), a human embryonic kidney cell line (293), a breast cell line (MCF7), and a brain cell line (SY5Y). All PCR products were electrophoresed on 1% agarose, transferred to nitrocellulose, and probed with an internal radiolabeled oligonucleotide specific for the predicted product. (C) Western analysis of NF-E4 in cell lines. Nuclear extract isolated from 293T (lane 1), COS (lane 2), K562 (lane 3), and HeLa (lane 4) cell lines was resolved on a 12% polyacrylamide gel, transferred to a PVDF membrane, probed with polyclonal anti-NF-E4 antiserum, and developed with ECL. Molecular size standards are indicated in kilodaltons. (D) NF-E4 protein in cord blood and bone marrow. CD34+ cells were isolated from fresh bone marrow (B.M; lane 2) or cord blood (C.B; lane 3) and cultured for 7 days (see Materials and Methods). Nuclear extracts were prepared from both samples and resolved by SDS-PAGE using a 12% gel. Crude K562 nuclear extract served as the control (lane 1). After transfer to PVDF, the samples were immunoblotted with anti-NF-E4 antiserum and developed with ECL. Positions of migration of NF-E4 and the molecular size standards (in kilodaltons) are indicated.
Enforced expression of NF-E4 in K562 cells induces fetal and embryonic globin gene expression.
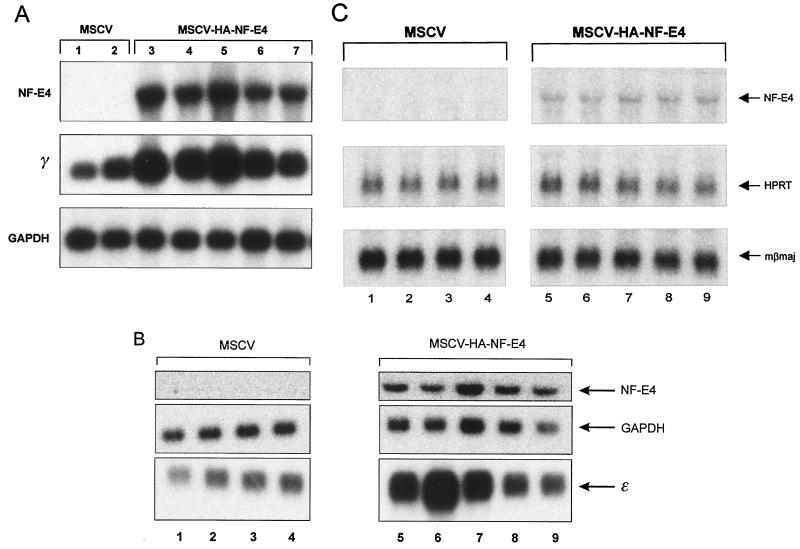
To examine the functional role of NF-E4 in globin gene expression, we used MSCV-HA-NF-E4. K562 cells were transduced with this vector or the parent GFP-containing vector (MSCV) and then sorted twice for green fluorescence by FACS and expanded in oligoclonal pools. All pools were subsequently shown to contain more than 99% GFP-positive cells by FACS analysis (data not shown). Northern analysis of pools derived from MSCV-HA-NF-E4-transduced cells showed a significant upregulation (5- to 10-fold as an average of all pools) of γ-gene expression compared to pools from the MSCV-transduced cells (Fig. 6A). Expression of the housekeeping gene (GAPDH) was unchanged between pools.
FIG. 6.
Enforced expression of NF-E4 induces γ- and ɛ-gene expression. (A) Northern analysis of K562 cell pools overexpressing NF-E4. K562 cells were transduced with either MSCV-HA-NF-E4 or MSCV alone, and GFP-positive cells were obtained by FACS. Cells were expanded, resorted, and cultured in oligoclonal pools; 10 μg of total RNA from five MSCV-HA-NF-E4 pools (lanes 3 to 7) or two MSCV pools (lanes 1 and 2) was analyzed with γ-gene and NF-E4 probes. GAPDH served as the control. (B) Enforced expression of NF-E4 induces ɛ-gene expression. K562 cells were transduced with either MSCV-HA-NF-E4 or MSCV, and GFP-positive cells were obtained by FACS. Cells were expanded, resorted, and cultured in oligoclonal pools; 10 μg of total RNA from four MSCV pools (lanes 1 to 4) or five MSCV-HA-NF-E4 pools (lanes 5 to 9) was analyzed with ɛ-gene and NF-E4 probes. GAPDH served as the control. (C) Northern analysis of MEL cell pools overexpressing NF-E4. MEL cells were transduced with either MSCV-HA-NF-E4 or MSCV alone, and GFP-positive cells were obtained by FACS. Cells were expanded, resorted, and cultured in oligoclonal pools; 10 μg of total RNA from four MSCV pools (lanes 1 to 4) or five MSCV-HA-NF-E4 pools (lanes 5 to 9) was analyzed with βmaj gene and NF-E4 probes. HPRT served as the control.
A previous report has suggested a potential role for the SSP in embryonic gene regulation (22). We therefore examined the effects of enforced expression of NF-E4 on ɛ-gene expression. Total RNA from oligoclonal pools of K562 cells transduced with either the MSCV (lanes 1 to 4) or MSCV-HA-NF-E4 (lanes 5 to 9) was analyzed by Northern blotting (Fig. 6B). Results comparable to those observed for γ-gene expression were obtained, with significant induction of ɛ-gene expression in the cells transduced with MSCV-HA-NF-E4. No evidence of β-gene activation was observed in any pool (data not shown).
To evaluate the specificity of NF-E4 activity, we transduced a murine erythroleukemia cell line (MEL) with either MSCV (lanes 1 to 4) or MSCV-HA-NF-E4 (lanes 5 to 9) and sorted for GFP expression. Oligoclonal pools were analyzed by Northern blotting. As shown in Fig. 6C, no change in murine βmaj expression was observed. Expression of murine ɛy and βH1 was undetectable in all clones (data not shown).
Enforced expression of NF-E4 in cord blood progenitors induces γ-gene and represses β-gene expression.
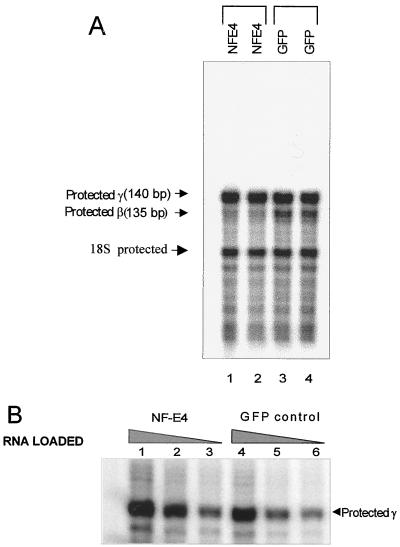
To extend the functional studies of NF-E4, we generated stable FLYRD18 producer cell lines containing either MSCV or MSCV-HA-NF-E4 (10). Supernatants from these lines were used to transduce CD34+ cells derived from human cord blood (see Materials and Methods). The cells were then cultured for 12 days in differentiation medium, and GFP- and glycophorin A-positive cells were separated by FACS. RNA was prepared from these cells and analyzed by RNase protection assay. As shown in Fig. 7A, the most striking difference between the MSCV-NF-E4 and control MSCV pools was the reduction in β-gene expression in the NF-E4-transduced pools. After normalization for the housekeeping gene 18S, the reduction in β-gene expression induced by NF-E4 was approximately 10-fold. As the signal detected with the γ-gene probe in this assay was intense in both MSCV and NF-E4 pools, we examined various dilutions of RNA to determine whether a difference was evident at lower concentrations. As shown in Fig. 7B, analysis in the linear portion of the assay revealed a twofold increase in γ-gene expression in pools transduced with MSCV-NF-E4 compared with those transduced with the control MSCV retrovirus.
FIG. 7.
Enforced expression of NF-E4 in cord blood progenitors induces γ-gene and represses β-gene expression. (A) RNase protection assays on RNA from transduced cord blood CD34+ cells. CD34+ cells were isolated from fresh cord blood and transduced with either MSCV-HA-NF-E4 or the MSCV generated in the FLYRD18 packaging cell line (see Materials and Methods). Cells were expanded in in vitro differentiation culture (59) for 12 days, and then GFP- and glycophorin A-positive cells were obtained by FACS; 1 μg of total RNA was used in each lane, and the predicted protected fragments for human γ- and β-globin are shown on the left. (B) Quantitation of γ-gene expression by RNase protection assay. Total RNA (lanes 1 and 4, 0.8 μg; lanes 2 and 5, 0.4 μg; lanes 3 and 6, 0.1 μg) from MSCV-HA-NF-E4 (lanes 1 to 3) or MSCV (lanes 4 to 6) transduced cord blood CD34+ cells (as detailed above) was used; the predicted protected fragment for human γ-globin and 18S is shown on the right.
DISCUSSION
In this report we detail the molecular cloning and characterization of human NF-E4, a novel gene encoding the tissue-restricted component of the SSP complex. The gene was isolated from a yeast two-hybrid screen of a K562 cell cDNA library using CP2, the previously identified ubiquitous component of the SSP, as the bait. As predicted from our previous studies (29), NF-E4 is essential for DNA binding of the SSP, as demonstrated by the disruption of the SSP-SSE complex induced by NF-E4 antiserum and the activation in the yeast one-hybrid assay induced by NF-E4. Based on our GST chromatographic assays and previous UV cross-linking data, it appears that the SSP is composed of two molecules of NF-E4 linked to a single molecule of CP2.
Analysis of the NF-E4 cDNA and protein sequence revealed no homology to known genes or ESTs. Specifically, no known DNA binding, protein dimerization, or transactivation domains were evident. Protein translation appears to commence at a CUG codon. This is supported by our in vitro transcription-translation assays, analysis of in vivo translation with retroviral vectors, and the molecular weight of the native protein. It is, however, conceivable that a very small region of additional 5′ coding sequence exists, despite the multiple 5′ RACE clones which terminated at the same nucleotide. Efforts to formally map the 5′ end of the transcript by RNase protection and primer extension were unsuccessful due to the rarity of the transcript in expressing cells (data not shown). Despite this, the NF-E4 clone that we have isolated appears to fulfill its predicted functional role.
Non-AUG initiation has been previously reported for a variety of mammalian proteins, most often from CUG codons (6, 33, 46). However, studies of some mammalian proteins and of viral mRNA in mammalian cells have also demonstrated initiation mediated by GUG, ACG, AUA, and AUU (38). Although our evidence for CUG initiation of native NF-E4 is indirect, it is significant that none of these alternate initiating codons are present either 5′ or within 125 bp 3′ of the CUG. It is also significant that, like many of the other factors with non-AUG-initiated isoforms, NF-E4 may also exist in a truncated form initiated from a downstream AUG (7, 44). This is evidenced by the smaller species observed in Western analyses and immunoprecipitation experiments with anti-NF-E4 antisera. Although it is possible that this smaller species represents a proteolytic cleavage product of NF-E4, the generation of a comigrating protein from an MSCV carrying the NF-E4 cDNA truncated to this ATG suggests that this species represents the product of alternate translation initiation (data not shown). The use of non-AUG initiation codons in many proteins plays a key regulatory role (7, 23, 34, 39, 41, 50, 55, 68). For example, the CUG- and AUG-initiated isoforms of the steroid receptor binding protein Bag-1 and the proto-oncogene products Int2 and Hck-1 differ in their subcellular localization (1, 36, 44). In addition, the CUG isoform of Bag-1 interacts with different protein partners and consequently has a unique functional role (17). Interestingly, the smaller NF-E4 peptide was immunoprecipitated with anti-NF-E4 antisera but not coimmunoprecipitated with anti-CP2 antisera. This suggests that there may be intrinsic functional differences between these species.
The demonstration of NF-E4 mRNA and/or protein in fetal liver, bone marrow, and cord blood raises the question of the developmental stage specificity of the SSP complex. This finding is analogous to the expression pattern observed for another stage-specific globin regulatory factor, EKLF, which is present at both mRNA and protein level in yolk sac, fetal liver, and adult bone marrow (12; Jane and Cunningham, unpublished). Despite this, β-gene expression is observed only in definitive erythroid cells, and mice nullizygous for EKLF demonstrate no abnormalities in primitive erythropoiesis (42, 48). The mechanism underlying this selectivity remains unknown, although the increase in γ-gene transcription observed in the fetal livers of EKLF−/− mice transgenic for the human globin locus suggests that it may be influenced by promoter competition (47, 66). Functionally, NF-E4 also displays a high degree of selectivity, as it induces fetal and embryonic but not adult globin gene expression. The lack of β-gene induction is observed in the context of K562 cells, in which constitutive β-gene expression is absent, and MEL cells, in which high levels of β-globin gene expression are observed. In contrast, induction of fetal and embryonic globin is observed only in cells in which these genes are normally transcribed. This suggests that the chromatin modifications associated with fetal-embryonic gene silencing cannot be altered simply by changing the transcription factor milieu. This observation is comparable to the effects of enforced expression of EKLF, which fails to induce β-gene expression in cells in which the adult globin gene is constitutively silent (12).
The suppression of β-gene and augmentation of γ-gene expression observed in primary cord blood progenitors are of interest, as this is a cell population in which γ- and β-globin are normally expressed concurrently. This finding suggests that in the context of promoter competition, the presence of NF-E4 is sufficient to alter the balance between the promoters, favoring transcription of the fetal gene. This finding is reminiscent of the studies of EKLF nullizygous mice carrying the human β-globin locus yeast artificial chromosome (β-YAC mice), in which enhanced γ-gene expression is observed in the fetal liver due to the diminution of effective competition from the inactive β promoter (47, 66). The difference in magnitude between β-globin suppression and γ-gene activation in the cord blood progenitors in the setting of enforced expression of NF-E4 is intriguing. One interpretation is that although the γ promoter has the competitive advantage over the β promoter in the presence of NF-E4, allowing its preferential interaction with the LCR, other factors necessary for optimal γ-gene expression are diminishing as the switch from fetal to adult globin expression progresses. This interpretation is consistent with our previous findings which suggest that binding of the SSP confers only weak transcriptional activation in the setting of promoter competition (28). Studies of the effects of enforced expression of NF-E4 in earlier developmental stages in the β-YAC transgenic line will further address this issue (18).
As low levels of fetal globin expression are detectable in adult bone marrow, the presence of some NF-E4 in this tissue is not surprising. However, the levels observed appear similar to those observed in cord blood, in which the expression of the γ genes is appreciably greater. This finding may indicate that another fetal factor such as FKLF is also required for the developmental specificity of γ-gene expression (5).
Our previous studies have suggested that the enhancer activity of the LCR is influenced by the developmentally specific factors EKLF and the SSP (2). This effect is not mediated by direct protein-protein interactions between the stage-specific promoter-bound factors and factors bound to the LCR (19). An alternate model is that EKLF and the SSP alter promoter structure, rendering it more amenable to the effects of the LCR enhanceosome. This model is supported by the loss of HS formation in the β promoter in EKLF nullizygous mice and by the recent demonstration of complex formation between EKLF and the SWI-SNF chromatin remodeling factors (4, 66). Studies addressing the role of NF-E4 in chromatin modification are in progress.
The ultimate goal of defining factors which activate fetal globin is their potential for therapeutic intervention in the hemoglobinopathies. Patients with β-thalassemia and sickle cell disease who also inherit genetic mutations which prolong fetal globin expression after birth have a significantly ameliorated clinical course (49). To date, three factors which augment γ-gene expression in cell lines have been identified. The first of these, the helix-loop-helix protein Id2, is ubiquitously expressed and likely to have pleiotropic effects on gene regulation (26). However, NF-E4 and FKLF have highly restricted patterns of expression and offer promise for both pharmacological manipulation and gene therapy. The ability of enforced expression of NF-E4 in cord blood progenitors to suppress β-gene expression may have significant implications in genetic therapy of sickle cell disease with the dual beneficial effects of enhanced fetal globin expression and reduction of βS synthesis. Studies of NF-E4 in mouse models of human hemoglobin switching (18) and hemoglobinopathies (9, 45, 52) represent the next step in the evaluation of this factor as a therapeutic tool.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Stuart Orkin, Art Nienhuis, Jerry Adams, and Glenn Begley for critical reading of the manuscript, Robert Hawley for the gift of the MSCV plasmid, Patrick Kelly and Derek Persons for the gift of the RD18 packaging cell line, and Amy McEwan and Helen Zogos for technical assistance. We also thank members of the Jane and Cunningham laboratories for helpful discussions and Art Nienhuis for continuing support.
This work was supported by the NHMRC of Australia, The Wellcome Trust (S.M.J.), the Anti-Cancer Council of Victoria (D.R.C.), NIH PO1 HL53749-03, Cancer Center Support CORE grant P30 CA 21765, the American Lebanese Syrian Associated Charities (ALSAC), and the Assisi Foundation of Memphis.
REFERENCES
- 1.Acland P, Dixon M, Peters G, Dickson C. The subcellular fate of the Int-2 oncoprotein is determined by choice of initiation codon. Nature. 1990;343:662–665. doi: 10.1038/343662a0. [DOI] [PubMed] [Google Scholar]
- 2.Amrolia P J, Cunningham J M, Jane S M. Maximal activity of an erythroid-specific enhancer requires the presence of specific protein binding sites in linked promoters. J Biol Chem. 1998;273:13593–13598. doi: 10.1074/jbc.273.22.13593. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K P, Lloyd J A, Ponce E, Crable S C, Neumann J C, Lingrel J B. Regulated expression of the human β-globin gene in transgenic mice requires an upstream globin or nonglobin promoter. Mol Biol Cell. 1993;4:1077–1085. doi: 10.1091/mbc.4.10.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodelling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 5.Asano H, Li X S, Stamatoyannopoulos G. FKLF, a novel Krüppel-like factor that activates human embryonic and fetal β-like globin genes. Mol Cell Biol. 1999;19:3571–3579. doi: 10.1128/mcb.19.5.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruening W, Pelletier J. A non-AUG translational initiation event generates novel WT1 isoforms. J Biol Chem. 1996;271:8646–8654. doi: 10.1074/jbc.271.15.8646. [DOI] [PubMed] [Google Scholar]
- 8.Choi O R B, Engel J D. Developmental regulation of β-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 9.Ciavatta D J, Ryan T M, Farmer S C, Townes T M. Mouse model of human β0-thalassemia: targeted deletion of the mouse βmaj and βmin-globin genes in embryonic stem cells. Proc Natl Acad Sci USA. 1995;92:9259–9263. doi: 10.1073/pnas.92.20.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosset F-L, Takeuchi Y, Battini J-L, Weiss R A, Collins M K L. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 12.Donze D, Townes T M, Bieker J J. Role of erythroid Kruppel-like factor in human γ- to β-globin switching. J Biol Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 13.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I K, Kennedy M, Keller G, Groudine M. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 14.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 15.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 16.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froesch B A, Takayama S, Reed J C. BAG-1L protein enhances androgen receptor function. J Biol Chem. 1998;273:11660–11666. doi: 10.1074/jbc.273.19.11660. [DOI] [PubMed] [Google Scholar]
- 18.Gaensler K M L, Kitamura M, Kan Y W. Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human β-globin locus in transgenic mice. Proc Natl Acad Sci USA. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallarda J L, Foley K P, Yang Z, Engel J D. The β-globin stage selector element factor is erythroid-specific promoter/enhancer binding protein NF-E4. Genes Dev. 1989;3:1845–1859. doi: 10.1101/gad.3.12a.1845. [DOI] [PubMed] [Google Scholar]
- 20.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 21.Gumucio D L, Blanchard-McQuate K L, Heilstedt-Williamson H, Tagle D A, Gray T A, Tarle S A, Gragowski L, Goodman M, Slightom J L, Collins F S. Gamma-globin gene regulation: evolutionary approaches. In: Stamatoyannopoulos G, Nienhuis A W, editors. The regulation of hemoglobin switching. Proceedings of the Seventh Conference on Hemoglobin Switching. Baltimore, Md: The Johns Hopkins University Press; 1991. pp. 277–289. [Google Scholar]
- 22.Gumucio D L, Shelton D A, Bailey W J, Slightom J L, Goodman M. Phylogenetic footprinting reveals unexpected complexity in trans factor binding upstream from the ɛ-globin gene. Proc Natl Acad Sci USA. 1993;90:6018–6022. doi: 10.1073/pnas.90.13.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hann S R, Sloan-Brown K, Spotts G D. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes Dev. 1992;6:1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- 24.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 25.Holmes M L, Haley J D, Cerruti L, Zhou W-L, Zogos H, Smith D E, Cunningham J M, Jane S M. Identification of Id2 as a globin regulatory protein by representational difference analysis of K562 cells induced to express γ-globin with a fungal compound. Mol Cell Biol. 1999;19:4182–4190. doi: 10.1128/mcb.19.6.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jane S M, Cunningham J M. Understanding fetal globin gene expression: a step towards effective HbF reactivation in hemoglobinopathies. Br J Hematol. 1998;102:415–422. doi: 10.1046/j.1365-2141.1998.00811.x. [DOI] [PubMed] [Google Scholar]
- 27.Jane S M, Gumucio D L, Ney P A, Cunningham J M, Nienhuis A W. Methylation enhanced binding of Sp1 to the stage selector element of the human γ-globin gene promoter may regulate developmental specificity of expression. Mol Cell Biol. 1993;13:3272–3281. doi: 10.1128/mcb.13.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jane S M, Ney P A, Vanin E F, Gumucio D L, Nienhuis A W. Identification of a stage selector element in the human γ-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the β-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jane S M, Nienhuis A W, Cunningham J M. Hemoglobin switching in man and chicken is mediated by a heteromeric complex between the ubiquitous transcription factor CP2 and a developmentally specific protein. EMBO J. 1995;14:97–105. doi: 10.1002/j.1460-2075.1995.tb06979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim C H, Heath C, Bertuch A, Hansen U. Specific stimulation of simian virus 40 late transcription in vitro by a cellular factor binding the simian virus 40 21-base-pair repeat promoter element. Proc Natl Acad Sci USA. 1987;84:6025–6029. doi: 10.1073/pnas.84.17.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kioussis D, Vanin E F, deLange T, Flavell R A, Grosveld F. β-Globin inactivation by DNA translocation in γβ-thalassemia. Nature. 1983;306:662–664. doi: 10.1038/306662a0. [DOI] [PubMed] [Google Scholar]
- 32.Kozak M. An analysis of 5′-non-coding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8145. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Downstream secondary structure facilitates recognition of initiation codons by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemaire P, Vesque C, Schmitt H, Stunnenberg H, Frank R, Charnay P. The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol Cell Biol. 1990;10:3456–3467. doi: 10.1128/mcb.10.7.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim L C, Swendeman S L, Sheffery M. Molecular cloning of the α-globin transcription factor CP2. Mol Cell Biol. 1992;12:828–835. doi: 10.1128/mcb.12.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lock P, Ralph S, Stanley E, Boulet I, Ramsay R, Dunn A R. Two isoforms of murine hck generated by utilization of alternative translational initiation codons exhibit different patterns of subcellular localization. Mol Cell Biol. 1991;11:4363–4370. doi: 10.1128/mcb.11.9.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozzio C B, Lozzio B B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 38.Mehdi H, Ono E, Gupta K C. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene. 1990;91:173–178. doi: 10.1016/0378-1119(90)90085-6. [DOI] [PubMed] [Google Scholar]
- 39.Mellentin J D, Smith S D, Cleary M L. Lyl-1, a novel gene altered by chromosomal translocation in T-cell leukemia, codes for a protein with a helix-loop-helix DNA binding motif. Cell. 1989;58:77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- 40.Moritz T, Dutt P, Xiao X, Carstanjen D, Vik T, Hanenberg H, Williams D A. Fibronectin improves transduction of reconstituting hematopoietic stem cells by retroviral vectors: evidence of direct viral binding to chymotryptic carboxy-terminal fragments. Blood. 1996;88:855–862. [PubMed] [Google Scholar]
- 40a.Morley B J, Abbott C A, Wood W G. Regulation of human fetal and adult globin genes in mouse erythroleukemia cells. Blood. 1991;78:1355–1363. [PubMed] [Google Scholar]
- 41.Nagpal S, Zelent A, Chambon P. RAR-beta 4, a retinoic acid receptor isoform is generated from RAR-beta 2 by alternative splicing and usage of a CUG initiator codon. Proc Natl Acad Sci USA. 1992;89:2718–2722. doi: 10.1073/pnas.89.7.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective hematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 43.Orkin S H. Regulation of globin gene expression in erythroid cells. Eur J Biochem. 1995;231:271–281. doi: 10.1111/j.1432-1033.1995.tb20697.x. [DOI] [PubMed] [Google Scholar]
- 44.Packham G, Brimmell M, Cleveland J L. Mammalian cells express two differently localised Bag-1 isoforms generated by alternative translation initiation. Biochem J. 1997;328:807–813. doi: 10.1042/bj3280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paszty C, Brion C M, Manci E, Witkowska H E, Stevens M E, Mohandas N, Rubin E M. Transgenic knockout mice with exclusively human sickle hemoglobin and sickle cell disease. Science. 1997;278:876–878. doi: 10.1126/science.278.5339.876. [DOI] [PubMed] [Google Scholar]
- 46.Peabody D S. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 47.Perkins A C, Gaensler K M L, Orkin S H. Silencing of human fetal globin expression is impaired in the absence of the adult β-globin gene activator protein, EKLF. Proc Natl Acad Sci USA. 1996;93:12267–12271. doi: 10.1073/pnas.93.22.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkins A C, Sharpe A H, Orkin S H. Lethal β-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 49.Poncz M P, Henthorn P, Stoekert C, Surrey S. Globin gene expression in hereditary persistence of fetal hemoglobin and δβ0-thalassemia. In: McLean N, editor. Oxford surveys of eukaryotic genes. Oxford, United Kingdom: Oxford University Press; 1989. pp. 163–203. [PubMed] [Google Scholar]
- 50.Prats H, Kaghad M, Prats A C, Klagsburn M, Lelias J M, Liauzun P, Chalon P, Tauber J P, Amalric F, Smith J A, Caput D. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rowley P T, Ohlsson-Wilhelm B M, Wisniewski L, Lozzio C B, Lozzio B B. K562 human leukemia cell passages differ in embryonic globin gene expression. Leuk Res. 1984;8:45–54. doi: 10.1016/0145-2126(84)90030-4. [DOI] [PubMed] [Google Scholar]
- 52.Ryan T M, Ciavatta D J, Townes T M. Knockout-transgenic mouse model of sickle cell disease. Science. 1997;278:873–876. doi: 10.1126/science.278.5339.873. [DOI] [PubMed] [Google Scholar]
- 53.Sabatino D E, Cline A P, Gallagher P G, Garrett L J, Stamatoyannopoulos G, Forget B G, Bodine D M. Substitution of the human β-spectrin promoter for the human Aγ-globin promoter prevents silencing of a linked human β-globin gene in transgenic mice. Mol Cell Biol. 1998;18:6634–6640. doi: 10.1128/mcb.18.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Saris C J M, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991;10:655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shirra M K, Zhu Q, Huang H-C, Pallas D, Hansen U. One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol Cell Biol. 1994;14:5076–5087. doi: 10.1128/mcb.14.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatoyannopoulos G, Nienhuis A W. Hemoglobin switching. In: Stamatoyannopoulos G, Nienhuis A W, Majerus P J, Varmus H, editors. The molecular basis of blood diseases. 2nd ed. Philadelphia, Pa: W. B. Saunders; 1994. pp. 107–156. [Google Scholar]
- 58.Starck J, Sarkar R, Romana M, Bhargava A, Scarpa A L, Tanaka M, Chamberlain J W, Weissman S M, Forget B G. Developmental regulation of human γ- and β-globin genes in the absence of the locus control region. Blood. 1994;84:1656–1665. [PubMed] [Google Scholar]
- 59.Sui X, Krantz S B, You M, Zhao Z. Synergistic activation of MAP kinase (ERK1/2) by erythropoietin and stem cell factor is essential for expanded erythropoiesis. Blood. 1998;92:1142–1149. [PubMed] [Google Scholar]
- 60.Tagle D A, Koop B F, Goodman M, Slightom J L, Hess D L, Jones R T. Embryonic ɛ and γ globin genes of a prosimian primate (Galago crassicaudatus) J Mol Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- 61.Taramelli R, Kioussis D, Vanin E F, Bartram K, Groffen J, Hurst J, Grosveld F G. γδβ-Thalassemias 1 and 2 are the result of a 100 kbp deletion in the human β-globin cluster. Nucleic Acids Res. 1986;14:7017–7029. doi: 10.1093/nar/14.17.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tuan D, Solomon W, Li Q, London I M. The “β-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uv A E, Thompson C R L, Bray S J. The Drosophila tissue-specific factor grainyhead contains novel DNA-binding and dimerization domains that are conserved in the human protein CP2. Mol Cell Biol. 1994;14:4020–4031. doi: 10.1128/mcb.14.6.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van der Ploeg L H, Konings A, Oort M, Roos D, Bernini L, Flavell R A. γβ-Thalassemia studies showing that deletion of the γ and δ genes influences β-globin expression in man. Nature. 1980;283:637–642. doi: 10.1038/283637a0. [DOI] [PubMed] [Google Scholar]
- 65.Vanin E F, Henthorn P S, Kioussis D, Grosveld F, Smithies O. Unexpected relationships between four large deletions in the human β-globin gene cluster. Cell. 1983;35:701–709. doi: 10.1016/0092-8674(83)90103-4. [DOI] [PubMed] [Google Scholar]
- 66.Wijgerde M, Gribnau J, Trimborn T, Nuez B, Philipsen S, Grosveld F, Fraser P. The role of EKLF in human β-globin gene competition. Genes Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 67.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 68.Xiao J H, Davidson I, Matthes H, Garnier J, Chambon P. Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 69.Yang Z, Engel J D. Biochemical characterisation of the developmental stage- and tissue-specific erythroid transcription factor, NF-E4. J Biol Chem. 1994;269:10079–10087. [PubMed] [Google Scholar]
- 70.Yoon J-B, Li G, Roeder R G. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol Cell Biol. 1994;14:1776–1785. doi: 10.1128/mcb.14.3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]