Abstract
Chicory (Cichorium intybus var. sativum) is an industrial crop species cultivated for the production of a fructose polymer inulin, which is used as a low‐calorie sweetener and prebiotic. Besides, inulin chicory taproots also accumulate sesquiterpene lactones (STLs). These are bitter tasting compounds, which need to be removed during inulin extraction, resulting in additional costs. In this work, we describe chicory lines where STL accumulation is almost completely eliminated. Genome editing using the CRISPR/Cas9 system was used to inactivate four genes that encode the enzyme that performs the first dedicated step in STL synthesis, germacrene A synthase (CiGAS). Chicory lines were obtained that carried null mutations in all four CiGAS genes. Lines lacking functional CiGAS alleles showed a normal phenotype upon greenhouse cultivation and show nearly complete elimination of the STL synthesis in the roots. It was shown that the reduction in STLs could be attributed to mutations in genetically linked copies of the CiGAS‐short gene and not the CiGAS‐long gene, which is relevant for breeding the trait into other cultivars. The inactivation of the STL biosynthesis pathway led to increase in phenolic compounds as well as accumulation of squalene in the chicory taproot, presumably due to increased availability of farnesyl pyrophosphate (FFP). These results demonstrate that STLs are not essential for chicory growth and that the inhibition of the STL biosynthesis pathway reduced the STL levels chicory which will facilitate inulin extraction.
Keywords: chicory, sesquiterpene lactones, germacrene A synthase, genome editing
Introduction
Cichorium species are perennial plants, which are grown both as vegetable and as an industrial crop. C. intybus is grown for many different applications and is divided into several different varieties according to their use (Street et al., 2013). C. intybus var. foliosum is cultivated as a vegetable, appreciated for its bitter taste in the regions of northern France, Belgium and the Netherlands as an etiolated compact leaf structure known as Belgian endive (also witloof) and in Italy as radicchio. The related species C. endivia is also consumed as a green leafy vegetable (endive). The taproot of another species, C. intybus var. sativum, is grown for an industrial application, the isolation of inulin (van Arkel et al., 2012). Inulin is a fructose polymer that is a water soluble dietary fibre according to the EU and CODEX definitions (EC 1169/2011, Alinorm 09/3/12, March 2009) and the FDA (Addition to FDA‐2016‐D‐3401, February 2018). It is fermented to short chain fatty acids by the gut bacteria contributing to local and whole body health (Ahmed and Rashid, 2019; Roberfroid, 2002; Roberfroid, 2007). It finds an application as a food fibre and low‐calorie sweetener in a wide variety of food products such as bakery and dairy products, confectionary, infant food and increasingly in cosmetic applications (MINTEL GNPD database, 2020), where enhancing a healthy skin microbiome builds on the well‐established gut microbiome research. The inulin market size is well over 100 000 ton with a yearly growth of approximately 4% (International, 2020). The chicory taproot is the primary source of inulin and with an average inulin content of 17% and a typical root yield of 45 tons per hectare, the global acreage of chicory is calculated to be over 13 000 ha (Wilson et al., 2004). Apart from inulin, the chicory root is also rich in sesquiterpene lactones (STLs), which are characteristic of the Asteraceae family. More than 5000 different STLs have so far been identified in the Asteraceae and are thought to have been a major factor in their widespread adaptation to different environments (Chadwick et al., 2013). STLs are synthesized throughout the plant but are particularly concentrated in the trichomes and the latex which is stored in laticifers. STLs have been shown to have a defence role in plants, ranging from allelopathic activity to protection against herbivorous insects of roots and flowers (Huber et al., 2016; Molinaro et al., 2016; Padilla‐Gonzalez et al., 2016; Prasifka et al., 2015). Some STLs exhibit medicinal properties such as anti‐inflammatory, anti‐cancer and analgesic activity and have therefore been explored in medicinal applications (Chadwick et al., 2013). The major STLs of chicory belong to the class of guaianolide sesquiterpene lactones and are thought to be derived from a single sesquiterpene, germacrene A (Figure 1) (de Kraker et al., 1998). Germacrene A does not accumulate in chicory but is efficiently converted by the cytochrome P450 enzymes germacrene A oxidase (GAO) and costunolide synthase (COS) to costunolide, a common precursor of guaianolide sesquiterpenes (Cankar et al., 2011; Ikezawa et al., 2011; Liu et al., 2011; Nguyen et al., 2010). Subsequently, biosynthetic enzymes modify costunolide (e.g. through oxidations, lactone ring closures and conjugations to oxalate, hydroxyphenyl acetate and/or glycosyl moieties) to diverse biochemical structures. In chicory, the most predominant STLs are lactucin, lactucopicrin and 8‐deoxylactucin in their oxalated forms (Sessa et al., 2000). In the industrial process, the very bitter tasting STLs from the chicory taproot are co‐extracted with inulin. This bitter taste hinders the broad food application of inulin, and therefore, additional purification steps are required, increasing cost of the process and eventually of inulin itself (Meyer and Blaauwhoed, 2009). Therefore, there is considerable interest in the development of chicory lines that are low in STL content, for both cost savings in inulin isolation and the production of a wide selection of less bitter leafy chicory varieties that could make chicory more suitable for other markets. Although there is some natural variation in STL content in chicory, variants of chicory which have considerably lower STL content have not yet been identified, and therefore, reduction in STL content by traditional breeding approaches is difficult.
Figure 1.
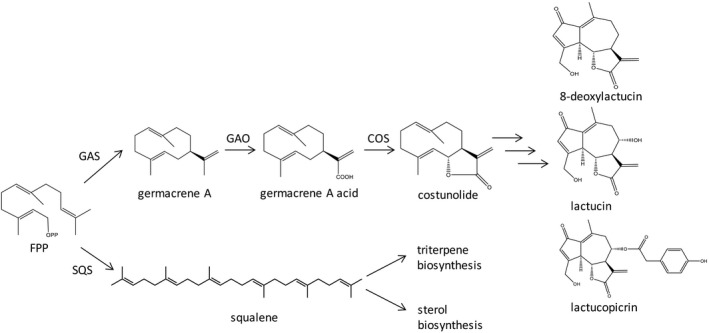
Biosynthetic pathway of chicory sesquiterpene lactones. COS, costunolide synthase; FPP, farnesyl pyrophosphate; GAO, germacrene A oxidase; GAS, germacrene A synthase; SQS, squalene synthase.
The genes underlying STL biosynthesis have been partially characterized. Genes encoding the enzyme catalysing the first step of the STL production, germacrene A synthase, have been identified (Bouwmeester et al., 2002; de Kraker et al., 1998). In chicory, four different germacrene A synthase genes (CiGAS) are present, one copy of GAS‐long (CiGAS‐L) and three physically linked copies of GAS‐short (CiGAS‐S1, CiGAS‐S2 and CiGAS‐S3) (Bogdanović et al., 2019). Expression analysis of the CiGAS genes has shown that both the CiGAS‐L and CiGAS‐short genes are expressed in chicory roots, while predominantly only the CiGAS‐L is expressed in chicory leaves (Bogdanović et al., 2019; Bouwmeester et al., 2002). Silencing of the CiGAS genes by a microRNA approach resulted in reduced expression of both the long and short germacrene A synthases and a partial decrease of STL levels (Bogdanovic et al., 2020). However, it is still not known which of the CiGAS genes play a role in different tissues and which CiGAS genes need to be inactivated before a strong effect on STL production can be observed. Genome editing in plants using CRISPR/Cas9 is well established and has been used in many plant species. The method involves expression of a targeting RNA (guide RNA) and bacterial endonuclease, which together introduce a DNA double‐strand break (DSB) at a target sequence, such as in an exon. Mis‐repair of the DSB(s) by the cellular DNA repair machinery results in small indels at the target site, which often disrupt the gene function. Due to the efficiency of the CRISPR/Cas9 system, it can also be exploited to inactivate multiple alleles of a gene family to identify phenotypes that, due to complementation, would not be apparent in individual mutants. Bernard et al. (2019 ) have recently shown that the CRISPR/Cas9 system efficiently introduces mutations in the C. intybus genome using either Agrobacterium rhizogenes mediated transformation or protoplast transfection. Therefore, the CRISPR/Cas9 system is applicable for creating mutations in the different copies of the chicory CiGAS genes. Successful gene editing of this gene has just recently been reported; however, the effect of the CiGAS gene on the secondary metabolism of chicory was not assessed (De Bruyn et al., 2020). In this work, chicory lines were produced in which the CiGAS genes were inactivated and STL accumulation was almost completely eliminated, which led to a corresponding overproduction of other secondary metabolites (phenolics and squalene). To our knowledge, this is the first example of an Asteraceae species where STLs were eliminated and as such is a valuable resource to study the role of STLs on plant growth and defence.
Results
Evaluating CRISPR/Cas9 mutagenesis of the chicory GAS genes
In the first step of STL production, germacrene A synthase (GAS) converts farnesyl pyrophosphate (FPP) to germacrene A. Four GAS genes have been identified in chicory, one copy of GAS‐long (CiGAS‐L) and three copies of GAS‐short (CiGAS‐S1, CiGAS‐S2, CiGAS‐S3) (Bogdanović et al., 2019; Bouwmeester et al., 2002). Gene expression data for GAS‐long and GAS‐short suggest that there is clear tissue specificity in the role of the different CiGAS isogenes (Bogdanović et al., 2019; Bouwmeester et al., 2002). CiGAS‐long is anticipated to be responsible for STL biosynthesis at least in leaves and possibly also in roots, while it is unclear what the contribution of the 3 CiGAS‐short genes would be. Possibly several CiGAS alleles must be mutated in a single line before an effect on the STL levels can be observed.
Mutagenesis of the CiGAS genes using CRISPR/Cas9 reagents was performed in chicory leaf protoplasts. The CRISPR/Cas9 reagents themselves were either expressed transiently from introduced plasmid constructs or introduced into the protoplasts as a ribonucleoprotein complex (RNP) made up of the SpCas9 protein and guide RNA. Individual protoplasts were then regenerated into mature plants with low levels of chimaerism. Chicory protoplasts were found to be very amenable to transformation and regeneration and showed rapid growth rates in tissue culture, making them very suitable for CRISPR/Cas9‐mediated mutagenesis. The transfection efficiency of chicory mesophyll protoplasts was assessed using a fluorescent reporter plasmid and PEG‐mediated transfection, and 58 ± 6% of the protoplasts displayed a signal. A number of guide RNAs were first tested for their ability to introduce mutations at five sequences in exon 4 of the CiGAS‐S1 gene which are conserved in the four CiGAS genes (Figure S1). Exon 4 encodes a part of the CiGAS active site based on its homology with the tobacco 5‐epi‐aristolochene synthase (TEAS) (Bouwmeester et al., 2002), for which both the crystal structure and the amino acids essential for activity are known, and therefore, any type of mutation in exon 4 can potentially disrupt gene function. Indel mutations can be characterized as those that change the protein sequence by altering the coding frame or those that add or delete codons without altering the frame itself (e.g. −3 bp, −6 bps). A change in the coding frame is likely to inactivate the protein, whereas deleting a small number of codons may have little effect on the phenotype. However, by ensuring that the CiGAS region targeted includes the enzyme active site it becomes more likely that in‐frame deletions also have a negative impact on enzyme activity. As the mutagenesis efficiencies of different guide RNAs cannot be predicted in silico beforehand, we first assessed the activity of five different guide RNAs targeting different positions in the CiGAS‐S1 exon 4 (Figure S1). Plasmids each carrying a different guide RNA expression cassette were each mixed with the SpCas9 expression plasmid and used for chicory leaf protoplast transfection. After 48 h, the protoplasts were used for genomic DNA isolation, the CiGAS‐S1 exon 4 target sites were amplified and these were then bulk sequenced to quantify the percentage of amplicons in each sample that contained CRISPR/Cas9‐mediated mutations. Based on these results, the guide RNA showing the highest activity (sgRNA1; Figure S1) was selected for further experiments. While exon 4 of CiGAS‐S1 and CiGAS‐S2 are nearly identical, some SNPs are present in exon 4 of the CiGAS‐S3 and CiGAS‐L genes, although all of the PAM sequences are conserved (Figure S1). Variants of sgRNA1, targeting the corresponding region in the CiGAS‐S3 and CiGAS‐L genes, were designed (sgRNA1.1 and sgRNA1.2). A single construct was then synthesized that expressed all three guide RNAs that target the respective CiGAS genes (Figure S2). To assess the activity of this construct for CiGAS mutagenesis, it was transfected to chicory protoplasts together with the SpCas9 expression construct and the mutations were then analysed as before by amplicon sequencing. The percentage of sequence reads derived from each CiGAS gene that contained mutations is shown in Table 1 and was found to be sufficient for mutagenesis experiments. For the RNP experiments, chicory protoplasts were transfected with complexes of SpCas9 and the most efficient guide, sgRNA1, identified in the previous screen. Two different versions of sgRNA1 were used, the first consisting of unmodified RNA and the second incorporating modified RNA nucleotides that improve sgRNA resistance to endogenous nucleases and thereby may improve the mutagenesis efficiency (Hendel et al., 2015; O'Reilly et al., 2019). In these RNP experiments, only the single sgRNA1 guide was used as simultaneous transfection of different RNPs with different guides is known to have a negative impact on the mutagenesis efficiency. Several SNPs are present between sgRNA1 and GAS‐S3 and GAS‐L, which may influence the mutagenesis efficiency at these targets. However, SpCas9 is able to tolerate a limited number of SNPs depending on their position. The RNPs containing either unmodified or modified sgRNA1 were transfected to chicory protoplasts and their mutagenesis efficiency at the GAS genes quantified by amplicon sequencing (Table 1). The RNP incorporating the modified sgRNA1 was clearly more efficient, although the SNP in GAS‐L significantly reduced the activity. Therefore, the efficiency of these RNPs was deemed to be satisfactory for further experiments.
Table 1.
Mutations found in protoplasts and regenerated shoots using the different mutagenesis methods
| Method | % indels in protoplasts | # regenerated shoots | % mutant CiGAS alleles in shoots | # shoots with null alleles in multiple CiGAS genes* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | L | S1 | S2 | S3 | L | 0 genes | 1 gene | 2 genes | 3 genes | 4 genes | ||
| Plasmid | 14 | 14 | 15 | 16 | 37 † | 78 | 78 | 80 | 75 | 8 | 0 | 1 | 3 | 25 |
| RNPs (+sgRNA) | 24 | 25 | 9 | 4 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| RNPs (modified sgRNAs) | 38 | 38 | 40 | 20 | 79 | 84 | 77 | 76 | 28 | 9 | 8 | 13 | 42 | 7 |
Putative null alleles, genes which contain either homozygous or biallelic mutations (either in or out of frame) which are both expected to eliminate gene function.
39% of the regenerated shoots were shown by PCR to have integrated copies of SpCas9 ORF in the genome.
Protoplasts were transfected with either the plasmid constructs or the RNP containing the modified sgRNA1, and the cells were maintained in tissue culture until they had formed calli which then regenerated into mature plants. The karyotype of the plants was then confirmed, and diploid rooted plants were then genotyped by Illumina sequencing to identify CiGAS mutants plants with indel mutations at the CiGAS genes (Table 1). Both mutagenesis methods generated lines with very high numbers of mutated alleles, confirming the high efficiency of the CRISPR/Cas9 system in chicory. Plants were then placed in classes based on the number of putative null alleles (indels in both copies of the target genes) they contained (Table 1). As argued above, we assumed that in frame indel mutations in the enzyme active site would also eliminate the protein function. As chicory is a self‐incompatible species that cannot be selfed, the effect of recessive mutations can only be determined in primary transformants when both copies of a gene are inactivated. We were able to identify lines with homozygous mutations in all four CiGAS genes as well as plants in which different combinations of the CiGAS genes had been mutated (Figure S3), generating both frame‐shift and in‐frame mutations in this gene. Therefore, next to previously used transient expression of CRISPR/Cas9 reagents in chicory protoplasts (Bernard et al., 2019; De Bruyn et al., 2020) also the transfection of SpCas9/sgRNA RNPs appears to be a very efficient method to introduce mutations in multiple targets simultaneously and create a pool of germplasm with a variety of functional and non‐functional alleles in multiple isogenes. While plasmid transfection is designed to provide transient expression of the CRISPR/Cas9 reagents for mutagenesis, the transfected plasmids may also integrate into the genome resulting in stable transformation. Indeed, we were able to detect SpCas9 sequences in 39% of the regenerated plants, indicating that chicory readily integrates exogenous plasmid DNA into its genome and confirming the previous findings (Bernard et al., 2019). The leaves of diploid plants carrying a spectrum of different homozygous CiGAS mutations and lacking integrated plasmid DNA were sampled in tissue culture, and a selection of lines was transferred to the greenhouse and assayed for STL content in roots. Regenerated lines that lacked CiGAS mutations served as wild‐type (WT) controls.
Inactivation of CiGAS leads to reduction in STLs in chicory leaves and roots
Sesquiterpene lactone content was determined in the leaves of 17 CiGAS mutant lines and 7 control plants that also were obtained via the regeneration process (Table 2). The genotypes of these lines are depicted in Figure S3. The major sesquiterpenes observed in the chicory leaves were lactucin, lactucopicrin and 8‐deoxylactucin, predominantly in their oxalated form. The level of STLs in the seven control lines was varied somewhat indicating that the regeneration process may have introduced some somaclonal variation in STL amount in the control plants. However, several lines containing mutations in the CiGAS genes showed a clear and strong reduction in the amount of STLs (Table 2). Multiple chicory lines that carried a mutation in the six CiGAS‐short alleles or all 8 CiGAS‐short and CiGAS‐long alleles were obtained in which nearly complete elimination of sesquiterpene lactones was observed. Although these lines are genotypically unique (Figure S3) as the CRISPR mediated cut is directed but the repair mechanism at that site is random, the mutations lead to the same biochemical phenotype. There appears to be a direct correlation between the identity of the functional CiGAS genes present and the levels of STLs produced. Several chicory lines carrying mutations in the three CiGAS‐short genes show elimination of terpenes to the same extent as when all 4 CiGAS genes, including also the CiGAS‐long gene, are mutated. Apparently, the CiGAS‐L gene hardly contributes to STL production in the leaves.
Table 2.
Terpene and phenolic composition of chicory leaves of WT and CiGAS chicory lines
| Line | CiGAS gene genotype | Sesquiterpene lactone and phenolic compound quantification | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | L | L µg/gFW | LP µg/gFW | 8‐DOL relative abundance | Lox relative abundance | LPox relative abundance | 8‐DOLox relative abundance | CA µg/gFW | CiA µg/gFW | 3,5‐DiCQA µg/gFW | |
| PL1 | W/W | W/W | W/W | W/W | 6.8 ± 2.1ab | 10.7 ± 3.2abc | 10.2 ± 2.8bc | 149.6 ± 55.4c | 320.2 ± 120.2b | 154.1 ± 50.4e | 95.0 ± 26.6 a | 1535.6 ± 55.7ab | 172.8 ± 9.9a |
| PL2 | W/W | W/W | W/W | W/W | 4.3 ± 0.0ab | 8.0 ± 0.5abc | 9.2 ± 0.1abc | 104.5 ± 7.6c | 259.2 ± 6.4b | 140.7 ± 9.9de | 131.7 ± 4.9 a | 1649.3 ± 58.0 ab | 227.9 ± 13.1a |
| PL3 | W/W | W/W | W/W | W/W | 5.9 ± 1.6ab | 12.4 ± 4.7bc | 9.2 ± 2.4abc | 118.3 ± 15.8c | 317.7 ± 65.4b | 126.8 ± 13.2de | 74.7 ± 4.3 a | 1608.1 ± 18.4ab | 164.0 ± 31.5a |
| PL4 | W/W | W/W | W/W | W/W | 9.9 ± 4.4b | 15.9 ± 7.5c | 19.4 ± 8.6c | 127.1 ± 26.3c | 293.8 ± 78.5b | 166.4 ± 37.6e | 138.7 ± 33.9 a | 1963.3 ± 152.8ab | 172.2 ± 58.8a |
| RN1 | W/W | W/W | W/W | W/W | 4.1 ± 0.7ab | 6.2 ± 0.9abc | 8.0 ± 1.8ab | 88.7 ± 14.4bc | 195.4 ± 24.9b | 112.6 ± 27.3de | 56.4 ± 23.9 a | 1320.3 ± 195.8ab | 122.5 ± 5.9a |
| RN2 | W/W | W/W | W/W | W/W | 4.7 ± 2.8ab | 7.2 ± 3.0abc | 7.3 ± 3.8ab | 117.5 ± 11.4c | 250.7 ± 0.2b | 127.6 ± 3.0de | 92.6 ± 9.7 a | 1531.1 ± 163.9ab | 152.4 ± 1.8a |
| RN3 | W/W | W/W | W/W | W/W | 3.0 ± 0.2ab | 3.8 ± 0.8ab | 3.0 ± 0.5ab | 120.2 ± 32.5c | 200.0 ± 77.3b | 79.9 ± 33.9 cd | 72.5 ± 5.7 a | 1417.1 ± 382.2ab | 109.1 ± 11.4a |
| RN4 | −5/−11 | −9/−9 | −7/ins | W/W | 0.2 ± 0.1a | 0.6 ± 0.5a | 0.3 ± 0.3ab | 0.1 ± 0.1a | 8.3 ± 7.5a | 1.2 ± 1.2a | 52.0 ± 3.7 a | 1360.2 ± 237.1ab | 164.8 ± 5.4a |
| RN5 | −12/−9 | W/W | W/W | W/W | 6.7 ± 4.5ab | 8.2 ± 4.2abc | 5.5 ± 3.3ab | 120.1 ± 12.1c | 221.4 ± 2.9b | 77.0 ± 6.0bcd | 109.6 ± 1.3 a | 1437.5 ± 100.5ab | 166.0 ± 31.2 a |
| RN6 | −19/−4 | W/W | −4/W | W/W | 6.1 ± 2.8ab | 11.1 ± 4.8abc | 8.7 ± 4.5ab | 114.7 ± 3.6c | 289.2 ± 5.8b | 115.4 ± 6.1de | 102.7 ± 13.2 a | 1773.4 ± 215.1ab | 227.6 ± 7.1a |
| RN7 | −4/−17 | −11/−11 | −2/−7 | W/W | 0.3 ± 0.1a | 0.7 ± 0.1a | 0.7 ± 0.2ab | 0.0 ± 0.0a | 16.3 ± 2.0a | 4.6 ± 0.9a | 92.6 ± 38.8 a | 1404.9 ± 157.7ab | 151.6 ± 51.4a |
| RN8 | −14/−7 | −13/−10 | −2/W | W/W | 2.0 ± 0.1a | 8.4 ± 1.0abc | 4.8 ± 1.2ab | 35.2 ± 5.1ab | 267.5 ± 0.3b | 79.8 ± 7.6 cd | 63.6 ± 7.2 a | 1720.2 ± 151.5ab | 156.3 ± 7.4a |
| RN9 | −9/−9 | −10/W | −4/−4 | W/W | 4.2 ± 2.2ab | 5.6 ± 2.6abc | 5.2 ± 2.4ab | 88.8 ± 15.1bc | 166.9 ± 30.3ab | 70.3 ± 18.1abcd | 100.4 ± 12.3 a | 1133.6 ± 198.0ab | 104.9 ± 12.2a |
| RN10 | −3/−5 | −6/−3 | −7/W | W/W | 0.2 ± 0.0a | 0.4 ± 0.1a | 0.3 ± 0.1ab | 0.0 ± 0.0a | 7.0 ± 1.8a | 1.3 ± 0.4a | 124.0 ± 56.6 a | 1615.3 ± 564.9ab | 222.3 ± 1.6a |
| RN11 | −7/−7 | −5/−5 | −2/−2 | W/W | 0.2 ± 0.0a | 0.1 ± 0.0a | 0.1 ± 0.1ab | 0.0 ± 0.0a | 0.4 ± 0.5a | 0.1 ± 0.1a | 96.5 ± 78.7 a | 967.9 ± 690.1a | 156.1 ± 119.7a |
| RN12 | −4/−7 | +1/−7 | −7/−16 | W/W | 0.3 ± 0.0a | 0.9 ± 0.2a | 0.6 ± 0.3ab | 0.0 ± 0.0a | 21.4 ± 10.4a | 4.3 ± 3.2a | 68.6 ± 23.4 a | 1428.9 ± 33.0ab | 201.2 ± 4.4a |
| RN13 | −11/−11 | −12/−6 | −9/−7 | W/W | 0.3 ± 0.0a | 0.5 ± 0.1a | 0.7 ± 0.3ab | 0.0 ± 0.0a | 9.2 ± 3.0a | 2.7 ± 0.8a | 142.6 ± 4.4 a | 1812.5 ± 350.9ab | 283.0 ± 3.5a |
| RN14 | −7/−7 | −9/−7 | −3/−3 | −11/W | 0.4 ± 0.2a | 0.7 ± 0.4a | 0.6 ± 0.4ab | 0.0 ± 0.0a | 9.5 ± 6.3a | 2.5 ± 2.3a | 89.7 ± 12.1 a | 1928.7 ± 386.8ab | 192.8 ± 7.0a |
| RN15 | W/−16 | W/W | W/−5 | W/W | 4.4 ± 1.2ab | 7.6 ± 3.9abc | 7.4 ± 2.1ab | 89.6 ± 0.8b | 191.6 ± 30.2b | 107.3 ± 18.1de | 94.8 ± 39.8 a | 1266.8 ± 250.1ab | 116.5 ± 35.3a |
| PL5 | −9/−12 | W/W | −9/−2 | −6/−6 | 5.8 ± 1.4ab | 9.2 ± 4.4abc | 9.3 ± 1.3abc | 99.1 ± 19.8b | 214.2 ± 82.0b | 101.7 ± 17.4de | 71.3 ± 16.2a | 1479.7 ± 113.9ab | 128.4 ± 43.2a |
| RN16 | −2/−11 | −2/−11 | −9/−7 | −7/−16 | 0.5 ± 0.4a | 0.7 ± 0.4a | 0.7 ± 0.5ab | 0.0 ± 0.0a | 16.9 ± 19.8a | 7.6 ± 9.4ab | 176.0 ± 119.5a | 2608.0 ± 1100.1b | 426.6 ± 473.0a |
| RN17 | −6/−5 | −10/−6 | −11/+1 | −8/−9 | 0.1 ± 0.0a | 0.1 ± 0.0a | 0.0 ± 0.0a | 0.0 ± 0.1a | 0.3 ± 0.2a | 0.0 ± 0.0a | 107.8 ± 59.0a | 1009.9 ± 158.6a | 61.6 ± 56.1a |
| PL6 | −1/−1 | −4/−4 | −2/−11 | −1/−5 | 0.4 ± 0.2a | 0.7 ± 0.5a | 0.9 ± 0.5ab | 0.0 ± 0.0a | 17.3 ± 17.5a | 8.9 ± 8.2abc | 98.2 ± 29.5a | 1747.7 ± 168.8ab | 296.8 ± 90.8a |
| PL7 | −3/−3 | −2/−2 | −4/−4 | −5/−5 | 0.6 ± 0.1a | 1.1 ± 0.2a | 1.1 ± 0.1ab | 0.0 ± 0.0a | 22.2 ± 13.6a | 7.4 ± 4.7ab | 97.9 ± 59.9a | 1992.1 ± 547.0ab | 228.2 ± 23.4a |
The mean ± SD is shown for duplicate measurements. The letters indicate significantly different groups according to ANOVA and the Tukey’s post hoc test (P < 0.05). CA, chlorogenic acid; CiA, chicoric acid; 3,5‐DiCQA, 3,5‐dicaffeoylquinic acid; 8‐DOL, 8‐deoxylactucin; 8‐DOLox, 8‐deoxylactucin oxalate; ins, large insertion; L, lactucin; Lox, lactucin oxalate; LP, lactucopicrin; LPox, lactucopicrin oxalate; W, wild‐type genotype.
Next, the STL levels were measured in the roots. To this purpose, five CiGAS mutant lines were selected and were, together with six WT lines, grown in the greenhouse for 5 months until a taproot of approximately 15 cm in length was formed and the STLs profiles of harvested roots were analysed. The profile of 5 selected CiGAS mutant lines and 6 WT lines and the profile of STLs in different CiGAS deletion lines resembled closely the leaf measurements (Table 3). Line RN12 carrying a frame‐shift mutation in all six CiGAS‐short genes and line PL6 carrying a frame‐shift mutation in all eight alleles of CiGAS‐short and CiGAS‐long genes both showed nearly complete elimination of sesquiterpene lactones in the root. This suggests that the CiGAS‐short genes are most important for sesquiterpene lactone production in both leaves and roots. This was not expected, since the CiGAS‐short genes are hardly expressed in leaves (Bogdanović et al., 2019; Bouwmeester et al., 2002). Given that a small amount of STLs is still detected in the leaves and roots of lines with biallelic mutations in all 4 CiGAS genes, we conclude that there might be another sesquiterpene synthase producing small amounts of germacrene A present in chicory. Plants lacking STLs were viable and showed no obvious morphological differences with the controls under used greenhouse conditions. Next the inulin profile of lines PL6 and RN12 was analysed by size exclusion chromatography. The polymerization degrees of the two CiGAS deletion lines PL6 and RN12 were similar as that of the control plant PL4. The weight average DPs of all lines were between 20.5 to 21.0 and number average DPs between 11.5 and 12.7 (Table 4, Figure S4). Polydispersity was calculated to describe the distribution of the molecular units (Mw/Mn), thus the higher the polydispersity value, the wider the distribution. This analysis indicated that the reduction in terpenes did not affect the inulin composition, under the applied growth conditions. Though a comprehensive phenotypic test in many physiologically different conditions should be performed to fully characterize the role of the STLs, these initial observations indicate that STLs do not play an essential role in chicory development and inulin biosynthesis. As STLs have been hypothesized to be involved in defence, it has yet to be seen whether these plants have a disadvantage during normal field cultivation.
Table 3.
Terpene and phenolic composition of roots of selected WT and CiGAS chicory lines
| Line | CiGAS gene genotype | Sesquiterpene lactone and phenolic compound quantification | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | L | L µg/gFW | LP µg/gFW | 8‐DOL relative abundance | Lox relative abundance | LPox relative abundance | 8‐DOLox relative abundance | CA µg/gFW | CiA µg/gFW | 3,5‐DiCQA µg/gFW | |
| PL1 | W/W | W/W | W/W | W/W | 51.2 ± 5.2e | 38.4 ± 4.4f | 29.3 ± 2.5e | 424.7 ± 26.6d | 569.0 ± 26.1e | 349.6 ± 24.4f | 197.1 ± 18.7ab | 231.6 ± 11.8ab | 239.1 ± 47.6abc |
| PL2 | W/W | W/W | W/W | W/W | 21.3 ± 4.3b | 13.3 ± 2.1b | 9.0 ± 1.6b | 215.5 ± 65.2b | 246.2 ± 75.0bc | 123.5 ± 42.2bc | 161.4 ± 60.2a | 194.7 ± 142.1a | 147.9 ± 45.1a |
| PL3 | W/W | W/W | W/W | W/W | 33.7 ± 4.4d | 26.1 ± 1.9e | 17.4 ± 1.4d | 330.7 ± 12.2c | 423.0 ± 15.5d | 248.1 ± 6.8e | 307.4 ± 9.9 cd | 235.2 ± 7.4ab | 357.8 ± 1.5 cd |
| PL4 | W/W | W/W | W/W | W/W | 22.1 ± 1.2bc | 17.1 ± 0.8bcd | 14.2 ± 0.7 cd | 253.2 ± 5.3bc | 349.5 ± 5.0 cd | 233.7 ± 2.3e | 189.1 ± 12.4ab | 297.5 ± 20.1ab | 248.6 ± 9.6abc |
| RN1 | W/W | W/W | W/W | W/W | 22.2 ± 0.1bc | 14.1 ± 1.3bc | 9.3 ± 0.4b | 194.0 ± 9.7b | 227.2 ± 19.1b | 117.0 ± 5.6bc | 218.6 ± 2.3abc | 273.6 ± 7.6ab | 408.4 ± 4.5de |
| RN2 | W/W | W/W | W/W | W/W | 27.0 ± 2.0bcd | 25.2 ± 1.9e | 15.6 ± 0.8d | 225.6 ± 7.1b | 348.7 ± 20.2 cd | 188.9 ± 11.8de | 266.7 ± 20.1bc | 247.8 ± 25.7ab | 256.0 ± 27.8abc |
| PL5 | −9/−12 | W/W | −9/−2 | −6/−6 | 32.1 ± 1.9 cd | 20.9 ± 1.7cde | 13.2 ± 1.4bcd | 273.7 ± 11.6bc | 342.5 ± 18.9 cd | 167.9 ± 7.7 cd | 185.9 ± 11.9ab | 143.3 ± 1.3a | 220.9 ± 3.9ab |
| RN6 | −19/−4 | W/W | −4/W | W/W | 30.9 ± 1.3bcd | 24.0 ± 0.5de | 13.4 ± 0.3bcd | 245.4 ± 3.1bc | 320.2 ± 3.8bcd | 155.8 ± 4.4bcd | 220.6 ± 3.0abc | 294.5 ± 26.1ab | 294.0 ± 45.3bcd |
| RN8 | −14/−7 | −13/−10 | −2/W | W/W | 31.4 ± 1.2bcd | 23.8 ± 1.7de | 10.4 ± 0.7bc | 216.3 ± 15.2b | 343.5 ± 29.0 cd | 102.9 ± 8.1b | 389.9 ± 25.7d | 565.2 ± 92.0 cd | 503.3 ± 51.6e |
| RN12 | −4/−7 | +1/−7 | −7/−16 | W/W | 2.5 ± 0.0a | 1.7 ± 0.1a | 1.1 ± 0.0a | 2.2 ± 3.2a | 13.8 ± 0.5a | 3.8 ± 0.0a | 527.5 ± 11.3e | 675.2 ± 12.6d | 892.8 ± 18.0f |
| PL6 | −1/−1 | −4/−4 | −2/−11 | −1/−5 | 4.5 ± 0.2a | 2.4 ± 0.1a | 2.5 ± 0.0a | 0.0 ± 0.0a | 23.9 ± 1.0a | 9.6 ± 0.3a | 655.5 ± 37.0f | 442.8 ± 21.0bc | 1201.1 ± 11.2g |
The mean ± SD is shown for duplicate measurements. The letters indicate significantly different groups according to ANOVA and the Tukey’s post hoc test (P < 0.05). CA, chlorogenic acid; CiA, chicoric acid; 3,5‐DiCQA, 3,5‐dicaffeoylquinic acid; 8‐DOL, 8‐deoxylactucin; 8‐DOLox, 8‐deoxylactucin oxalate; L, lactucin; Lox, lactucin oxalate; LP, lactucopicrin; LPox, lactucopicrin oxalate; W, wild‐type genotype.
Table 4.
Inulin polymerisation degrees (DP) of WT and CiGAS KO lines PL6 and RN12. Polydispersity (Mw/Mn)
| Line | Mn (Daltons) | Mw (Daltons) | Polydispersity | Weight average DP | Number average DP |
|---|---|---|---|---|---|
| PL4 | 2050 | 3366 | 1.6 | 20.8 | 12.7 |
| PL6 | 1868 | 3316 | 1.8 | 20.5 | 11.5 |
| RN12 | 1995 | 3394 | 1.7 | 21.0 | 12.3 |
Inactivation of GAS activity leads to overproduction of squalene in chicory taproot
Inactivation of the CiGAS genes may influence the accumulation of other metabolites in the terpene pathway and therefore a hexane extract of chicory roots of six WT and five CiGAS mutant lines was analysed by GC‐MS analysis. In root tissues in mutant chicory lines displaying the highest reduction in STLs (RN12 and PL6), other than trace amounts of farnesene and farnesol, no accumulation of monoterpenes or sesquiterpenes was observed as compared to WT lines. We did not observe accumulation of germacrene A or costunolide, which are common guaianolide STL precursors. However, a large new peak was detected in the chromatogram of lines RN12, PL6 and RN8 at the retention time of 26.7 min (Figure 2). This compound was identified as squalene by comparison of the retention time and mass spectrum to the authentic standard of squalene (Figure S5). The amount of squalene accumulating in the root was quantified at 147 ± 4, 124 ± 3 and 30 ± 1 µg/gFW in chicory lines RN12, PL6 and RN8, respectively. RN12 and PL6 are the lines with strongest STLs reduction in the roots while RN8 did not show statistically significant STL reduction (Table 3). No squalene was observed in chicory root extracts of lines PL5 and RN6, nor in the extract of the six WT chicory roots.
Figure 2.
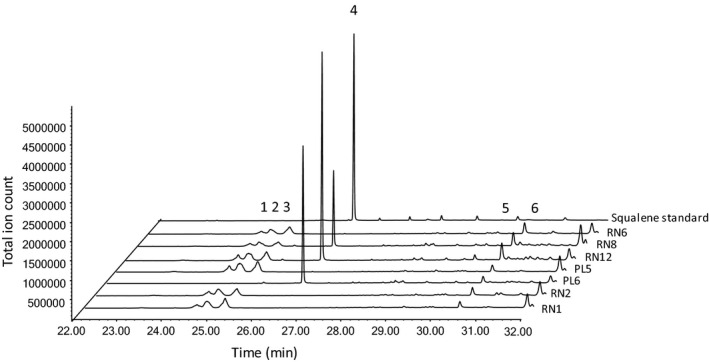
GC‐MS chromatogram of chicory root extracts. Chromatograms of two WT (RN1, RN2) and 5 CiGAS deletion lines (PL5, PL6, RN6, RN8, RN12) are shown. Peak 1 – 3: acetylated triterpenes (C32H52O2, MW = 468), Peak 4 – squalene, peak 5: stigmasterol, peak 6: sitosterol. Squalene accumulation was detected in lines PL6, RN12 and RN8.
Squalene is a precursor for the biosynthesis of triterpenes and phytosterols. WT chicory roots accumulate small amounts of acetylated‐triterpenes (Figure 2). Upon comparison of the WT plants to the CiGAS deletion lines, no increase in the amount of triterpenes was observed. The accumulation of phytosterols sitosterol, campesterol and stigmasterol in CiGAS deletion lines was next compared with the WT chicory plants. Sitosterol was the major observed sterol in the root tissue of WT chicory plants. The amount of sitosterol was not statistically significantly increased in CiGAS deletion lines. The amount of stigmasterol and campesterol was below 5 μg/g FW for both WT and KO lines and could not be quantified. In contrast to squalene, the changes in phytosterols are small and not clearly correlated with STL levels.
The GC‐MS analysis revealed that squalene accumulation in the leaves of the chicory lines was very low and was only observed in line PL6 at 13 ± 2 μg/g FW. None of the other KO lines showed any increased squalene accumulation in the leaves. No additional accumulation of monoterpenes, sesquiterpenes, triterpenes or sterols beyond WT levels was observed in the leaves of chicory CiGAS deletion lines.
GAS inactivation leads to an increase of phenolic compounds in chicory roots
The PDA spectrum of chicory methanolic extract was examined at the wavelength of 320 nm for detection of phenolic compounds and corresponding compounds were identified in the LC‐MS chromatogram. In chicory root tissues, 3,5‐dicaffeoylquinic acid, chicoric acid and chlorogenic acid were observed as major phenolic compounds (Tables 2 and 3) in accordance to previous findings (Legrand et al., 2016). Surprisingly, a statistically significant increase in phenolic compounds was observed in the CiGAS deletion lines PL6 and RN12 in the root tissue. Lines PL6 and RN12 were subjected to antioxidant activity analyses, and their radical scavenging activities were compared with control line PL4. Antioxidant activity showed a clear positive correlation with phenolic compound concentration and negative correlation with sesquiterpene lactones in both roots and leaves (Figure S6). Correlation coefficients with IC50 values were R 2 = 0.996…0.997 for sesquiterpene lactones and R 2 = −0.807… −0.986 for phenolic compounds. No statistically significant increase in phenolic compound was observed in the leaves.
Discussion
In this report, we have used the CRISPR/Cas9 system to inactivate all of the CiGAS genes in chicory resulting in a strong reduction in the levels of STL production throughout the plant. During inulin extraction from chicory roots, the STLs are also co‐isolated, but they need to be removed because of their strong bitter taste which impairs the quality of the inulin. This results in a more intense process consuming significantly more energy, and giving additional costs for their removal. The CiGAS mutant lines described here no longer produce significant amounts of STLs and could therefore contribute to simplify the inulin extraction process substantially.
The CRISPR/Cas9 system combined with efficient protoplast regeneration is a powerful tool to elucidate gene function. Chicory protoplasts divide robustly and quickly, enabling the production of mutant lines within 5 to 6 months. As the lines are derived from individual cells, we observe very low levels of chimerism that enables screening to be performed on the primary regenerants. In most cases, the regenerated plants contained mutations in both alleles of a gene, avoiding the need for selfing to produce homozygous mutants, which is particularly cumbersome in a perennial species such as chicory that requires long periods of vernalization and the creation of mutations in two compatible lines. Additionally, as demonstrated by this study, the use of gRNA‐Cas9 complex is advantageous since no foreign DNA is introduced as only transient RNA and protein are used to produce mutations. Other approaches for gene down‐regulation in chicory, such as RNAi, have previously been used. Bogdanovic et al. (2020) used microRNA to down‐regulate the expression of the CiGAS‐short and CiGAS‐long genes, but this approach gave only partial silencing and led to very variable levels of STL production compared with this study.
The chicory genome contains four copies of the CiGAS gene. Three copies of the short CiGAS gene were mapped to a single locus on linkage group LG3, while the CiGAS‐long gene was found to be located in linkage group LG9 (Bogdanović et al., 2019; Cadalen et al., 2010). The relative contributions of the CiGAS‐short and CiGAs‐long enzymes on STL production were unknown before this study. Differences in the STL levels in the various gene‐edited lines shed light on the relative importance of the CiGAS‐short and CiGAS‐long genes for STL production. It is clear that CiGAS‐short genes account for the majority of STL production, while CiGAS‐long (CiGAS‐L) does not appear to play a major role. Based on previous reports, both the CiGAS‐S1 and CiGAS‐L genes are expressed in the roots, increasing in the cortex and the epidermis where the levels of STLs are known to be the highest. In contrast, only the CiGAS‐L gene is expressed in chicory leaves (Bogdanović et al., 2019; Bouwmeester et al., 2002). In both tissues, these genes also seem to be expressed in all cell types and in general CiGAS‐L is expressed (one‐ to threefold) at a higher level (Bogdanović et al., 2019). Based on the expression pattern, we expected that CiGAS‐L would account for the majority of the CiGAS activity in chicory, particularly in the leaves. However, the analysis of gene‐edited lines generated in this study shows that the CiGAS‐short genes are responsible for STL production in the leaves even though they do not appear to be expressed in this tissue. The CiGAS‐L gene seems to play a minimal role in STL production, even though it is expressed throughout the plant. Therefore, the exact role of CiGAS‐L in the plant, and its role in STL accumulation in chicory is not yet clear. The CiGAS‐L proteins may be produced under specific environmental challenges, and therefore, it would be interesting to repeat the STL analysis on these mutant lines grown under different growth conditions. In plants where all 8 alleles of the CiGAS genes were mutated, a residual amount of STLs was still observed indicating that yet another terpene synthase from chicory might produce small amounts of germacrene A. It is known that sesquiterpene synthases often produce multiple products and therefore often produce in addition to their major products, several other different sesquiterpenes from the common FPP substrate (Degenhardt et al., 2009; Durairaj et al., 2019). As STLs account for the bitterness of chicory, these results provide an important insight into which genes are important to fine‐tune the chicory taste profile of vegetable chicory for the consumer. The CiGAS‐short genes which have a major contribution in the biosynthesis of chicory STLS are co‐localized in the chicory genome which would offer advantages for chicory breeding programmes aimed to reduce STLs to facilitate inulin extraction or modulate bitterness of leafy chicory varieties such as Belgian endive, radicchio or endive. These results also raise some interesting questions on the role of STLs in the Asteraceae. STL production and diversification are thought to underlie the ability of this group of plants to colonize new habitats, and although STLs may be important in particular environments these results indicate that they are not essential for chicory growth and development, at least not under the conditions applied in this study.
Surprisingly, CiGAS inactivation also led to significant increase in phenolic compounds in chicory roots, which are biosynthesized via an unrelated pathway. A similar finding was reported upon silencing of other terpene synthase genes (TPS9 and TPS12) in tomato where up‐regulation of several genes involved in flavonoid biosynthesis was demonstrated (Coppola et al., 2018). Similarly, a systems biology study of the terpenoid and shikimate pathway in basil showed that differential gene expression at major metabolic branch points is responsible for controlling the overall production of phenylpropanoid versus terpenoid constituents in the glandular trichomes of the different basil lines (Xie et al., 2008). It seems that also in chicory the regulation of terpene and shikimate pathways is interdependent. Although the phenolic compounds increase the antioxidant activity, their contribution to bitter taste needs to be evaluated and they may still need to be removed during inulin processing.
We found that the roots of chicory lines with reduced levels of STLs showed a corresponding increase in the production of squalene. Squalene (C30) is synthesized by the condensation of two molecules of FPP (C15) through the action of the enzyme squalene synthase (SQS). This enzyme is located in the membrane of the endoplasmic reticulum, and squalene is the first product in the sterol synthesis pathway, leading to the production of triterpenes and plant sterols, such as sitosterol, campesterol and stigmasterol. Apparently, inactivation of CiGAS terpene synthase led to an accumulation of FPP in the cytosol which is then available to squalene synthase for squalene production. This is somewhat surprising as normally FFP levels are tightly regulated. In plants, post‐translational feedback regulation by FPP controls degradation of the rate‐limiting enzyme of the mevalonate pathway hydroxymethylglutaryl CoA reductase (HMGR), and therefore, we would expect an increase in FPP levels to decrease the flux through this pathway (Gardner and Hampton, 1999). Alternatively, reduction in the CiGAS activity might have caused a compensatory effect on the overexpression of the squalene synthase. Silencing of a different terpene synthase (amorphadiene synthase) in Artemisia annua did not result in increased accumulation of squalene (Catania et al., 2018). On the other hand, sesquiterpene biosynthesis and squalene biosynthesis have been shown to be linked in Nicotiana benthamiana, where silencing of squalene synthase led to enhanced yields of sesquiterpenes (Cankar et al., 2015). Accumulation of squalene may be specific for laticifer‐rich chicory roots as we also did not observe increased accumulation of squalene in the leaves of the mutant lines.
This study clearly demonstrates that the CRISPR/Cas9 system is an efficient system for the manipulation of biochemical pathways in chicory. Inactivation of STL production yielded chicory lines which have hitherto not been achieved by traditional breeding, from which inulin can be more cost‐effectively extracted, opening the possibility that this health‐promoting dietary fibre can be used in a wider range of food products.
Experimental procedures
CRISPR/Cas9 reagent design
Five guide RNAs (sgRNAs) targeting different positions in exon 4 of the CiGAS‐S1 gene were designed and synthesized on plasmids under control of the A. thaliana U6 promoter (NCBI Genbank accession numbers MH350853–MH350858). For SpCas9 expression, a plasmid was made carrying the A. thaliana codon‐optimized SpCas9 ORF with a C‐terminal NLS driven by the constitutive parsley ubiquitin promoter. After testing for activity, three guides were selected for the mutagenesis experiments and these were synthesized together on a single plasmid separated by tRNA sequences and expressed using the A. thaliana U6 promoter.
Isolation and transfection of chicory protoplasts
Cichorium intybus subsp. intybus var. sativum cultivar Orchies was used in the study. Orchies is an economically relevant inulin producing cultivar. In vitro clone C37 shows typical characteristics of the Orchies cultivar (European Union Community Plant Variety Office, 2005) and was selected due to high transformation and regeneration capacity. Protoplast isolation, transfection and culture were performed as previously described (Deryckere et al., 2012; Frearson et al., 1973; Kao and Michayluk, 1975; Negrutiu et al., 1987; Nenz et al., 2000) with several modifications. In vitro shoot cultures of Cichorium intybus var. sativum (Orchies C37) were maintained on MS20 medium with 0.8% agar in high plastic jars at 16/8 h photoperiod under 100 µmol m−2 s−1 PPFD at 25 °C and 60–70% RH. Young leaves (2–3) were harvested, placed in a dish containing 5 mL CPW9M medium (Frearson et al., 1973) and were gently sliced perpendicularly to the mid nerve to ease the penetration of the enzyme mixture. Sliced leaves (approximately 0.2–0.3 g fwt) were transferred to a dish containing 20 mL CPW9M and an enzyme mixture (1% (w/v) Cellulase Onozuka RS, 0.2% (w/v) Macerozyme Onozuka R10). Digestion was carried out at room temperature for 14–16 h, in the dark. The protoplasts were filtered through a 50 µm nylon sieve and then harvested by centrifugation for 5 min at 85 ×g. Protoplasts were then resuspended in 1 mL CPW9M medium and then added to a tube containing a mix of 87% CPW15S and 13% CPW9M. This was then centrifuged for 10 min at 85 ×g at room temperature. Live protoplasts were then harvested from the interface layer and transferred to a fresh tube and mixed with 11 mL CPW9M. The protoplast density was determined in a haemocytometer. To assess the chicory protoplast transformation efficiency, different concentrations of a fluorescent reporter plasmid (35S::3xYFP) were transfected. This plasmid DNA was mixed with 0.25 × 106 protoplasts in a total volume of 250 µL MaMg medium and 250 µL PEG solution (400 g/L polyethylene glycol4000, Sigma‐Aldrich #81240; 70 g/L mannitol; 0.1 m Ca(NO3)2) was then added. The transfection was allowed to take place for 20 min at room temperature followed by the addition of 5 mL 0.275 m Ca(NO3)2 solution, which was thoroughly but gently mixed in. The protoplasts were harvested by centrifugation for 5 min at 85 ×g and resuspended in 2 mL K1Cg culture medium. Fluorescent protoplasts were quantified using flow cytometry (Accuri C6, BD). The highest number of fluorescent protoplasts was detected after transfection of 60 µg plasmid DNA, and the experiment was repeated with six different batches of protoplasts to assess the robustness of the transfection.
For the plasmid approach, 60 µg of the SpCas9 expression plasmid and 20 µg of a plasmid carrying a sgRNA cassette were mixed in a 50 µl final volume. For the RNP approach, the sgRNA1 guide was generated in vitro. The unmodified sgRNA1 was generated using an oligonucleotide and the EnGen sgRNA Synthesis kit (New England Biolabs, E3322V). The protected version of sgRNA1 contained three phosphorothioate linkages and 2’‐O‐methyl RNA modifications at both ends and was synthesized (www.synthego.com). Then, 10 µg of guide RNA was combined with 10 µg SpCas9 (New England Biolabs, M0646T) in a 20 µL volume. For the transfections, the CRISPR reagents were then introduced into chicory protoplasts using PEG transfection as described above. When protoplasts were used for sequencing experiments, they were maintained in the culture medium for 48 h and then harvested for genomic DNA isolation. For regeneration, the protoplasts were centrifuged at 85 ×g for 5 min at room temperature and then resuspended at a density of 0.25 × 106 cells in 5 mL 9 m medium. An equal volume of alginate solution was added dropwise and mixed thoroughly, and 1ml of the mixture was layered on a Ca‐agar plate (5 cm dish), dispersing the mixture evenly over the whole plate surface to form a disc. The alginate was allowed to polymerize for one hour and was then transferred to a 5 mL culture dish containing 1 mL K1Cg medium. After 7 days of culture in the dark at 28 °C, the liquid culture medium was replaced with 4 mL K5CgK medium and the discs were cultured for a further 14 days using the same conditions. The discs were cut into 5 mm broad strips and transferred to 9 cm plates with B5g‐10‐0,2‐SP‐NB medium, two discs per plate. These were incubated at 25 °C in the dark for three weeks to form microcalli. For each experiment, approximately 500 microcalli were picked with tweezers and transferred to MS10‐IB plates and incubated at 25 °C under low light for the first week followed by full light for the remainder of the regeneration. Calli were transferred to fresh MS10‐IB medium every 3–4 weeks until regeneration. Overall, more than 80% of the calli developed shoots and once a healthy shoot had been harvested the callus was discarded. The developing shootlets were rooted on MS20 medium, karyotyped and then genotyped. Plants obtained by plasmid transfection were marked with the code PL, and plants obtained by the RNP approach were marked with code RN. The regenerated plants lacking CiGAS mutations were used as controls. Regenerated in vitro plants were grown under the same conditions as the plants used for protoplast isolation. Some wild‐type plants regenerated from untreated Orchies 37 protoplasts varied in size, which was attributed to somaclonal variation induced by regeneration itself. However, most regenerated plants developed normally in tissue culture. Leaves of 2 clones for each genotype of well‐established plants in tissue culture were sampled for metabolomics analysis for 17 GAS mutants and 7 WT lines. A selection of five WT and five GAS mutant lines were transferred to the greenhouse and were grown in pots for a period of 5 months under standard greenhouse conditions during the summer period, until taproots of approximately 15 cm in length developed which were sampled in duplicate.
Genotyping chicory plants
Genomic DNA was isolated from protoplasts or regenerated chicory plants using the Maxwell Plant DNA kit (Promega), and the target sites in each gene were then amplified separately using specific primers (Table S1). A nested PCR was done on each PCR product using the appropriate forward and reverse primers, and a final third PCR was performed with barcoded Illumina primers to enable later identification of the sequences. All of these PCR products were then pooled and paired‐end sequenced on an Illumina MiSeq apparatus. The sequences were analysed for the presence of indel mutations at the target sites.
Analysis of sesquiterpene lactones and phenolic compounds by LC‐MS
Chicory leaf and root material (100 mg) was frozen and powdered in liquid nitrogen. Extraction was performed using 77% methanol containing formic acid (0.1%), and the samples were then vortexed, sonicated for 15 min and then centrifuged at 21 000 g at room temperature. The clear supernatant was transferred to a fresh vial and used for LC–MS analysis. LC‐MS analysis was performed using the LC‐PDA‐LTQ‐Orbitrap FTMS system (Thermo Scientific), which consist of an Acquity UPLC (H‐Class) with Acquity elambda photodiode array detector (220–600 nm) connected to a LTQ/Orbitrap XL hybrid mass spectrometer equipped with an electrospray ionizator (ESI). The injection volume was 5 μL. Chromatographic separation was on a reversed phase column (Luna C18/2,3 μ, 2.0 × 150 mm; Phenomenex, Torrance, CA, USA) at 40 °C. Degassed eluent A [ultra‐pure water: formic acid (1000:1, v/v)] and eluent B [acetonitrile:formic acid (1000:1, v/v)] were used at a flow rate of 0.19 mL min−1. A linear gradient from 5 to 75% acetonitrile (v/v) in 45 min was applied, which was followed by 15 min of washing and equilibration. FTMS full scans (m/z 90.00–1350.00) were recorded with a resolution of 60 000. The samples were analysed for the presence of six STLs (lactucin, lactucin 15‐oxalate, 8‐deoxylactucin, 8‐deoxylactucin 15‐oxalate, lactucopicrin and lactucopicrin 15‐oxalate). Quantification of the STL and phenolic compounds was performed by the integration of the selective ion peak (5 ppm) in Excalibur software (Thermo Fischer Scientific, Waltham, MA, USA). Quantification of metabolites was performed by comparison to a standard curve prepared from authentic standards of chicoric acid (Sigma Aldrich, Saint Louis, MO, USA), 3,5‐dicaffeoylquinic acid, chlorogenic acid (Sigma), lactucin (Extrasynthese) and lactucopicrin (Extrasynthese). Other compounds for which authentic standards are not available were relatively quantified and relative abundance is presented. Data were subjected to statistical analysis using SPSS software (version 23 for Windows; IBM, Armonk, NY, USA) for the analysis of variance (ANOVA). Tukey’s post hoc test (P > 0.05) was used to analyse differences between lines.
GC‐MS analysis of chicory root and leaf extracts
Chicory root and leaf material (300 mg) were frozen and powdered in liquid N2. The samples were then extracted with 1.5 mL of hexane: ethyl acetate mixture (v/v 85:15). The samples were sonicated for 15 min and centrifuged for 10 min at 1200 g. The extracts were dried over a Na2SO4 column prepared in a glass wool plugged glass pipette. Analytes from 1 μL samples were separated using a gas chromatograph (5890 series II; Hewlett‐Packard, Palo Alto, CA, USA) equipped with a 30 m × 0.25 mm, 0.25 mm film thickness column (ZB‐5; Phenomenex) using helium as carrier gas at flow rate of 1 mL/min. The injector was used in splitless mode with the inlet temperature set to 250 °C. The initial oven temperature of 45 °C was increased after 1 min to 310 °C at a rate of 10 °C/min and held for 5 min at 300 °C. The GC was coupled to a mass‐selective detector (model 5972A; Hewlett‐Packard), scanning from 45 to 500 atomic mass units. Experimental samples were compared with authentic standards of squalene (Sigma‐Aldrich, Saint Louis, MO, USA), campesterol (Sigma‐Aldrich, Saint Louis, MO, USA), stigmasterol (Extrasynthese) and sitosterol (Extrasynthese) for verification.
Analyses of inulin by size exclusion chromatography (SEC)
The molar mass measurements of inulin containing samples were performed with SEC using alkaline eluent. The lyophilized chicory root samples were dissolved into 0.1 m NaOH with the final concentration of 1 mg/mL. Samples were filtered (0.45 µm) prior the measurement. The SEC measurements were performed in 0.1 m NaOH as an eluent (pH 13, 0.5 mL/min, T = 25 °C) using PSS MCX 1000 & 100 000 Å columns with a pre‐column. The elution curves were detected using Waters 2410 RI detector. The molar mass distributions (MMD) were calculated against 8 × pullulan (6100–708 000 g/mol) standards, using Waters Empower 3 software. The data of inulin polymerization degree (DP) were expressed as weight average DP Eq. (1) and number average DP Eq. (2) using molecular weight of dehydrated glucose (162 g/mol):
| (1) |
| (2) |
where Mw is weight average molar mass (ΣM·w/Σw) and Mn is number average molar mass (ΣM·n/Σn).
Radical scavenging activity
Antioxidant activity of selected GAS‐edited chicory root and leaf samples was evaluated using the DPPH radical scavenging method as described in Shimamura et al., (2014) with modifications. Lyophilized root or leaf samples were disrupted using steel beads and a Retsch mill (MM301, Haan, Germany) and extracted with methanol (50 mg/mL) for 30 min in magnetic stirring. After centrifugation (700 g, 10 min), the supernatant was collected and diluted with methanol (roots 1:5, 1:10, 1:25, 1:50, 1:100, 1:125 and leaves 1:5, 1:10, 1:25, 1:50, 1:100, 1:200, 1:250). The DPPH solution (23.8 mg of DPPH/100 mL of methanol) was diluted 1:5 with methanol, and 0.1 mL of the diluted solution was pipetted on multiwell plate. Diluted extracts (0.1 mL) were added to DPPH solution. Samples were incubated at room temperature in the dark for 30 min. The absorbance of the samples was measured at 515 nm in a Multiskan spectrophotometer (EX 355; Thermo Fischer). The EC50 absorbance value was determined for trolox (6‐hydroxy‐2,5,7,8‐tetramethylchroman‐2‐carboxylic acid) within concentration levels 0–12.9 µg/mL, and the radical scavenging activity of the samples was expressed as EC50 concentrations (mg/mL). Thus, the higher the EC50 value, the lower the antioxidant activity.
Conflict of interest
The authors have submitted a patent application (No. WO2021122982) regarding the reduction in terpenes via gene editing of the GAS gene.
Author contributions
KC, PB, JB, IM, MB and DB devised the study. KC, PB, RS, JH and SH performed the experiments and analysed the data. KC, PB and DB drafted the manuscript. All authors reviewed the manuscript and agreed to the final version of the manuscript.
Supporting information
Figure S1 Design and efficiency of guide RNAs for CiGAS.
Figure S2 Sequence of the cassette carrying three guide RNAs.
Figure S3 Mutations in the CiGAS genes of regenerated chicory plants.
Figure S4 Molar mass distributions of inulin (in 0.1 m NaOH) against Pullulan standards for chicory lines PL4, PL6 and RN12.
Figure S5 Comparison of mass spectrum of (a) authentic squalene standard to (b) the mass spectrum of squalene produced by chicory.
Figure S6 Correlation of radical scavenging activity (IC50 values) and total content of sesquiterpene lactones and phenolic compounds in chicory lines PL4, PL6 and RN12.
Table S1 Primers used for genotyping.
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 760891 (H2020‐NMBP‐BIOTEC‐07‐2017: New Plant Breeding Techniques (NPBT) in molecular farming: Multipurpose crops for industrial bioproducts). Matthew de Roode is acknowledged for his valuable contribution on market data and uses of inulin. Tuuli Teikari and Bert Schipper are acknowledged for skillful technical assistance. We thank Atte Mikkelson for inulin analyses.
Cankar, K. , Bundock, P. , Sevenier, R. , Häkkinen, S. T. , Hakkert, J. C. , Beekwilder, J. , van der Meer, I. M. , de Both, M. and Bosch, D. (2021) Inactivation of the germacrene A synthase genes by CRISPR/Cas9 eliminates the biosynthesis of sesquiterpene lactones in Cichorium intybus L. Plant Biotechnol. J., 10.1111/pbi.13670
References
- Ahmed, W. and Rashid, S. (2019) Functional and therapeutic potential of inulin: a comprehensive review. Crit. Rev. Food Sci. Nutr. 59, 1–13. [DOI] [PubMed] [Google Scholar]
- van Arkel, J. , Vergauwen, R. , Sévenier, R. , Hakkert, J.C. , van Laere, A. , Bouwmeester, H.J. , Koops, A.J. et al. (2012) Sink filling, inulin metabolizing enzymes and carbohydrate status in field grown chicory (Cichorium intybus L.). J. Plant Physiol. 169, 1520–1529. [DOI] [PubMed] [Google Scholar]
- Bernard, G. , Gagneul, D. , Alves Dos Santos, H. , Etienne, A. , Hilbert, J.‐L. and Rambaud, C. (2019) Efficient genome editing using CRISPR/Cas9 technology in chicory. Int. J. Mol. Sci. 20, 1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović, M. , Cankar, K. , Todorović, S. , Dragicević, M. , Simonović, A. , van Houwelingen, A. , Schijlen, E. et al. (2019) Tissue specific expression and genomic organization of bitter sesquiterpene lactone biosynthesis in Cichorium intybus L. (Asteraceae). Ind. Crops Prod. 129, 253–260. [Google Scholar]
- Bogdanovic, M. , Cankar, K. , Dragicevic, M. , Bouwmeester, H. , Beekwilder, J. , Simonovic, A. and Todorovic, S. (2020) Silencing of germacrene A synthase genes reduces guaianolide oxalate content in Cichorium intybus L . GM Crops Food, 11, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester, H.J. , Kodde, J. , Verstappen, F.W. , Altug, I.G. , de Kraker, J.W. and Wallaart, T.E. (2002) Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol. 129, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn, C. , Ruttink, T. , Eeckhaut, T. , Jacobs, T. , de Keyser, E. , Goosens, A. and van Laere, K. (2020) Establishment of CRISPR/Cas9 genome editing in Witloof (Cichorium intybus var. foliosum). Front. Genome Ed. 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadalen, T. , Morchen, M. , Blassiau, C. , Clabaut, A. , Scheer, I. , Hilbert, J.L. , Hendriks, T. et al. (2010) Development of SSR markers and construction of a consensus genetic map for chicory (Cichorium intybus L.). Mol. Breeding, 25, 699–722. [Google Scholar]
- Cankar, K. , van Houwelingen, A. , Bosch, D. , Sonke, T. , Bouwmeester, H. and Beekwilder, J. (2011) A chicory cytochrome P450 mono‐oxygenase CYP71AV8 for the oxidation of (+)‐valencene. FEBS Lett. 585, 178–182. [DOI] [PubMed] [Google Scholar]
- Cankar, K. , Jongedijk, E. , Klompmaker, M. , Majdic, T. , Mumm, R. , Bouwmeester, H. , Bosch, D. et al. (2015) (+)‐Valencene production in Nicotiana benthamiana is increased by down‐regulation of competing pathways. Biotechnol. J. 10, 180–189. [DOI] [PubMed] [Google Scholar]
- Catania, T.M. , Branigan, C.A. , Stawniak, N. , Hodson, J. , Harvey, D. , Larson, T.R. , Czechowski, T. et al. (2018) Silencing amorpha‐4,11‐diene synthase genes in Artemisia annua Leads to FPP accumulation. Front. Plant Sci. 9, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick, M. , Trewin, H. , Gawthrop, F. and Wagstaff, C. (2013) Sesquiterpenoids lactones: benefits to plants and people. Int. J. Mol. Sci. 14, 12780–12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola, M. , Cascone, P. , Bossi, S. , Corrado, G. , Garonna, A.P. , Maffei, M. , Rao, R. et al. (2018) TPS genes silencing alters constitutive indirect and direct defense in tomato. Int. J. Mol. Sci. 19(9), 2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt, J. , Kollner, T.G. and Gershenzon, J. (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry, 70, 1621–1637. [DOI] [PubMed] [Google Scholar]
- Deryckere, D. , Eeckhaut, T. , van Huylenbroeck, J. and van Bockstaele, E. (2012) Low melting point agarose beads as a standard method for plantlet regeneration from protoplasts within the Cichorium genus. Plant Cell Rep. 31, 2261–2269. [DOI] [PubMed] [Google Scholar]
- Durairaj, J. , di Girolamo, A. , Bouwmeester, H.J. , de Ridder, D. , Beekwilder, J. and van Dijk, A.D. (2019) An analysis of characterized plant sesquiterpene synthases. Phytochemistry, 158, 157–165. [DOI] [PubMed] [Google Scholar]
- European Union Community Plant Variety Office . 2005. Protocol for distinctness, uniformuity and stability tests Cichorium intybus L. partim, industrial chicory CPVO‐TP/172/2. https://cpvo.europa.eu/sites/default/files/documents/cichorium_172‐2.pdf. Accessed on 01 December 2005.
- Frearson, E.M. , Power, J.B. and Cocking, E.C. (1973) The isolation, culture and regeneration of Petunia leaf protoplasts. Dev. Biol. 33, 130–137. [DOI] [PubMed] [Google Scholar]
- Gardner, R.G. and Hampton, R.Y. (1999) A highly conserved signal controls degradation of 3‐hydroxy‐3‐methylglutaryl‐coenzyme A (HMG‐CoA) reductase in eukaryotes. J. Biol. Chem. 274, 31671–31678. [DOI] [PubMed] [Google Scholar]
- Hendel, A. , Bak, R.O. , Clark, J.T. , Kennedy, A.B. , Ryan, D.E. , Roy, S. , Steinfeld, I. et al. (2015) Chemically modified guide RNAs enhance CRISPR‐Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, M. , Epping, J. , Schulze Gronover, C. , Fricke, J. , Aziz, Z. , Brillatz, T. Swyers, M. et al. (2016) A latex metabolite benefits plant fitness under root herbivore attack. PLoS Biol. 14, e1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa, N. , Gopfert, J.C. , Nguyen, D.T. , Kim, S.U. , O'Maille, P.E. , Spring, O. and Ro, D.K. (2011) Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio‐ and stereoselective hydroxylations in sesquiterpene lactone metabolism. J. Biol. Chem. 286, 21601–21611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International, E . 2020. portal.euromonitor.com. Accessed on 29 May 2020.
- Kao, K.N. and Michayluk, M.R. (1975) Nutritional requirements for growth of Vicia hajastana cells and protoplasts at a very low population density in liquid media. Planta, 126, 105–110. [DOI] [PubMed] [Google Scholar]
- de Kraker, J.W. , Franssen, M.C. , de Groot, A. , Konig, W.A. and Bouwmeester, H.J. (1998) (+)‐Germacrene A biosynthesis. The committed step in the biosynthesis of bitter sesquiterpene lactones in chicory. Plant Physiol. 117, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand, G. , Delporte, M. , Khelifi, C. , Harant, A. , Vuylsteker, C. , Morchen, M. , Hance, P. et al. (2016) Identification and characterization of five BAHD acyltransferases involved in hydroxycinnamoyl ester metabolism in chicory. Front. Plant Sci. 7, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Majdi, M. , Cankar, K. , Goedbloed, M. , Charnikhova, T. , Verstappen, F.W. , de Vos, R.C. et al. (2011) Reconstitution of the costunolide biosynthetic pathway in yeast and Nicotiana benthamiana . PLoS One, 6, e23255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, D. and Blaauwhoed, J.P. (2009) Handbook of hydrocolloids: inulin. In Woodhead Publishing Series in Food Science, Technology and Nutrition ( Phillips, G.O. and Williams, P.A. , eds), (2nd edn), pp. 829–848. Cambridge, England: Woodhead Publishing Limited. [Google Scholar]
- MINTEL GNPD database . www.mintel.com. Accessed on 29 May 2020.
- Molinaro, F. , Monterumici, C.M. , Ferrero, A. , Tabasso, S. and Negre, M. (2016) Bioherbicidal activity of a germacranolide sesquiterpene dilactone from Ambrosia artemisiifolia L. J Environ. Sci. Health B, 51, 847–852. [DOI] [PubMed] [Google Scholar]
- Negrutiu, I. , Shillito, R. , Potrykus, I. , Biasini, G. and Sala, F. (1987) Hybrid genes in the analysis of transformation conditions: I. Setting up a simple method for direct gene transfer in plant protoplasts. Plant Mol. Biol. 8, 363–373. [DOI] [PubMed] [Google Scholar]
- Nenz, E. , Varotto, S. , Lucchin, M. and Parrini, P. (2000) An efficient and rapid procedure for plantlet regeneration from chicory mesophyll protoplasts. Plant Cell Tissue Organ. Cult. 62, 85–88. [Google Scholar]
- Nguyen, D.T. , Gopfert, J.C. , Ikezawa, N. , Macnevin, G. , Kathiresan, M. , Conrad, J. , Spring, O. et al. (2010) Biochemical conservation and evolution of germacrene A oxidase in asteraceae. J. Biol. Chem. 285, 16588–16598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly, D. , Kartje, Z.J. , Ageely, E.A. , Malek‐Adamian, E. , Habibian, M. , Schofield, A. , Barkau, C.L. et al. (2019) Extensive CRISPR RNA modification reveals chemical compatibility and structure‐activity relationships for Cas9 biochemical activity. Nucleic Acids Res. 47, 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla‐Gonzalez, G.F. , dos Santos, F.A. and da Costa, F.B. (2016) Sesquiterpene lactones: more than protective plant compounds with high toxicity. Crit. Rev. Plant Sci. 35, 0735–2689. [Google Scholar]
- Prasifka, J.R. , Spring, O. , Conrad, J. , Cook, L.W. , Palmquist, D.E. and Foley, M.E. (2015) Sesquiterpene lactone composition of wild and cultivated sunflowers and biological activity against an insect pest. J. Agric. Food Chem. 63, 4042–4049. [DOI] [PubMed] [Google Scholar]
- Roberfroid, M.B. (2002) Functional foods: concepts and application to inulin and oligofructose. Br. J. Nutr. 87(Suppl 2), S139–S143. [DOI] [PubMed] [Google Scholar]
- Roberfroid, M.B. (2007) Inulin‐type fructans: functional food ingredients. J. Nutr. 137, 2493S–2502S. [DOI] [PubMed] [Google Scholar]
- Sessa, R.A. , Bennett, M.H. , Lewis, M.J. , Mansfield, J.W. and Beale, M.H. (2000) Metabolite profiling of sesquiterpene lactones from Lactuca species. Major latex components are novel oxalate and sulfate conjugates of lactucin and its derivatives. J. Biol. Chem. 275, 26877–26884. [DOI] [PubMed] [Google Scholar]
- Shimamura, T. , Sumikura, Y. , Yamazaki, T. , Tada, A. , Kashiwagi, T. , Ishikawa, H. , Matsui, T. et al. (2014) Applicability of the DPPH assay for evaluating the antioxidant capacity of food additives ‐ inter‐laboratory evaluation study. Anal. Sci. 30, 717–721. [DOI] [PubMed] [Google Scholar]
- Street, R.A. , Sidana, J. and Prinsloo, G. (2013) Cichorium intybus: traditional uses, phytochemistry, pharmacology, and toxicology. Evid. Based Complement. Alternat. Med 2013. Article ID 579319, 13. 10.1155/2013/579319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.G. , Smith, J.A. and Yonts, C.D. (2004) Chicory root yield and carbohydrate composition is influenced by cultivar selection, planting, and harvest date. Crop Sci. 44, 748–752. [Google Scholar]
- Xie, Z. , Kapteyn, J. and Gang, D.R. (2008) A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J. 54, 349–361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Design and efficiency of guide RNAs for CiGAS.
Figure S2 Sequence of the cassette carrying three guide RNAs.
Figure S3 Mutations in the CiGAS genes of regenerated chicory plants.
Figure S4 Molar mass distributions of inulin (in 0.1 m NaOH) against Pullulan standards for chicory lines PL4, PL6 and RN12.
Figure S5 Comparison of mass spectrum of (a) authentic squalene standard to (b) the mass spectrum of squalene produced by chicory.
Figure S6 Correlation of radical scavenging activity (IC50 values) and total content of sesquiterpene lactones and phenolic compounds in chicory lines PL4, PL6 and RN12.
Table S1 Primers used for genotyping.


