Abstract
Invasive fungal infections caused by yeasts of the genus Candida carry high morbidity and cause systemic infections with high mortality rate in both immunocompetent and immunosuppressed patients. Resistance rates against antifungal drugs vary among Candida species, the most concerning specie being Candida auris, which exhibits resistance to all major classes of available antifungal drugs. The presently available identification methods for Candida species face a severe trade-off between testing speed and accuracy. Here, we propose and validate a machine-learning approach adapted to Raman spectroscopy as a rapid, precise, and labor-efficient method of clinical microbiology for C. auris identification and drug efficacy assessments. This paper demonstrates that the combination of Raman spectroscopy and machine learning analyses can provide an insightful and flexible mycology diagnostic tool, easily applicable on-site in the clinical environment.
Keywords: Raman imaging, Raman spectroscopy, Candida auris, machine-learning, glucans, ergosterol
Introduction
Invasive candidiasis, a life-threatening opportunistic infection, nowadays represents a major cause of morbidity and mortality (Méan et al., 2008). In this pathological context, the emergence of multidrug species in the genus Candida is spread worldwide and poses severe public health implications. Candida albicans has long been the predominant species, but a recent shift has occurred toward non-albicans species with reduced susceptibility to antifungal agents. Candida auris was first identified and classified in the Makimura’s research group in 2009 (Sato et al., 2009). The multidrug-resistant C. auris resists commonly used drugs and often eludes a correct identification (Chaabane et al., 2019). In a lack of prompt and reliable microbiological identifications, the spread of infection by C. auris is unpredictable, and the situation is further exacerbated by unreported cases. Achieving an appropriate management of invasive candidiasis by C. auris entirely depends on two distinct factors: a rapid identification of species and a timely selection of an effective drug.
Speciation of Candida species can be performed by a number of different techniques, which are classified into two main categories: phenotypic/biochemical and molecular tests. Tests belonging to the former category, i.e., germ tube test, chlamydospore formation test, sugar assimilation, and sugar fermentation tests, commonly exploit directly observable characteristics. The germ tube test is the fastest method, but it only provides a limited range of speciation (C. albicans and Candida dubliniensis vs. all other Candida species) and is prone to false positive results (Crist et al., 1996). In the case of identification failure by the germ tube test, chlamydospore formation test or sugar assimilation and fermentation tests are usually performed. However, these methods are labor intensive (although recently being conspicuously automatized) and time consuming (3–14 days) (Chander, 2009). Newer commercially available tests, which include CHROMagarTM, Candida Plus (Mulet Bayona et al., 2020; Borman et al., 2021), API systems, and Vitek 2 ID system, offer more rapid and/or cost effective choices. However, none of the above-mentioned tests is capable to differentiate C. auris from other Candida yeast species (CDC, 2019). On the other hand, molecular tests specifically targeting drug-resistant C. auris have been developed, which are based on quantitative polymerase chain reaction (qPCR), real-time polymerase chain reaction (PCR), or loop-mediated isothermal amplification (LAMP) (Yamamoto et al., 2018; Camp et al., 2020). The assay time for these tests is in the order of 1 to few hours, excluding the time required for DNA extraction. Therefore, they are quite time consuming, despite being highly reliable. As a separate type of approach, the matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) method has the unique advantage of enabling the analysis from aliquots of positive blood culture bottles after a short time needed for protein extraction. MALDI-TOF MS analyses require times in the order of 90s to identify the fungus developed on the surface of a solid. However, detections from biological fluids (i.e., from positive haemoculture bottles) might take about 30 min. The main advantage of this method is its capacity of identifying all pathogens included in the database, including C. auris (Bal and McGill, 2018; Camp et al., 2020). Methods of genomic analysis have revealed fundamental aspects of Candida species and opened the path to gene deletion technologies. These technologies have been adapted to track C. auris and to screen it for drug resistance, mating, and virulence (Grahl et al., 2017). Among these methods, the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 genome modification systems, which was originally expression-constructed for C. albicans species, has recently been modified to enable genetic analyses of C. auris (Munoz et al., 2018). Precisely sequencing genomes from different Candida clades and species requires access to genomic sequence data with long lists of genes taking significant time to be analyzed (Skrzypek et al., 2018). Overall, the presently available identification methods for Candida species face a severe trade-off between testing speed and accuracy.
Different from genomics, Raman spectroscopy follows a structural analytical path to identify different yeast species. Resembling a molecular fingerprint, the Raman spectrum provides highly specific chemical information without requiring exogenous probes. Combined with high-resolution hyperspectral imaging techniques, Raman spectroscopy can be used as a molecular microscopy tool that enables in situ label-free screening of yeast cells (Noothalapati et al., 2016). The availability of a large number of spectra from Raman imaging then provides the opportunity to apply chemometric methods to yeast speciation. Chemometrics coupled with Raman analyses and imaging has so far been successfully employed to screen the quality of pharmaceutical products (Neuberger and Neususs, 2015), to unfold polymorphic transitions (Farias and Carneiro, 2014), to discriminate oral (Zhu et al., 2007) and pathogenic bacteria (Ho et al., 2019), and to identify biological contamination (Roesch et al., 2005a). Raman spectroscopy has already been applied to characterize Candida isolates and related biofilms in order to link the observed Raman spectroscopic characteristics to their genomic diversity (Samek et al., 2014; Chouthai et al., 2015; Potocki et al., 2019; Rebrosova et al., 2019); yet, such analyses have not been performed on multidrug-resistant C. auris isolates. Here, we present a preliminary study that combines Raman spectroscopy and imaging with chemometrics to develop a rapid and accurate algorithm for differentiating C. albicans from C. auris. Despite the presented Raman analyses being limited to only two (out of five known) C. auris clades, they suggest that the presented Raman platform might allow for a fast and accurate identification of C. auris species within minutes, thus potentially reducing healthcare costs and antibiotic misuse.
Experimental Procedures
Candida Species
Candida albicans ATCC® 90028 (C. albicans) cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA, United States). The following C. auris isolates were used: the LSEM 3673 (belonging to the South African Clade III) and the LSEM 0643 (or JCM15448T; belonging to the East Asian Clade II). The two latter clades were provided by Teikyo University. A brain heart infusion (BHI) medium was prepared by adding 25 g of BHI broth (Nihon Pharmaceutical Co., Ltd., Tokyo, Japan) into 1 L of distilled water and successively boiling the mixture. After sterilizing in an autoclave at 121°C for 15 min, the mixture was poured into 35 mm glass bottom dish (MatTek Life Sciences, MA, United States). The C. auris clades were cultured in SD agar at 36°C for 24 h under atmospheric pressure before Raman spectroscopic measurements. The C. albicans clade was cultured in RPMI 1640 Media (Nacalai Tesque, Inc., Kyoto, Japan) supplemented with 10% FCS for 24 h at 37°C in 35 glass bottom dishes.
In situ Raman Spectroscopy
Raman spectra were collected in situ on as-cultured single isolate C. albicans and two isolates C. auris (Clades II and III) living yeast cells. Spectra were obtained using a dedicated instrument (LabRAM HR800, Horiba/Jobin-Yvon, Kyoto, Japan) operating with a 20× optical lens with the spectroscope set in confocal mode. The spectroscope was equipped with a holographic notch filter enabling efficient and high spectrally resolved acquisitions. Excitation was made with a 532 nm solid-state laser source operating at 10 mW. A spectral resolution of better than 1 cm–1 was achieved by using an internal reference (neon emission) to calibrate the spectrometer. The Raman scattered light was monitored by a single monochromator connected with an air-cooled charge-coupled device (CCD) detector (Andor DV420-OE322; 1024 × 256 pixels). The acquisition time for a single spectrum was typically 10 s. Thirty spectra were collected at different locations over an area of ∼2 mm2 for each clade and averaged in order to obtain a representative spectrum for each clade.
Reference Raman spectra were collected on pure compounds and stored in a purposely built library. The reference library compilation contained more than 40 elementary compounds (simply referred to as the “library,” henceforth), including polysaccharides (e.g., chitin, β-1,3-glucans, β-1,6-glucans, and α-1, 3-glucans), mono- and disaccharides (e.g., trehalose, β-D-glucose, D-dextrose), lipids (e.g., triolein, trilinolein, 1,2-dipalmitoyl-L-α-lecithin), polyols [e.g., D-(+)-arabitol and L-(−)-arabitol], and other key molecules such as adenine, ergosterol, and glycine. The spectra from the pure compounds were collected with a highly resolved spectrometer (T-64000; Jobin-Ivon/Horiba Group, Kyoto, Japan) equipped with a nitrogen-cooled CCD detector (CCD-3500V, Jobin-Ivon/Horiba Group, Kyoto, Japan). The excitation source in these latter experiments was a 514 nm line of an Ar-ion laser operating with a nominal power of 200 mW. The spectral resolution was better than 1 cm–1.
Raman imaging of C. albicans cells was obtained using a dedicated Raman device (RAMANtouch, Nanophoton Co., Minoo, Osaka, Japan) operated in microscopic measurement mode with confocal imaging capability in two dimensions. This Raman microscope can achieve ultra-fast simultaneous image acquisition of up to 400 spectra. The spectroscope was specially designed to be compatible with cells’ life. It used an excitation source of 532 nm. The spectral resolution was ∼2 cm–1 (spectral pixel resolution equal to 0.3 cm–1/pixel) with an accuracy in laser spot location of 100 nm. Raman hyperspectral images were generated using commercially available software (Raman Viewer, Nanophoton Co., Minoo, Osaka, Japan). With the purpose of avoiding possible distortions, the Raman images were generated using intensity ratios in the normalized spectra. In order to minimize errors related to spectral resolution and possible shifts in band position, we used the average intensity of the pixels at the band nominal location ±3 pixels, rather than single pixel intensity.
Machine Learning Algorithm for Spectral Deconvolution
The experimentally obtained Raman spectra were preliminary treated with a baseline subtraction procedure and then automatically deconvoluted into a series of Voigtian sub-bands. Baseline subtraction and deconvolution procedure were performed using options available in commercial software (LabSpec 4.02, Horiba/Jobin-Yvon, Kyoto, Japan). The software applied a polynomial-fitting criterion for baseline subtraction. All spectra were analyzed for their relative intensity after normalization to the glucose ring signal (seen at 478 cm–1). The average spectra, Sav(ν), was then fitted with an automatic solver, which exploited a linear polynomial expression of Voigtian functions, V(Δν, σ, γ); with ν, Δν, σ, and γ representing the Raman frequency, the shift in frequency from each sub-band’s maximum (ν0), the standard deviation of each Gaussian component, and the full-width at half-maximum of the Lorentzian component, respectively. A working algorithm was applied to match the experimental spectrum upon searching for the minimum value of the following equation:
| (1) |
where the index i locates each compound in a series of n contributing to the overall spectrum, and the index j locates the Voigtian sub-bands of a series of m in the Raman spectrum of each compound of an n series. A computer program was built up for locating a selected series of Voigtian sub-bands from pre-selected compounds belonging to the library, including mono-, di-, and polysaccharides, specific lipids, polyols, and other key molecules, pre-selected according to previously published literature on the structure of Candida species. Upon operating a pre-selection of the component molecules from the library, the algorithm located the best fit to the experimental spectra. The computational procedure preserved relative intensities (βij), spectral positions (ν0), and full-width at half-maximum (σ and γ) values for the individual sub-bands of the deconvoluted spectra from each elementary compound (i.e., within ±3 cm–1, considering the resolution of the spectrometer and the possibility of slight alterations in molecular structure). The adopted criteria for the selection of band positions, relative intensity, and bandwidths provide a number of mathematical constraints that allow a univocal deconvolution of the experimental spectra. When specific sub-bands were located, which the solver could not fit according to the pre-selected library, the software labeled them as unknown and required adding a new compound from the library in the pre-selection. Then, a search for additional compounds is launched in the library to match the unknown band following the same criteria as described above. Upon adjusting for the overall intensity contribution (αi) of each elementary compound within the given constraints, the software located a best fit for the experimental spectrum.
The output of this newly developed machine-learning computational program was threefold: (1) it automatically screened the experimental spectra and proposed a deconvolution by best fitting the average spectrum based on Eq. 1; (2) it indicated the main molecules that contributed the observed intensity of each sub-band; and (3) it located sub-bands whose signal intensity was contributed at >90% by a single reference molecule.
Regarding the details of the machine learning algorithm and related training/validation procedures, we used a K-means clustering algorithm based on sets of 4 × 104 spectra obtained from Raman imaging using arrays of 100 arrays of 400 spectra. On all investigated samples, the algorithm was able to identify the main cluster related to the presence of different Candida species. C. auris presented several distinct traits, including that related the ergosterol/D-arabitol peak at ∼715 cm–1, as discussed later in the experimental results.
Chemometric Analysis
Statistical analyses and visualization of the large Raman data sets obtained during high-resolution imaging were performed according to principal component analysis (PCA) (Wold et al., 1987). The PCA analysis was carried out on the Origin software platform (OriginLab® Co., Northampton, MA, United States). PCA enabled summarizing the information with a set of two “summary indices,” referred to as principal components PC1 and PC2, which clearly differentiated the Raman spectra of different Candida clades. In the specific case of Candida analyses, coupling PCA with a machine-learning algorithm of spectral deconvolution (as shown in the previous section) allowed extraction and interpretation of the maximum relevant chemical information that unequivocally differentiates Candida species and helped in understanding their chemical characteristics.
Experimental Results
Average Raman Spectra From Different Candida Species
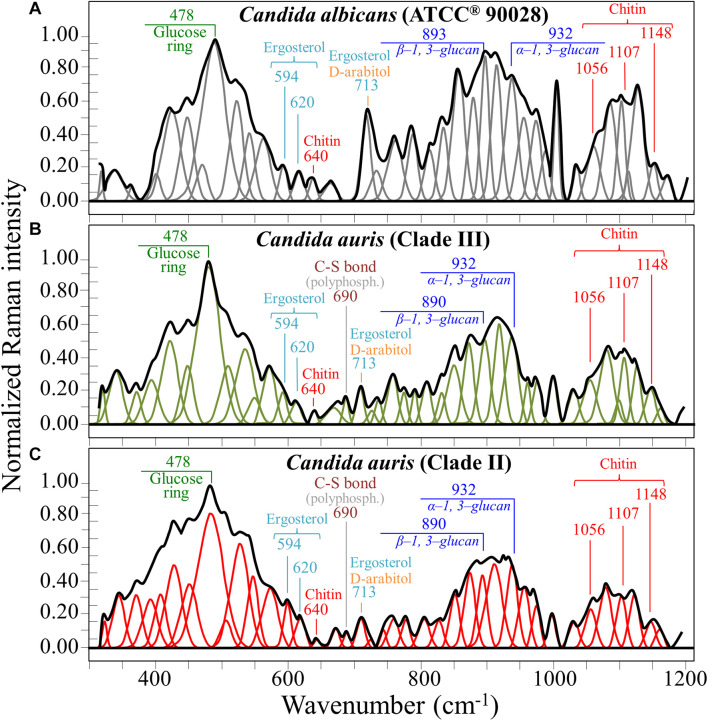
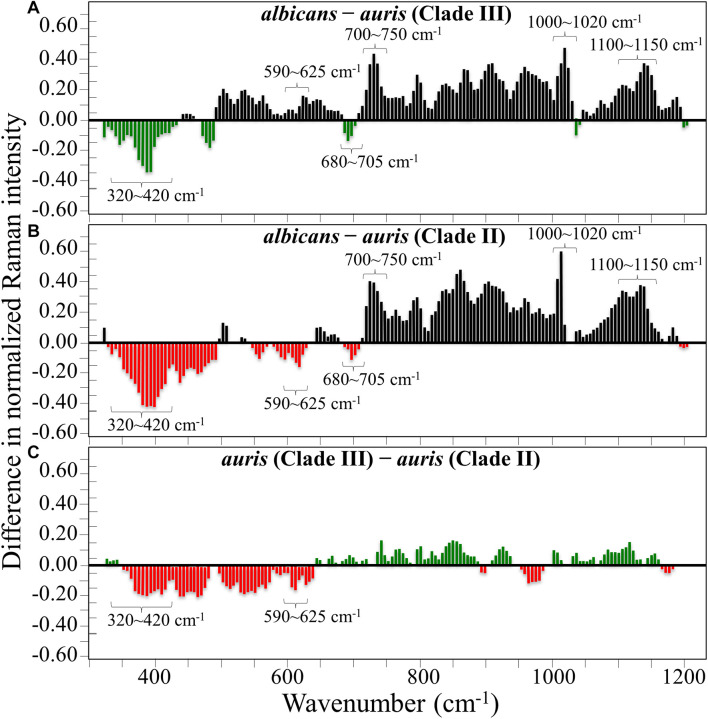
Figures 1A–C show the average Raman spectra of C. albicans, C. auris (Clade III), and C. auris (Clade II), respectively, as collected in the wavenumber interval 300–1200 cm–1. The spectra were first normalized to their common glucose ring band centered at ∼478 cm–1 and then deconvoluted into a series of Voigtian sub-bands by means of the machine-learning algorithm described in section “Chemometric Analysis.” The initial choice of molecular components included β-1, 3-glucans, α-1, 3-glucans, chitin, ergosterol, D-arabitol, trehalose, and adenine. A comparison among the collected (average) spectra revealed a number of vibrational bands common to all Candida species, but also bold differences in relative intensity and missing vibrational signals. Such differences are important because they locate specific fingerprints that enable the identification of predominant chemical patterns in different species. In an effort to better visualize the differences, spectral subtractions were performed as shown in Figure 2 (cf. labels). Bold differences in band relative intensities were noticed both at low and high wavenumbers. In order to facilitate the interpretation of these differences, reference Raman spectra are shown for four reported main constituents of yeast cells as collected on pure compounds. These spectra belong to a library of Raman spectra, which was built for elementary molecules. Spectra from β-1, 3-glucans and α-1, 3-glucans are shown in Figures 3A,B, respectively (cf. also structures in Figures 3C,D, respectively). Figures 4A–D show reference spectra/structures of ergosterol and chitin, respectively. At the lower measured wavenumbers (i.e., 320–420 cm–1), both C. auris species showed stronger Raman signals as compared to C. albicans (cf. Figures 2A,B). In yeasts, these wavenumbers mainly correspond to ring deformation and skeletal vibrations in glucans and chitin (De Gussem et al., 2005). The most prominent band in this low-wavenumber interval, which is located at ∼440 cm–1, included signal contributions from α-1, 3-glucans, chitin, and ergosterol (cf. Figures 3, 4), which were hard to separate. On the other hand, the machine-learning algorithm (i.e., Eq. 1) located two bands (at 594 and 620 cm–1) as mainly (>95%) contributed by ergosterol. Upon comparing the sum of the relative intensities of these two bands (cf. Figures 1, 2) in different clades, the highest fraction of ergosterol appeared to occur in C. auris (Clade II) (0.50) and the lowest in C. auris (Clade III) (0.34), with the C. albicans lying in the middle (0.41). Note that also the intense band at ∼713 cm–1 is contributed by ergosterol, but it contains additional (non-negligible) contributions from D-arabitol, chitin, and adenine. Accordingly, it could hardly serve as a fingerprint for ergosterol alone. Spectral differences in ergosterol bands are important since it has recently been suggested that mutations in Candida species could cause depletion and alteration of the ergosterol composition with an impact on antifungal resistance (Kean and Ramage, 2019). This point will be discussed later.
FIGURE 1.
Average Raman spectra of (A) C. albicans (ATCC® 90028), (B) C. auris (Clade III), and (C) C. auris (Clade II) in the wavenumber interval 300–1200 cm–1; spectra are normalized to the glucose ring band at ∼478 cm–1 and deconvoluted into Voigtian sub-bands by means of the machine-learning algorithm described in section “Chemometric Analysis.” The wavenumbers and the assignments of the main bands discussed in the text are given with labels in inset.
FIGURE 2.
Results of subtraction of the average normalized spectra in Figure 1: (A) subtraction of the C. auris (Clade III) spectrum from the C. albicans spectrum; (B) subtraction of the C. auris (Clade II) spectrum from the C. albicans spectrum; (C) subtraction of the C. auris (Clade II) spectrum from the C. auris (Clade III) spectrum.
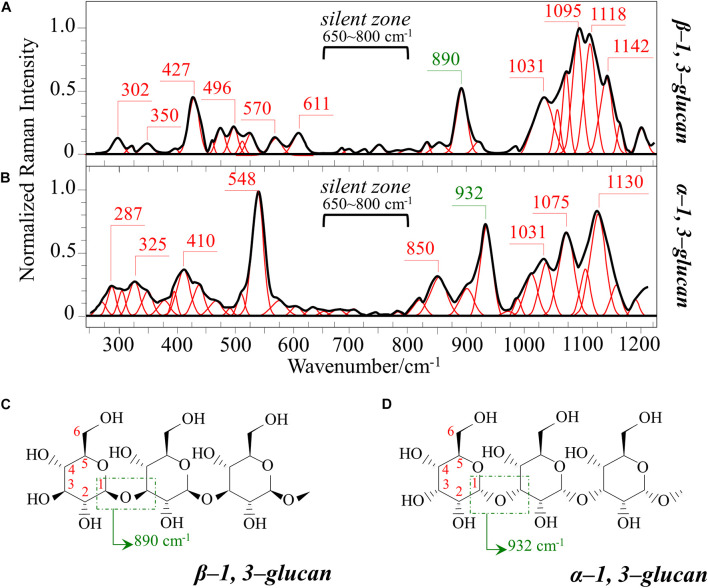
FIGURE 3.
Reference Raman spectra of: (A) β-1, 3-glucans and (B) α-1, 3-glucans (frequencies in cm–1 of the main bands given by labels in inset); (C,D) the respective glucan structures (cf. labels in inset). The spectra are deconvoluted in series of Voigtian bands.
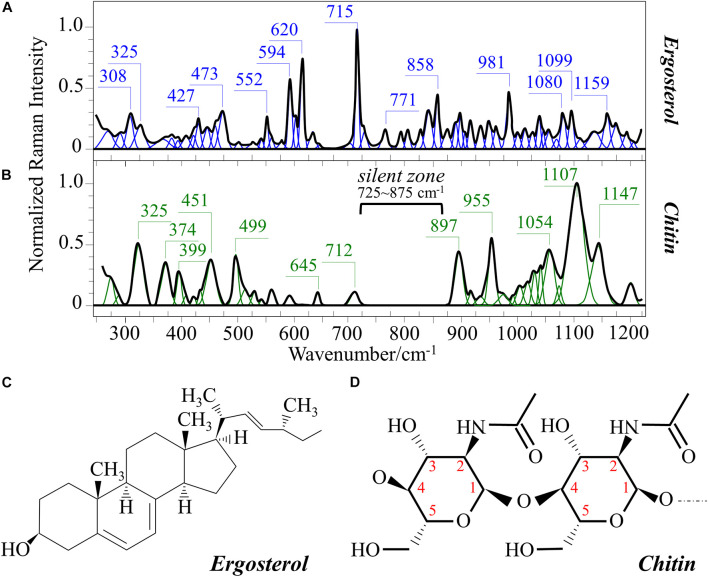
FIGURE 4.
Reference Raman spectra of: (A) ergosterol and (B) chitin (frequencies in cm– 1 of the main bands given in inset); (C,D) give the respective molecular structures (cf. labels in inset). The spectra are deconvoluted in series of Voigtian bands.
The wavenumber zone at 680–705 cm–1, which was another additional (narrow) interval of stronger Raman intensity for both C. auris species (cf. Figure 2), corresponded to a well distinct band centered at ∼690 cm–1. This band, which was only present in C. auris species (cf. Figure 1), was located by means of the machine-learning algorithm as one requiring the choice of an additional compound from the library, besides the pre-selected components given above. As a matter of fact, in this wavenumber interval, both glucans and chitin present either a silent zone or very weak signals (cf. Figures 3A,B and 4B, respectively), and ergosterol shows no Raman bands (cf. Figure 4A). Therefore, the C. auris band centered at ∼690 cm–1 should belong to a different compound. One possible candidate in assigning this band are S-containing amino acids, namely, methionine and cysteine, because the C–S bond stretching vibration scatters in this zone (Diaz Fleming et al., 2009). The observed band frequency corresponds to C–S stretching in methionine and/or cysteine (in gauche and trans configurations, respectively). In favor of this hypothesis is the fact that the Raman intensity is generally strong for bonds involving sulfur even when only small fractions of S-containing amino acids are present. The C. auris Clade III displayed a Raman intensity at 690 cm–1 60% higher than the C. auris Clade II (cf. Figures 1B,C). In a recent paper by Costa et al. (2015) mechanisms of flucytosine resistance in Candida glabrata were found to link with sixteen genes related to nitrogen metabolism. These included genes involved with biosynthesis of methionine (MET2 and MET7), among other genes related to proline, tyrosine/phenylalanine, and leucine productions. Based on this finding, it was suggested a role of methionine fractions included in the external membrane proteins in the drug-resistance behavior of different Candida species. In this context, note also that the relative intensity of the Raman band at ∼1002 cm–1 (related to phenylalanine ring vibrations) was much more pronounced in the spectrum of C. albicans as compared to those of both the investigated C. auris isolates (cf. Figures 1, 2). Methionine serves as the major biological methyl donor in methylation reactions, which are essential for the biosynthesis of phospholipids, proteins, DNA, and RNA (Fontecave et al., 2004). Similar to the case of subgingival bacteria (Gosschalk et al., 2020), increased virulence could also be related to S-containing transpeptidase enzymes that covalently attach proteins to peptidoglycan precursors. Bednarova et al. (2012) assigned a 688 cm–1 band (symmetric P–O–P stretching) to polyphosphates contained in yeast vacuoles. This band was one of a couple with a threefold stronger band centered at ∼1154 cm–1 (symmetric PO2 stretching), which was indicated as a marker for vacuole formation. The presence of vacuoles is a sign of adaptation of C. albicans to environmental changes due to severe nutrition limitations, and a sign of its reaction against host immune defense (Kim and Sudbery, 2011). The function of vacuoles is to store amino acids, ions, and metabolites that serve in pH homeostasis and osmoregulation, while also playing a crucial role in breaking peptide links and recycling nutrients (Li and Kane, 2009). In the Raman spectra of Figure 1, a band at ∼690 cm–1 is completely missing in C. albicans (cf. Figure 1A) and only present in the two C. auris isolates (Figures 1B,C). A band at 1154 cm–1 is also seen in the spectra of both C. auris clades (and missing in the C. albicans isolate), but only as a weak shoulder. The 1154 cm–1 band in the spectrum of polyphosphate pure compound is about three times more intense than that at 690 cm–1 (Bednarova et al., 2012), while the intensity of the shoulder band seen at 1154 cm–1 in C. auris spectra is about a half of the intensity of the 690 cm–1 one. Note also that different types of Candida cells were analyzed after being cultured in nutritional medium and not in a stress state. Moreover, secretion of phospholipase and protease enzymes in C. auris has been reported to be generate at significantly lower levels as compared to C. albicans (Larkin et al., 2017). For all the above reasons, we believe that the correct assignment of the 690 cm–1 band is C–S bond stretching.
The membrane of Candida species includes both α- and β-glucans polysaccharides, which only differ in their C–O–C bond configurations at the anomeric C1 position (cf. Figures 3C,D). Such difference in bond configuration is reflected in fine morphological differences of the Raman spectrum in correspondence of the wavenumber interval 850–950 cm–1, which is sensitive to glycosidic linkages (Noothalapati et al., 2016). Note that the overall relative intensity of the average Raman spectra recorded in this spectral zone was clearly higher in C. albicans as compared to Clades II and III of C. auris (cf. Figures 2A,B). Additional spectral differences arise in the region of C–H equatorial bending vibrations, which are found at 880–910 and 830–870 cm–1 in the cases of β- and α-anomer, respectively (Cael et al., 1974; Corbett et al., 1991). Examination of band overlap with other biomolecules revealed that the deconvoluted sub-band at ∼890 cm–1 can be taken as a marker for β-1, 3-glucans, while the antisymmetric ring vibration seen at 932 cm–1 is a fingerprint for α-1, 3-glucans in C. auris. Note that also the signal at ∼850 cm–1 is contributed by α-1, 3-glucans, but it strongly overlaps with a main band from tyrosine (in-plane phenol ring vibrations) and cannot easily be singled out. The above choice of the 932 cm–1 marker is in line with previously published studies of α-1, 3-glucans (Synytsya et al., 2009; Mikkelsen et al., 2010). According to the above spectral choices, the fractional ratio of α- to β-glucans of the yeast wall structure can be computed for different Candida species as: 0.83, 1.03, and 1.09 for C. albicans, C. auris (Clade III), and C. auris (Clade II), respectively. In summary, both C. auris clades possessed lower total amounts of glucans as compared to the C. albicans isolate, but they were richer in α-1, 3-glucans.
In the Raman spectra of polysaccharides, the region representing C–C and C–O symmetrical stretching corresponds to the interval 950–1200 cm–1. Although all polysaccharides show a number of structural similarities (e.g., the silent zone at intermediate frequencies, 725–875 cm–1; cf. Figure 4B), distinctions can yet be made in the C–C/C–O spectral region. This is the case of chitin (Figure 4D), which is structurally different due to the presence of an amide group at the C2 position to replace an OH group (De Gussem et al., 2005). Due to this structural feature, the chitin spectrum shows peculiar bands at 955, 1107, and 1149 cm–1, in addition to other spectral features at the lower frequencies of 325 and 645 cm–1 (cf. Figure 4B). Upon machine-learning examination of the Candida spectra in Figure 1, the triplet at 1056, 1107, and 1148 cm–1 and the low-frequency band at 640 cm–1 were located as non-overlapping chitin bands and indicated as fingerprints for chitin since they conspicuously preserved both relative intensities and band morphology characteristics of the elementary compound. Building upon this automatic indicator, we compared the relative intensities of chitin-related bands (with respect to the 478 cm–1 glucose ring band) among different Candida clades and found that C. albicans was richer in chitin by ∼31 and 50% as compared to Clades III and II C. auris, respectively. Since chitin adds rigidity and structural support to the yeast cell walls, C. albicans appeared to possess the most rigid cell-wall structure, while C. auris Clade II is the most flexible one.
Candida albicans cells experience elevated chitin content in their wall as a consequence of an adaptive (remodeling) mechanism to maintain intact (i.e., not permeable) the cell walls and to fix resistance to specific drugs that inhibit synthesis of β-1, 3-glucans (i.e., echinocandins) (Ibe and Munro, 2021). The same chitin-enrichment remodeling effect was also found in C. auris species; however, the extent of such effect was significantly less pronounced (Lara-Aguilar et al., 2021). On the other hand, a higher flexibility of yeast cell walls, which is dictated by the interplay between β-1, 3-glucans and chitin, is also a sign of cellular adaptation to environmental change (Munro, 2013), since it confers resistance to osmotic stress (Ene et al., 2015). Regarding the origin of the osmotic stress resistance, the current understanding links it to the circumstance of yeast cells entering the bloodstream and becoming suddenly exposed to glucose. Such sudden exposure imposes to develop a mechanism of osmotic stress resistance (Van Ende et al., 2019). C. auris exploits different strategies to resist various kinds of external stress and to promote virulence as compared to C. albicans (Day et al., 2018). Raman spectroscopy reveals here large differences in the structure of yeast cell walls between unstressed C. albicans and C. auris clades, while much less pronounced differences were found between different C. auris clades. The detected spectral differences suggest that the wall structures of C. auris clades underwent permanent compositional changes to improve cell wall elasticity, notwithstanding the yeast capacity to manipulate the structure of their walls in a dynamic response to environmental perturbations and to display fast structural realignments to increase survival probability following osmotic shock.
Raman Imaging of Living Yeast Cells
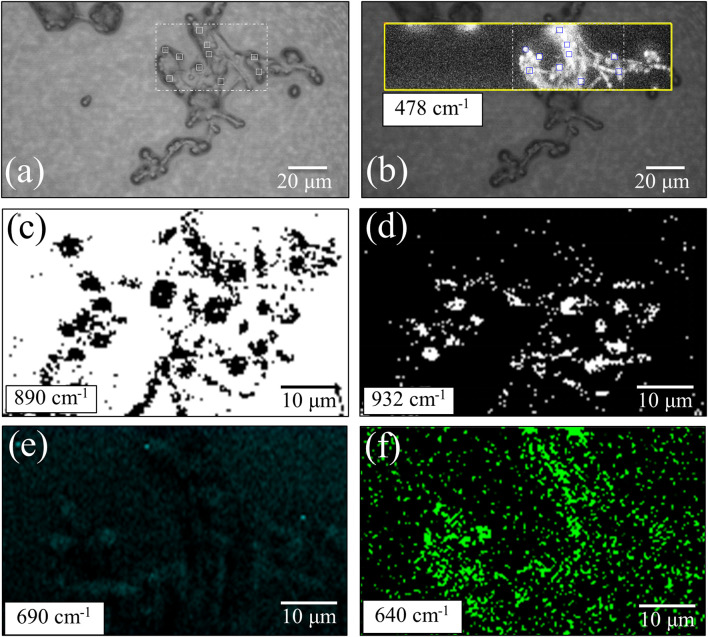
The machine-learning analyses on average Raman spectra shown in the previous section revealed significant differences in yeast wall composition, including lipids and polysaccharides, and a possible variation in concentration of S-containing amino acids (present both intracellular and in the external membrane proteins). In order to spatially locate the differences and to statistically substantiate the findings, we performed extensive Raman imaging on living cultures of the three investigated Candida isolates (the total number of spectra collected per each species is in the order of 105), as shown in Figures 5–7. Figure 5 shows an optical micrograph (a) of C. albicans culture and spatially resolved Raman maps (taken in the square region) of signals at 478 cm–1 (b), 890 cm–1 (c), 932 cm–1 (d), 690 cm–1 (e), and 640 cm–1 (f), representing the entirety of glucose rings in membrane polysaccharides, β-1, 3-glucans, α-1, 3-glucans, S-containing amino acids both intracellular and in membrane mannoproteins, and chitin, respectively. The comparison between the maps of β-1, 3-glucans and α-1, 3-glucans [black spots on white background and white spots on black backgrounds in (c) and (d), respectively] makes quantitative the ratio between these two polysaccharides in the C. albicans isolate, which was the richest in β-1, 3-glucans among the investigated species. The fractional ratio computed from black and white spots related to the same area indicated an α/β glucan-ratio ∼0.81, which is in good agreement with the 0.83 value computed from band-intensity ratio in the average spectrum in Figure 1A. Also the conspicuous lack of 690 cm–1 signal from C–S bond stretching, already noticed in the average spectrum in Figure 1A, was confirmed in Raman mapping experiments (cf. Figure 5e). The chitin spots in the C. albicans map in Figure 5f were quite pronounced in correspondence of the cells, but they also appeared in minor fractions in neighboring areas.
FIGURE 5.
(a) Optical micrograph of cultured C. albicans cells; spatially resolved Raman maps in: (b) glucose rings in membrane polysaccharides at 478 cm–1 (in the yellow square region), (c) β-1, 3-glucans (in the white broken-line square region) at 890 cm–1 (black spots on white background), (d) α-1, 3-glucans (in the broken-line square region) at 932 cm–1 (white spots on black background), (e) S-containing amino acids (in the white broken-line square region) at 690 cm–1, and (f) chitin at 640 cm–1 (in the white broken-line square region). The 10 smaller square areas located in a,b represent the regions used for PCA analyses.
FIGURE 7.
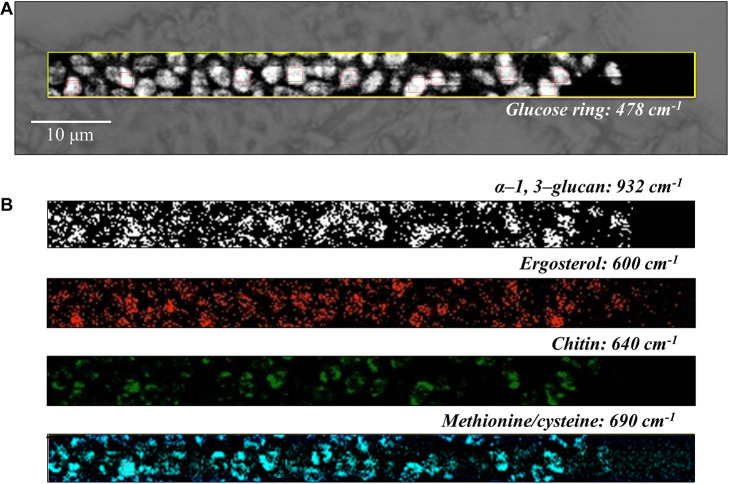
(A) Optical micrograph and Raman map of glucose rings in membrane polysaccharides at 478 cm–1 (in the yellow square region) of C. auris (Clade II) culture; in (B) from top to bottom (cf. labels), α-1, 3-glucans at 932 cm–1 (white spots on black background), ergosterol at 600 cm–1, chitin at 640 cm–1, and S-containing amino acids at 690 cm–1. The maps in (B) correspond to the same square region in (A). The 10 smaller square areas located in (A) represent the regions used for PCA analyses.
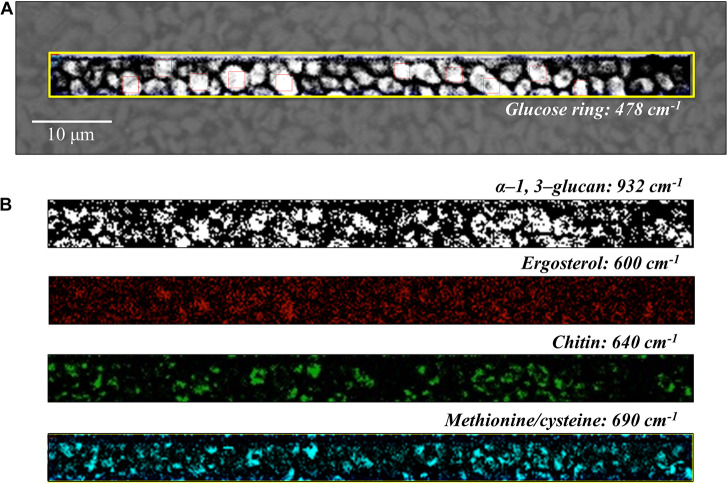
Spatially resolved Raman images of C. auris Clades III and II are shown in Figures 6, 7, respectively. Glucose rings in membrane polysaccharides (478 cm–1), α-1, 3-glucans (932 cm–1), chitin (640 cm–1), and S-containing amino acids in membrane mannoproteins (690 cm–1) are shown in sections (a) and (b) of each figure (cf. labels). Both C. auris clades were confirmed to be richer in α-1, 3-glucans (932 cm–1 white spots on black background) and methionine (690 cm–1) as compared with C. albicans. On the other hand, the ergosterol map showed a detectably higher spot concentration in the C. auris Clade II as compared to Clade III (cf. maps at ∼600 cm–1 in Figures 6, 7). Finally, the consistency between average spectra and Raman imaging experiments could be confirmed with comparing the chitin maps at ∼640 cm–1; Raman imaging confirmed that the C. auris Clade II clade was the least chitin rich among the three Candida isolates investigated (cf. Figures 5–7).
FIGURE 6.
(A) Optical micrograph and Raman map of glucose rings in membrane polysaccharides at 478 cm–1 (in the yellow square region) of C. auris (Clade III) culture; in (B) from top to bottom (cf. labels), α-1, 3-glucans at 932 cm–1 (white spots on black background), ergosterol at 600 cm–1, chitin at 640 cm–1, and S-containing amino acids at 690 cm–1. The maps in (B) correspond to the same square region in (A). The 10 smaller square areas located in (A) represent the regions used for PCA analyses.
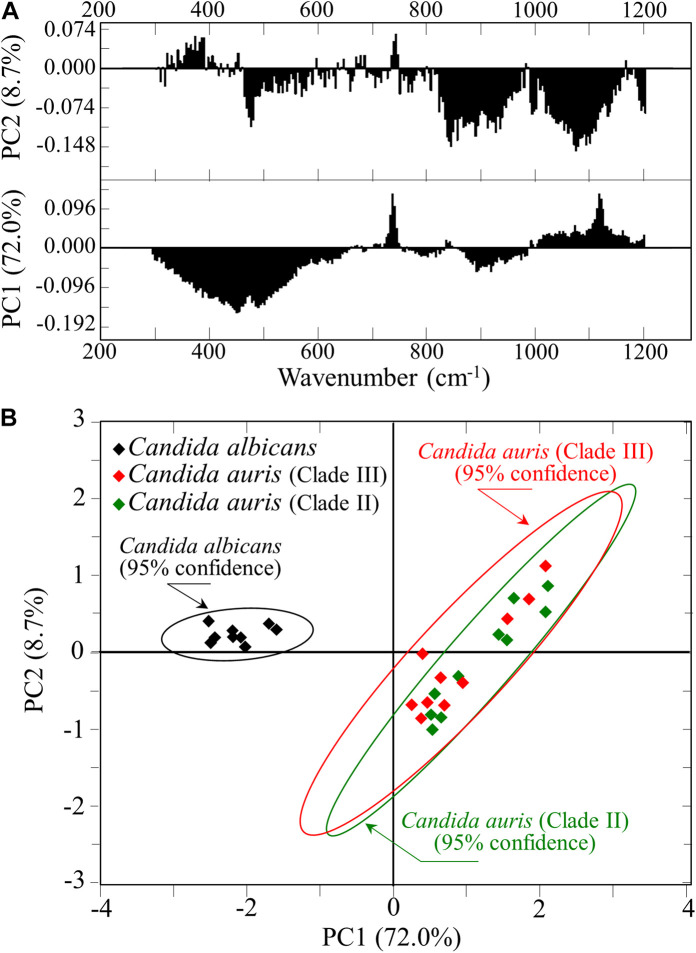
Statistical Principal Component Analyses
For each Candida clade investigated, 10 locations were selected as shown by square insets [cf. section (a) of Figures 5–7]. Each location was ∼20 μm2 in size and contained 100 spectra, which were in turn averaged to obtain a single spectrum for each location. Figure 8A shows the first and second principal components (PC1 and PC2, respectively) of a PCA analysis conducted on the Raman spectra representative of the above-mentioned 10 selected locations from each map of the three isolates of two Candida species investigated. As seen in Figure 8B, C. albicans vs. C. auris data sets displayed as well separated (95% confidence) upon plotting the loading vectors PC1 and PC2 for the entire spectral region 300–1200 cm–1. However, the PCA analysis also pointed out that it was not possible to distinguish between the two C. auris clades. The present application of the PCA statistical method, which reduces the dimensionality of the Raman data matrix to only two orthogonal variables, gives clear evidence that it is possible to identify C. albicans and C. auris by discrimination between their Raman spectra.
FIGURE 8.
(A) First and second principal components (PC1 and PC2, respectively) of a PCA analysis of Raman spectra at the 10 selected locations from maps of C. albicans and two C. auris clades (Figures 5–7); (B) plot of the loading vectors PC1 and PC2 for the spectral region 300–1200 cm–1.
As a final observation with reference to Figure 1, one could note that the Raman spectra of C. albicans and C. auris are markedly different, so that they could be differentiated at a glance even without applying specific algorithms such as machine learning. For example, the relative intensities of the ergosterol peak at ∼713 cm–1 or the phenylalanine peak at ∼1000 cm–1 could be taken as fingerprints for C. albicans with respect to C. auris clades. However, besides adding computational exactness and exhaustiveness to the spectral differentiation procedure, the machine-learning algorithm enables to exactly locate the contributions made by different molecular constituents to the structure of different Candida species. This approach could ultimately enable quantifying both membrane fluidity/elasticity and antifungal resistance of different mutants.
Discussion
Structural Peculiarities of Candida auris Revealed by Raman Analyses
The Raman analyses presented in this study have located (with statistical relevance) some fundamental differences in the structures of C. albicans and C. auris. Specifically, two main differences were related to the polysaccharide structure of the yeast cell membrane, as follows:
-
(i)
Both C. auris Clades II and III contained similar fractional ratios of α- to β-glucans, which were 20–30% higher than that measured in C. albicans.
-
(ii)
C. albicans was richer in chitin by ∼31 and 50% as compared to C. auris Clades III and II, respectively.
The two above characteristics suggest that both C. auris clades possess cell walls less permeable to external agents and structurally more flexible. α-1, 3-glucans are insoluble in water due to the presence of strong hydrogen bonds, and their accumulation in the yeast cell walls was shown to protect the yeast cells from antifungal agents and degrading enzymes encountered during infection (Fujikawa et al., 2009). A wall structure “engineered” with larger amounts of α-1, 3-glucans could be linked to the fact that the majority of clinical C. auris species display resistance to the main classes of antifungals (azoles, polyenes, or echinocandins) (Cernakova et al., 2021). The observed higher fractions of α-1, 3-glucans in both studied C. auris clades also likely relate to their reported strong biofilm-mediated resistance (Kean and Ramage, 2019). Note also that the α-1, 3-glucans are more abundant in pathogenic yeasts, and are reported to be needed for their normal morphology and virulence (Ruiz-Herrera and Ortiz-Castellanos, 2019). Regarding the resistance of C. auris to osmotic stress, this characteristic is related to a high flexibility of the yeast cell walls. This, in turn, is related to the lower fraction of chitin contained in the cell walls. According to Ene et al. (2015) the β-glucan/chitin interlinked composition of the yeast cell walls inner matrix is key in modulating elasticity and strength. The ability of Candida species to display rapid structural realignments in the compositional fractions of glucans vs. chitin greatly impacts survivorship following osmotic shock. C. auris was found clearly more resistant to the oxidative stress imposed by H2O2 than C. albicans, and displayed higher levels of resistance to the cationic stress imposed by either sodium or calcium chloride, but it also proved much less capable to adapt to acidic or alkaline pH environments than C. albicans (Day et al., 2018). Conversely, C. albicans was found capable to undergo cell wall remodeling in acidic environment, which resulted in a promptly enhanced chitin contents (Sherrington et al., 2017). All these specific characteristics, which relate to both composition and (chemical vs. mechanical) resistance of the yeast cell walls, illustrate the unique stress resistance profile of C. auris as compared to other Candida species. As a matter of fact, the trade off between cell-wall flexibility and chemical stability depends on the type of polysaccharides.
Regarding the structure of cell-wall lipids, the Raman experiments revealed that the C. auris Clade II possessed the highest fraction of ergosterol while the C. auris Clade III the lowest. It has been recently clarified that mutations in Candida species can cause depletion and alteration of the ergosterol composition (Kean and Ramage, 2019). Alterations in the ergosterol pathway, which in turn depend on ergosterol gene mutations, are considered the main responsible for resistance to amphotericin B (Kordalewska et al., 2018). Ergosterol gene mutations lead to reduced ergosterol levels in the yeast plasma membrane, which in turn increases the resistance to polyenes because polyene-based drugs specifically target ergosterol by binding, to form pores or simply sequestrating it to induce membrane disruption (Mesa-Arango et al., 2012; Frias-De-Leon et al., 2020).
An additional spectral difference among the three studied Candida isolates was the Raman intensity at ∼690 cm–1, which was assigned to C–S bond vibrations in specific rotamers of S-containing amino acids (i.e., methionine and cysteine). At this specific frequency, the C. auris Clade III displayed a Raman signal 60% higher than the C. auris Clade II, while no signal could be detected in the C. albicans species. Methionine affects morphogenesis and virulence through the protein kinase A, a pathway that is also involved in cellular processes related to cell wall integrity and drug sensitivity (Biswas et al., 2007; Schrevens et al., 2018; Kuplinska and Rzad, 2021). Both methionine and cysteine are also important biofilm inducers (Schrevens et al., 2018). C. albicans was found capable to synthesize methionine (Prasannan et al., 2009) and to interconvert cysteine and methionine through the trans-sulfuration pathway (Thomas and Surdin-Kerjan, 1997; Schrevens et al., 2018). Therefore, the lack of 690 cm–1 Raman signal in the C. albicans spectrum can only be attributed to one specific missing rotamer or to definite physiological circumstances of the cultured yeast cells, rather than to an intrinsic structural characteristic.
Importance of the Raman Approach to Candida Species Differentiation
The Raman method is widely applied in cell biology and microbiology with several different purposes, including identification of different types of cell (e.g., cancer and stem cells) (Chan et al., 2006; Kast et al., 2008; Leslie et al., 2012), differentiation between Gram-positive and Gram-negative bacteria (Paret et al., 2010), tracking down of mutant viruses (Yeh et al., 2020), and characterization of environmentally and physiologically driven changes in cellular and microbial metabolism (Pezzotti, 2021; Pezzotti et al., 2021a,b). Raman molecular fingerprints have also been used for the identification of yeasts and fungi (Roesch et al., 2005b). In this study, we have applied machine-learning-supported Raman method and succeeded in differentiating between C. albicans and C. auris clades. From a general viewpoint, the present findings are important because they show that creating a reference Raman library for clinically important Candida pathogens may provide a tool for rapid identification of pathogens in clinical practice. For the specific case studied here, the possibility of locating the presence of C. auris clades with a conspicuously reduced identification time could allow a prompt selection of the most appropriate cure. In line with the present state of the art in deep learning applied to biospectroscopy and biospectral imaging (He et al., 2021), we have proposed here a reliable and versatile Raman method to locate subtle variations and hidden features within big data collected on different Candida clades. Once translated into clinical practice, the speed of the presented Raman approach will provide a significant advantage for all those patients at immunological risk, who have limited ability to fight invasive fungal infections.
In this context, it is important to note that the MALDI-TOF MS method is already replacing traditional methods for identifying pathogens (including yeasts and fungi) in clinical practice (Rychert, 2019). This technique, which relies on measuring microbial proteins for discriminating different species, can already exploit a wide database; it has also been shown to be faster and more accurate than conventional biochemical methods for definitive identifications of Candida species (Bal and McGill, 2018; Camp et al., 2020). The Raman spectroscopic method, which we newly apply here to C. auris, is also a suitable tool for fast on-site identifications of Candida species. It cannot yet rely on a complete database, but it has a tangible advantage with respect to the MALDI-TOF MS method: it concurrently gives precise information on polysaccharides, protein, lipids, and DNA structures, with respect to their fractions and molecular symmetry. While exploiting this peculiarity, the Raman method can be applied in time-lapse mapping experiments on living cells to follow up in real time the metabolic reactions of the Candida species to environmental stress. The Raman findings could become clinically important to obtain molecular-scale evidences in drug development as, for example, in understanding why echinocandin and amphotericin B treatments frequently lead to therapeutic failures among patients suffering from invasive C. auris infections. Such in situ studies are presently ongoing.
Limitations of This Study
At the present time, five different C. auris clades have been discovered. However, the present basic study yet addresses Raman experiments on only one isolate each from South African and East Asian clades. Accordingly, the present results do not allow us to unequivocally conclude that the presented Raman approach will be valid in general. While it has worked well in speciation of C. albicans and C. auris, additional work is needed to check whether misidentifications could occur when other Candida species are also considered. It should also be noted that different C. auris clades possess different degrees of virulence (Borman et al., 2016; Forgacs et al., 2020) and different in vitro susceptibility toward different antifungal agents (Szekely et al., 2019; Kovacs et al., 2021; Papp et al., 2021). Therefore, characterizations of all clades are mandatory to confirm whether or not the present Raman spectroscopic approach could be extended to and give unequivocal molecular-scale information about all other C. auris clades. Encouraging results were obtained and we are presently completing a number of additional Raman characterizations on different clades/isolates. The results will be published in a forthcoming report. Moreover, samples taken on-site from patients should also be examined in future experiments in order to test the actual suitability of Raman classifications in the clinical practice.
Conclusion
Differences in Raman fingerprints of C. albicans and C. auris yeasts belonging to different clades were studies using a double approach based on the analysis of average spectra collected with a resolution 20× and maps of 105 spectra collected with high spatial resolution. The former approach unfolded several spectral signatures traceable to specific structural features peculiar to different Candida species. The latter approach, while confirming the characteristics retrieved from average spectra, allowed the application of a chemometric method based on PCA to mechanistically distinguish between C. albicans and C. auris. We succeeded in unequivocally distinguishing between these two different types of Candida yeasts, the proposed identification procedure accomplishing both accuracy and testing speed. Moreover, the results we obtained in classification of different Candida species by using machine learning analyses of Raman spectroscopic data are relevant to practical treatments because they reveal structural details that might relate to both virulence and drug resistance characteristics. Future experiments should be conducted to provide assessments of a larger number of Candida species in order to build up a comprehensive library. The present findings justify the design and development of a rapid, low-cost, portable Raman system for application as a precise, fast, and reliable diagnostic tool in the clinical practice.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
GP, TY, NK, OM, and IN contributed to conception and design of the study. MK, TAs, TaN, NM, TAd, and EO performed the analyses. WZ organized the database. EM performed the statistical analysis. GP wrote the first draft of the manuscript. TeN and KM wrote sections of the draft. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The Raman experiments with Aramis equipment were conducted at the Kyoto Municipal Institute of Industrial Technology and Culture (Shimogyo-ku, Kyoto, Japan).
References
- Bal A. M., McGill M. (2018). Rapid species identification of Candida directly from blood culture broths by Sepsityper-MALDI-TOF mass spectrometry: impact on antifungal therapy. J. R. Coll. Phys. Edinb. 48 114–119. 10.4997/JRCPE.2018.203 [DOI] [PubMed] [Google Scholar]
- Bednarova L., Palacky J., Bauerova V., Hruskova-Heidingsfeldova O., Pichova I., Mojzes P. (2012). Raman microspectroscopy of yeast vacuoles. Spectrosc. Int. J. 27 503–507. 10.1002/tcr.201200008 [DOI] [PubMed] [Google Scholar]
- Biswas S., Van Dijck P., Datta A. (2007). Environmental sensing and signal transduction pathways regulating morphopathogenic determinants. Microbiol. Mol. Biol. Rev. 71 348–376. 10.1128/MMBR.00009-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A. M., Fraser M., Johnson E. M. (2021). CHROMagarTM Candida Plus: a novel chromogenic agar that permits the rapid identification of Candida auris. Med. Mycol. 59 253–258. 10.1093/mmy/myaa049 [DOI] [PubMed] [Google Scholar]
- Borman A. M., Szekely A., Johnson E. M. (2016). Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. Clin. Sci. Epidem. 1 e189–e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cael J. J., Koenig J. L., Blackwell J. (1974). Infrared and Raman spectroscopy of carbohydrates: Part IV. Identification of configuration- and conformation-sensitive modes for D-glucose by normal coordinate analysis. Carbohydr. Res. 32 79–91. [Google Scholar]
- Camp I., Spettel K., Willinger B. (2020). Molecular methods for the diagnosis of invasive candidiasis. J. Fungi 6:101. 10.3390/jof6030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2019). Centers for Disease Control and Prevention Identification of Candida auris. Available online at: https://www.cdc.gov/fungal/candida-auris/recommendations.html. (accessed July 5, 2021). [Google Scholar]
- Cernakova L., Roudbary M., Bras S., Tafaj S., Rodrigues C. F. (2021). Candida auris: a quick review on identification, current treatments, and challenges. Int. J. Mol. Sci. 22:4470. 10.3390/ijms22094470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabane F., Graf A., Jequier L., Coste A. T. (2019). Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front. Microbiol. 10:2788. 10.3389/fmicb.2019.02788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. W., Taylor D. S., Zwerdling T., Lane S. M., Ihara K., Huser T. (2006). Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 90 648–656. 10.1529/biophysj.105.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander J. (2009). A Textbook of Medical Mycology, 3rd Edn. New Delhi: Mehta Publishers, 266–290. [Google Scholar]
- Chouthai N. S., Shah A. A., Salimnia H., Palyvoda O., Devpura S., Klein M., et al. (2015). Use of Raman spectroscopy to decrease time for identifying the species of Candida growth in cultures. Avicenna J. Med. Biotech. 7 45–48. [PMC free article] [PubMed] [Google Scholar]
- Corbett E. C., Zichy V., Goral J., Passingham C. (1991). Fourier transform Raman studies of materials and compounds of biological importance-II. The effect of moisture on the molecular structure of the alpha and beta anomers of D-glucose. Spectrochim. Acta Part A: Mol. Spectrosc. 47 1399–1411. 10.1016/0584-8539(91)80231-7 [DOI] [Google Scholar]
- Costa C., Ponte A., Pais P., Santos R., Cavalheiro M., Yaguchi T., et al. (2015). New mechanisms of Flucytosine resistance in C. glabrata unveiled by a chemogenomics analysis in S. cerevisiae. PLoS One 10:e0135110. 10.1371/journal.pone.0135110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist A. E., Dietz T. J., Kampschroer K. (1996). Comparison of the MUREX C. albicans, albicans-sure, and bacticard Candida test kits with the germ tube test for presumptive identification of Candida albicans. J. Clin. Microbiol. 34 2616–2618. 10.1128/jcm.34.10.2616-2618.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A. M., McNiff M. M., da Silva Dantas A., Gow N. A. R., Quinn J. (2018). Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere 3 e506–e518. 10.1128/mSphere.00506-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gussem K., Vandenabeele P., Verbeken A., Moens L. (2005). Raman spectroscopic study of Lactarius spores (Russulales. Fungi). Spectrochim. Acta Part A 61 2896–2908. 10.1016/j.saa.2004.10.038 [DOI] [PubMed] [Google Scholar]
- Diaz Fleming G., Finnerty J. J., Campos-Vallette M., Celis F., Aliaga A. E., Fredes C., et al. (2009). Experimental and theoretical Raman and surface-enhanced Raman scattering study of cysteine. J. Raman Spectrosc. 40 632–638. 10.1002/jrs.2175 [DOI] [Google Scholar]
- Ene I. V., Walker L. A., Schiavone M., Lee K. K., Martin-Yken H., Dague E., et al. (2015). Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6:e00986. 10.1128/mBio.00986-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias M., Carneiro R. (2014). Simultaneous quantification of three polymorphic forms of carbamazepine in the presence of excipients using Raman spectroscopy. Molecules 19 14128–14138. 10.3390/molecules190914128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M., Atta M., Mulliez E. (2004). S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29 243–249. 10.1016/j.tibs.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Forgacs L., Borman A. M., Prepost E., Toth Z., Kardos G., Kovacs R., et al. (2020). Comparison of in vivo pathogenicity of four Candida auris clades in a neutropenic bloodstream infection murine model. Emerg. Microb. Infect. 9 1160–1169. 10.1080/22221751.2020.1771218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-De-Leon M. G., Hernandez-Castro R., Vite-Garin T., Arenas R., Bonifaz A., Castanon-Olivares L., et al. (2020). Antifungal resistance in Candida auris: molecular determinants. Antibiotics 9:568. 10.3390/antibiotics9090568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa T., Kuga Y., Yano S., Yoshimi A., Tachiki T., Abe K., et al. (2009). Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73 553–570. 10.1111/j.1365-2958.2009.06786.x [DOI] [PubMed] [Google Scholar]
- Gosschalk J. E., Chang C., Sue C. K., Siegel S. D., Wu C., Kattke M. D., et al. (2020). A cell-based screen in Actinomyces oris to identify sortase inhibitors. Sci. Rep. 10:8520. 10.1038/s41598-020-65256-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N., Demers E. G., Crocker A. W., Hogan D. A. (2017). Use of RNA–protein complexes for genome editing in non-albicans Candida species. mSphere 2:e00218-17. 10.1128/mSphere.00218-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Yan S., Lyu D., Xu M., Ye R., Zheng P., et al. (2021). Deep learning for biospectroscopy and biospectral imaging: state-of-the-art and perspectives. Anal. Chem. 93 3653–3665. 10.1021/acs.analchem.0c04671 [DOI] [PubMed] [Google Scholar]
- Ho C.-S., Jean N., Hogan C. A., Blackmon L., Jeffrey S. S., Holodniy M., et al. (2019). Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 10:4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibe C., Munro C. A. (2021). Fungal cell wall: an underexploited target for antifungal therapies. PloS Pathog. 17:e1009470. 10.1371/journal.ppat.1009470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast R. E., Serhatkulu G. K., Cao A., Pandya A. K., Dai H., Thakur J. S., et al. (2008). Raman spectroscopy can differentiate malignant tumors from normal breast tissue and detect early neoplastic changes in a mouse model. Biopolym. 89 235–241. 10.1002/bip.20899 [DOI] [PubMed] [Google Scholar]
- Kean R., Ramage G. (2019). Combined antifungal resistance and biofilm tolerance: the global threat of Candida auris. mSphere 4:e00458-19. 10.1128/mSphere.00458-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Sudbery P. (2011). Candida albicans, a major human fungal pathogen. J. Microbiol. 49 171–177. 10.1007/s12275-011-1064-7 [DOI] [PubMed] [Google Scholar]
- Kordalewska M., Lee A., Park S., Berrio I., Chowdhary A., Zhao Y., et al. (2018). Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob. Agents Chemother. 62 e00238–18. 10.1128/AAC.00238-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs R., Toth Z., Locke J. B., Forgacs L., Kardos G., Nagy F., et al. (2021). Comparison of in vitro killing activity of rezafungin, anidulafungin, caspofungin, and micafungin against four Candida auris clades in RPMI-1640 in the absence and presence of human serum. Microorganisms 9:863. 10.3390/microorganisms9040863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuplinska A., Rzad K. (2021). Molecular targets for antifungals in amino acid and protein biosynthetic pathways. Amino Acids 53 961–991. 10.1007/s00726-021-03007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Aguilar V., Rueda C., Garcia-Barbazan I., Varona S., Monzon S., Jimenez P., et al. (2021). Adaptation of the emerging pathogenic yeast Candida auris to high caspofungin concentrations correlates with cell wall changes. Virulence 12 1400–1417. 10.1080/21505594.2021.1927609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin E., Hager C., Chandra J., Mukherjee P. K., Retuerto M., Salem I., et al. (2017). The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob. Agents Chemother. 61 2396–2316. 10.1128/AAC.02396-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie D. G., Kast R. E., Poulik J. M., Rabah R., Sood S., Auner G. W., et al. (2012). Identification of pediatric brain neoplasms using Raman spectroscopy. Pediatr. Neurosurg. 48 109–117. 10.1159/000343285 [DOI] [PubMed] [Google Scholar]
- Li S. C., Kane P. M. (2009). The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta 1793 650–663. 10.1016/j.bbamcr.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méan M., Marchetti O., Calandra T. (2008). Bench-to-bedside review: Candida infections in the intensive care unit. Crit. Care 12:204. 10.1186/cc6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Arango A. C., Scorzoni L., Zaragoza O. (2012). It only takes one to do many jobs: amphotericin B as antifungal and immunomodulatory drug. Front. Microbiol. 3:286. 10.3389/fmicb.2012.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M. S., Jespersen B. M., Moller B. L., Laerke H. N., Larsen F. H., Engelsen S. B. (2010). Comparative spectroscopic and rheological studies on crude and purified soluble barley and oat β-glucan preparations. Food Res. Int. 43 2417–2424. 10.1016/j.foodres.2010.09.016 [DOI] [Google Scholar]
- Mulet Bayona J. V., Salvador García C., Tormo Palop N., Gimeno Cardona C. (2020). Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagarTM Candida for the detection of Candida auris in surveillance samples. Diagn. Microbiol. Infect. Dis. 98:115168. 10.1016/j.diagmicrobio.2020.115168 [DOI] [PubMed] [Google Scholar]
- Munoz J. F., Gade L., Chow N. A., Loparev V. N., Juieng P., Berkow E. L., et al. (2018). Genomic insight into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 9:5346. 10.1038/s41467-018-07779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro C. A. (2013). Chitin and glucan, the yin and yang of the fungal cell wall, implications for antifungal drug discovery and therapy. Adv. Appl. Microbiol. 83 145–172. 10.1016/B978-0-12-407678-5.00004-0 [DOI] [PubMed] [Google Scholar]
- Neuberger S., Neususs C. (2015). Determination of counterfeit medicines by Raman spectroscopy: systematic study based on a large set of model tablets. J. Pharm. Biomed. Anal. 112 70–78. 10.1016/j.jpba.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Noothalapati H., Sasaki T., Kaino T., Kawamukai M., Ando M., Hamaguchi H.-O., et al. (2016). Label-free chemical imaging of fungal spore walls by Raman microscopy and multivariate curve resolution analysis. Sci. Rep. 6:27789. 10.1038/srep27789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp Z., Borman A. M., Forgacs L., Kovacs R., Toth Z., Chun-Ju C., et al. (2021). Unpredictable in vitro killing activity of amphotericin B against four Candida auris clades. Pathogens 10:990. 10.3390/pathogens10080990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paret M. L., Sharma S. K., Green L. M., Alvarez A. M. (2010). Biochemical characterization of gram-positive and gram-negative plant-associated bacteria with micro-Raman spectroscopy. Appl. Spectrosc. 64 433–441. 10.1366/000370210791114293 [DOI] [PubMed] [Google Scholar]
- Pezzotti G. (2021). Raman spectroscopy in cell biology and microbiology. J. Raman Spectrosc. 10.1002/jrs.6204 [DOI] [Google Scholar]
- Pezzotti G., Asai T., Adachi T., Ohgitani E., Yamamoto T., Kanamura N., et al. (2021a). Antifungal activity of polymethyl methacrylate/Si3N4 composites against Candida albicans. Acta Biomater. 126 259–276. 10.1016/j.actbio.2021.03.023 [DOI] [PubMed] [Google Scholar]
- Pezzotti G., Boschetto F., Ohgitani E., Fujita Y., Zhu W., Marin E., et al. (2021b). Silicon nitride: a potent solid-state bioceramic inactivator of ssRNA viruses. Sci. Rep. 11:2977. 10.1038/s41598-021-82608-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L., Depciuch J., Kuna E., Worek M., Lewinska A., Wnuk M. (2019). FTIR and Raman spectroscopy-based biochemical profiling reflects genomic diversity of clinical Candida isolates that may be useful for diagnosis and targeted therapy of candidiasis. Int. J. Mol. Sci. 20:988. 10.3390/ijms20040988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasannan P., Suliman H. S., Robertus J. D. (2009). Kinetic analysis of site-directed mutants of methionine synthase from Candida albicans. Biochem. Biophys. Res. Commun. 382 730–734. 10.1016/j.bbrc.2009.03.098 [DOI] [PubMed] [Google Scholar]
- Rebrosova K., Siler M., Samek O., Ruzicka F., Bernatova S., Jezek J., et al. (2019). Identification of ability to form biofilm in Candida parapsilosis and Staphylococcus epidermidis by Raman spectroscopy. Future Microbiol. 14 509–518. 10.2217/fmb-2018-0297 [DOI] [PubMed] [Google Scholar]
- Roesch P., Harz M., Schmitt M., Peschke K.-D., Ronneberger O., Burkhardt H., et al. (2005a). Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71 1626–1637. 10.1128/AEM.71.3.1626-1637.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch P., Harz M., Schmitt M., Popp J. (2005b). Raman spectroscopic identification of single yeast cells. J. Raman Spectrosc. 36 377–379. 10.1002/jrs.1312 [DOI] [Google Scholar]
- Ruiz-Herrera J., Ortiz-Castellanos L. (2019). Cell wall glucans of fungi. A review, Cell Surf. 5:100022. 10.1016/j.tcsw.2019.100022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychert J. (2019). Benefits and limitations of MALDI-TOF Mass Spectrometry for the identification of microorganisms. J. Infectiology 2 1–5. 10.29245/2689-9981/2019/4.1142 32681212 [DOI] [Google Scholar]
- Samek O., Mlynarikova K., Bernatova S., Jezek J., Krzyzanek V., Siler M., et al. (2014). Candida parapsilosis biofilm identification by Raman spectroscopy. Int. J. Mol. Sci. 15 23924–23935. 10.3390/ijms151223924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. (2009). Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 53 41–44. 10.1111/j.1348-0421.2008.00083.x [DOI] [PubMed] [Google Scholar]
- Schrevens S., Van Zeebroeck G., Riedelberger M., Tournu H., Kuchler K., Van Dijck P. (2018). Methionine is required for cAMP-PKA mediated morphogenesis and virulence of Candida albicans. Mol. Microbiol. 108 258–275. 10.1111/mmi.13933 [DOI] [PubMed] [Google Scholar]
- Sherrington S. L., Sorsby E., Mahtey N., Kumwenda P., Lenardon M. D., Brown I., et al. (2017). Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 13:e1006403. 10.1371/journal.ppat.1006403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek M. S., Binkley J., Sherlock G. (2018). Using the Candida genome database. Methods Mol. Biol. 1757 31–47. 10.1007/978-1-4939-7737-6_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synytsya A., Mickova K., Synytsya A., Jablonsky I., Spevacek J., Erban V., et al. (2009). Glucans from fruit bodies of cultivated mushrooms Pleurotus ostreatus and Pleurotus eryngii: Structure and potential prebiotic activity. Carbohydr. Polym. 76 548–556. 10.1016/j.carbpol.2008.11.021 [DOI] [Google Scholar]
- Szekely A., Borman A. M., Johnson E. M. (2019). Candida auris isolates of the Southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J. Clin. Microbiol. 57 e02055–18. 10.1128/JCM.02055-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Surdin-Kerjan Y. (1997). Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61 503–532. 10.1128/.61.4.503-532.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ende M., Wijnants S., Van Dijck P. (2019). Sugar sensing and signaling in Candida albicans and Candida glabrata. Front. Microbiol. 10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S., Esbensen K., Geladi P. (1987). Principal component analysis. Chemometr. Intell. Lab. Syst. 2 37–52. [Google Scholar]
- Yamamoto M., Alshahni M. M., Tamura T., Satoh K., Iguchi S., Kikuchi K., et al. (2018). Rapid Detection of Candida auris based on loop-mediated isothermal amplification (LAMP). J. Clin. Microbiol. 56 e00591–18. 10.1128/JCM.00591-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.-T., Gulino K., Zhang Y., Sabestien A., Chou T.-W., Zhou B., et al. (2020). A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. U.S.A. 117 895–901. 10.1073/pnas.1910113117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Quivey R. G., Jr., Berger A. J. (2007). Raman spectroscopic measurement of relative concentrations in mixtures of oral bacteria. Appl. Spectrosc. 61 1233–1237. 10.1366/000370207782597021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.