Abstract
To evaluate the racial and ethnic differences in prevalence of germline pathogenic variants (PVs) and the effect of race and ethnicity on breast cancer (BC) risk among carriers, results of multigene testing of 77 900 women with BC (non-Hispanic White [NHW] = 57 003; Ashkenazi-Jewish = 4798; Black = 6722; Hispanic = 5194; and Asian = 4183) were analyzed, and the frequency of PVs in each gene were compared between BC patients (cases) and race- and ethnicity-matched gnomAD reference controls. Compared with NHWs, BRCA1 PVs were enriched in Ashkenazi-Jews and Hispanics, whereas CHEK2 PVs were statistically significantly lower in Blacks, Hispanics, and Asians (all 2-sided P < .05). In case-control studies, BARD1 PVs were associated with high risks (odds ratio > 4.00) of BC in Blacks, Hispanics, and Asians; ATM PVs were associated with increased risk of BC among all races and ethnicities except Asians, whereas CHEK2 and BRIP1 PVs were associated with increased risk of BC among NHWs and Hispanics only. These findings suggest a need for personalized management of BC risk in PV carriers based on race and ethnicity.
Germline pathogenic variants (PVs) in breast cancer (BC) predisposition genes are detected in approximately 10% of women with BC undergoing multigene panel testing (MGPT) (1,2). However, these estimates have been derived predominantly from the non-Hispanic White (NHW) population. Studies of other populations have been limited by sample size (3-5). A better understanding of racial- and ethnic-specific genetic risk for BC associated with PVs is expected to improve targeting of genetic testing and impact management of carriers from other populations.
Clinical genetic testing results for 12 established BC predisposition genes (ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, PALB2, PTEN, RAD51C, RAD51D, TP53) from 77 900 women with BC (NHW = 57 003, Ashkenazi-Jewish = 4798, Black = 6722, Hispanic = 5194, Asian = 4183) (Supplementary Tables 1 and 2, available online) referred to Ambry Genetics between March 2012 and December 2016 for MGPT were used to evaluate associations between PVs in individual genes and BC in each population. The details of patient ascertainment, race and ethnicity classification, germline sequencing, variant classification, and statistical analyses are described in the Supplementary Methods (available online). Enrichment of PVs in commonly mutated genes among different racial and ethnic groups was assessed relative to NHW while adjusting for family history of breast and ovarian cancer, age at diagnosis, and tumor estrogen receptor (ER) status. Likelihood ratio tests were used to estimate P values for the pairwise comparisons. All tests were 2-sided, and a P value less than .05 was considered statistically significant.
The pooled frequency of PVs in BC predisposition genes was 8.7% for NHW, 7.5% for Ashkenazi-Jews, 9.7% for Blacks, 9.9% for Hispanics, and 7.5% for Asians (Table 1), with corresponding variants of uncertain significance rates of 16.1%, 13.7%, 26.6%, 20.8%, and 29.0%, respectively (Supplementary Table 3, available online). The PV frequency in the Ashkenazi-Jewish population may have been slightly lower because of screening for BRCA1 or BRCA2 founder PVs over the past 20 years. Nonetheless, BRCA1 and BRCA2 had the highest frequency of PVs in all populations, similar to prior studies (1,2,5-9). PVs in PALB2 were also consistently observed at approximately 1% among NHWs, Blacks, Hispanics, and Asians.
Table 1.
Frequency and relative enrichment of pathogenic variants (PVs) in breast cancer predisposition genes in racial and ethnic groups relative to non-Hispanic Whites
| Gene | NHW | Ashkenazi-Jewish |
Black |
Hispanic |
Asian |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PV (%) | PV (%) | OR (95% CI)a |
P b | PV (%) | OR (95% CI)a |
P b | PV (%) | OR (95% CI)a |
P b | PV (%) | OR (95% CI)a |
P b | |
| ATM | 434 (1.3) | 19 (1.0) |
0.98 (0.54 to 1.62) |
.94 | 33 (0.8) |
0.70 (0.44 to 1.05) |
.11 | 29 (0.9) |
0.71 (0.42 to 1.14) |
.18 | 16 (0.6) |
0.51 (0.26 to 0.9) |
.03 |
| BARD1 | 73 (0.2) | 2 (0.1) | ND | ND | 18 (0.5) | ND | ND | 14 (0.5) | ND | ND | 7 (0.3) | ND | ND |
| BRCA1 | 747 (1.7) | 65 (2.6) |
2.58 (1.86 to 3.51) |
5.2 x 10-9 | 161 (2.9) |
0.99 (0.79 to 1.22) |
.89 | 127 (3.1) |
1.39 (1.08 to 1.78) |
.01 | 73 (2.2) |
1.31 (0.95 to 1.75) |
.08 |
| BRCA2 | 867 (2.0) | 46 (1.9) |
1.12 (0.76 to 1.6) |
.54 | 161 (2.9) |
1.21 (0.97 to 1.49) |
.09 | 107 (2.6) |
1.27 (0.98 to 1.63) |
.07 | 73 (2.2) |
1.08 (0.78 to 1.44) |
.64 |
| BRIP1 | 104 (0.3) | 3 (0.2) | ND | ND | 16 (0.4) | ND | ND | 9 (0.3) | ND | ND | 1 (0.0) | ND | ND |
| CDH1 | 27 (0.1) | 0 (0.0) | ND | ND | 5 (0.1) | ND | ND | 3 (0.1) | ND | ND | 4 (0.1) | ND | ND |
| CHEK2 c | 568 (1.7) | 23 (1.2) |
0.64 (0.34 to 1.09) |
.13 | 10 (0.2) |
0.17 (0.07 to 0.33) |
3.2 x 10-6 | 13 (0.4) |
0.36 (0.18 to 0.62) |
8.4 x 10-4 | 6 (0.2) |
0.14 (0.04 to 0.34) |
1.3 x 10-4 |
| PALB2 | 303 (0.9) | 6 (0.3) |
0.57 (0.22 to 1.17) |
.17 | 50 (1.1) |
1.29 (0.9 to 1.81) |
.15 | 37 (1.1) |
1.57 (1.03 to 2.29) |
.02 | 33 (1.2) |
1.69 (1.08 to 2.52) |
.02 |
| PTEN | 28 (0.1) | 0 (0.0) | ND | ND | 8 (0.1) | ND | ND | 4 (0.1) | ND | ND | 2 (0.1) | ND | ND |
| RAD51C | 65 (0.2) | 2 (0.1) | ND | ND | 10 (0.3) | ND | ND | 13 (0.5) | ND | ND | 2 (0.1) | ND | ND |
| RAD51D | 29 (0.1) | 4 (0.2) | ND | ND | 6 (0.2) | ND | ND | 2 (0.1) | ND | ND | 2 (0.1) | ND | ND |
| TP53 | 81 (0.2) | 2 (0.1) | ND | ND | 14 (0.2) | ND | ND | 11 (0.3) | ND | ND | 10 (0.3) | ND | ND |
| Totald, % | 8.7 | 7.5 | — | — | 9.7 | — | — | 9.9 | — | — | 7.5 | — | — |
Enrichment of PVs in commonly mutated genes among racial and ethnic groups relative to non-Hispanic White adjusted for family history of breast and ovarian cancer, age at diagnosis, and tumor estrogen receptor status. CI = confidence interval; ND = not determined because of insufficient number (<5) of PVs across all races and ethnicities; NHW = non-Hispanic White; OR = odds ratio; — = Not Applicable.
Likelihood ratio test for pairwise comparison relative to non-Hispanic White.
Missense and low penetrance PVs in CHEK2 were excluded from analyses (see Supplementary Methods, available online).
Total is the sum of PV frequency across all breast cancer predisposition genes.
Compared with NHW, BRCA1 PVs were statistically significantly (P < .05) enriched in Ashkenazi-Jewish and Hispanic women, and PALB2 PVs were enriched in Hispanic and Asian women, whereas CHEK2 PVs were statistically significantly less frequent (<1%) among Black, Hispanic, and Asian women (Table 1). The c.1100delC PV accounted for the majority of CHEK2 PVs in NHWs, Ashkenazi-Jews, and Blacks but was only observed in 3 Hispanics and not in Asians. Among self-reported Ashkenazi-Jews, 92 of the 111 (82.8%) BRCA1 and BRCA2 PVs were due to founder PVs (10). In Hispanics, c.2167_2168delAT accounted for 8 of the 37 (21.6%) PALB2 PVs, and c.577C>T accounted for 6 of the 13 (46.1%) RAD51C PVs. Frequencies of PVs in other candidate breast cancer predisposition genes are shown in Supplementary Table 4 (available online).
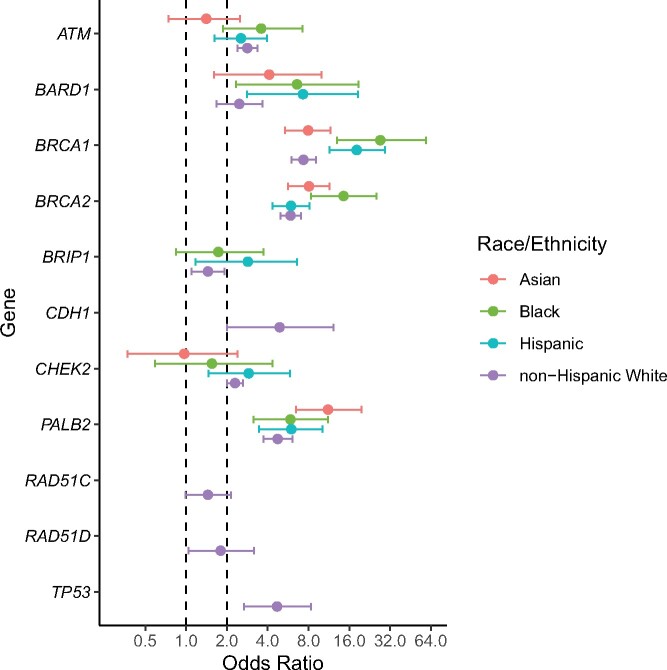
Associations between PVs in each gene and BC risk by race and ethnicity were assessed in case-control analysis by comparing frequencies of PVs in BC patients (cases) with race- and ethnicity-matched gnomAD (v2.1.1) reference controls using the Fisher exact test and adjusting for multiple comparisons (false discovery rate = 0.1) (11). PVs in BRCA1, BRCA2, and PALB2 were associated with a statistically significant high risk (odds ratio [OR] > 4.00) of BC in all populations (Figure 1;Supplementary Table 5, available online). PVs in BARD1 were associated with a high risk of BC in Blacks, Hispanics, and Asians, whereas the risk was only moderately elevated (OR = 2.00-4.00) in NHW. ATM PVs were associated with moderately elevated risk among all races and ethnicities except the Asian population. CHEK2 and BRIP1 PVs were associated with an increased risk of BC among NHWs and Hispanics only. Furthermore, in all racial and ethnic groups, BRCA1, BRCA2, and PALB2 PVs were associated with a statistically significant high risk of both ER-positive and ER-negative BC, whereas PVs in BARD1 were associated with statistically significant high risk of ER-negative BC only (Supplementary Tables 6 and 7, available online).
Figure 1.
Forest plot for case-control comparisons of frequency of pathogenic variants between breast cancer cases and ethnicity- and race-matched reference controls in gnomAD. Plots show odds ratios and 95% confidence intervals for each gene in 4 common race and ethnicity groups. Odds ratios are not shown for genes with less than 5 pathogenic variants in cases or controls. Dashed lines are presented at odds ratios of 1 and 2, the latter representing the threshold at which pathogenic variants in a gene may be considered to confer moderate risk of breast cancer.
The odds ratios were subsequently combined with age-adjusted race- and ethnicity-specific incidence rates from the Surveillance, Epidemiology, and End Results program to estimate lifetime absolute risks (up to age 85 years) of BC associated with PVs in each gene. BRCA1 PVs were associated with high (>50%) lifetime risk of BC across all populations, whereas BRCA2 and PALB2 PVs were associated with high risks in selective populations. In particular, lifetime risk of BC for BRCA1 carriers was 85% in Blacks compared with 59% in NHWs and 50% in Asians (Supplementary Table 8, available online), although this needs to be interpreted with caution because of the higher proportion of triple-negative cases among Black women. Lifetime risk of BC for ATM carriers ranged from 12% in Asians to 32% in Blacks, whereas for CHEK2 carriers, risks ranged from 9% in Asians to 26% in NHWs. The observed PVs are listed in Supplementary Table 9 (available online).
Although the frequency of PVs in BC predisposition genes is approximately 10% across racial and ethnic groups, the findings from this study suggest that race and ethnicity may be a modifying factor for gene-specific BC risk. For instance, CHEK2 carriers among Blacks and Asians did not have an increased risk of BC, with c.1100delC primarily accounting for the differences in frequency of PVs (12); ATM carriers did not have an increased risk of BC among Asians; and BARD1 PVs were associated with high risk of BC (ER negative) in Blacks, Hispanics, and Asians but only moderate risks in the NHW population. Differences in gene-specific BC risk by race and ethnicity underscore the need for more personalized screening and management recommendations in current guidelines (13) for carriers of PVs in specific genes in minority populations.
Despite including a large sample of racially and ethnically diverse populations, this study has limitations. First, this was a study of patients who underwent MGPT at a clinical testing laboratory and was not a population-based study. Therefore, it is unclear if the differences in PV frequencies across racial and ethnic populations were influenced by genetic testing favoring high-risk groups in some minority populations, given that non-White populations are less likely to undergo genetic testing (14-16). Second, clinical variables and ethnicity and race data were not comprehensively verified. Third, not all patients underwent genetic testing using the same multigene panel. Finally, the lack of information on age at testing or family history in gnomAD may have influenced the results from case-control analysis. However, overall, we confirm the utility of MGPT in identifying clinically actionable PVs in patients of different racial and ethnic backgrounds and provide tools for discussing race- and ethnicity-based probability of carrying PVs. We share these findings with the goal of improving the use of genetic testing in populations facing health inequalities and to support efforts to better serve minority populations as a collective of health-care providers.
Funding
This study was supported in part by NIH grants R01CA116167, R01CA176785, R01CA192393, R01CA225662, and R35CA253187; an NIH Specialized Program of Research Excellence (SPORE) in Breast Cancer [P50CA116201]; and the Breast Cancer Research Foundation.
Notes
Role of the funder: The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: HL, NN, SG, AY, BTD, ECC, TP, and JSD are employees of Ambry Genetics Incorporated. FJC reports receipt of personal fees from Ambry Genetics and Qiagen. All other authors have no other conflicts of interest to disclose.
Role of the authors: Siddhartha Yadav: Contributor Information: Formal analysis; Investigation; Writing – original draft; Writing – review & editing. Holly LaDuca: Data curation; Investigation; Resources; Supervision; Writing – original draft; Writing – review & editing. Eric Polley: Data curation; Formal analysis; Supervision; Writing – original draft; Writing – review & editing. Chunling Hu: Data curation; Formal analysis; Writing – original draft; Writing – review & editing. Nancy Niguidula: Data curation; Writing – original draft; Writing – review & editing. Hermela Shimelis: Data curation; Writing – original draft; Writing – review & editing. Jenna Lilyquist: Data curation; Writing – original draft; Writing – review & editing. Jie Na: Formal analysis; Writing – original draft; Writing – review & editing. Kun Lee: Data curation; Writing – original draft; Writing – review & editing. Stephanie Gutierrez: Data curation; Writing – original draft; Writing – review & editing. Amal Yussuf: Contributor Information: Data curation; Writing – original draft; Writing – review & editing. Steven Hart: Data curation; Formal analysis; Visualization; Writing – original draft; Writing – review & editing. Brigette Davis: Resources; Supervision; Writing – original draft; Writing – review & editing. Elizabeth Chao: Resources; Writing – original draft; Writing – review & editing. Tina Pesaran: Contributor Information: Data curation; Project administration; Writing – original draft; Writing – review & editing. David Goldgar: Contributor Information: Conceptualization; Writing – original draft; Writing – review & editing. Jill Dolinsky: Conceptualization; Resources; Supervision; Writing – original draft; Writing – review & editing. Fergus J Couch: Conceptualization; Formal Analysis; Investigation; Resources; Supervision; Writing - original draft; Writing - review and editing.
Prior presentation: This study was presented as a poster discussion at the 2019 Annual Meeting of American Society of Clinical Oncology (ASCO) in Chicago, Illinois, on May 13, 2019.
Data Availability
The data underlying this article were provided by Ambry Genetics by permission. Data will also be shared on request to the corresponding author with permission of Ambry Genetics.
Supplementary Material
References
- 1. Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3(9):1190-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34(13):1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynce F, Graves KD, Jandorf L, et al. Genomic disparities in breast cancer among Latinas. Cancer Control. 2016;23(4):359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ding YC, Steele L, Chu LH, et al. Germline mutations in PALB2 in African-American breast cancer cases. Breast Cancer Res Treat. 2011;126(1):227-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Churpek JE, Walsh T, Zheng Y, et al. Inherited predisposition to breast cancer among African American women. Breast Cancer Res Treat. 2015;149(1):31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ricker C, Culver JO, Lowstuter K, et al. Increased yield of actionable mutations using multi-gene panels to assess hereditary cancer susceptibility in an ethnically diverse clinical cohort. Cancer Genetics. 2016;209(4):130-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong ESY, Shekar S, Met-Domestici M, et al. Inherited breast cancer predisposition in Asians: multigene panel testing outcomes from Singapore. NPJ Genomic Med. 2016;1(1):15003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang YA, Jian JW, Hung CF, et al. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer. 2018;18(1):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silva FC, Lisboa BC, Figueiredo MC, et al. Hereditary breast and ovarian cancer: assessment of point mutations and copy number variations in Brazilian patients. BMC Med Genet. 2014;15(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levy-Lahad E, Catane R, Eisenberg S, et al. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet. 1997;60(5):1059-1067. [PMC free article] [PubMed] [Google Scholar]
- 11. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 1995;57(1):289-300. [Google Scholar]
- 12. Evans DG, Eccles DM, Rahman N, et al. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41(6):474-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NCCN Clinical Practice Guidelines in Oncology. Genetic/Familial High-Risk Assessment: Breast, Ovarian and Pancreatic Version 1.2021. https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf. Accessed October 29, 2020. [DOI] [PubMed]
- 14. McCarthy AM, Bristol M, Domchek SM, et al. Health care segregation, physician recommendation, and racial disparities in BRCA1/2 testing among women with breast cancer. J Clin Oncol. 2016;34(22):2610-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hann KEJ, Freeman M, Fraser L, et al. ; for the PROMISE study team. Awareness, knowledge, perceptions, and attitudes towards genetic testing for cancer risk among ethnic minority groups: a systematic review. BMC Public Health. 2017;17(1):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cruz-Correa M, Perez-Mayoral J, Dutil J, et al. ; on behalf of the Puerto Rico Clinical Cancer Genetics Consortia. Clinical cancer genetics disparities among Latinos. J Genet Counsel. 2017;26(3):379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article were provided by Ambry Genetics by permission. Data will also be shared on request to the corresponding author with permission of Ambry Genetics.