Abstract
The normal gut microbiome modulates host enterocyte metabolism and shapes local and systemic immunity. Accumulation of urea and other waste products in chronic kidney disease induces gut dysbiosis and intestinal wall inflammation (leaky gut). There are decreased numbers of bacteria that generate short-chain fatty acids, which are an important nutrient source for host enterocytes and also contribute to regulation of the host immune system. Anaerobic proteolytic bacteria that express urease, uricase and indole and p-cresol enzymes, such as Enterobacteria and Enterococci, are increased. Microbial-derived uremic toxins such as indoxyl sulfate and trimethylamine N-oxide contribute to the pathophysiology of immune-related kidney diseases such as diabetic nephropathy, lupus nephritis and immunoglobulin A (IgA) nephropathy. Animal and clinical studies suggest potential benefits of dietary and probiotic interventions in slowing the progression of immune-related kidney diseases.
Keywords: chronic kidney disease, diabetic nephropathy, gut microbiome, IgA nephropathy, lupus nephritis
THE GUT MICROBIOME IN HEALTH
The healthy adult gastrointestinal tract harbors ∼100 trillion microbes including 2000–4000 species, both aerobic and anaerobic. This is 10-fold greater than the number of cells in the human body [1, 2]. The genome of the gut microbiome encompasses 3.3 million genes, which is at least 150-fold larger than the human genome [3]. Bacteria are the most abundant (>50 phyla); archaea and eucarya are also present, and microbial density is the highest in the colon [4]. The anaerobic phyla Bacteroidetes (genus-level examples include Bacteroides, Prevotella) and Firmicutes (Clostridium, Eubacterium, Roseburia and Ruminococcus) contribute >90% of bacterial species [3]. Although the diversity of the human gut microbiota is influenced by gender, ethnicity, immune status, nationality, age, diet, geographic location, alcohol and drug consumption, and smoking [5, 6], >50% of healthy individuals share the same 75 bacterial species [3]. Bacterial abundance and diversity increase from the stomach (102–104 cells/mL) to the colon (>1012 cells/mL) as oxygen tension declines [7].
Gut microbes evolved symbiotically and confer important benefits to the host. The microbiota is involved in gut motility, modulates energy harvesting and absorption of complex carbohydrates, and is a source of micronutrients including short-chain fatty acids (SCFAs), vitamins such as B vitamins and vitamin K6, and amino acids such as lysine and threonine [8, 9]. SCFAs include acetate, butyrate, propionate and d-lactate, and are derived from fermentation of starch in the colon by Bacteroides; host enterocytes derive 60–70% of their energy from SCFA oxidation [10]. SCFAs from gut microbiota metabolism are present in hepatic, portal and peripheral blood, and influence lipid, glucose and cholesterol metabolism in various tissues [10]. SCFAs bind and activate G-protein coupled receptors free fatty acid receptor 2 (FFAR2, also called GPR 41) and free fatty acid receptor 3 (FFAR3, also called GPR43) expressed on immune cells, endocrine cells, enterocytes, adipose tissue and the autonomic nervous system, and modulate host energy homeostasis [2].
The gut microbiota offers direct protection against pathogens via production of antibiotics and bacteriocins. The gut mucosal barrier, consisting of a mucus layer, defensins and lectins, shields the epithelium from direct contact with microbiota [11]. Nonetheless, intestinal microbes modulate development of local as well as systemic immunity (Figure 1), shaping the composition of T cell and natural killer cells subsets as discussed below.
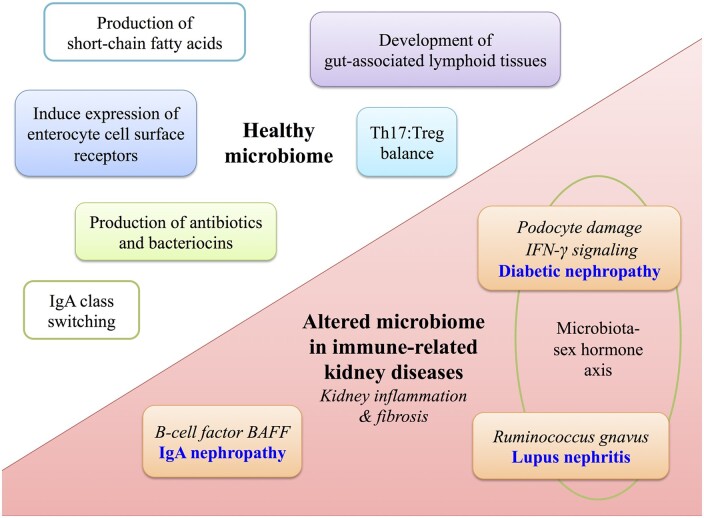
FIGURE 1.
Role of the gut microbiome in health and in autoimmune kidney diseases. Upper left: the healthy gut microbiome shapes local and systemic immunity including production of antibiotics and bacteriocins, induction of surface pattern recognition receptors on host intestinal cells and production of SCFAs. SCFAs provide a nutrient source to host intestinal cells and also modulate immune function via induction of natural killer cells and expansion of Tregs. The microbiota also modulates development of gut-associated lymphoid tissues (Peyer’s patches, mesenteric lymph nodes), shapes the balance between cytotoxic Th17 and Treg cells and induces IgA class switching. Lower right: the microbiome is altered in CKD and bacterial-derived uremic toxins translocate across the leaky gut and induce systemic effects including kidney inflammation and fibrosis. In diabetic nephropathy, the toxin phenyl sulfate induces podocyte damage and proteinuria. There is a microbiota–sex hormone axis whereby transfer of microbiota from male to female mice increases IFN-γ signaling and reduced incidence of diabetes. In lupus nephritis, higher gut abundance of Ruminococcus gnavus strain 2 is associated with more severe lupus nephritis kidney pathology. A microbiota–sex hormone axis is also observed, whereby female mice have more severe disease. Mouse studies indicate that overexpression of BAFF, a B-cell factor crucial for IgA synthesis, develop a commensal flora-dependent IgA nephropathy.
GUT MICROBIOTA SHAPE THE HOST IMMUNE SYSTEM
The normal microbial flora colonizes the intestinal tract after birth and modulates antigen-responsiveness of the lymphoid tissues [12]. Evidence from germ-free mice indicates that the gut microbiota is involved in the development of gut-associated lymphoid tissues including Peyer’s patches and mesenteric lymph nodes, antibody production [immunoglobulin A (IgA) class switching] and expression of surface pattern recognition receptors on intestinal epithelial cells [13].
Two T-cell subsets, regulatory T cells (Tregs) and T helper 17 (Th17) cells are enriched in the intestinal lamina propria and play critical roles in host defense and homeostasis. Th17 cells produce interleukin (IL)-17 and IL-22, two cytokines that stimulate the intestinal epithelium to produce anti-microbial peptides; Th17 cells are also critical for recruitment of neutrophils to eliminate bacteria that have traversed the epithelium [12]. If the latter is left unchecked, then there can be excessive gut wall inflammation with tissue destruction. Treg cells in turn are critical in providing immunologic self-tolerance countering Th17 effects, to avoid excessive inflammation; the gut microbiota shapes this balance between effector and Tregs [12]. SCFAs produced by Bacteroides also contribute to immune system activation through (i) neutrophil chemotaxis, (ii) induction of natural killer cells and (iii) differentiation and proliferation of Tregs (butyrate is the most potent SCFA in Treg expansion) [14, 15].
GUT DYSBIOSIS IN CHRONIC KIDNEY DISEASE
Disruptions in the gut microbiome have been implicated in progression of numerous conditions including chronic kidney disease (CKD), inflammatory bowel disease, metabolic disorders, cardiovascular diseases, cancer and allergic disorders [1, 16]. Simenhoff et al. [17] performed endoscopy studies in the 1970s and were the first to demonstrate markedly altered gut flora in CKD patients. They noted that antibiotic treatment altered the microbiome, decreased serum levels of amine toxins and patients reported improved mentation [18]. The duodenum and jejunum, which are only lightly colonized in the healthy individual, become intensely colonized by aerobic and anaerobic bacteria in CKD patients [17].
As further evidence of gut dysbiosis, gas chromatography studies have shown significantly altered exhaled breath gases in CKD rats [19] and end-stage kidney disease (ESKD) patients [20] compared with healthy controls. It has been proposed that influx of urea and other retained toxins in CKD, from the bloodstream into the gut lumen, selects for microbes that express urease, uricase and indole and p-cresol-forming enzymes [21, 22]. These changes in the gut flora are exacerbated by the CKD diet, which by virtue of being low in potassium and phosphorus equates to a diet low in fermentable plant fiber and poor in symbiont-rich cheese/yogurt [16]. The fiber-poor CKD diet also impacts microbial production of beneficial SCFAs; transit of plant-derived resistant starches to the colon is necessary for fermentation by Bacteroides to release hydrogen, carbon dioxide, alcohol and SCFAs [7]. Indeed, fecal analysis has demonstrated that dialysis patients show decreased numbers of bacteria that are able to produce the SCFA butyrate [22].
In an earlier study, our group isolated stool microbial DNA from ESKD patients on hemodialysis and a group of age-, sex- and ethnicity-matched healthy individuals. Via a phylogenic microarray technique, we demonstrated highly significant differences in the abundance of >200 bacterial operational taxonomic units (OTUs) belonging to 23 bacterial families between the ESKD and the healthy control groups; of note, there were lower numbers of Lactobacillaceae and Prevotellaceae families (both are considered normal colonic microbiota) and 100 times higher numbers of Enterobacteria and Enterococci anaerobic species (which are normally present in lower proportion) [21]. In order to isolate the effects of uremia from those of comorbid conditions, medications, diet and other inter-individual variations, the stool microbiome was studied in rats 8 weeks after 5/6 nephrectomy or sham operation. The CKD rats showed significant differences in the abundance of 175 bacterial OTUs, thus confirming the effect of CKD on gut dysbiosis [21]. In a subsequent study, stool microbial genomic analyses confirmed that hemodialysis patients demonstrated expansion of bacteria that express urease, uricase and p-cresyl- and indole-forming enzymes concurrent with depletion of the bacteria that produce SCFAs [22]. Among the 19 microbial families that were dominant in ESKD patients, 12 possessed urease (Alteromonadaceae, Cellulomonadaceae, Clostridiaceae, Dermabacteraceae, Enterobacteriaceae, Halomonadaceae, Methylococcaceae, Micrococcaceae, Moraxellaceae, Polyangiaceae, Pseudomonadaceae and Xanthomonadaceae), five possessed uricase (Cellulomonadaceae, Dermabacteraceaea, Micrococcaceae, Polyangiaceae and Xanthomonadaceae) and three possessed indole and p-cresol-forming enzymes (Clostridiaceae, Enterobacteriaceae and Verrucomicrobiaceae). Lactobacillaceae and Prevotellaceae, two species that generate the SCFA butyrate, were among the four microbial families that were depleted in ESKD patients [22].
Gut dysbiosis has similarly been reported in pediatric ESKD patients. Crespo-Salgado et al. examined the gut microbiome in pediatric patients undergoing hemodialysis or peritoneal dialysis; increased Bacteroidetes with decreased Proteobacteria was noted in pediatric hemodialysis patients, while increased Enterobacteriaceae and decreased Firmicutes and Actinobacteria was noted in pediatric peritoneal dialysis patients [23]. Increased numbers of glucose-fermentable bacteria Enterobacteriaceae in the setting of peritoneal dialysis was attributed to intestinal absorption of glucose from intraperitoneal dialysate [23]. It is important to note that despite the accumulating data regarding gut dysbiosis in animals and humans with CKD, it remains unknown to what degree these differences in the microbial subpopulations change microbial function and contribute to disease states.
In tandem with gut dysbiosis, there is widespread intestinal inflammation and increased gut permeability in CKD, with systemic consequences. Vaziri et al. described chronic inflammation throughout the gastrointestinal tract—extending from esophagus to large bowel—in chronic hemodialysis patients on autopsy studies performed in the 1980s [24]. In the early 1990s, studies utilizing polyethylene glycols of various molecular weights demonstrated increased permeability of the intestinal wall in CKD patients and rats [16]. To date, two mechanisms have been described that led to tight junction breakdown [16, 25]. Urea diffuses from the blood into the gut lumen and is metabolized by gut bacterial urease to ammonia. Ammonia is subsequently hydrolyzed into caustic ammonium hydroxide, which erodes the epithelial barrier and stimulates influx of inflammatory leukocytes. This triggers the second mechanism whereby local cytokine production induces retraction and endocytosis of transcellular tight junction proteins including claudins and occludin. Animal studies have confirmed that CKD is associated with depletion of gut epithelial tight junction proteins [26, 27].
This leaky gut barrier facilitates systemic translocation of gut bacterial DNA and products of bacterial protein catabolism; the latter generate uremic toxins [e.g. indoxyl sulfate, p-cresyl sulfate, indole-3 acetic acid, trimethylamine N-oxide (TMAO) and phenylacetylglutamine], which are detectable in the blood of CKD and ESKD patients [1]. Tryptophan is metabolized into indole by intestinal bacteria and is subsequently sulfated in the liver to form indoxyl sulfate. p-cresol/p-cresyl sulfate is derived from phenylalanine and tyrosine; p-cresol is conjugated by intestinal microbes to p-cresyl sulfate (the main circulating toxin) and p-cresyl glucoronidate (which is metabolized to p-cresyl glucuronide in the liver). TMAO is derived from bacterial metabolism of quaternary amines, such as phosphatidylcholine or L-carnitine which generates trimethylamine, with subsequent conversion to TMAO by flavin monooxygenase enzymes in the liver.
A definitive study by Aronov et al. [28] confirmed the colonic origin of microbial-derived uremic toxins (including p-cresyl sulfate and indoxyl sulfate) by mass spectroscopy analysis of plasma solutes from ESKD patients who had undergone total colectomy. Compared with ESKD and control subjects who had intact colons, the colectomized ESKD individuals had significantly lower or absent plasma levels of >30 solutes [28]. Nearly all of these compounds were significantly lower in healthy individuals, suggesting that they represented uremic solutes. These findings provide irrefutable evidence for colonic microbes being the source of numerous uremic solutes—many of which have yet to be identified.
GUT DYSBIOSIS AND IMMUNE-RELATED KIDNEY DISEASES
Gut-derived uremic toxins have been shown to promote kidney inflammation and fibrosis in animal models including 5/6 nephrectomized rats and ApoE-knockout mice [1]. Below we summarize evidence known to date about gut microbiome interactions with immune-related kidney diseases such as diabetic nephropathy, lupus nephritis and IgA nephropathy (Table 1 and Figure 1). Other end-organ dysfunction (cardiovascular, hematologic, neurohormonal, etc.) induced by systemic effects of gut-derived uremic toxins from the altered microbiome has been discussed in previous reviews [1, 2, 16, 25].
Table 1.
Changes in the gut microbiota in CKD and in immune-related kidney diseases
| Chronic disease state | Increased genera | Decreased genera |
|---|---|---|
| CKD | Adult dialysis patients: Enterobacteriaceae, Clostridia, Bacteroidia and Pseudomonadaceae Pediatric dialysis patients: Enterobacteriaceae, Bacteroidia | Adult dialysis patients: Bifidobacteriaceae, Lactobacillaceae and Prevotellaceae Pediatric dialysis patients: Proteobacteria, Firmicutes and Actinobacteria |
| Diabetic nephropathy | Type 1 diabetes mellitus: Bacteroides, Blautia, Rikenellaceae, Ruminococcus, Streptococcus, Clostridium perfringens, Veillonella and Fusobacteria Type 2 diabetes mellitus: Escherichia, Prevotella, Clostridium bolteae, Clostridium symbiosum, Clostridium ramosum, Clostridium hathewayi, Betaproteobacteria and Desulfovibrio | Type 1 diabetes mellitus: Lactobacillaceae, Bifidobacterium, Prevotella, Staphylococcus, Lachnospiraceae, Veillonellaceae, Coprococcus eutactus, Dialister invisus, Roseburia faecis, Faecalibacterium prausnitzii, Clostridium clostridioforme, Blautia coccoides, Pseudobutyrivibrio and Akkermansia muciniphila Type 2 diabetes mellitus: Bifidobacterium, Lactobacillaceae, Eubacterium rectale, Faecalibacterium, Roseburia intestinalis, Clostridiales, Akkermansia muciniphila (Verrucomicrobiaceae) and Streptococcus |
| Lupus nephritis | Lachnospiraceae, Lactobacillaceae, Bacteroides and RG | Firmicutes |
| IgA nephropathy | Sutterellaceae, Enterobacteriaceae and Firmicutes (latter includes genera/species of Ruminococcaceae, Lachnospiraceae, Eubacteriaceae and Streptococcaeae) | Clostridium, Lactobacillus, Enterococcus and Bifidobacterium |
DIABETIC NEPHROPATHY
In a study of 14 patients with biopsy-proven diabetic nephropathy, Tao et al. reported significant differences in expression of the Escherichia-Shigella and Prevotella genera compared with diabetic individuals who did not have kidney disease [29]. Dietary habits were not a determining factor, as household contacts of the diabetic patients had similar microbiota composition to non-household healthy controls. The investigators proposed that increased levels of Eschericha-Shigella contribute to the pathophysiology of diabetes by promoting breakdown of the gut epithelial barrier and by producing ethanol, which leads to disordered fatty acid metabolism [29]. Alterations in Lactobacillus and Bifidobacterium genera are common trend seen across proteinuric kidney diseases (Table 1) [30]. As further evidence of the important role of the gut microbiota in kidney disease, frequent use of antibiotics (which disrupts the balance of normal intestinal flora) has been shown to correlate with severity of diabetic nephropathy in patients with Type 1 diabetes [31].
Kikuchi et al. demonstrated that phenyl sulfate, derived from gut microbial metabolism of tyrosine, contributes to podocyte damage and albuminuria in animal CKD models [32]. They generated transgenic rats overexpressing the organic anion transporting polypeptide transporter SLCO4C1 in the proximal tubule to amplify renal excretion of uremic toxins; transgenic rats with streptozotocin-induced diabetes had less proteinuria than wild-type diabetic rats. Further investigations were done in diabetic mouse models (db/db and KKAy mice on high-fat diet), whereby oral administration of phenyl sulfate induced albuminuria and podocyte damage. Inhibition of tyrosine phenol-lyase, a bacterial enzyme responsible for phenol synthesis from dietary tyrosine before it is metabolized into phenyl sulfate in the liver, reduced albuminuria in diabetic mice. Kikuchi et al. also reported findings from the urinary biomarker for continuous and rapid progression of diabetic nephropathy patient cohort whereby baseline blood phenyl sulfate levels predict 2-year progression of albuminuria in patients with baseline microalbuminuria [32]. Similar to the autoimmunity in systemic lupus (discussed below), there appears to be a microbiota–sex hormone axis in Type 1 diabetes; transfer of microbiota from male to female mice was shown to raise testosterone and reduce diabetes incidence in female animals, partly due to enhanced Type I interferon (IFN)-γ signaling [33]. p-cresyl sulfate and TMAO via inducing accelerated aging pathways also likely contribute to the microvascular complications of diabetes including nephropathy [34]. Diabetes is independently associated with higher free p-cresol serum concentrations after adjustment for kidney function [35], suggesting that this gut-derived microbial toxin may be a shared pathologic factor in both diabetes and CKD.
LUPUS NEPHRITIS
Systemic lupus erythematosus (SLE) is a complex autoimmune disease associated with excessive signaling of RNA-sensing Toll-like receptor 7 and Type I IFNs. Investigations of gut microbiota using animal SLE models have generated varying results. In MRL/lpr lupus-prone mice, severity of lupus disease indices correlated positively with increased levels of Lachnospiraceae in gut microflora but was inversely correlated with Lactobacillaceae (which was decreased in the gut microbiome) [36]. In contrast, the NZB/W F1 mouse SLE model demonstrates increased abundance of gut Lactobacillaceae and this increase was reversed by dexamethasone [37]. Fecal samples from SLE patients have demonstrated increased Lactobacillus species, and decreased amount of Firmicutes relative to Bacteroidetes [38–41]. Recently, Azzouz et al. reported that the abundance of gut Ruminococcus gnavus (RG) was associated with lupus disease activity score (Systemic Lupus Erythematosus Disease Activity Index score) and correlated inversely with C3 and C4 complement levels [42]. Furthermore, highest levels of serum anti-RG antibodies against the RG-2 strain (not RG-1) were detected in those with proliferative lupus nephritis (including Classes III and IV). The findings were validated in two small independent SLE cohorts that include both men and women of diverse ethnic background [42].
The exact molecular pathways linking gut microbiota to SLE and lupus nephritis are not fully elucidated. Emerging data from animal and human studies support several mechanisms including translocation of microbial antigens from the leaky gut, molecular mimicry and the microbiota–sex hormone axis. First, SLE patients exhibit limited gut microbiota diversity and gut-derived pathobionts including Enterococcus gallinarum are detectable in mesenteric lymph nodes and the liver [43]. Animal studies show that gut-derived E. gallinarum reaches the liver and induces development of lupus autoantibodies, partly via activating the aryl hydrocarbon receptor-CYP1A1 pathway triggering Th17 cell activation and anti-dsDNA antibody production [43]. Second, molecular mimicry of human autoantigens by bacterial orthologs such as 60 kDa Ro protein (Ro60) triggers cross-reactive T- and B-cell autoimmune responses [40]. Third, the microbiota–sex hormone axis has been implicated similar to that observed in Type 1 diabetes pathophysiology. In mouse SLE models, gut microbiota differ between female and male lupus-prone mice and the female mice have more severe disease [36]; castration of male mice reversed this difference indicating an androgen-dependent pathway [33].
IgA NEPHROPATHY
IgA nephropathy patients show increased stool Sutterellaceae, Enterobacteriaceae and Firmicutes, while levels of Clostridium, Enterococcus, Bifidobacterium and Lactobacillus are decreased, compared with healthy controls [44]. IgA nephropathy is characterized by circulating immune complexes containing aberrantly glycosylated IgA1, and prominent mesangial IgA deposition in the kidneys. Recent data suggest that gut-associated lymphoid tissue plays a major role in the development of IgA nephropathy. Genome-wide association studies showed that most loci associated with the risk of IgA nephropathy are also associated with inflammatory bowel disease, suggesting that disruption of the intestinal barrier contributes to disease pathogenesis [45]. This is further supported by data from experimental models. For example, mice overexpressing BAFF, a B-cell factor crucial for IgA synthesis, develop a commensal flora-dependent IgA nephropathy [46].
PRE- AND PROBIOTICS IN CKD AND AUTOIMMUNE DISEASES
Prebiotics (nondigestible food ingredients that can stimulate growth and/or activity of beneficial gut bacteria) and probiotics (living organisms ingested via food or supplements that can improve the health of the host) are being actively studied as approaches to dampen chronic inflammation and improve quality of life in CKD patients (Table 2). Our group previously reported that feeding uremic rats with high amylose-resistant starch (a prebiotic) improved creatinine clearance and reduced kidney inflammation and fibrosis [48]. Resistant starches transit to the colon undigested and are metabolized by bacteria to SCFAs, which, as discussed above, are important nutrients to enterocytes. A recent randomized controlled trial of 46 hemodialysis patients in Iran noted that a diet enriched with amylose (HAM-RS2 biscuits) for 8 weeks significantly reduced serum tumor necrosis factor (TNF)-α, IL-6 and urea with patients reporting less constipation [49]. Total serum antioxidant activity and hs-CRP levels were unchanged compared with placebo (wheat-flour biscuits). Other small trials in ESKD patients utilizing oligofructose–inulin or resistant starch supplementation have demonstrated significant reduction in circulating levels of gut-derived uremic toxins such as indoxyl sulfate and p-cresyl sulfate [50, 51].
Table 2.
Studies of prebiotics and probiotics in immune-related kidney diseases
| Chronic disease state | Intervention | n per group | Outcomes |
|---|---|---|---|
| Animal studies | |||
| Rat adenine CKD model [46] Prebiotic study | High amylose maize resistant starch (HI-MAIZE 260) | CTL n = 6 CKD n = 9 CKD-HI-MAIZE n = 9 | Improved expression of epithelal tight junction proteins in the colon; reduced kidney inflammation and fibrosis; improved creatinine clearance |
| Mouse MRL/lpr lupus nephritis model [47] Probiotics study | Mix of five Lactobacillus strains | PBS control n = 7 Probiotics n = 7 | Improved gut epithelial barrier, decreased IgG2a, increased IL-10 and improved Treg–Th17 balance in the kidneys at 7 weeks. Benefits seen only in female mice and castrated male mice, suggesting modulation of the microbiota by sex hormones |
| Human studies | |||
| Chronic hemodialysis patients in Iran [48] Prebiotic study | Amylose resistant starch (HAM‐RS2) | Wheat-flour control n = 23 HAM-RS2 n = 23 | Decreased serum levels of TNF‐α, IL‐6 and malondialdehyde at 4 weeks Decreased constipation No significant difference in serum IL‐1β, hs‐CRP and total antioxidant activity |
| Chronic hemodialysis patients in Belgium [49] Prebiotic study | Oligofructose-enriched inulin (ORAFTI®Synergy 1, Tienen, Belgium) | n = 22 All patients subjected to escalating dose regimen | Serum p-cresyl sulfate significantly reduced at 4 weeksNo change in indoxyl sulfate |
| Chronic hemodialysis patients in USA [50] Prebiotic study | High-amylose corn starch (Hi-maize 260) | CTL n = 20 Hi-maize n = 20 | Decreased serum levels of indoxyl sulfate at 6 weeksNo change in p-cresol sulfate |
| Chronic hemodialysis patients in USA [51] Probiotics study | Renadyl formulation: 30 billion CFU of S. thermophilus KB 19, L. acidophilus KB 27, and B. longum KB 31 | n = 22 Placebo-controlled crossover study | No significant change in uremic toxin levels or quality of life scores at 6 months |
| Predialysis CKD Stages 4–5 patients in Australia [52] Symbiotic study | Prebiotics: combination of high-molecular weight inulin (inulin high performance), fructo-oligosaccharides and galacto-oligosaccharides Probiotics: nine different strains across the Lactobacillus, Bifidobacteria and Streptococcus genera | n = 31 Placebo-controlled crossover study | Serum p-cresyl sulfate decreased at 6 weeks. Increased stool Bifidobacterium and decreased Ruminococcaceae No change in indoxyl sulfate, endotoxin level, serum inflammatory biomarkers (IL-1β, IL-6, IL-10 and TNF-α), oxidative stress biomarkers (F2-isoprostanes and glutathione peroxidase), urinary Kim-1 or proteinuria Increase in albuminuria of 38 mg/24 h |
| Non-obese patients with Type 2 diabetes mellitus in Iran [53] Symbiotic study | Prebiotic: Fructo-oligosaccharide Probiotics: Lactobacillus, Bifidobacterium and Streptococus thermophiles | Placebo n = 35 Symbiotic therapy n = 35 | Significant decrease in HbA1c and albuminuria at 9 weeksNo change in urea, creatinine or lipid profile |
Probiotics have been tested in CKD with the goal of producing a less pathogenic microflora and thus reduce generation of uremic toxins. However, a randomized controlled trial with the Renadyl formulation of Streptococcus thermophilus, Lactobacillus acidophilus and Bifidobacterium longum in 22 patients with CKD Stages 3 and 4 failed to reduce plasma concentrations of uremic toxins and did not improve quality of life parameters [52]. The lack of benefit with probiotics may be explained by persistent uremia-induced changes in the gut biochemical environment combined with dietary restrictions and medications, which create an unfavorable milieu for the symbiotic microbiota. Thus, attempts to restore the desired microbiome without simultaneously improving the gut’s biochemical milieu can be futile. To address this deficit, the Synbiotics Easing Renal Failure by Improving Gut Microbiology (SYNERGY) trial examined the combination of pro- and prebiotic therapy (symbiotic formulation) >6 weeks in predialysis CKD patients and noted decreased serum p-cresyl sulfate and improvement in gut microbiome diversity; however, systemic markers of inflammation and proteinuria were not changed [54]. Studies of longer duration are needed to define clinical impact of pre- and probiotics in the CKD population. We also caution that choice of probiotic microbe is important; inclusion of bacteria that express urease with the intention to metabolize gut urea may be misguided since the downstream products ammonia and ammonium hydroxide can promote intestinal wall inflammation, as discussed above [16, 25].
Probiotics have been studied in CKD-related autoimmune diseases including Type 1 diabetes mellitus and SLE. In nonobese diabetic mice, oral administration of probiotic enriched in Lactobacillaceae prevented development of overt Type 1 diabetes, decreased IL-1β and increased CD103+ tolerogenic dendritic cell differentiation in the gut [55]. The TEDDY study evaluated the effects of early probiotic supplementation in children with genetic risk factors for Type 1 diabetes [53]. This multicenter prospective study involved 7473 children aged 4–10 years. Early probiotic exposure within 27 days of life correlated with decreased risk of islet cell autoimmunity. In a randomized controlled trial of 70 nonobese Iranian adults with Type 2 diabetes, symbiotic therapy prevented increase in microalbuminuria and Hgb A1c, while there was no effect on markers of kidney function [47].
Studies targeting gut microbiome dysbiosis in SLE have been restricted to animal models. In MRL/lpr lupus-prone mice, Lactobacillus probiotic treatment reduced proteinuria and kidney disease activity (assessed on biopsy samples) and prolonged survival [56]. Lactobacillus supplementation restored IL-10 production and decreased circulating IgG2a, an antibody isoform that tends to deposit in the kidneys of MRL/lpr mice. Flow cytometry studies of kidney tissue demonstrated that Lactobacillus skewed the Treg–Th17 balance toward a less inflammatory Treg phenotype. The findings were sex-dependent; benefits of Lactobacillus supplementation were observed in female and castrated male mice, but not in noncastrated male mice [56], consistent with a microbiota–sex hormone axis (discussed above). To date, clinical trials using probiotics as adjuvant therapy in SLE patients are lacking.
MEDICATION EFFECTS ON THE GUT MICROBIOTA
Disease-specific and immunomodulatory medications can impact the gut microbiome. Metformin, an established oral hypoglycemic agent, has been reported to increase beneficial gut bacteria that produce SCFAs [57]. The newer sodium-glucose cotransporter 2 (SGLT2) inhibitors were reported in animal investigations to alter the gut microbiome and decrease serum levels of indoxyl and p-cresyl sulfates [58, 59]; however, a randomized trial in patients with Type 2 diabetes found no change in microbiome composition with the SGLT2 inhibitor dapagliflozin [60]. Steroids are a common treatment for lupus nephritis and IgA nephropathy; dexamethasone treatment in NZB/W F1 lupus-prone mice was shown to increase gut microbial diversity while lowering abundance of pathogenic Lactobacillaceae [37]. Conversely, animal studies examining transplant immunosuppression medications (including prednisolone, tacrolimus, sirolimus or everolimus, mycophenolate mofetil) have demonstrated significant decrease in bacterial diversity leading to a more catabolic microbial profile [61, 62]. The combination of prednisolone, tacrolimus and mycophenolate mofetil induced variable changes in the fecal microbiota; of concern, a dramatic increase in uropathogenic Escherichia coli strain 536 was noted [62]. In a small trial of 19 kidney transplant recipients, fecal abundance of Faecalibacterium prausnitzii was significantly higher in patients who required dose escalation of tacrolimus to achieve therapeutic blood levels [63]. While evidence in this area is still exploratory, the data suggest that alterations to the gut microbiome with immunomodulatory medications can in turn affect host response to therapy for autoimmune-related kidney diseases.
CONCLUSIONS
The gut microbiome is altered in CKD with lower numbers of Lactobacillaceae and Prevotellaceae families and higher concentrations of anaerobic species that express urease, uricase, and p-cresyl- and indole-forming enzymes. Production of SCFAs falls, depriving host enterocytes of an important nutrient source and immune-modulating signal. Microbe-derived uremic toxins such as indoxyl sulfate, p-cresyl sulfate and TMAO induce systemic inflammation and widespread end-organ damage including pathogenesis of immune-related CKD states such as diabetic nephropathy, IgA nephropathy and lupus nephritis. Additional studies are needed to better understand gut dysbiosis in these disease states, and to examine potential benefits of gut-targeted interventions to restore a balanced microbiome in these patients.
FUNDING
W.L.L. received research funding from NIH NINDS R01 NS20989 and AHA 17IRG33410803.
AUTHORS’ CONTRIBUTIONS
W.L.L. and Y.C. drafted the manuscript and N.D.V. participated in manuscript revision.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest. This paper has not been published previously in whole or part.
REFERENCES
- 1. Lau WL, Savoj J, Nakata MB. et al. Altered microbiome in chronic kidney disease: systemic effects of gut-derived uremic toxins. Clin Sci (Lond) 2018; 132: 509–522 [DOI] [PubMed] [Google Scholar]
- 2. Jazani NH, Savoj J, Lustgarten M. et al. Impact of gut dysbiosis on neurohormonal pathways in chronic kidney disease. Diseases 2019; 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qin J, Li R, Raes J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Selber-Hnatiw S, Rukundo B, Ahmadi M. et al. Human gut microbiota: toward an ecology of disease. Front Microbiol 2017; 8: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capurso G, Lahner E.. The interaction between smoking, alcohol and the gut microbiome. Best Pract Res Clin Gastroenterol 2017; 31: 579–588 [DOI] [PubMed] [Google Scholar]
- 6. Ursell LK, Clemente JC, Rideout JR. et al. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol 2012; 129: 1204–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anders HJ, Andersen K, Stecher B.. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int 2013; 83: 1010–1016 [DOI] [PubMed] [Google Scholar]
- 8. Hooper LV, Midtvedt T, Gordon JI.. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 2002; 22: 283–307 [DOI] [PubMed] [Google Scholar]
- 9. Burkholder PR, McVeigh I.. Synthesis of vitamins by intestinal bacteria. Proc Natl Acad Sci USA 1942; 28: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. den Besten G, van Eunen K, Groen AK. et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 2013; 54: 2325–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hooper LV, Littman DR, Macpherson AJ.. Interactions between the microbiota and the immune system. Science 2012; 336: 1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cerf-Bensussan N, Eberl G.. The dialog between microbiota and the immune system: shaping the partners through development and evolution. Semin Immunol 2012; 24: 1–2 [DOI] [PubMed] [Google Scholar]
- 13. Round JL, Mazmanian SK.. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim CH, Park J, Kim M.. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation. Immune Netw 2014; 14: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei B, Wingender G, Fujiwara D. et al. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J Immunol 2010; 184: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lau WL, Kalantar-Zadeh K, Vaziri ND.. The gut as a source of inflammation in chronic kidney disease. Nephron 2015; 130: 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simenhoff ML, Saukkonen JJ, Burke JF. et al. Bacterial populations of the small intestine in uremia. Nephron 1978; 22: 63–68 [DOI] [PubMed] [Google Scholar]
- 18. Simenhoff ML, Saukkonen JJ, Burke JF. et al. Amine metabolism and the small bowel in uraemia. Lancet 1976; 308: 818–821 [DOI] [PubMed] [Google Scholar]
- 19. Meinardi S, Jin KB, Barletta B. et al. Exhaled breath and fecal volatile organic biomarkers of chronic kidney disease. Biochim Biophys Acta 2013; 1830: 2531–2537 [DOI] [PubMed] [Google Scholar]
- 20. Lee HJ, Pahl MV, Vaziri ND. et al. Effect of hemodialysis and diet on the exhaled breath methanol concentration in patients with ESRD. J Ren Nutr 2012; 22: 357–364 [DOI] [PubMed] [Google Scholar]
- 21. Vaziri ND, Wong J, Pahl M. et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int 2013; 83: 308–315 [DOI] [PubMed] [Google Scholar]
- 22. Wong J, Piceno YM, Desantis TZ. et al. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am J Nephrol 2014; 39: 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crespo-Salgado J, Vehaskari VM, Stewart T. et al. Intestinal microbiota in pediatric patients with end stage renal disease: a Midwest Pediatric Nephrology Consortium study. Microbiome 2016; 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaziri ND, Dure-Smith B, Miller R. et al. Pathology of gastrointestinal tract in chronic hemodialysis patients: an autopsy study of 78 cases. Am J Gastroenterol 1985; 80: 608–611 [PubMed] [Google Scholar]
- 25. Vaziri ND, Zhao YY, Pahl MV.. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol Dial Transplant 2016; 31: 737–746 [DOI] [PubMed] [Google Scholar]
- 26. Vaziri ND, Yuan J, Rahimi A. et al. Disintegration of colonic epithelial tight junction in uremia: a likely cause of CKD-associated inflammation. Nephrol Dial Transplant 2012; 27: 2686–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vaziri ND, Yuan J, Nazertehrani S. et al. Chronic kidney disease causes disruption of gastric and small intestinal epithelial tight junction. Am J Nephrol 2013; 38: 99–103 [DOI] [PubMed] [Google Scholar]
- 28. Aronov PA, Luo FJ, Plummer NS. et al. Colonic contribution to uremic solutes. J Am Soc Nephrol 2011; 22: 1769–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tao S, Li L, Liu Y. et al. Understanding the gut-kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: an analysis of the gut microbiota composition. Acta Diabetol 2019; 56: 581–592 [DOI] [PubMed] [Google Scholar]
- 30. Kanbay M, Onal EM, Afsar B. et al. The crosstalk of gut microbiota and chronic kidney disease: role of inflammation, proteinuria, hypertension, and diabetes mellitus. Int Urol Nephrol 2018; 50: 1453–1466 [DOI] [PubMed] [Google Scholar]
- 31. Simonsen JR, Harjutsalo V, Järvinen A. et al. Bacterial infections in patients with type 1 diabetes: a 14-year follow-up study. BMJ Open Diab Res Care 2015; 3: e000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kikuchi K, Saigusa D, Kanemitsu Y. et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat Commun 2019; 10: 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yurkovetskiy L, Burrows M, Khan AA. et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013; 39: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fernandes R, Viana SD, Nunes S. et al. Diabetic gut microbiota dysbiosis as an inflammaging and immunosenescence condition that fosters progression of retinopathy and nephropathy. Biochim Biophys Acta Mol Basis Dis 2019; 1865: 1876–1897 [DOI] [PubMed] [Google Scholar]
- 35. Meijers BK, Claes K, Bammens B. et al. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin J Am Soc Nephrol 2010; 5: 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang H, Liao X, Sparks JB. et al. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol 2014; 80: 7551–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo XM, Edwards MR, Mu Q. et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol 2017; 84: e02288–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zegarra-Ruiz DF, El Beidaq A, Iñiguez AJ. et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe 2019; 25: 113–127.e116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He Z, Shao T, Li H. et al. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog 2016; 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greiling TM, Dehner C, Chen X. et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med 2018; 10: eaan2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Meulen TA, Harmsen HJM, Vila AV. et al. Shared gut, but distinct oral microbiota composition in primary Sjögren’s syndrome and systemic lupus erythematosus. J Autoimmun 2019; 97: 77–87 [DOI] [PubMed] [Google Scholar]
- 42. Azzouz D, Omarbekova A, Heguy A. et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis 2019; 78: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manfredo Vieira S, Hiltensperger M, Kumar V. et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018; 359: 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Angelis M, Montemurno E, Piccolo M. et al. Microbiota and metabolome associated with immunoglobulin A nephropathy (IgAN). PLoS One 2014; 9: e99006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiryluk K, Li Y, Scolari F. et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 2014; 46: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fagarasan S, Kawamoto S, Kanagawa O. et al. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol 2010; 28: 243–273 [DOI] [PubMed] [Google Scholar]
- 47. Ebrahimi ZS, Nasli-Esfahani E, Nadjarzade A. et al. Effect of symbiotic supplementation on glycemic control, lipid profiles and microalbuminuria in patients with non-obese type 2 diabetes: a randomized, double-blind, clinical trial. J Diabetes Metab Disord 2017; 16: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vaziri ND, Liu SM, Lau WL. et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS One 2014; 9: e114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tayebi Khosroshahi H, Vaziri ND, Abedi B. et al. Effect of high amylose resistant starch (HAM-RS2) supplementation on biomarkers of inflammation and oxidative stress in hemodialysis patients: a randomized clinical trial. Hemodial Int 2018; 22: 492–500 [DOI] [PubMed] [Google Scholar]
- 50. Meijers BK, De Preter V, Verbeke K. et al. Cresyl sulfate serum concentrations in haemodialysis patients are reduced by the prebiotic oligofructose-enriched inulin. Nephrol Dial Transplant 2010; 25: 219–224 [DOI] [PubMed] [Google Scholar]
- 51. Sirich TL, Plummer NS, Gardner CD. et al. Effect of increasing dietary fiber on plasma levels of colon-derived solutes in hemodialysis patients. Clin J Am Soc Nephrol 2014; 9: 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Natarajan R, Pechenyak B, Vyas U. et al. Randomized controlled trial of strain-specific probiotic formulation (Renadyl) in dialysis patients. Biomed Res Int 2014; 2014: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uusitalo U, Liu X, Yang J. et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr 2016; 170: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rossi M, Johnson DW, Morrison M. et al. Synbiotics easing renal failure by improving gut microbiology (SYNERGY): a randomized trial. Clin J Am Soc Nephrol 2016; 11: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dolpady J, Sorini C, Di Pietro C. et al. Oral probiotic VSL#3 prevents autoimmune diabetes by modulating microbiota and promoting indoleamine 2,3-dioxygenase-enriched tolerogenic intestinal environment. J Diabetes Res 2016; 2016: 7569431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mu Q, Zhang H, Liao X. et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017; 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vallianou NG, Stratigou T, Tsagarakis S.. Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones (Athens) 2019; 18: 141–144 [DOI] [PubMed] [Google Scholar]
- 58. Mishima E, Fukuda S, Kanemitsu Y. et al. Canagliflozin reduces plasma uremic toxins and alters the intestinal microbiota composition in a chronic kidney disease mouse model. Am J Physiol Renal Physiol 2018; 315: F824–F833 [DOI] [PubMed] [Google Scholar]
- 59. Lee DM, Battson ML, Jarrell DK. et al. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc Diabetol 2018; 17: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van Bommel EJM, Herrema H, Davids M. et al. Effects of 12-week treatment with dapagliflozin and gliclazide on faecal microbiome: results of a double-blind randomized trial in patients with type 2 diabetes. Diabetes Metab 2019. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 61. Bhat M, Pasini E, Copeland J. et al. Impact of immunosuppression on the metagenomic composition of the intestinal microbiome: a systems biology approach to post-transplant diabetes. Sci Rep 2017; 7: 10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tourret J, Willing BP, Dion S. et al. Immunosuppressive treatment alters secretion of ileal antimicrobial peptides and gut microbiota, and favors subsequent colonization by uropathogenic Escherichia coli. Transplantation 2017; 101: 74–82 [DOI] [PubMed] [Google Scholar]
- 63. Lee JR, Muthukumar T, Dadhania D. et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One 2015; 10: e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]