Summary
The source–sink relationship determines the overall agronomic performance of rice. Cloning and characterizing key genes involved in the regulation of source and sink dynamics is imperative for improving rice yield. However, few source genes with potential application in rice have been identified. Glucan, Water‐Dikinase 1 (GWD1) is an essential enzyme that plays a pivotal role in the first step of transitory starch degradation in source tissues. In the present study, we successfully generated gwd1 weak mutants by promoter editing using CRISPR/Cas9 system, and also leaf‐dominant overexpression lines of GWD1 driven by Osl2 promoter. Analysis of the gwd1 plants indicated that promoter editing mediated down‐regulation of GWD1 caused no observable effects on rice growth and development, but only mildly modified its grain transparency and seed germination. However, the transgenic pOsl2::GWD1 overexpression lines showed improvements in multiple key traits, including rice yield, grain shape, rice quality, seed germination and stress tolerance. Therefore, our study shows that GWD1 is not only involved in transitory starch degradation in source tissues, but also plays key roles in the seeds, which is a sink tissue. In conclusion, we find that GWD1 is an ideal biotechnological target with promising potential for the breeding of elite rice cultivars via genetic engineering.
Keywords: GWD1, Osl2 promoter, rice yield, eating and cooking quality, seed germination, stress tolerance
Introduction
Rice, one of the most important staple crops in the world, serves as the primary source of carbohydrates and nutrition for more than half of the world’s population. Improving rice yield is essential to prevent a global food crisis. Grain yield in rice mainly consists of three major factors; the number of grains per panicle, the 1000‐grain weight and the number of panicles per unit area (Ren et al., 2020). Recently, the number of cloned genes that directly contribute to rice yield has increased. For example, a number of genes related to grain size have been successfully cloned and characterized, such as GS3 and qTGW3/TGW3 (Hu et al., 2018; Li et al., 2019; Mao et al., 2010; Ying et al., 2018). In addition, some key genes that control plant architecture in rice have also been cloned and studied, including IPA1 and DEP1. A point mutation in IPA1 resulted in an ideal rice plant that had a reduced number of tillers and increased lodging resistance, which acted to enhance grain yield (Jiao et al., 2010; Wang et al., 2018 Science). Mutations in DEP1 promote cell division and increase panicle density, branch number and grain number per panicle, thereby increasing rice yield by 15% to 20% (Huang et al., 2009). Moreover, some of these genes also showed interactions, and they act coordinately to regulate rice yield, thus forming a complex regulatory network (Liu et al., 2018b; Sun et al., 2018).
Nevertheless, grain yield in rice is not only affected by genes that are directly related to sink strength, such as seed size. Other factors involved in source capacity and flow fluence also play important roles in determining grain filling which, in turn, is vital to rice yield (Li et al., 2018a; Liang et al., 2001). Thus, modulating expression of sink‐related genes has only a limited contribution to final rice yield, which could be one major reason why most cloned yield‐related genes have not been widely applied in modern rice breeding practice. It is generally thought that harmonizing the source, sink and flow of rice is a prerequisite for high yield potential (Peng et al., 2000; Zhai et al., 2020). However, most progress on cloning and characterizing source‐related genes comes from studies in the model dicotyledonous plant Arabidopsis thaliana. These studies have shown how chloroplasts fix carbon during photosynthesis and then transport the fixed carbon between mesophyll and phloem parenchyma cells in the form of soluble sugar, and the subsequent synthesis and degradation of soluble sugar (Sonnewald and Kossmann, 2013; Abt and Zeeman 2020). In leaf tissues, transitory starch is synthesized during the light period and degraded at night. During the first 2 h in the darkness, the process is accelerated and then proceeds at a constant rate. At the end of the light phase, the leaf starch content reaches its peak, while at the end of the dark phase, leaf transitory starch is almost all degraded. The diurnal starch synthesis and products of its degradation supply plants continually with carbohydrates necessary for metabolism (Streb and Zeeman 2012). Nevertheless, it remains mostly unknown whether the regulation of transitory starch metabolism in source organs will affect sink organ activity and the final yield in rice, a major crop and model monocotyledonous plant species.
Starch phosphorylation is known to be the only covalent modification that occurs naturally in the process of transitory starch degradation. It is important to understand the effect and function of starch phosphorylation during starch metabolism. Studies have shown that appropriate phosphorylation of starch can improve its function and change certain physicochemical properties, which meet the requirements of certain industrial production and processing applications (Singh et al., 2003; Wiesenborn et al., 1994). In addition, the phosphorylation of starch can increase the specific surface area of starch granules and make the starch more digestible and softer in texture, thus meeting the requirements of consumers and the food processing industry (Mahlow et al., 2016). In the late 1990s, the R1 gene was cloned and shown to be related to starch phosphorylation in potato (Lorberth et al., 1998). The protein encoded by the R1 gene was then found to be an α‐glucan hydration dikinase (GWD), which can catalyse the reaction of α‐glucan and adenosine triphosphate (ATP) with water to generate α‐glucan phosphate monoester, adenosine monophosphate (AMP) and orthophosphate (Ritte et al., 2002). The GWD gene is present in many plants, but the evolutionary history and enzyme functions are not identical (Muyiwa et al., 2020). In Arabidopsis, GWD1 phosphorylates glucosyl residues of amylopectin at the C6 position (Ritte et al., 2002); then, GWD3/PWD further phosphorylates a different glucosyl residue at the C3 position (Baunsgaard et al., 2005; Kötting et al., 2005). Either the mutation of GWD1 or GWD3/PWD caused excessive transitory starch accumulation in plant leaves (Yu et al., 2001; Baunsgaard et al., 2005). GWD catalyses starch phosphorylation (Blennow and Engelsen 2010) which in turn regulates starch degradation (Mikkelsen et al., 2005) and biosynthesis (Skeffington et al., 2014; Xu et al., 2017). In cassava (Manihot esculenta), GWD1 is essential for characterization of morphogenesis and the turnover of transitory starch in leaves, and it contributes to source/sink strength, thus affecting storage root growth (Zhou et al., 2017). A previous report also showed that excessive starch accumulated in the leaves of the GWD1 rice mutant LSE1, but it had no significant effect on rice vegetative growth; however, the grain yield was notably decreased due to fewer panicles per plant, smaller and less ripened grains in mutant plants during reproductive growth (Hirose et al., 2013). Chen et al. (2017) overexpressed the potato GWD1 gene under control of an endosperm‐specific D‐hordein promoter in rice, and the transgenic plants produced highly phosphorylated rice starch. Interestingly, endosperm‐specific down‐regulation of GWD expression in wheat increased the grain biomass (Ral et al., 2012). Therefore, GWD could be an ideal target for improving both grain yield and starch quality in crops via genetic engineering.
In addition to the careful selection of suitable target genes, the choice of the appropriate promoter is also crucial in genetic engineering‐mediated crop improvement. In general, gene promoters are generally classified into three types based on their expression mode and functions and include constitutive, tissue‐specific and inducible promoters. Tissue‐specific promoters are characterized by certain temporal and spatial expression patterns. Target genes can be specifically expressed in a certain type of tissue or organ, avoiding the negative effects of over‐accumulated proteins in unintended tissues, thus achieving precise expression control of the target gene in the expected time, position and even quantity (Yi et al., 2010; Jeong and Jung 2015). Tissue‐specific promoters are of great significance for improving desirable agronomic traits and stress tolerance in crops via genetic engineering. Although a number of tissue‐specific promoters have been successfully identified and used, such as the rice Glutelin promoters (Li et al., 2018b; Qu et al., 2008), the number of tissue‐specific promoters is still far from adequate for research and crop breeding applications.
When rice leaves undergo senescence, the assimilation ability and conversion efficiency of transitory starch gradually decreases, resulting in reduced glucose and stored starch, and a reduction in the final grain yield. To investigate the function of GWD1 and evaluate its potential for application in rice breeding programmes, our plan was to overexpress GWD1 in rice leaves, particularly to enhance its expression in the senescent leaves in order to promote the conversion efficiency of photosynthate from senescent leaves to developing seeds. Therefore, we chose the Osl2 promoter, because its expression is mainly observed in the leaves, and the expression is also induced by leaf senescence (Ansari et al., 2005; Lee et al., 2001). In addition, we were inspired by two recent reports by direct editing Wx gene promoter to moderately down‐regulation of its expression, thus fine‐tuning of the amylose content of rice and improved rice quality (Huang et al., 2020; Zeng et al., 2020). Therefore, we also generated weak gwd1 mutants via CRISPR/Cas9‐mediated gene edition of GWD1 core promoter. Therefore, we were able to comprehensively evaluate the effects of GWD1 on rice yield, quality, seed germination and stress tolerance by using these GWD1‐related rice materials in the present study.
Results
Expression analysis of GWD1 and Osl2 in rice
First, the expression pattern of GWD1 was determined via qRT‐PCR analysis. The highest expression levels of GWD1 were observed in leaf blades of both young seedlings and mature plants (Figure S1), which is consistent with previous reports (Hirose et al., 2013). Second, we also examined the expression pattern of Osl2. Considering that Osl2 is mainly expressed in rice leaves and its expression is specifically enhanced during rice leaf senescence, we intended to use its promoter to drive the expression of GWD1 in rice, thus promoting the expression of GWD1 in rice leaves during the late stage of grain filling. Our qRT‐PCR results showed that Osl2 is primarily expressed in rice leaves, and its expression in stems, sheaths, roots and developing seeds is low (Figure 1a). Specifically, the expression of Osl2 in rice leaves gradually increased as the seeds developed (Figure 1b). Therefore, the Osl2 promoter was suitable to drive the expression of GWD1 in rice and should help to enhance the translocation of photosynthetic product from source to sink tissues during the later stage of grain filling.
Figure 1.
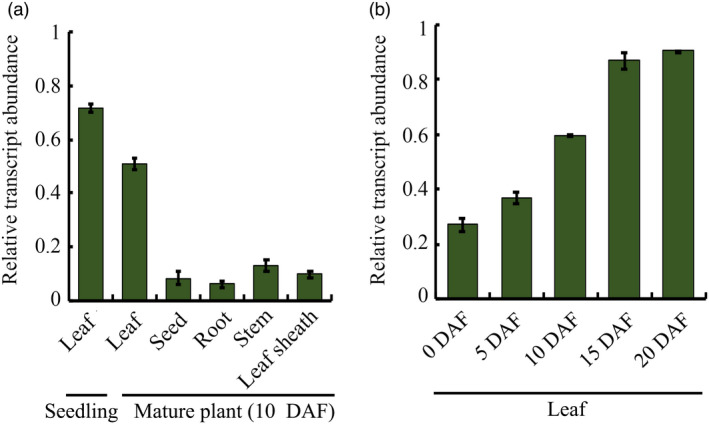
Spatial and temporal expression analysis of Osl2 in rice. (a) Expression analysis of Osl2 in different tissues of rice. (b) Expression of Osl2 in rice leaves at different stages of senescence. DAF, day after flowering. The rice Actin gene was the internal control for normalization of gene expression. Error bars represent SDs. Each experiment was repeated at least three times
Generation of GWD1 overexpression lines and gwd1 mutants
We constructed the pOsl2::GWD vector and transformed the japonica rice variety 9983 via Agrobacterium‐mediated transformation (Figure 2a). Genotyping analysis identified more than 10 positive, independent transgenic lines (Figure 2b). Then, we planned to select representative lines for further analysis based on the following criteria. First, the line should only contain single copy of the T‐DNA. Second, it should be homozygous and the expression of GWD1 is notably increased in the leaves. The plants from each target T2 rice line were genotyped by PCR, and the line was homozygous if all the genotyped plants from the line were positive. Finally, its general phenotype should be consistent with most other positive homozygous lines. Therefore, three representative homozygous lines expressing pOsl2::GWD (Lines 1, 2 and 3, T2 generation) were selected. The results indicated that the expression of GWD1 was mainly enhanced in the leaves of pOsl2::GWD1 transgenic plants, and no significant differences were observed in the seeds of the transgenic plants compared to the wild‐type (WT) control (Figure 2e; Figure S2), suggesting that the Osl2 promoter successfully directed dominant expression of GWD1 in rice leaves. Next, the GWD1 core promoter, 200bp upstream of translation initiation codon (ATG), was analysed for the presence of potential cis‐acting motifs using the PLACE database. Th result showed that one TATA box, one TATCCAY motif, one pyrimidine box and four GATA boxes were included in the core promoter (Figure 2c; Table S1). Among them, TATCCAY motif and pyrimidine box are responsible for sugar repression (Mena et al., 2002; Morita et al., 1998; Toyofuku et al., 1998) while GATA box is required for high level, light regulated and tissue‐specific expression (Reyes et al., 2004). Therefore, a specific target site close to four clustered cis‐elements was selected for CRISPR/Cas9‐mediated editing using CRISPR‐GE (https://bio.tools/crispr‐ge). Then, the GWD1‐Cas9 plasmid was constructed and transformed into the japonica rice variety ‘Zhonghua11’ (ZH11), and seven independent transgenic rice lines were generated. Genotyping and sequencing analysis allowed us to select two homozygous gwd1 mutant lines without T‐DNA insertions in their genomes. These were designated gwd1‐1 and gwd1‐2 and carried a 1 bp (G) deletion and a 1 bp (C) insertion in the target region respectively (Figure 2d; Figure S3). Then, qRT‐PCR was performed to assess the effect of promoter mutation on GWD1 expression. The result indicated that the transcription abundance of both two gwd1 mutants was about 50% of that of the WT control (Figure 2f).
Figure 2.
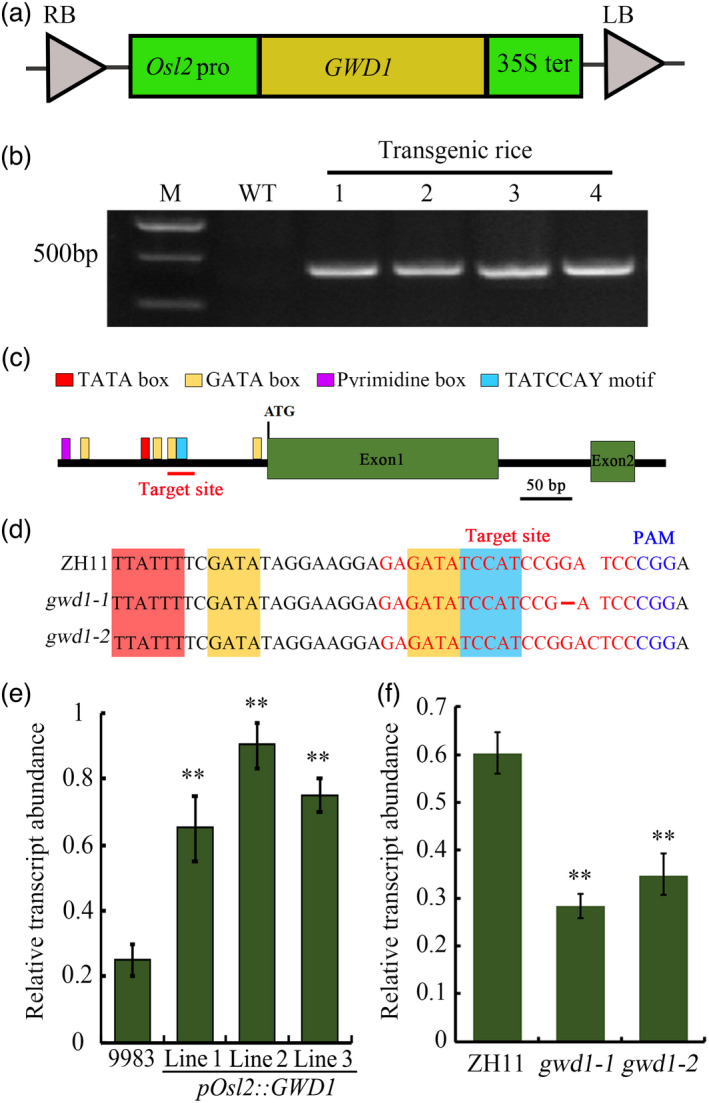
Generation and identification of the pOsl2::GWD1‐overexpressing transgenic rice plants and gwd1 knock‐down mutants. (a) Schematic diagram showing the structure of the pOsl2::GWD1 transformation construct. Osl2 pro, Osl2 promoter; 35S term, CaMV 35S terminator. (b) PCR analysis of total DNA from representative pOsl2::GWD1 transgenic rice lines and the WT. (c) Analysis of the potential cis‐elements in the GWD1 core promoter. (d) DNA sequences around the editing site and identification of gwd1 mutants. The guide RNA sequence used in CRISPR/Cas9‐mediated gene editing of GWD1 is highlighted in red, and the protospacer adjacent motif (PAM) is highlighted in blue. The deletion in gwd1‐1 is indicated by a hyphen. Expression analysis of GWD1 in the leaves of pOsl2::GWD1 overexpression lines (e) and gwd1 mutants (f). Error bars represent the SDs. Each experiment was repeated at least three times. **P < 0.01 (Student’s t‐test)
The GWD1 mutation suppresses starch degradation in rice leaves
Since GWD1 catalyses starch phosphorylation and promotes its degradation, down‐regulation or mutation of GWD1 will cause over‐accumulation of starch in plant leaves. Although iodine staining is not quantitative, comparison with wild‐type plants at specific times during the diurnal cycle allows the identification of plants with less or more starch accumulated than normal (Caspar et al., 1991; Streb and Zeeman 2012). For example, the end of the dark cycle is an ideal time point of distinguishing the difference in starch accumulation between gwd1 mutant and the wild‐type control. Then, we first examined starch accumulation in 14‐day‐old seedlings of gwd1 mutant at the end of the dark cycle. Iodine staining showed that gwd1 mutant seedlings had dark staining starch in the leaves, while the WT control did not show any observable staining (Figure 3), suggesting that a large amount of starch remained in the rice leaves after the dark cycle. Since the transcription of GWD1 was increased in the pOsl2::GWD1 transgenic lines, we speculated that the efficiency of transitory starch degradation could be accelerated in pOsl2::GWD1 plants. Therefore, we stained the leaves of both pOsl2::GWD1 rice and its WT control during the light cycle. However, no difference in staining was observed between seedlings of the GWD1 overexpression lines and the WT control (Figure S4), suggesting that the mild difference of starch content in the pOsl2::GWD1 rice and the WT control was indistinguishable.
Figure 3.

Iodine staining of ZH11 (WT) and gwd1‐1 mutant seedlings for the presence of starch in the leaves
GWD1 overexpression increases plant height and 1000‐grain weight in rice
We examined agronomic traits in the selected pOsl2::GWD1 transgenic lines and the gwd1 mutants. In general, plant height was increased in the GWD1 overexpression line compared to the WT (Figure 4a, b). Moreover, both 1000‐grain weight and grain yield per plant in the pOsl2::GWD1 transgenic rice plants showed about 10% increase while the seed setting rate, panicle number per plant and grain number per panicle were unchanged (Figure 4d; Figure S5a‐c). No significant difference was observed between the gwd1 mutant plants and the WT in all traits tested (Figure 4e‐h), which was different from the reported GWD1 null mutant (Hirose et al., 2013), suggesting the strategy to fine‐tune GWD1 expression via editing its core promoter is effectual.
Figure 4.
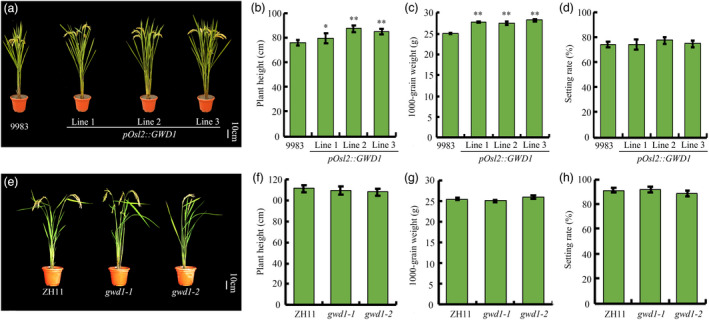
Characterization of the pOsl2::GWD1 transgenic lines and the gwd1 knock‐out mutants. Phenotypes of mature plants (a), plant height (b), 1000‐grain weight (c), and seed setting rate (d) of the three pOsl2::GWD1 transgenic rice lines and the WT japonica line 9983 control. Mature plant phenotypes (e), plant height (f), 1000‐grain weight (g), and seed setting rate (h) of the two gwd1 mutant lines and the WT control. Error bars represent the SDs. Each experiment was repeated at least three times. **P < 0.01 (Student’s t‐test)
The effect of GWD1 on grain size and transparency
The result of grain shape analysis showed that GWD1 overexpression noticeably increased grain length but reduced grain width (Figure 5a‐c); thus, the pOsl2::GWD1 grains were more slender than those from WT plants. The rice grains were then dehusked and polished, and the rice appearance was evaluated using a rice quality detector (SC‐E, Wanshen, China). The data showed that GWD1 overexpression had no effect on rice chalkiness (Figure 5d‐f). Interestingly, although the declined GWD1 expression did not alter grain shape of gwd1 mutants (Figure 5g‐i), it significantly reduced the degree of chalkiness and the percentage of chalky rice grains (Figure 5j‐i), thus improved rice appearance quality. Therefore, GWD1 can either alter grain shape or transparency in rice depending on its expression level and pattern.
Figure 5.
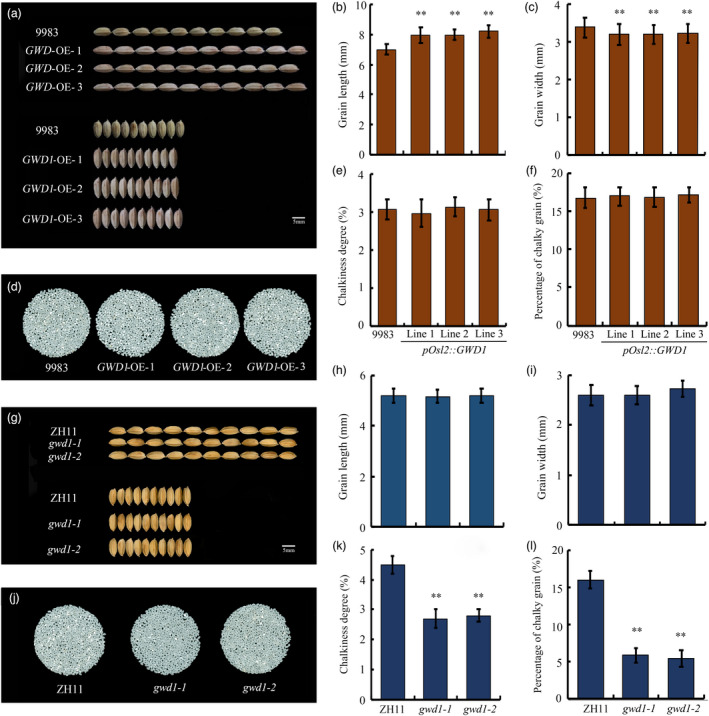
The effects of modulating GWD1 expression on grain shape and transparency in rice. Mature paddy rice grains of japonica cultivar 9983 and the pOsl2::GWD1 transgenic lines: grain shape (a), length (b), and width (c). Appearance (d), degree of chalkiness (e), and chalky rice grain percentage (f) of milled rice from 9983 and the three pOsl2::GWD1 transgenic lines. Mature paddy rice grains from japonica cultivar ZH11 and the two gwd1 knock‐out mutants: grain shape (g), length (h), and width (i). Appearance (j), degree of chalkiness (k), and chalky rice grain percentage (l) of milled rice from ZH11 and the gwd1 mutants. Each experiment was repeated at least three times. Error bars represent the SDs; **P < 0.01 (Student’s t‐test)
Examination of rice grain physicochemical characteristics in the GWD1‐related materials
In addition to evaluating the yield‐related traits and appearance quality of rice from the pOsl2::GWD1 transgenic and gwd1 mutant plants, we further examined the physicochemical properties of the grain, which are closely related to the eating and cooking qualities (ECQs). The analyses showed that GWD1 overexpression or mutation had no significant effect on either the apparent amylose content (AAC) or gel consistency (GC) of the rice (Figure 6a‐d). We also look at the thermodynamic properties; differential scanning calorimeter (DSC) analysis showed that rice from the gwd1 mutant had similar gelatinization curves to rice from the control ZH11, but GWD1 overexpression slightly increased the onset temperature (To), peak temperature (Tp) and conclusion temperature (Tc) of gelatinization when compared with the WT control (Figure 6e, f; Table S2). Finally, a Rapid Visco Analyzer (RVA) assay was performed to evaluate the pasting properties of rice flour in order to assess its apparent viscosity changes during heating and cooling, which will predict the texture of cooked rice. The result showed that the values for peak viscosity (PKV), hot paste viscosity (HPV) and cool paste viscosity (CPV) were all elevated. Most importantly, the breakdown viscosity (BDV) was notably increased while the setback viscosity (SBV) was obviously decreased (Figure 6g), suggesting that the ECQ of the rice from the pOsl2::GWD1 transgenic plants was improved. This conclusion was further confirmed by the taste value result, which indicated that the taste value was also increased in the pOsl2::GWD1 rice (Table S3). In contrast, the PKV, HPV, CPV and BDV of rice flour from gwd1 mutant plants were all slightly decreased, while the SBV was increased (Figure 6h), implying the ECQ of gwd1 rice was somewhat decreased.
Figure 6.
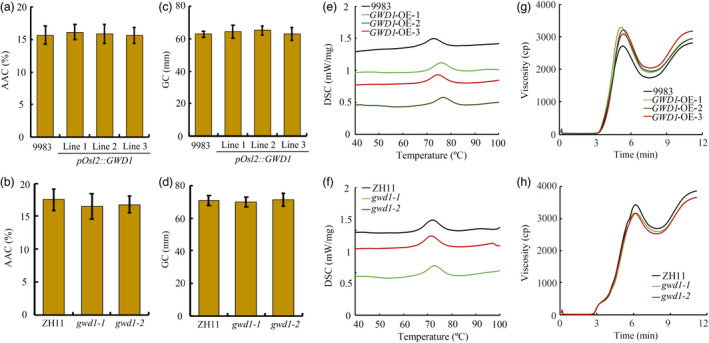
Physicochemical qualities of flours made from mature seeds of the GWD1‐related rice materials and the corresponding WT controls. (a, b) Apparent amylose content (AAC), (c, d) gel consistency (GC), (e, f) gelatinization and (g, h) Rapid Visco Analyzer (RVA) spectra of the three pOsl2::GWD1 transgenic lines and the two gwd1 knock‐out mutant lines respectively. Error bars represent the SDs
GWD1 is a positive regulator of rice seed germination and stress tolerance
Seed germination is not only a key event in plant reproduction and development, but it is also critical for crop production. Therefore, the effect of GWD1 overexpression or knock‐down mutation on rice seed germination was also investigated. The analysis showed that GWD1 overexpression increased germination while the gwd1 mutation decreased rice seed germination (Figure 7a, b; Figure S6a‐d). We next used the starch board test and enzyme activity test to evaluate the α‐amylase activity of GWD1‐related materials during germination. The results indicated that there is no significant difference in α‐amylase activity between rice from transgenic pOsl2::GWD1 plants compared to the WT control (Figure 7c, e, g). However, the α‐amylase activity was strikingly decreased in gwd1 mutants in both tests (Figure 7d, f, h). All of the above analyses demonstrated that GWD1 plays a positive role in regulating rice seed germination. Moreover, seed germination is precisely controlled by various environmental cues and endogenous hormones. Environmental stresses can suppress rice seed germination and cause yield losses. Therefore, it is essential to evaluate seed germination in the GWD1‐related materials in response to stress. The test results showed that germination in the GWD1 overexpression lines was less sensitive while the gwd1 mutants were more sensitive to both salt stress and drought stress (Figure 8a‐d). Therefore, GWD1 can promote rice seed germination under either normal or stress conditions, at least partially by modulating the α‐amylase activity. Since the promotional effect of GWD1 overexpression on rice drought resistance was more remarkable, we further evaluated the response of pOsl2::GWD1 transgenic seedlings to drought stress. The results indicate that GWD1‐overexpressing seedlings are more resistant to PEG treatment than are the WT control seedlings. In addition, almost all of the pOsl2::GWD1 transgenic seedlings recovered, while the WT seedlings died 36 h after the PEG‐6000 was removed (Figure 8e).
Figure 7.
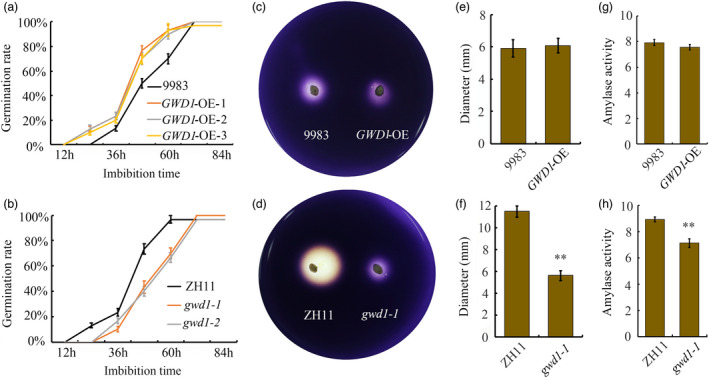
GWD1 is a positive regulator of seed germination in rice. Germination rates of the three pOsl2::GWD1 transgenic rice lines (a) and the two gwd1 mutants (b) and their WT controls. Representative photos of the starch board test of s pOsl2::GWD1 transgenic line (c) and a gwd1 mutant (d) and the respective control lines 9983 and ZH11. Diameters of the colourless halos produced by the grain samples from the pOsl2::GWD1 transgenic line (e) and the gwd1‐1 mutant (f). α‐amylase activity in the grains of the pOsl2::GWD1 transgenic line (g) and the gwd1‐1 mutant (h). Each experiment was repeated at least three times. Error bars represent the SDs. **P < 0.01 (Student’s t‐test)
Figure 8.
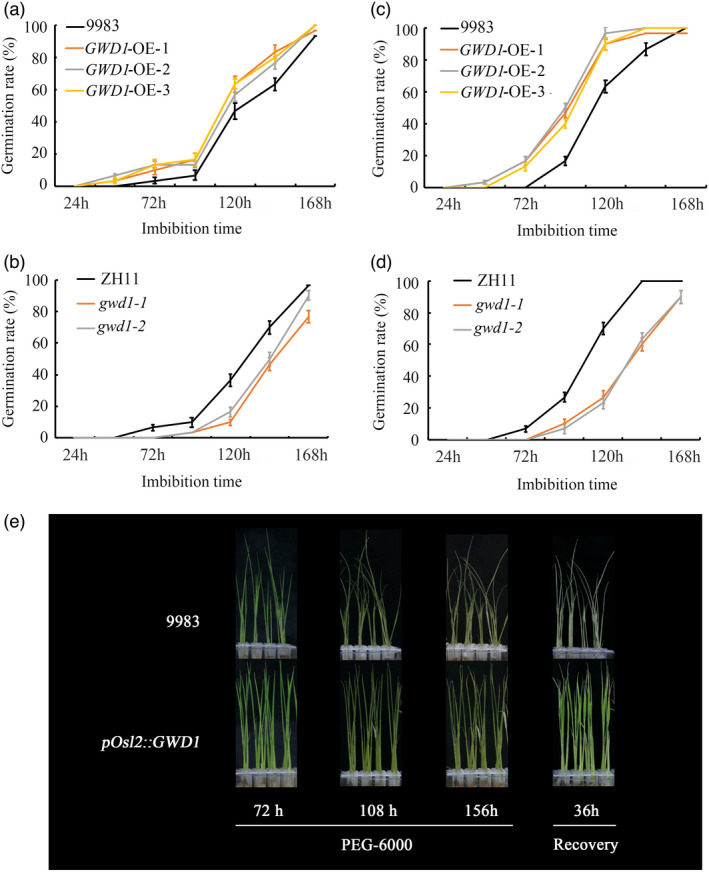
GWD1 overexpression increases resistance to abiotic stress in rice. Germination rates of the three pOsl2::GWD1 transgenic rice lines (a) and the two gwd1 mutants (b) in response to salt treatment (200 mM NaCl). Germination rates of the pOsl2::GWD1 transgenic lines (c) and the gwd1 mutants (d) in response to 30% PEG‐6000 treatment. (e) The pOsl2::GWD1 transgenic seedlings were more resistant to PEG treatment than the WT control
Discussion
Starch is an α‐D‐glucan that consists of a mixture of two polymers, amylose and amylopectin. The composition and structure of starch in the endosperm are key determinants of rice grain quality. Therefore, the expression and function of starch biosynthesis‐related genes in rice endosperm have been extensively studied (Butardo et al., 2019; Pandey et al., 2012). Some genes, such as Wx, are considered to be the key determinants of rice quality (Huang et al., 2020; Zhang et al., 2019). However, studies of genes involved in starch metabolism in rice source organs, such as leaves, are still rare. The starch phosphorylation reaction catalysed by GWD is the beginning of the degradation of transitory starch produced by photosynthesis. GWD, a key enzyme with essential roles in the first step of transitory starch degradation, plays important roles in both spore‐forming plants and angiosperms (Mahlow et al., 2016; Muyiwa et al., 2020). GWD has been studied in a number of plant species, including Arabidopsis, potato, rice, maize and wheat (Betti et al., 2016; Bowerman et al., 2016; Hirose et al., 2013; Malinova et al., 2018; Xu et al., 2017). In most of these studies, the expression of GWD was down‐regulated and consequently resulted in a starch‐excess phenotype accompanied by growth retardation, demonstrating the positive roles played by GWD in enhancing the degradation of transitory starch and in plant growth.
In addition, the effects of GWD on the phosphorylation, structure and degradation of starch have also been investigated (Chen et al., 2017). However, there are few comprehensive studies of the effects of GWD on important economic traits of crops. A GWD RNAi construct driven by the constitutive Ubiquitin promoter was introduced and expressed in maize, which resulted in an increased leaf starch content with no change in plant biomass (Weise et al., 2012). In rice, the gwd1 mutant LSE1 was generated by Tos17 insertion and about one third of the CDS of GWD1 was deleted. Therefore, there is no functional GWD1 protein in the LSE1 rice, which led to the hyperaccumulation of transitory starch in leaves but had no notable effects on rice growth. However, LSE1 rice had smaller grains, fewer panicles per plant and lower percentage of ripened grains, thus leading to a 20∼40% reduction in rice yield (Hirose et al., 2013). In wheat, a different conclusion was reached; down‐regulation of GWD1 specifically in the endosperm via RNAi promoted both vegetative biomass and grain yield in wheat (Bowerman, et al., 2016; Ral et al., 2012), which could be due to the use of an endosperm‐specific promoter. Therefore, using a tissue‐specific promoter, instead of a constitutive promoter, would be more practical for improving target crop traits because the plants would experience fewer adverse side effects. In our research, we intensively studied the effects of GWD1 on rice growth and grain yield in both GWD1 overexpression transgenic lines and in weak gwd1 mutant plants. It is worth noting that the GWD1 gene in the overexpression lines was driven by the Osl2 promoter, which imparted a leaf‐specific and senescence‐inducible expression pattern. Our results demonstrate that tissue‐specific overexpression of GWD1 enhanced several yield‐related traits in rice, such as plant height, 1000‐grain weight and grain yield per plant (Figure 4a‐c, Figure S5c). As to the gwd1 mutants, the mutation site was located between the TATA box and the initiation codon ATG of GWD1. Although the mutations did not directly modify any known motifs, the expression of GWD1 was notably declined. We speculated that the destruction of unknown motifs or violation of spacing constraints may lead to GWD1 expression change in gwd1 mutants. In fact, a previous report indicated that single‐nucleotide deletions at any position between the TATA box and the initiator sharply reduced transcription of target genes (Patwardhan et al., 2009). Moreover, two recent studies in rice also reported similar findings to us. Huang et al. (2020) reported that editing a target site in the core promoter of Wx gene, without alteration of the TATA box or other known motifs, caused distinct change in Wx expression. In another study, Zeng et al. (2020) also generated a number of mutated Wx alleles by modifying its promoter, some of which only had a fragment deletion without known motifs included or even only contained a single‐nucleotide change. Nevertheless, the expression of Wx was significantly declined in these lines. In the present study, the mutation in the core promoter of GWD1 had milder effects on rice growth and development than that of the reported GWD1 null mutant LSE1. For example, the yield‐related traits of gwd1 mutant were unchanged in the present study, but the grain yield of LSE1 mutant was declined by more than 20% (Figure 4e‐h; Hirose et al., 2013). We infer that modulation of GWD1 expression by editing its core promoter did not affect its protein structure and functions, thus causing no observable negative effects on both rice growth and development, including plant height, seed size, setting rate and 1000‐grain weight. In brief, the existence of functional GWD1 protein or not is the major cause that led to different yield performance of LSE1 rice and gwd1 mutants.
Although knock‐out of the target gene is useful in studying its functions in the lab and sometimes it could also improve certain traits of crops, it often accompanies with a series of other negative side effects. That is one important reason why most cloned yield‐related genes have not been successfully applied in crop production. On the other hand, we could fine‐tune the expression of the target gene by promoter editing using CRISPR/Cas9 technology, without damaging its protein product, thus leading to less negative side effects on plant growth and development. Our study demonstrated that the strategy of editing the core promoter of target gene is effectual and applicable in modern breeding programmes. Moreover, we also studied the quality of the rice harvested from the GWD1‐related materials in detail, including both appearance quality and ECQ. The shape of rice grains from the GWD1 overexpression lines was slender (Figure 5a‐c), meeting the requirements of seed shape in modern rice breeding programmes. Moreover, its pasting properties were modified, with increased BDV and decreased SBV (Figure 6g), which resulted in improved apparent viscosity and texture of the cooked rice. Nevertheless, the effects of the gwd1 mutation on grain yield and quality were milder. No notable change was observed in grain size and rice ECQ (Figure 5g‐i; Figure 6b, d, f, h). However, the degree of chalkiness and the percentage of chalky grains in the gwd1 mutants were significantly reduced (Figure 5j‐l). Therefore, GWD1 overexpression driven by the Osl2 promoter improved multiple agronomic traits related to both rice yield and quality, exhibiting potential for application in rice breeding programmes. On the contrary, the down‐regulation of GWD1 expression by promoter editing caused over‐accumulation of starch in the leaves and a reduction in grain chalkiness without any visible changes in other agronomic traits. The grain chalkiness traits are closely related to rice appearance quality and milling quality. In the market, the transparent rice is high in price and welcomed by consumers. From an industrial standpoint, it is critical to maximize the milling recovery of whole grain polished rice. Therefore, the rice processing enterprises will prefer transparent rice grains because they are not easy to be broken during milling process, thus increasing head rice yield.
Source–sink communication is critical in almost every stage of plant growth and development (Li et al., 2018a). Various factors, especially sugar allocation and phytohormones, are involved in the regulation of the source–sink relationship (Chen et al., 2019; Mathan et al., 2021; Réthoré et al., 2020; Zhang et al., 2021). As mentioned above, modulating the expression of the leaf‐dominant gene GWD1 not only affected starch accumulation in the source organ leaves, but also changed sink organ seed‐related traits, including grain shape, rice transparency and physicochemical characteristics. Another important issue related to source–sink communication is seed germination. Seed germination, the beginning of a new life cycle in plants, involves a series of complex physiological and biochemical reactions, including degradation of seed storage reserves. A recent report showed that the metabolism of glutelin, the major storage protein in rice seeds, is involved in phytohormone‐regulated seed germination in rice (Xiong et al., 2021). Whether changes in the level of starch, another major component of the rice seed, also affects seed germination is still largely unknown. Since GWD1 is the primary enzyme responsible for starch phosphorylation, an integral step in starch degradation, GWD1‐related genetic materials are appropriate for studying the effect of starch metabolism on seed germination. However, there are only three published studies, in wheat, barley and tomato, that mention the effect of GWD on germination. In wheat, endosperm‐specific suppression of GWD expression caused a slight reduction in germination rate (Ral et al., 2012). In barley, GWD overexpression had combination effects on both starch granule structure and amylase activities, thus affecting cereal grain germination and seedling establishment (Shaik et al., 2014). In tomato, the gwd mutant legwd::Ds, caused by insertion of the transposon Ds, showed excess accumulation of starch in the pollen and reduced pollen germination, which led to male gametophytic lethality (Nashilevitz et al., 2009). However, no such data have been reported from rice. In the present study, we have demonstrated that GWD1 is a positive regulator of seed germination in rice by using both GWD1 overexpression lines and gwd1 mutants. Overexpression of GWD1 promoted seed germination, while the gwd1 mutation suppressed seed germination, which was due at least in part to changes in the activity of α‐amylase (Figure 7a‐h). Although both starch metabolism and the levels of soluble sugars in plants contribute to alleviation of the damaging effects of stress (Krasensky and Jonak 2012; Siddiqui et al., 2020), direct evidence confirming the role of GWD in the stress response is rare. A single study in Arabidopsis showed that mutation of GWD promoted electrolyte leakage and reduced sugar accumulation and plant survival rate under freezing stress (Yano et al., 2005). In our study, we directly demonstrated that GWD1 enhances tolerance to abiotic stress during both seed germination and seedling growth stages in rice (Figure 8a‐e), thus further adding to our knowledge of the role of GWD in mediating the plant stress response.
In conclusion, leaf‐specific overexpression or knocking down GWD1 expression in rice allowed us to comprehensively analyse the function and effects of GWD1 on plant growth, rice yield, grain quality, seed germination and the stress response. Significantly, the gwd1 weak mutants generated by promoter editing caused no observable effects on rice growth and development, but only mildly modified its grain appearance quality and seed germination. Nevertheless, pOsl2::GWD1 transgenic rice plants exhibited improvements in multiple key traits, including increased 1000‐grain weight, improved grain shape and rice quality, and increased seed germination and stress tolerance. Therefore, our study reveals that GWD1 is not only directly involved in the degradation of transitory starch in rice leaves, but also plays positive roles in a number of other growth and developmental events in rice. Therefore, GWD1 is an ideal biotechnological target for rice breeding programmes. The combination of GWD1 with an appropriate promoter, such as the Osl2 promoter, will make impressive contributions towards potential improving rice yield, grain quality and stress tolerance. Moreover, targeted modification of gene core promoter is also promising in fine‐tuning of the expression of target genes. Considering the wide distribution of the GWD gene in plant species, a similar strategy could also be applied to the improvement of other crops.
Materials and methods
Plant materials and growth conditions
Two japonica rice varieties were used in this study, 9983 and ‘Zhonghua11’ (ZH11). 9983 was used for transgenic GWD1 overexpression, and ‘Zhonghua11’ was used for CRISPR/Cas9‐mediated knock‐out of GWD1. All rice plants were grown in the field at Yangzhou University (Yangzhou, Jiangsu Province, China) in the summer, using the same climate and crop management conditions. Three replicate plots were used in the experiments, and the plots were arranged in a randomized block pattern.
Promoter analysis and generation of GWD1 knock‐down mutant
To identify major motifs within the core promoter of GWD1, 200 bp DNA sequence upstream of the GWD1 translation initiation codon ATG was scanned in PLACE database (http://www.dna.affrc.go.jp/PLACE/). To generate the gwd1 knock‐down mutant, a specific target site guide RNA sequence (5’‐GAGATATCCATCCGGATCC‐3’) located between TATA box and the ATG of GWD1 was selected and cloned into the sgRNA‐Cas9 expression plasmid (Wang et al., 2019), which was then transformed into the japonica rice cultivar ZH11 (Slamet‐Loedin et al., 2014). Then, DNA fragment containing the GWD1 target site was amplified by PCR using the primer pair GWD1MT‐F and GWD1MT‐R (Table S4) and then used for sequencing. The plants with mutation were selected, and their seeds were grown into the next generation plants. Then, the homozygous rice mutants were identified by sequencing. Next, the Cas9 gene, located inside the T‐DNA region, was identified by PCR using primer Cas9‐F and Cas9‐R (Table S4). The transgene‐free homozygous mutants were selected for following analysis.
Generation and identification of GWD1 overexpression rice lines
The GWD1 coding sequence and the Osl2 promoter were amplified and cloned into the binary vector pCAMBIA1300 to generate the pOsl2::GWD1 expression vector. This construct was then successfully introduced into the genome of the rice variety 9983 via Agrobacterium‐mediated transformation (Slamet‐Loedin et al., 2014). Genomic DNA PCR analysis was performed to identify the positive transgenic plants (T0 generation) by GWD1OE‐F and GWD1OE‐R primers (Table S4). Then, the single copy transgenic lines (T1 generation) were selected based on genotyping result and Mendel’s law of segregation. Next, the seeds of six positive plants from each selected T1 line were collected, respectively, and used to produce corresponding T2 progeny lines. Finally, total 20 plants from each T2 rice line were further genotyped by PCR to determine whether the line was homozygous.
Agronomic trait investigations of the GWD1‐related rice materials
The main agronomic traits for rice were recorded after seed maturity. Plant height refers to the length from top of the rice main panicle to the field ground. Seed setting rate is the ratio of full‐filled grains to total grains of the panicle. Totally, 10 identical main panicles were collected from the three replicate plots of the same rice sample. The number of total grains and the full‐filled grains of each panicle was counted and recorded, respectively, for calculating the seed setting rate. Then, these full‐filled mature seeds were air‐dried for subsequent analyses of 1000‐seed weight, grain morphology and rice quality. About 200 rice grains or polished seeds were measured, respectively, for seed size or chalkiness using a grain appearance analyzer ScanMaker (Microtek, Shanghai, China). The data for each sample represent the mean of three plots.
Total RNA extraction and qRT‐PCR gene expression analysis
Rice tissue samples of ZH11 for spatial expression analysis of Osl2 and GWD1 were collected from 14‐day‐old whole seedlings or the leaf, seed, root, stem and leaf sheath of mature rice plant (10 DAF). Leaf samples for temporal expression analysis of Osl2 were collected from ZH11 mature rice plants 0, 5, 10, 15 and 20 DAF respectively. Leaf samples for expression analysis of GWD1 in its transgenic rice or mutants were from homozygous plants 10 DAF (T2 generation). Then, total RNA was extracted from the collected rice samples using the RNAsimple Total RNA Kit (Tiangen, Beijing, China), after which it was treated with DNase I (Qiagen, Hilden, Germany) to remove residual genomic DNA. 1 μg samples of total RNA were reverse‐transcribed using the SuperScript™ First‐Strand cDNA Synthesis System (Invitrogen, Carlsbad, CA). qRT‐PCR assays were then performed on a MyiQ Real‐Time system (Bio‐Rad, CA) using Actin as the reference gene for normalization of gene expression. The primer pairs used for expression analysis of Osl2 and GWD1 were Osl2qRT‐F/Osl2qRT‐R and GWD1qRT‐F/GWD1qRT‐R respectively. Each experiment consisted of three biological replicates, and the primer sequences are given in Supplementary Table S4.
Iodine staining of starch in rice seedlings
Rice seeds were sterilized with 70% ethanol, washed twice with Milli‐Q water and allowed to imbibe in water for four days for germination. The germinated seeds were transferred to a PCR plate with the bottom cut out and then cultured in water in a climate‐controlled chamber (12‐h light and 12‐h dark at a temperature of 26 °C), and the water was changed every 48 h. 14‐day‐old seedlings at the end of the dark cycle (gwd1 mutant) or six hours after the beginning of the light cycle (pOsl2::GWD1) were collected for starch staining with potassium iodide as previously described (Yu et al., 2001).
Analysis of seed quality using Grain analyzer and preparation of rice flour
Mature rice seeds were air‐dried, dehusked and subsequently polished using a grain polisher (Kett, Tokyo, Japan). The seed quality of polished rice was measured using the Grain analyzer (Perten IM9500, Sweden). Then, the polished grains were further ground into powder using a FOSS 1093 Cyclotec Sample Mill (Tecator, Höganäs, Sweden) with a 0.5 mm sieve. The rice powder samples were then stored in sealed bags under refrigeration at 4 °C until they were used for analysis.
Analysis of the physicochemical characteristics of rice
Apparent amylose content (AAC) was determined using the iodine colorimetric method as described previously (Tan et al., 1999). Gel consistency (GC) was determined as previously described (Zhu et al., 2010). A differential scanning calorimeter (DSC 200 F3, Netzsch Instruments NA LLC; Burlington, MA) was used to evaluate starch gelatinization and retrogradation temperatures as described (Zhang et al., 2017). Rapid Visco Analyzer (RVA) profiles were determined using the Rapid Viscosity Analyzer (RVA) (Techmaster, Newport Scientific, Warriewood, Australia) (Li et al., 2018b). All tests were performed in triplicate.
Seed germination analysis
Plump and uniform rice seeds were selected, dehulled, and sterilized with 70% ethanol and washed twice with Milli‐Q water. The sterilized seeds were transferred to a sterile petri dish and allowed to germinate in the dark in a climate‐controlled incubator at a temperature of 26 °C and a relative humidity of 70%.
Enzyme activity analysis
Seeds were imbibed at 28 °C for 72 h, and starch was extracted from the germinating seeds. Starch hydrolase was then used to hydrolyse the starch to produce reducing sugars, which react with 3,5‐dinitrosalicylic acid to produce a brownish red compound with an absorption peak at 540 nm. Amylase activity was determined by measuring the increase in the rate of absorbance at 540 nm (Liu et al., 2018a).
Starch degradation experiment
Seeds of the GWD1 overexpression and mutant lines as well as their respective WT controls were sterilized, and half‐seeds without embryos were placed on a 2% agar plate containing 2% potato dextrose agar. The plates were then incubated in the dark at 28 °C for four days, after which they were stained with iodine solution for five minutes and subsequently photographed (Liu et al., 2018a). The diameters of at least 20 halos for each rice sample were measured by using ImageJ software.
Stress treatment analysis
For the analysis of seed germination under abiotic stress conditions, seeds were exposed to either 200 mM NaCl or 30% PEG‐6000 treatments. Germination rates were recorded every day. Each seed germination assay included at least three independent biological replicates, and each replicate contained 30 germinated seeds (Li et al., 2020). For PEG treatment at the seedling stage, 2‐week‐old seedlings of the pOsl2::GWD1 transgenic line and the WT control were treated with 30% PEG for seven days, after which they were transferred to fresh water without PEG to allow for recovery. Photographs of seedlings were taken at 72 h, 108 h and 156 h after PEG treatment and at 36 h after recovery.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
Q.F.L. and Q.Q.L. conceived the project. Z.W., K.W., M.X., J.W., X.F. and L.H. performed the experiments and analysed the data. C.Z. and D.Z. participated in the design and coordination of the study. Z.W. and Q.F.L. wrote and revised the manuscript. All authors read and approved the final version of the manuscript.
Supporting information
Figure S1. Expression of the GWD1 gene in six tissues of rice.
Figure S2. Expression analysis of GWD1 in the developing seeds of pOsl2::GWD1 transgenic rice plants.
Figure S3. Sequencing result of ZH11 control and gwd1 mutant.
Figure S4. Iodine staining of seedlings of the pOsl2::GWD1 transgenic line and the WT control 9983.
Figure S5. Analysis of several yield‐related traits of pOsl2::GWD1 transgenic rice.
Figure S6. Shoot lengths of germinated seeds of GWD1‐related materials at 96 h after imbibition (HAI).
Table S1. The putative cis‐regulatory motifs in the key region of GWD1 promoter.
Table S2. Thermal properties of GWD1‐related rice samples as determined by differential scanning calorimetry (DSC).
Table S3. Data of rice grain quality were analysed by Grain analyzer.
Table S4. Names and sequences of oligonucleotide primers used in this study.
Acknowledgements
This work was financially supported by the Ministry of Science and Technology of China (2016YFD0100902 and 2016ZX08009003‐004), the National Natural Science Foundation of China (31825019), the Science Fund for Distinguished Young Scholars of Jiangsu Province (BK20200045) and the Programs (SWYY‐154, PAPD and Qinglan Project) from Jiangsu Government.
Wang, Z. , Wei, K. , Xiong, M. , Wang, J.‐D. , Zhang, C.‐Q. , Fan, X.‐L. , Huang, L.‐C. , Zhao, D.‐S. , Liu, Q.‐Q. and Li, Q.‐F. (2021) Glucan, Water‐Dikinase 1 (GWD1), an ideal biotechnological target for potential improving yield and quality in rice. Plant Biotechnol. J., 10.1111/pbi.13686
References
- Abt, M.R. and Zeeman, S.C. (2020) Evolutionary innovations in starch metabolism. Curr. Opin. Plant Biol, 55, 109–117. [DOI] [PubMed] [Google Scholar]
- Adegbaju, M S. , Morenikeji, O B. , Borrego, E J. , Hudson, A O. and Thomas, B N. (2020) Differential evolution of α‐glucan water dikinase (GWD) in plants. Plants, 9, 1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari, M.I. , Lee, R.H. and Chen, S.C.G. (2005) A novel senescence‐associated gene encoding γ‐aminobutyric acid (GABA):pyruvate transaminase is upregulated during rice leaf senescence. Physiol. Plantarum, 123, 1–8. [Google Scholar]
- Baunsgaard, L. , Lütken, H. , Mikkelsen, R. , Glaring, M.A. , Pham, T.T. and Blennow, A. (2005) A novel isoform of glucan, water dikinase phosphorylates pre‐phosphorylated α‐glucans and is involved in starch degradation in Arabidopsis. Plant J, 41, 595–605. [DOI] [PubMed] [Google Scholar]
- Betti, C. , Vanhoutte, I. , Coutuer, S. , De Rycke, R.M. , Mishev, K. , Vuylsteke, M. , Aesaert, S. et al. (2016) Sequence‐specific protein aggregation generates defined protein knockdowns in plants. Plant Physiol, 171, 773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow, A. and Engelsen, S.B. (2010) Helix‐breaking news: fighting crystalline starch energy deposits in the cell. Trends Plant Sci, 15, 236–240. [DOI] [PubMed] [Google Scholar]
- Bowerman, A.F. , Newberry, M. , Dielen, A.‐S. , Whan, A. , Larroque, O. , Pritchard, J. , Gubler, F. et al. (2016) Suppression of glucan, water dikinase in the endosperm alters wheat grain properties, germination and coleoptile growth. Plant Biotechnol. J, 14, 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo, V.M. Jr , Sreenivasulu, N. and Juliano, B.O. (2019) Improving rice grain quality: state‐of‐the‐art and future prospects. Methods Mol. Biol, 1892, 19–55. [DOI] [PubMed] [Google Scholar]
- Caspar, T. , Lin, T.P. , Kakefuda, G. , Benbow, L. , Preiss, J. and Somerville, C. (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol, 95, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Li, G. , Islam, M.R. , Fu, W. , Feng, B. , Tao, L. and Fu, G. (2019) Abscisic acid synergizes with sucrose to enhance grain yield and quality of rice by improving the source‐sink relationship. BMC Plant Biol, 19, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Sun, X. , Zhou, X. , Hebelstrup, K.H. , Blennow, A. and Bao, J. (2017) Highly phosphorylated functionalized rice starch produced by transgenic rice expressing the potato GWD1 gene. Sci. Rep, 7, 3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T. , Aoki, N. and Harada, Y. (2013) Disruption of a rice gene forα‐glucan water dikinase, OsGWD1, leads to hyperaccumulation of starch in leaves but exhibits limited effects on growth. Front. Plant Sci, 4, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Lu, S.‐J. , Wang, M.‐J. , He, H. , Sun, L.E. , Wang, H. , Liu, X.‐H. et al. (2018) A novel QTL qTGW3 encodes the GSK3 / SHAGGY‐like kinase OsGSK5 / OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant, 11, 736–749. [DOI] [PubMed] [Google Scholar]
- Huang, L. , Li, Q. , Zhang, C. , Chu, R. , Gu, Z. , Tan, H. , Zhao, D. et al. (2020) Creating novel Wx alleles with fine‐tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J, 18, 2164–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Qian, Q. , Liu, Z. , Sun, H. , He, S. , Luo, D.A. , Xia, G. et al. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet, 41, 494–497. [DOI] [PubMed] [Google Scholar]
- Jeong, H.J. and Jung, K.H. (2015) Rice tissue‐specific promoters and condition‐dependent promoters for effective translational application. J. Integr. Plant Biol, 57, 913–924. [DOI] [PubMed] [Google Scholar]
- Kötting, O. , Pusch, K. , Tiessen, A. , Geigenberger, P. , Steup, M. and Ritte, G. (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol, 137, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky, J. and Jonak, C. (2012) Drought, salt, and temperature stress‐induced metabolic rearrangements and regulatory networks. J. Exp. Bot, 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.H. , Wang, C.H. , Huang, L.T. and Chen, S.C. (2001) Leaf senescence in rice plants: cloning and characterization of senescence up‐regulated genes. J. Exp. Bot, 52, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Li, N. , Xu, R. and Li, Y. (2019) Molecular networks of seed size control in plants. Annu. Rev. Plant Biol, 70, 435–463. [DOI] [PubMed] [Google Scholar]
- Li, P. , Chang, T. , Chang, S. , Ouyang, X. , Qu, M. , Song, Q. , Xiao, L. et al. (2018a) Systems model‐guided rice yield improvements based on genes controlling source, sink, and flow. J. Integr. Plant Biol, 60, 1154–1180. [DOI] [PubMed] [Google Scholar]
- Li, Q.F. , Yu, J.W. , Lu, J. , Fei, H.Y. , Luo, M. , Cao, B.W. , Huang, L.C. et al. (2018b) Seed‐specific expression of OsDWF4, a rate‐limiting gene involved in brassinosteroids biosynthesis, improves both grain yield and quality in rice. J. Agric. Food Chem, 66, 3759–3772. [DOI] [PubMed] [Google Scholar]
- Li, Q.‐F. , Zhou, Y.U. , Xiong, M. , Ren, X.Y. , Han, L.I. , Wang, J.D. , Zhang, C.Q. et al. (2020) Gibberellin recovers seed germination in rice with impaired brassinosteroid signalling. Plant Sci, 293, 110435. [DOI] [PubMed] [Google Scholar]
- Liang, J. , Zhang, J. and Cao, X. (2001) Grain sink strength may be related to the poor grain filling of indica‐japonica rice (Oryza sativa) hybrids. Physiol. Plant, 112, 470–477. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Xia, W. , Li, H. , Zeng, H. , Wei, B. , Han, S. and Yin, C. (2018a) Salinity inhibits rice seed germination by reducing α‐amylase activity via decreased bioactive gibberellin content. Front. Plant Sci, 9, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Han, R. , Wu, K. , Zhang, J. , Ye, Y. , Wang, S. , Chen, J. et al. (2018b) G‐protein βγ subunits determine grain size through interaction with MADS‐domain transcription factors in rice. Nat. Commun, 9, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorberth, R. , Ritte, G. , Willmitzer, L. and Kossmannet, J. (1998) Inhibition of a starch‐granule‐bound protein leads to modified starch and repression of cold sweetening. Nat. Biotechnol, 16, 473–477. [DOI] [PubMed] [Google Scholar]
- Mahlow, S. , Orzechowski, S. and Fettke, J. (2016) Starch phosphorylation: insights and perspectives. Cell Mol. Life Sci, 73, 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinova, I. , Mahto, H. , Brandt, F. , AL‐Rawi, S. , Qasim, H. , Brust, H. , Hejazi, M. and et al. (2018) EARLY STARVATION1 specifically affects the phosphorylation action of starch‐related dikinases. Plant J, 95, 126–137. [DOI] [PubMed] [Google Scholar]
- Mao, H. , Sun, S. , Yao, J. , Wang, C. , Yu, S. , Xu, C. , Li, X. and et al. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U. S. A, 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan, J. , Singh, A. and Ranjan, A. (2021) Sucrose transport and metabolism control carbon partitioning between stem and grain in rice. J. Exp. Bot, 72, 4355–4372. [DOI] [PubMed] [Google Scholar]
- Mena, M. , Cejudo, F.J. , Isabel‐Lamoneda, I. and Carbonero, P. (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin‐responsive genes in barley aleurone. Plant Physiol, 130, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, R. , Mutenda, K. , Mant, A. , Schürmann, P. and Blennow, A. (2005) Alpha‐glucan, water dikinase (GWD): A plastidic enzyme with redox‐regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci, 102, 1785–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, A. , Umemura, T. , Kuroyanagi, M. , Futsuhara, Y. , Perata, P. and Yamaguchi, J. (1998) Functional dissection of a sugar‐repressed alpha‐amylase gene (Ramy1A) promoter in rice embryos. FEBS Lett, 423, 81–85. [DOI] [PubMed] [Google Scholar]
- Nashilevitz, S. , Melamed‐Bessudo, C. , Aharoni, A. , Kossmann, J. , Wolf, S. and Levy, A.A. (2009) The legwd mutant uncovers the role of starch phosphorylation in pollen development and germination in tomato. Plant J, 57, 1–13. [DOI] [PubMed] [Google Scholar]
- Pandey, M.K. , Rani, N.S. , Madhav, M.S. , Sundaram, R.M. , Varaprasad, G.S. , Sivaranjani, A. , Bohra, A. et al. (2012) Different isoforms of starch‐synthesizing enzymes controlling amylose and amylopectin content in rice (Oryza sativa L.). Biotechnol. Adv, 30, 1697–1706. [DOI] [PubMed] [Google Scholar]
- Patwardhan, R.P. , Lee, C. , Litvin, O. , Young, D.L. , Pe'er, D. and Shendure, J. (2009) High‐resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol, 27, 1173–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, S. , Laza, R. , Visperas, R. , Sanico, A. , Cassman, K.G. and Khush, G. (2000) Grain yield of rice cultivars and lines developed in the Philippines since 1966. Crop Sci, 40, 307–314. [Google Scholar]
- Qu, L.Q. , Xing, Y.P. , Liu, W.X. , Xu, X.P. and Song, Y.R. (2008) Expression pattern and activity of six glutelin gene promoters in transgenic rice. J. Exp. Bot, 59, 2417–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ral, J.P. , Bowerman, A.F. , Li, Z. , Sirault, X. , Furbank, R. , Pritchard, J.R. , Bloemsma, M. et al. (2012) Down‐regulation of Glucan, Water‐Dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnol. J, 10, 871–882. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Li, Y. , He, G. and Qian, Q. (2020) Multifloret spikelet improves rice yield. New Phytol, 225, 2301–2306. [DOI] [PubMed] [Google Scholar]
- Réthoré, E. , Ali, N. , Yvin, J.C. and Hosseini, S.A. (2020) Silicon regulates source to sink metabolic homeostasis and promotes growth of rice plants under sulfur deficiency. Int. J. Mol. Sci, 21, 3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.C. , Muro‐Pastor, M.I. and Florencio, F.J. (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol, 134, 1718–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte, G. , Lloyd, J.R. , Eckermann, N. , Rottmann, A. , Kossmann, J. and Steup, M. (2002) The starch‐related R1 protein is an alpha‐glucan, water dikinase. Proc. Natl. Acad. Sci. U. S. A, 99, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik, S.S. , Carciofi, M. , Martens, H.J. , Hebelstrup, K.H. and Blennow, A. (2014) Starch bioengineering affects cereal grain germination and seedling establishment. J. Exp. Bot, 65, 2257–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, H. , Sami, F. and Hayat, S. (2020) Glucose: sweet or bitter effects in plants‐a review on current and future perspective. Carbohydr. Res, 487, 107884. [DOI] [PubMed] [Google Scholar]
- Singh, N. , Singh, J. , Kaur, L. , Singh Sodhi, N. and Singh Gill, B. (2003) Morphological, thermal and rheological properties of starches from different botanical sources. Food Chem, 81, 219–231. [Google Scholar]
- Skeffington, A.W. , Graf, A. , Duxbury, Z. , Gruissem, W. and Smith, A.M. (2014) Glucan, water dikinase exerts little control over starch degradation in Arabidopsis leaves at night. Plant Physiol, 165, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamet‐Loedin, I.H. , Chadha‐Mohanty, P. and Torrizo, L. (2014) Agrobacterium‐mediated transformation: rice transformation. Methods Mol. Biol, 1099, 261–271. [DOI] [PubMed] [Google Scholar]
- Sonnewald, U. and Kossmann, J. (2013) Starches–from current models to genetic engineering. Plant Biotechnol. J, 11, 223–232. [DOI] [PubMed] [Google Scholar]
- Streb, S. and Zeeman, S.C. (2012) Starch metabolism in Arabidopsis. The Arabidopsis Book, 10, e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. , Wang, L. , Mao, H. , Shao, L. , Li, X. , Xiao, J. , Ouyang, Y. et al. (2018) A G‐protein pathway determines grain size in rice. Nat. Commun, 9, 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y.F. , Li, J.X. , Yu, S.B. , Xing, Y.Z. , Xu, C.G. and Zhang, Q. (1999) The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet, 99, 642–648. [DOI] [PubMed] [Google Scholar]
- Toyofuku, K. , Umemura, T. and Yamaguchi, J. (1998) Promoter elements required for sugar‐repression of the RAmy3D gene for alpha‐amylase in rice. FEBS Lett, 428, 275–280. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Liu, Q. , Shen, Y. , Hua, Y. , Wang, J. , Lin, J. , Wu, M. et al.(2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol, 37, 283–286. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018) A single transcription factor promotes both yield and immunity in rice. Science, 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Weise, S.E. , Aung, K. , Jarou, Z.J. , Mehrshahi, P. , Li, Z. , Hardy, A.C. , Carr, D.J. and et al. (2012) Engineering starch accumulation by manipulation of phosphate metabolism of starch. Plant Biotechnol. J, 10, 545–554. [DOI] [PubMed] [Google Scholar]
- Wiesenborn, D.P. , Orr, P.H. , Casper, H.H. and Tacke, B.K. (1994) Potato starch paste behavior as related to some physical/chemical properties. J. Food Sci, 59, 644–648. [Google Scholar]
- Xiong, M. , Chu, L. , Li, Q. , Yu, J. , Yang, Y. , Zhou, P. , Zhou, Y. et al. (2021) Brassinosteroid and gibberellin coordinate rice seed germination and embryo growth by regulating glutelin mobilization. Crop J, 10.1016/j.cj.2020.11.006. [DOI] [Google Scholar]
- Xu, X. , Dees, D. , Dechesne, A. , Huang, X.F. , Visser, R.G.F. and Trindade, L.M. (2017) Starch phosphorylation plays an important role in starch biosynthesis. Carbohydr. Polym, 157, 1628–1637. [DOI] [PubMed] [Google Scholar]
- Yano, R. , Nakamura, M. , Yoneyama, T. and Nishida, I. (2005) Starch‐related alpha‐glucan/water dikinase is involved in the cold‐induced development of freezing tolerance in Arabidopsis. Plant Physiol, 138, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, N. , Kim, Y.S. , Jeong, M.‐H. , Oh, S.‐J. , Jeong, J.S. , Park, S.‐H. , Jung, H. et al. (2010) Functional analysis of six drought‐inducible promoters in transgenic rice plants throughout all stages of plant growth. Planta, 232, 743–754. [DOI] [PubMed] [Google Scholar]
- Ying, J.‐Z. , Ma, M. , Bai, C. , Huang, X.‐H. , Liu, J.‐L. , Fan, Y.‐Y. and Song, X.‐J. (2018) TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant, 11, 750–753. [DOI] [PubMed] [Google Scholar]
- Yu, T.S. , Kofler, H. , Husler, R.E. , Hille, D. , Flügge, U.I. , Zeeman, S.C. and Smith, A.M. (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell, 13, 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, D. , Liu, T. , Ma, X. , Wang, B. , Zheng, Z. , Zhang, Y. , Xie, X. et al. (2020) Quantitative regulation of Waxy expression by CRISPR/Cas9‐based promoter and 5'UTR‐intron editing improves grain quality in rice. Plant Biotechnol. J, 18, 2385–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, L. , Wang, F. , Yan, A. , Liang, C. , Wang, S. , Wang, Y. and Xu, J. (2020) Pleiotropic effect of GNP1 underlying grain number per panicle on sink, source and flow in rice. Front. Plant Sci, 11, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Chen, S. , Ren, X. , Lu, Y. , Liu, D. , Cai, X. , Li, Q. et al. (2017) Molecular structure and physicochemical properties of starches from rice with different amylose contents resulting from modification of OsGBSSI activity. J. Agric. Food Chem, 65, 2222–2232. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Zhu, J. , Chen, S. , Fan, X. , Li, Q. , Lu, Y. , Wang, M. et al. (2019) Wx(lv), the ancestral allele of rice waxy gene. Mol. Plant, 12, 1157–1166. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Peng, K. , Cui, F. , Wang, D. , Zhao, J. , Zhang, Y. , Yu, N. et al. (2021) Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J, 19, 335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , He, S. , Naconsie, M. , Ma, Q. , Zeeman, S.C. , Gruissem, W. and Zhang, P. (2017) Alpha‐glucan, water dikinase 1 affects starch metabolism and storage root growth in cassava. Sci. Rep, 7, 9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.J. , Liu, Q.Q. , Sang, Y.J. , Gu, M.H. and Shi, Y.C. (2010) Underlying reasons for waxy rice flours having different pasting properties. Food Chem, 120, 94–100. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression of the GWD1 gene in six tissues of rice.
Figure S2. Expression analysis of GWD1 in the developing seeds of pOsl2::GWD1 transgenic rice plants.
Figure S3. Sequencing result of ZH11 control and gwd1 mutant.
Figure S4. Iodine staining of seedlings of the pOsl2::GWD1 transgenic line and the WT control 9983.
Figure S5. Analysis of several yield‐related traits of pOsl2::GWD1 transgenic rice.
Figure S6. Shoot lengths of germinated seeds of GWD1‐related materials at 96 h after imbibition (HAI).
Table S1. The putative cis‐regulatory motifs in the key region of GWD1 promoter.
Table S2. Thermal properties of GWD1‐related rice samples as determined by differential scanning calorimetry (DSC).
Table S3. Data of rice grain quality were analysed by Grain analyzer.
Table S4. Names and sequences of oligonucleotide primers used in this study.


