Plant height and branch angle are the key factors of rapeseed plant architecture that affect plant density and lodging, which are crucial for rapeseed yield. In China, ˜80% of growing areas are the semi‐winter type (Bol‐type), with taller plants that are easily affected by lodging and limit mechanical harvesting, especially of hybrids. Breeders have, therefore, been seeking an ‘ideotype’ for rapeseed breeding, deeming that semi‐dwarf and compact architecture could benefit yield (Fu and Zhou, 2013; Li et al., 2019). However, no optimized plant architecture has been identified in rapeseed germplasm. In this study, we used the CRISPR/Cas9 gene editing system to knock out the BnaA03.BP gene [a homolog of Arabidopsis BREVIPEDICELLUS (BP)] creating a novel germplasm for optimizing rapeseed plant architecture.
BP encodes a knotted1‐like homeobox gene that plays a key role in the regulation of leaf morphogenesis and pedicel bending in Arabidopsis (Lincoln et al., 1994; Venglat et al., 2002). Blast analysis identified two close homologs of BP in the rapeseed genome (ZS11A03G024840 and ZS11C03G030900), which we named BnaA03.BP and BnaC03.BP. The two BnaBPs are highly conserved, sharing 87.5% and 86.5% identity with BP at the amino acid level respectively. In addition, the two BnaBP homologs share 98.1% identity in their nucleotide sequences with each other. RNA‐seq and qRT‐PCR analysis showed that transcripts of both BnaBPs were more abundant in wild type (WT) stems, and both genes displayed similar expression patterns (Figure 1a). To explore the roles of BnaBP genes, we first over‐expressed BnaA03.BP in rapeseed (862, a spring variety) under control of the ubi promoter by Agrobacterium‐mediated transformation. Nine positive transgenic lines were obtained. None showed changes in leaf morphogenesis, but two lines exhibited dwarfism, erect axillary buds, downward siliques and reduced fertility (Figure 1b). Further, qRT‐PCR analysis showed that transcript levels of both BnaA03.BP and BnaC03.BP were significantly reduced in these two lines relative to WT, indicating co‐suppression occurred in these lines (Figure 1c; Krol et al., 1990). These morphological changes in plant height, pedicel bending and fertility changes are phenotypic of the bp mutants (Venglat et al., 2002), suggesting that the two BnaBPs probably have similar functions as BP in regulating stem elongation and pedicel bending. Significantly, the erect axillary buds that occurred in these transgenic plants provided strong evidence that down‐regulation of BnaBP genes could decrease the branch angle to create more compact plants.
Figure 1.
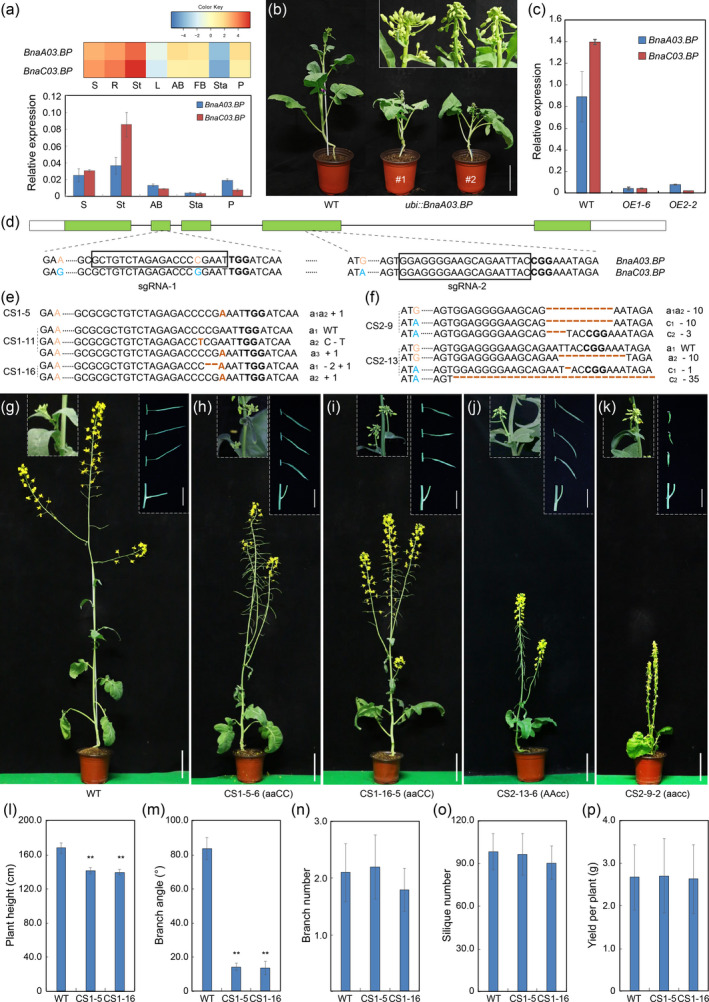
CRISPR/Cas9‐targeted mutagenesis of the BnaBP genes confers semi‐dwarf and compact architecture in rapeseed. (a) Heat‐map and qRT‐PCR analysis of the expression of BnaBP genes in various tissues of wild type (WT). Error bars ± standard deviations (n = 3). AB, axillary bud; FB, flower bud; L, leaf; P, pistil; R, root; S, seedling; St, stem; Sta, stamen. The BnaTMA7 gene was used as an internal control. (b) Morphological comparison of WT and ubi: BnaA03.BP transgenic lines at bolting stage. Bar = 15 cm. (c) qRT‐PCR of BnaBPs in pedicels from 6‐week‐old T2 plants. The BnaTMA7 gene was used as an internal control. (d) CRISPR/Cas9 sgRNA‐1 targets the second exon of BnaA03.BP and sgRNA‐2 targets the fourth exon of both BnaBPs. The orange or blue colours indicate the SNP upstream or on the sgRNA target to distinguish the homologous sequences. The protospacer adjacent motif (PAM) is indicated in bold. The box indicates the target sequences. (e–f) Sequencing of the BnaBP sites targeted by sgRNA‐1/2. Brown colours and hyphens in target sequences indicate insertions and deletions respectively. a1, a2, c1 and c2 indicate the four BnaBP alleles respectively. a3 indicates the chimeric allele. The remaining editing events are not shown in this figure. (g–k) Morphological comparison of WT, BnaA03.BP (aaCC), BnaC03.BP (AAcc) and BnaBPs (aacc) homozygous mutant plants at the reproductive stage. Bars = 15 cm. Dashed regions from left to right and top to bottom are as follows: morphology of axillary buds, pedicel and branch angle. Bars = 5 cm. (l–p) Statistical analysis of plant height, branch angle, branch number, silique number and yield per plant in WT, CS1‐5‐6‐2 and CS1‐16‐5‐1 homozygous plants in the green house. Error bars ± standard deviation (n = 10). Student’s t‐test was used for statistical analysis (*P ≤ 0.05; **P ≤ 0.01).
CRISPR/Cas9‐mediated gene editing has been proved to be a useful tool to generate gene mutations in rapeseed (Wu et al., 2020; Yang et al., 2017). To further verify these phenotypic changes resulting from changes in BnaBP gene expression, we designed two sgRNAs and constructed CRISPR/Cas9 vectors to knock them out. SgRNA‐1 (20 bp) targets a specific region within the second exon of BnaA03.BP to edit it individually, and sgRNA‐2 (19 bp) was designed to target a conserved region in the fourth exons of BnaA03.BP and BnaC03.BP to knock out both homologs (Figure 1d). The two Cas9‐sgRNA‐1/2 (CS1/2) vectors were individually introduced into rapeseed by Agrobacterium transformation. With CS1, editing events including nucleotide insertions, deletions and substitutions occurred in 19 of the 24 positive T0 lines (Figure 1e). Most of the mutations caused frameshifts in the coding region, leading to a premature stop codon. Notably, the majority of mutation events were a single‐nucleotide insertion (A nucleotide). With CS2, 17 positive lines were obtained, nine of which simultaneously edited both BnaA03.BP and BnaC03.BP. In these edited lines, relatively large deletions (7‐35 bps) that lost the PAM sequence (NGG) occurred (Figure 1f). Similarly, most of these mutations also caused frameshifts predicted to result in truncated proteins.
During vegetative growth, the leaf morphology of mutant lines was indistinguishable from WT. At the reproductive stage, as expected, the plants with homozygous or bi‐allelic mutations in the BnaA03.BP gene (aaCC) displayed similar mutant phenotypes, showing semi‐dwarf, erect axillary buds and slightly drooping siliques (e.g. CS1‐5/16), while the remaining heterozygous plants resembled wild type (Figure 1g–i). Strikingly, in the CS2 T0 generation, four lines were homozygous mutations, which simultaneously knocked out both BnaBP genes. These exhibited more severe phenotypes than those of the BnaA03.BP mutants, showing extremely dwarf, smaller branch angles and severely drooping and short siliques with sterility, which were consistent with the homozygous plants (aacc) from the self‐pollinated progeny of CS2‐9 (Figure 1k). In addition, the homozygous plants that only knocked out the BnaC03.BP gene (AAcc) from the self‐pollinated progeny of CS2‐13 heterozygous plants displayed similar phenotypes to CS1‐5 (aaCC), but were shorter in height (Figure 1j). Combining the expression levels of the two BnaBP genes with the phenotypic observations, we suggest that both of the homologs participate in the regulation of plant height, branch angle and pedicel bending with a dosage effect, but have redundant roles in controlling plant fertility.
Theoretically, knocking out the BnaA03.BP genes individually could obtain more moderate phenotypes with potential uses in rapeseed breeding. We next selected non‐transgenic plants (T‐DNA free) with mutant phenotypes in CS1‐5/16 segregating populations. The Cas9‐induced mutations in BnaBP.A03 were clearly heritable. In the T3 generation, we investigated several agronomic traits including plant height, branch angle, branch number, silique number and yield per plant at the mature stage. The plant height was about 15.8%–16.9% shorter in CS1‐5 and CS1‐16 compared with WT (Figure 1l); the branch angle was significantly decreased from 84° (WT) to 14° in CS1‐5/16 plants (Figure 1m), whereas there were no significant changes in the branch number, silique number and yield per plant between WT and the mutant plants (Figure 1n–p). These results collectively demonstrated that knock out of BnaA03.BP can optimize rapeseed plant architecture with potential for dense planting. Moreover, no nonsense mutations were found in any BnaA03.BP exons in our 531 rapeseed germplasm accessions, suggesting gene editing is the most efficient way to create new germplasm in rapeseed.
In summary, we designed sgRNAs to edit the two rapeseed BnaBP genes using the CRISPR/Cas9 system and analysed phenotype variations. Our results showed that knocking out BnaA03.BP genes individually could obtain semi‐dwarf and compact plant architecture without any other inferior traits. We also obtained stably inherited mutant lines that eliminated T‐DNA in the segregating offspring, and backcrossed these mutations into a widely planted variety (Zhong shuang11, a semi‐winter type) for further field agronomic trait characterization in the future. This is the first report of using CRISPR/Cas9 technology to create semi‐dwarf and compact inflorescence germplasm resources in rapeseed. Our study provides new insights into potential strategies for optimizing rapeseed plant architecture.
Conflict of interest
The authors declare no competing financial interest.
Author contributions
M.Z. and W.H. designed the experiments; S.F., L.Z., M.T., Y.C. and J.L. performed the experiments; H.L. analysed the genome and sequencing data; S.F. and M.Z. wrote the manuscript and J.L., W.T., H.W., W.H. and M.Z. revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31801402) and the Key Research Program & Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS‐ZDRW202105).
Fan, S. , Zhang, L. , Tang, M. , Cai, Y. , Liu, J. , Liu, H. , Liu, J. , Terzaghi, W. , Wang, H. , Hua, W. and Zheng, M. (2021) CRISPR/Cas9‐targeted mutagenesis of the BnaA03.BP gene confers semi‐dwarf and compact architecture to rapeseed (Brassica napus L.). Plant Biotechnol. J., 10.1111/pbi.13703
Contributor Information
Wei Hua, Email: huawei@oilcrops.cn.
Ming Zheng, Email: zhengming@caas.cn.
References
- Fu, T. and Zhou, Y. (2013) Progress and future development of hybrid rapeseed in China. Eng. Sci. 11, 13–18. [Google Scholar]
- Krol, A. , Mur, L. , Beld, M. , Mol, J. and Stuitje, A. (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell, 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Li, J. , Song, J. , Zhao, B. , Guo, C. , Wang, B. , Zhang, Q. et al. (2019) An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol. 222, 837–851. [DOI] [PubMed] [Google Scholar]
- Lincoln, C. , Long, J. , Yamaguchi, J. , Serikawa, K. and Hake, S. (1994) A knotted1‐like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell, 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat, S.P. , Dumonceaux, T. , Rozwadowski, K. , Parnell, L. , Babic, V. , Keller, W. , Martienssen, R. et al. (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl Acad. Sci. USA, 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Chen, C. , Xian, G. , Liu, D. , Lin, L. , Yin, S. , Sun, Q. et al. (2020) Engineering herbicide‐resistant oilseed rape by CRISPR/Cas9‐mediated cytosine base‐editing. Plant Biotechnol. J. 18, 1857–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Zhu, K. , Li, H. , Han, S. , Meng, Q. , Khan, S.U. , Fan, C. et al. (2017) Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]


