Highlights
-
•
Gastric, microbial and plant-based enzymes were used to produce Amaranth protein hydrolysates (APHs).
-
•
APHs displayed enhanced cholesterol esterase (CEase) and pancreatic lipase (PL) inhibitory activities.
-
•
Bromelain generated hydrolysates showed the highest CEase and PL inhibitory activity.
-
•
FPFPPTLGY, FGAPR, and FPFVPAPT were predicted as potential PL inhibitors and FPFVPAPT as CEase inhibitor.
Keywords: Amaranth hydrolysates, Bioactive peptides, Pancreatic lipase, Cholesterol esterase, HPLC
Abstract
Human diet is undergoing a shift towards plant-based diet as a sustainable source of protein compared to animal-derived protein. In this study, cholesterol esterase (CEase) and pancreatic lipase (PL) inhibitory activities of amaranth protein hydrolysates (APHs) were studied. Bromelain, chymotrypsin, and actinase E were used for generating APHs at 2, 4 & 6 h of hydrolysis. Higher PL inhibiting potential were observed in bromelain-derived APHs (IC50 = 0.38–0.66 mg/mL) in comparison to intact amaranth proteins (IC50 = 3.93 mg/mL). Bromelain-4 h hydrolysates (AB4) demonstrated significant inhibitory potential for both CEase (IC50 = 0.47 mg/mL) and PL (IC50 = 0.48 mg/mL) activity. Peptide identification in AB-4 hydrolysate revealed that among 17 bioactive peptides, three peptides (FPFPPTLGY, FGAPR, and FPFVPAPT) were predicted as potential PL inhibitors and only one peptide (FPFVPAPT) was predicted as CEase inhibitor based on the number of substrate binding sites on active site of the enzymes. This is the first study providing insights into amaranth protein derived bioactive peptide possessing CEase and LIP inhibitory potential.
Introduction
The rapid increase in the occurrence of hypercholesterolemia, a metabolic disorder that is primarily characterized by elevated levels of triglycerides and cholesterol in the blood plasma, is becoming a public health concern globally. These metabolic conditions lead to considerable number of death across the world (Su et al., 2016). Hypercholesterolemia plays a major role in the development of several cardiovascular diseases, and also, increases the risk for other complications such as hypertension, obesity, cancer, and diabetes. Cholesterol esterase (CEase) and pancreatic lipase (PL) are the two specific enzymes that are often associated with the development of these metabolic disorders (Adisakwattana, Intrawangso, Hemrid, Chanathong, & Mäkynen, 2012). Even though, different synthetic drugs such as statins, niacin, ezetimibes and fibrates with effective CEase and PL inhibition activity have been developed for the management of hypercholesterolemia and obesity (Chiou et al., 2006, Lunagariya et al., 2014), but long-term intake of these drugs has been associated with undesirable side effects on human health. In addition to these side effects, these medicines are also not affordable for a lot of people in developing countries. Considering these concerns and limitations with synthetic drugs, it is important to identify natural compounds with anti- hypercholesterolemic properties (Birari and Bhutani, 2007, Mudgil et al., 2019a).
Currently, plant-based proteins and their derived bioactive peptides are gaining immense importance from sustainability and food security point of view. Plant species with salt and draught tolerance and possessing high nutritious value are being cultivated in many regions of the world (Soriano-Santos & Escalona-Buendía, 2015). Amaranth, a pseudo-cereal, possesses high quality protein (13–19% of protein with 90% of digestibility), and balanced amino acids profile high in lysine, which is deficient in other cereals (Kurek, Karp, Wyrwisz, & Niu, 2018). It also lacks an allergenic protein (gluten/gliadin) making it more suitable as a celiac diet (Kurek et al., 2018). Several compounds found in amaranth are considered to provide considerable benefits to human health such as decreasing plasma cholesterol levels, stimulating the immune system, exerting an antitumor activity, reducing blood glucose levels, and improving conditions of hypertension and anemia (Soriano-Santos & Escalona-Buendía, 2015). Peptides resulting from unprocessed amaranth protein hydrolysis have previously demonstrated antioxidant capacity, antihypertensive, anticarcinogenic, and antidiabetic potential (Montoya-Rodríguez, De Mejía, Dia, Reyes-Moreno, & Milán-Carrillo, 2014). A recent review by (Soriano Santos et al., 2015) and (Tovar-Pérez, Lugo-Radillo, & Aguilera-Aguirre, 2019) presented amaranth proteins and their hydrolysates with different bioactive properties including antidiabetic, antioxidant, antihypertensive, antithrombotic, immunomodulatory, and anti-inflammatory activities.
Recently, there has been an increasing interest in the investigation of plant protein-derived bioactive peptides (BAPs) such as soybean, cowpea, lupin, etc. for functional food development. For example, lupin derived BAPs provided a cholesterol-lowering effect upon digestibility and bio-availability of protein isolate (Martins et al., 2005). Another interesting investigation on the peptides derived from incomplete digestion of soybean protein showed a maximum CEase inhibition activity and minimum accumulation of cholesterol in the cell (Bhargava, Shukla, & Ohri, 2007). In another study centered on the PL inhibition of common beans protein isolates, a reduction in fat absorption and cholesterol level was observed (Frota, Mendonça, Saldiva, Cruz, & Arêas, 2008). Jakubczyk, Karaś, Złotek, & Szymanowska, 2017) identified eight peptides from fermented beans that inhibited lipase activity.
Previous studies have shown cholesterol-lowering effect of amaranth derived bioactive peptides (BAPs). For example, Mendonça, Saldiva, Cruz, & Arêas, 2009) reported a reduced concentration of cholesterol in plasma, and this was attributed to the incomplete in-vitro gastro-intestinal digestion of derived Amaranth BAPs. Soares, Mendonça, De Castro, Menezes, & Arêas, 2015) identified three peptides with 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMG-CoA reductase) activity, a key enzyme in cholesterol biosynthesis from amaranth proteins upon simulated gastric and pancreatic digestion. Regardless of the numerous reports on the health benefits of amaranth derived bioactive peptides, research focusing on the CEase and PL inhibitory activity of amaranth protein hydrolysates is still very scarce or non-existing. Moreover, sequence identification and molecular binding mechanism of CEase and PL inhibiting bioactive peptide generated from amaranth protein hydrolysates has not been carried till now. Therefore, it is critical to investigate the potential of amaranth derive protein hydrolysates and bioactive peptides for their possible role in inhibiting key enzyme (CEase and PL) involved in hypercholesteremia. Furthermore, it is important to identify the peptides from amaranth protein that are responsible for inhibiting CEase and PL. Hence, this present work aims to provide new insights into CEase and PL inhibitory potentials of amaranth protein hydrolysates produced via enzymatic (bromelain, chymotrypsin, and actinase E) hydrolysis. Furthermore, the bioactive peptides in the selected hydrolysate were identified using LC-MS-QTOF and molecular binding mechanism of peptides with CEase and PL were elucidated using in silico structural activity relationship (SAR) via molecular docking.
Material and methods
Materials
All enzymes such as pancreatic lipase (EC 3.4.23.1, source: porcine pancreas; L3126: 100–500 units/mg protein), cholesterol esterase (EC-3.1.1.13, source: porcine pancreas; 26745: 32 units/mg solid), bromelain (EC 3.4.22.33, source: pineapple stem; B4882: 3-7 units/mg solid), α-chymotrypsin (EC 3.4.21.1, source: bovine pancreas; C4129 40 units/mg solid), and actinase E from Streptomyces griseus (EC 3.4.21.81; P5147: ≥3.5 units/mg solid), p-nitrophenyl butyrate (PNPB), were obtained from Sigma Aldrich (St. Louis, MO). Additional solvents and chemicals of analytical grades were also purchased from Sigma Aldrich (St. Louis, MO). Organic Amaranth (Amaranthus cruentus) Cream-brown colored grains (Infinity Foods, West Sussex, UK, Country of Origin: Peru) were procured from a local supermarket (Al Ain, Abu Dhabi, United Arab Emirates) in different batches and mixed together to be used as a consolidated sample.
Preparation of amaranth proteins isolates (API) and production of amaranth protein hydrolysates (APHs)
Amaranth seeds were first dried in a hot air oven (40 ◦C for 3 h), milled into a fine powder using IKA A11 basic analytic grinder (Guangzhou, China), and sieved through a screen with 3 μm pore size fitted to a sieve shaker OCTAGON 200CL (Endecotts Ltd., London, UK). The procedure described by Mudgil, Omar, Kamal, Kilari, and Maqsood (2019b) was followed for isolation of amaranth proteins. Amaranth flour was defatted by solvent extraction using hexane, followed by dispersion in deionized water and pH adjustment to 11 to promote protein solubilization using 1 mol L−1 NaOH. The aqueous dispersion was stirred at 300 rpm, 25 °C for 4 h and then centrifuged at the rate of 9000×g for 20 min, at 4 °C. The proteins from the aqueous supernatant were precipitated by addition of 1 mol·L−1 HCl to settle pH at 4.5, the precipitated proteins were isolated by centrifugation. Protein pellet thus recovered was re-suspended in water (1:5, w/v), and the pH was increased to 7.0 with 1 mol·L − 1 NaOH, and finally lyophilized. Separated amaranth proteins (4% w/v) were hydrolyzed with three enzymes (bromelain, chymotrypsin and actinase E) at the optimum pH for each enzyme and hydrolysis was carried out for 2, 4, and 6 h. A two-factor (enzyme and hydrolysis time) by three-level (2, 4, and 6 h) factorial design was used to design the experiment, which generated 9 different hydrolysates. At the end of each hydrolysis time, the enzymatic reaction was stopped by heating the hydrolysate samples in a water bath (100 °C for 5 min). Resultant hydrolysates were centrifuged at a speed of 10,000×g for 15 min at 4 °C, followed by collection of supernatants which was referred to as amaranth protein hydrolysates (APHs). Protein content in the hydrolysates obtained was measured using bicinchoninic acid assay following the protocol as described by (Nongonierma et al., 2019). Hydrolysates samples were collected and stored at − 20 °C until further analysis was carried out within 2 weeks. Selected APHs were lyophilized and processed for peptide sequencing and identification. Non-hydrolyzed amaranth protein isolate served as control. Each hydrolysate was produced in triplicates.
Determination of the degree of hydrolysis (DH)
The degree of hydrolysis via determination of free amino group content (AN) was measured among the amaranth protein hydrolysates (APHs) using the O-phthaldialdehyde method as previously described (Ashraf et al., 2021). Reaction mixture was read with a microplate reader (Multiskan Sky, Thermo Fisher Scientific, Waltham, MA) at wavelength of 340 nm. A standard curve of tryptone was prepared along with samples and used to calculate the free amino nitrogen content of the samples and the AN was calculated using Equation [1]:
where AN1 and AN2 are the free amino group contents of the amaranth proteins before and after hydrolysis, respectively. Degree of hydrolysis was performed on each batch of hydrolysates in triplicate.
Profiling of peptide by reverse Phase-Ultra performance liquid chromatography (RP-UPLC) technique
The methodology of (Nongonierma & FitzGerald, 2012) was slightly modified and employed to analyze the peptide profile of APHs using the RP-UPLC (UltiMate 3000, Thermoscientific, Germering, Germany). Briefly, samples were mixed with mobile phase containing 0.1%TFA in HPLC grade acetonitrile (1:1v/v) (Solvent A). The mixture was vortexed vigorously for 5 min and then filtered through 0.45 μm syringe filters (Millipore Corp., Bedford, MA, USA). Amaranth proteins and derived hydrolysates were separated through a 2.1 mm × 100 mm, 1.7 μm Acquity UPLC C18 BEH column (Waters, Milford, MA, USA) at a flow rate of 0.3 mL min−1 at 30 °C. Solvent B comprised of 0.05% (v/v) TFA in 60% HPLC grade ACN in water, and amaranth proteins and derived peptides were eluted using a linear gradient of solvent B from 0 to 80% for 30 min. The eluent absorbance was read at 220 nm.
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoretic pattern of API and APHs were determined by SDS-PAGE under reducing condition following the method described by Kamal et al. (2021).
Inhibition of cholesteryl esterase (CEase) by AP and APHs
The CEase inhibition was determined as per the method described by Mudgil et al. (2019a) with slight modification. APHs samples (50 μL) were incubated with 5 mM p-nitrophenyl butyrate (50 μL) in sodium phosphate buffer (100 mM; pH 7.2 containing 100 mM NaCl) in a 96-well microplate. The reaction was initiated by addition of 50 μL of pancreatic CEase (5 μg/mL) in each well and incubated for 30 min at 37 °C. After the incubation, the reaction was terminated by adding 1 M HCl. The p-nitrophenol released from enzymatic hydrolysis of p-nitrophenyl butyrate was analysed at 405 nm using a microplate reader and the percent CEase inhibition activity was determined using the equation below:
where A (control): absorbance in the presence of enzyme without the test sample, B (control blank): absorbance without enzyme and test sample, C (reaction): absorbance with enzyme and test sample, D (reaction blank): absorbance with test sample without enzyme. The IC50 (half-maximal inhibitory concentration) was determined from a curve by plotting the concentration of the test sample (0.25–100 mg/mL) against the percentage inhibition and expressed in mg (equivalent protein)/mL.
Pancreatic lipase (PL) inhibition assay
Pancreatic lipase inhibitory of APHs was determined as per the method of Jafar, Kamal, Mudgil, Hassan, and Maqsood (2018). Briefly, 25 μL p-nitrophenyl butyrate (5 mM) was added to 50 μL of different APHs samples in a 96 well microplate reader containing sodium phosphate buffer (100 mM, pH 7.2, containing 100 mM of NaCl) and pre-incubated at 37℃ for 5 min. The reaction was initiated by the addition of 20 μL of PL (50 mg/mL) and the final volume was adjusted to 150 μL with assay buffer. The reaction plate was then incubated for 30 min at 37 °C and the absorbance of released p-nitrophenyl produced for each test sample was recorded at 405 nm using a microplate reader (Epoch 2, BioTek, VT, USA). The % pancreatic lipase inhibition was determined using the following equation:
where A (control): absorbance in the presence of enzyme without the test sample, B (control blank): absorbance without enzyme and test sample, C (reaction): absorbance with enzyme and test sample, D (reaction blank): absorbance with test sample without enzyme. IC50 value as mg protein equivalent were determined using a curve by plotting the concentration of the test sample (0.25 to 100 mg/mL) against the percentage inhibition and expressed in mg (equivalent protein)/mL.
Identification of peptide by liquid chromatography mass spectrometry of quadrupole Time-of-Flight (LC-MS QTOF) and in silico peptide analysis
The method reported by Sarah et al. (2016) was adopted for the identification of peptides in APHs (AB4) using LC-MS-QTOF instrument (Agilent 6520). The test samples (1 mg) were solubilized in a 1 mL of deionized water containing 0.1 % formic acid, whereas the mobile phases used were deionized water containing 0.1 % formic acid (solvent A) and LCMS grade acetonitrile containing 0.1 % formic acid (solvent B). Peptides were separated on an Advance Bio Peptide Map, C18 column (2.1 × 100 mm, 2.7 μm particles; Agilent) at a flow rate of 15 μL/min. Peptides were analyzed using the electrospray ionization-quadrupole time-of-flight system (ESI-QTOF, Agilent 6520) with the setting of: (a) mass range: 100–2000 m/z, (b) collision energy: 6 V/100 Da (offset −2), (c) ion source voltage: 3.5 kV. Mass spectrometry data were subjected to Peaks studio version 6.0 with average local confidence (ALC) positioned above 80 %. From the obtained sequences, a predictive analysis of bioactivity was carried out on Peptide Ranker web server available at http://distilldeep.ucd.ie/PeptideRanker/. Peptides with a score of more than 0.80, based on the Peptide Ranker score were selected for novelty check using a database search software (i.e. BIOPEP, PeptideDB, SwePep, and EROP-Moscow) (Mooney, Haslam, Pollastri, Shields, & Kurgan, 2012). The molecular mechanism of enzyme inhibition by selected peptides was established by using in-silico molecular interaction carried out on the web-based program PepSite 2 (Trabuco, Lise, Petsalaki, & Russell, 12012, 2012). Hence, the porcine PL (PDB code: 1PCO) and CE (PDB code: 1AQL-b) crystal structures were downloaded from RCSB Protein Data Bank. The most potent bioactive peptide was selected according to the significance of binding at < 5% p-value and potential binding sites. The physico-chemical properties such as Mass, Isoelectric point (pI), Net charge, Hydrophobicity of selected peptides with potent PL and CEase inhibitory activities were calculated and structure of peptides were drawn using online available tool PepDraw available at http://www.tulane.edu/~biochem/WW/PepDraw/.
Statistical analysis
All the hydrolysates were generated in three separate batches and each hydrolysate was analyzed in duplicate giving us a total of three replicates with six analytical measurements for each hydrolysate unless otherwise specified. The analysis of variation between different treatments was analyzed using SPSS version 24.0 (SPSS, IBM, USA). The significant difference between the CEase and PL inhibitory IC50 values was analyzed using the General Linear Model and compared using Tukey’s test at a 5% significance level.
Results and discussion
Determination of the DH by amino nitrogen values (AN values)
The results pertaining to the effect of different enzymes (bromelain, chymotrypsin and actinase E) for different time periods (2, 4, and 6 h) on the degree of protein hydrolysis are presented in Table 1. The results obtained for degree of hydrolysis as determined through increase in free amino nitrogen content revealed that free amino content (AN) of 9 hydrolysates produced through bromelain, chymotrypsin and actinase E hydrolysis varied between 166.68 and 732.49 µg NH3/mL−1 for AP2 and AB6, respectively (Table 1). Significant differences (p < 0.05) were seen for the AN content when the time of hydrolysis was increased from 2 h to 6 h for most of the hydrolysates. The variation observed in AN content could be due to enzyme specificity which showed a different behavior as the duration of hydrolysis progressed. These results are in corroboration with those obtained by (Montoya-Rodríguez, Milán-Carrillo, Reyes-Moreno, & De Mejía, 2015), where hydrolysis of amaranth protein by pepsin and pancreatin showed gradual increase in extent of hydrolysis with time. Overall, free AN content of the hydrolysates showed an increasing trend with increase in time of hydrolysis. It was also observed that bromelain and chymotrypsin enzyme were more active in hydrolyzing amaranth proteins while actinase E enzyme was found to be weak in degrading amaranth proteins. The variation associated with degree of hydrolysis among different hydrolysates produced using different enzymes is usually associated with difference in enzyme specificity for different protein substrates and reaction rates (Al-Shamsi, Mudgil, Hassan, & Maqsood, 2018)). Similar findings were reported by Mudgil et al. (2019b) for quinoa and amaranth derived hydrolysates. Chymotrypsin which is categorized as a well known serine protease catalyzes the hydrolysis of peptide bonds, on the carboxyl side of bulky aromatic side chains (Tyr, Phe, Trp), specifically next to hydrophobic residues. Actinase E (Protease from S. griseus), is a mixture of many proteases (including five serine-type proteases, two Zn2+ -endopeptidases, two Zn2+ -leucine aminopeptidases, and one Zn2+ -carboxypeptidase) will cleave peptide bonds from both the N- and C-terminus and within the peptide to theoretically yield a mixture of all possible fragments. In addition, chymotrypsin-induced amaranth protein hydrolysis mimics the gastrointestinal digestion of ingested food. As indicated by Fritz, Vecchi, Rinaldi, and Añón (2011) 7S globulins and 11S acidic subunits of amaranth protein are considered more susceptible to enzymatic hydrolysis, whereas, 11S basic subunit shows resistance to degradation initially but degrades gradually as the time of hydrolysis progresses. The initial increase in the degree of hydrolysis after 2 h could be assigned to the degradation of 7S and 11S acidic subunits, while at around 6 h of hydrolysis to 11S basic subunits. Previous studies have reported a DH of approximately 51.1% after performing gastrointestinal simulation of amaranth protein isolate using pepsin and pancreatin (Sabbione, Nardo, Añón, & Scilingo, 2016). Another study by the same authors reported 25.8% DH of APH after successive hydrolysis with alcalase and trypsin (Sabbione, Scilingo, & Añón, 2015). Furthermore, chymotrypsin was able to hydrolyze quinoa proteins to a higher degree after 4 and 6 h (86.0–87.5%) while bromelain showed highest degree of hydrolysis for amaranth proteins hydrolyzed for 6 h (47.2% DH) (Mudgil et al., 2019b). Overall, bromelain and chymotrypsin displayed higher degree of hydrolysis compared to actinase E enzyme.
Table 1.
IC50 values of lipase and cholesterol esterase inhibitory activities of amaranth protein hydrolysates generated by bromelain, chymotrypsin, and actinase E after 2, 4 and 6 h and degree of hydrolysis as the amino group content (AN).
| Sample | AN (µg NH3 mL−1) | Inhibitory potential-IC50 values (mg/mL) |
|
|---|---|---|---|
| CE | PL | ||
| API | 1.50 ± 0.06b | 3.93 ± 0.33 e | |
| AB2 | 568.32 ± 12.1 e | 0.50 ± 0.01 a | 0.66 ± 0.02 ab |
| AB4 | 535.09 ± 4.36 d | 0.47 ± 0.001 a | 0.48 ± 0.001 a |
| AB6 | 732.49 ± 4.2g | 0.53 ± 0.001 a | 0.38 ± 0.001 a |
| AC2 | 502.02 ± 9.26c | 1.35 ± 0.03b | 2.48 ± 0.05 d |
| AC4 | 597.64 ± 3.22f | 10.5 ± 0.13 e | 1.08 ± 0.01b |
| AC6 | 632.55 ± 3.07g | ND | 0.81 ± 0.01 ab |
| AE2 | 166.68 ± 5.16 a | 4.49 ± 0.04 d | 7.34 ± 0.04f |
| AE4 | 275.53 ± 6.47b | 3.24 ± 0.06c | 2.88 ± 0.06 d |
| AE6 | 290.69 ± 4.67b | 3.33 ± 0.08c | 1.89 ± 0.06c |
Note: API: Amaranth: Intact amaranth protein isolate; AB2, AC2 and AE2 denotes bromelain, chymotrypsin and actinase E 2 h generated amaranth protein hydrolysates; AB4, AC4 and AP4 denotes bromelain, chymotrypsin and actinase E 4 h amaranth protein hydrolysate;; AB6, AC6 and AA6 denotes bromelain, chymotrypsin and actinase E 6 h amaranth protein hydrolysate;, respectively. ND: not detected.
Values are mean ± SD. Different letter in the column indicates significant difference among the values.
Characterization of APHs by reverse phase-ultra performance liquid chromatography (RP-UPLC) and sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) technique
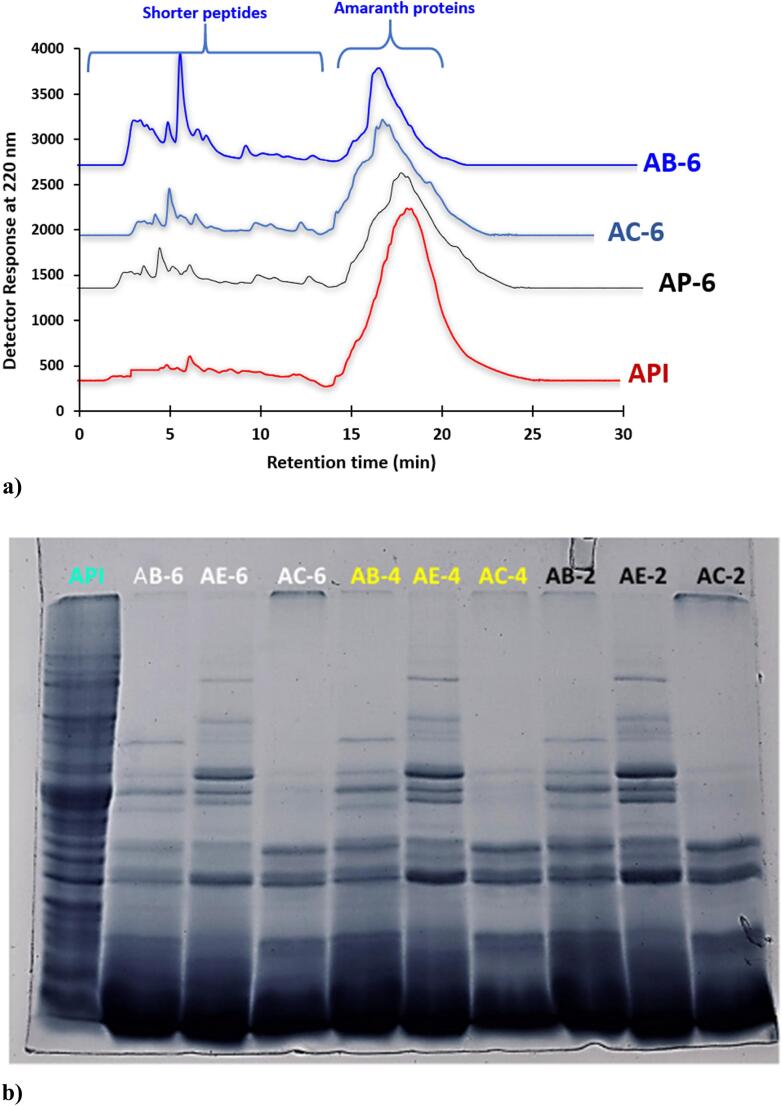
API and APHs were subjected to RP-UPLC analysis to monitor the hydrolysis of intact protein into peptides eluted over a retention time of 30 mins (Fig. 1a). Intact API (red line) displayed a broad intense protein peak eluting between 14 and 22 of mins with few small peaks toward low retention time 5–10 mins. Upon hydrolysis by bromelain, chymotrypsin and actinase E for 6 h, the intensity of major protein peak decreased with elution of newer small peptide peaks in the range of 3 to 12 mins of retention time. AB-6 hydrolysates displayed higher intensity of peptide peaks toward retention time ranging from 3 to 7 min, this indicates AB-6 h hydrolysate was extensively hydrolyzed compared to AC-6 h and AP-6 h hydrolysates. This was also reflected in the higher degree of hydrolysis value for AB-6 (732.49 ± 4.2 µg NH3 g−1) compared to AC-6 h (632.55 ± 3.07 µg NH3 g−1) and AP-6 h (290.69 ± 4.67 µg NH3 g−1) hydrolysates (Table 1). Peptide peaks in all the hydrolysates were eluted toward lower retention time on a reverse-phase C18 column which indicate wide range of hydrophobicity of the peptides. This also corroborated with the predicted hydrophobicity values of the selected peptides in hydrolysate AB-4 which shows hydrophobicity values in the positive value ranging from +4.34 Kcal * mol -1 to +9.79 Kcal * mol−1 (Table 4). Gu et al. (2021) also reported that hydrolysate fraction from the millet proteins showed majority of the peptide peaks eluted toward lower retention time ranging from 2 to 14 min indicating high polarity of the peptides eluted first. Overall, the peptide profile of APHs displayed lower intensity of the main intact protein peak with generation of new peptide peaks towards low retention time.
Fig. 1.
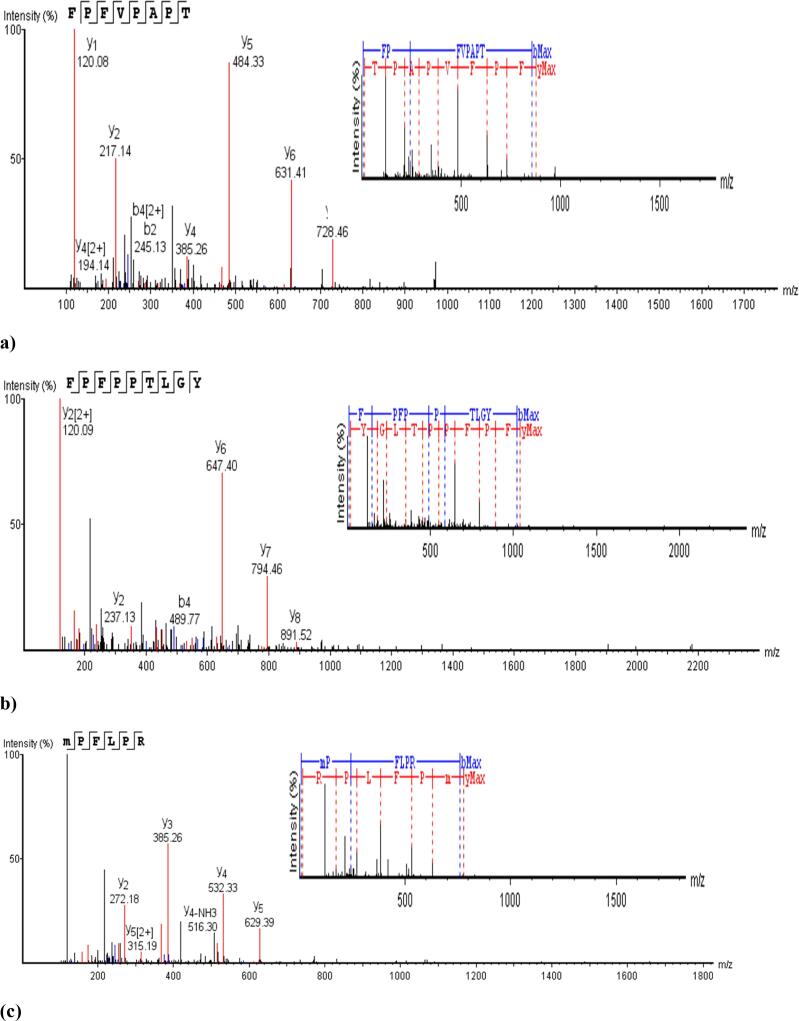
(a) RP_UPLC chromatogram of APHs produced by bromelain, chymotrypsin, and actinase E after 6 h, and (b) electrophoretic protein pattern of APHs generated after 2, 4 and 6 h. For Keynotes, please see foot note of Table 1.
Table 4.
Physico-chemical properties of selected peptides from AB4 hydrolysate predicted to be potent PL and CEase inhibitors (Data obtained from PepDraw webiste (http://www.tulane.edu/~biochem/WW/PepDraw/).
| Sequence | Mass | Isoelectric point (pI) | Net charge | Hydrophobicity | Structure | Activity |
|---|---|---|---|---|---|---|
| FPFVPAPT | 874.4575 | 5.32 | 0 | +5.19 Kcal * mol -1 | PL and CE inhibition | |
| MPFLPR | 759.4090 | 10.88 | +1 | +6.36 Kcal * mol -1 | 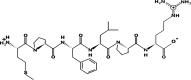 |
PL inhibition |
| FPFVGP | 662.3418 | 5.19 | 0 | +5.45 Kcal * mol -1 |  |
PL inhibition |
| FPFPPTLGY | 1037.5206 | 5.40 | 0 | +4.34 Kcal * mol -1 | PL inhibition | |
| FGAPR | 546.2907 | 10.90 | +1 | +9.79 Kcal * mol -1 | 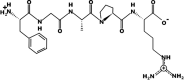 |
PL inhibition |
*Hotspots that could inhibit the activity of cholesterol esterase if bounded by the peptide.
API and APHs were characterized by using sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) as shown in Fig. 1b. Intact protein (API) displayed dense prominent bands over a wide range of molecular weight. Major protein bands detected in electrophoretic pattern of amaranth proteins have been reported to be globulin, albumin, glutelins, prolamin (Kamal et al., 2021, Sandoval-Sicairos et al., 2021). The protein profile of the amaranth reported in this study was similar to that of previously reported by Martínez and Anon (1996). When amaranth proteins were subjected to hydrolysis by different enzymes, disappearance or lightening of major protein bands were noticed in all the hydrolysates. Interestingly, actinase E generated hydrolysates showed dense and intact band which were not found in the APHs generated by bromelain and chymotrypsin, indicating that actinase E derived hydrolysates did not underwent high degree of hydrolysis when compared to other two types of hydrolysates. This was further supported by the lower degree of hydrolysis in actinase E derived APHs (166.68–290.69 µg NH3 mL−1) compared to those of bromelain (568.32–732.49 µg NH3 mL−1) and chymotrypsin (502.02–632.55 µg NH3 mL−1generated APHs (P < 0.05). When the time of hydrolysis increased from 2 to 6 h, slight degradation of the protein band was noticed after 4 h while considerable degradation was noticed after 6 h of hydrolysis, which corroborates with increase in amino nitrogen value (AN values) from 2 to 6 h of hydrolysis in all APHs (Table 1). All the APHs derived from different enzymes showed a long thick smear toward the bottom of the gel indicating that low molecular weight peptides were retained toward the end of the gel which could not separate further. Similar findings were also reported by Mudgil et al. (2019b) in quinoa protein hydrolysates generated by bromelain, chymotrypsin and actinase E. Sandoval-Sicairos et al. (2021) also reported that sequential hydrolysis (pepsin and pancreatin) of germinated and non-germinated amaranth seed flour showed disappearance of the major proteins and retention of the low molecular weight band towards the end of the gel.
Inhibition of cholesterol esterase (CEase) activity
Inhibition of cholesterol esterase is the prime target in the treatment of hypercholesterolemia via regulation of plasma cholesterol level (Gururaja, Mundkinajeddu, Dethe, Sangli, Abhilash, & Agarwal, 2015). Cholesterol and free fatty acids are liberated within the intestinal lumen after hydrolysis of long-chain fatty acid esters of cholesterol by pancreatic cholesterol esterase also known as a bile salt-dependent lipase. The process of hydrolysis of cholesterol ester into cholesterol by cholesterol esterase in the intestinal lumen is required for absorption because cholesterol ester is not directly absorbed by the intestinal epithelial cells (Gururaja et al., 2015). Therefore, inhibition of CEase is required to retard the production and absorption of cholesterol.
The inhibition of cholesterol esterase by intact amaranth protein (API) and APHs generated with three different enzymes at different time intervals are depicted in Table 1. Intact API revealed the CEase inhibitory potential with an IC50 value of 1.50 mg/mL, whereas the more effective inhibition was displayed by bromelain hydrolysates at 4 h hydrolysis (AB4) with the lowest IC50 value of 0.47 mg/mL. Among all the hydrolysates, bromelain showed the maximum inhibitory potential of cholesterol esterase (0.47–0.53 mg/mL). The chymotrypsin derived APHs resulted in decreasing CEase inhibitory potential with the increase of hydrolysis time from 2 to 4 h (IC50 values of 1.35 to 10.5 mg/mL, respectively). This could be because of the degradation of CEase inhibitory peptides to inactive peptides which could not bind on the active site of CEase. Actinase E derived APHs showed least inhibition of CEase with IC50 value ranging from 4.49 to 3.24 mg/mL
Till date only few scientific studies were published reporting data on the hypocholesterolemic effect of amaranth proteins, where it was shown to lower the blood cholesterol levels as depicted in the clinical studies on humans (Maier, Turner, & Lupton, 2000). Soares et al. (2015) explored the peptides from amaranth proteins as inhibitors of HMG-CoA reductase suggesting its hypocholesterolemic effect. Moreover Mendonça et al. (2009) demonstrated hypocholesterolemic effect in hamsters by the intake of amaranth protein which led to a remarkable decrease in total/HDL-cholesterol ratio. The authors strongly speculate that the activity may be attributed due to peptides that are likely to be produced by partial protein digestion and may affect cholesterol absorption in the gut. There are no studies available to date on amaranth protein hydrolysates' activity towards cholesterol esterase inhibition under in-vitro studies. This study may stand as a reference for future researchers on providing the knowledge of amaranth protein hydrolysates that could generate the anti-hypercholesterolemic peptides.
Inhibition of pancreatic lipase (PL)
The key enzyme for lipid digestion and absorption is pancreatic lipase that hydrolyzes the triacylglycerols present in the gut. The inhibitors of pancreatic lipase plays a role to limit intestinal fat absorption and are proved as useful medication in controlling hyperlipidemia and offers themselves as promising anti-obesity agents (Gholamhoseinian, Shahouzehi, & Sharifi-Far, 12010, 2010). The IC50 values of amaranth protein (non-hydrolysate), amaranth protein hydrolysates (AB2-6; AC2-6 and AP2-6) for porcine pancreatic lipase inhibitory activity are documented in Table 1. The highest inhibitory activity was observed in bromelain derived 6 h hydrolysates (AB6) (IC50 value = 0.38 mg/mL). Actinase E derived APHs showed the lowest efficacy in inhibiting the PL which is in agreement with CEase inhibition wherein lowest inhibition was shown by actinase E derived APHs. Decrease in lipase inhibitory IC50 values with increase in hydrolysis time was noticed for all the three enzymatic hydrolysates that indicate the increase in lipase inhibitory activity with increase in time of hydrolysis. The significant (p < 0.05) inhibitory potential for bromelain hydrolysates by the low IC50 values ranged between 0.38 and 0.66 mg/mL. Lowest CEase and PL inhibition was demonstrated by the APHs which had lowest DH, which indicates that low degree of hydrolysis might have not been sufficient to release peptide which could bind on to the active site of CEase and PL. Our results are in corroboration with those of (Velarde-Salcedo, González de Mejía, & Barba de la Rosa, 2012) who reported that amaranth proteins hydrolysates have shown anti-lipidemic activity by reducing fat accumulation in mouse adipocyte cultures. Previous studies on amaranth derived proteins and protein hydrolysates have been mostly carried out in in-vivo model system, however very limited scientific data has been reported on proteins and peptides that could inhibit pancreatic lipase activity in in vitro conditions (Gargouri et al., 1984, Jafar et al., 2018, Mudgil et al., 2018). Recently, Awosika and Aluko (2019) have reported pancreatic lipase inhibitory activity of pea protein hydrolysates generated by alcalase, chymotrypin, pepsin and trypsin. It was reported that the trypsin (IC50 value of 3.95 ± 0.04 mg mL − 1) and alcalase (IC50 3.98 ± 0.4 mg mL − 1) generated pea protein hydrolysates had the lowest IC50 values, indicating higher pancreatic lipase inhibitory potential than the chymotrypsin and pepsin derived hydrolysate (Awosika & Aluko, 2019). Interesting, the lipase inhibitory IC50 value (0.38–2.88 mg/mL) obtained in this study for all the amaranth protein hydrolysates except actinase E-2 h hydrolysates were lower than those reported in the study of Awosika and Aluko (2019), indicating higher potential of amarath protein hydrolysates compared to pea protein hydrolysates. Overall, lipase inhibiting properties of different protein hydrolysates is least explored (Awosika and Aluko, 2019, Gil-Rodríguez and Beresford, 2021, Mudgil et al., 2018). The result of the current study indicates that the bromelain generated hydrolysates with lowest lipase inhibitory IC50 values could be used as natural inhibitors of pancreatic lipase and thus can play a role in obesity management.
Identification of CEase and PL inhibitory peptides from selected amaranth protein hydrolysate
Amaranth protein hydrolysate produced by bromelain at 4 h hydrolysis time (AB4) was selected for further peptide identification using LC-MS QTOF based on the highest CEase inhibitory activities and are shown in Table 1. Overall, a total of 17 peptides identified in sample AB4 having a peptide ranker score of 0.8 and above were subjected to in-silico structure activity relationship with the target enzymes through Peptide Ranker web server available at http://distilldeep.ucd.ie/PeptideRanker/ (Table 2). Table S1 shows a complete list of peptides (116 peptides) generated from AB4 hydrolysate.
Table 2.
Identification of the peptides in the selected amaranth protein hydrolysate AB4 and their binding potential on CE (1AQL-b) as protein receptor.
| Peptide sequence | Peptide Ranker score1 | PepSite2 P value2 | Reactive residues in peptide | Bound residues of CE |
|---|---|---|---|---|
| FPFPR | 0.988236 | 0.005523 | F1, P2, F3, P4 | Phe235, Trp236, His283, Tyr284 |
| DFPF | 0.987642 | 0.0107 | D1, F2, P3, F4 | Phe235, Trp236, His283, Tyr284 |
| PLMF | 0.979313 | 0.08692 | P1, L2, F4 | Phe235, Trp236, His283 |
| MPFPR | 0.967245 | 0.003743 | M1, P2, F3, R5 | Tyr7, Phe12 |
| FPFLPR | 0.96619 | 0.0948 | F1, P2, F3, L4 | Phe235, Trp236, His283, Tyr284 |
| MPFLPR | 0.93323 | 0.04707 | M1, P2, F3, L4 | Tyr7, Phe12 |
| FPFVGP | 0.928911 | 0.09026 | F1, P2, F3, V4 | Phe235, Trp236, His283, Tyr284 |
| MRAF | 0.919531 | 0.004841 | M1, R2, A3, F4 | Tyr7, Phe12 |
| CCPDGL | 0.918875 | 0.1911 | C2, P3, D4 | Tyr7, Phe12 |
| ML | 0.894564 | 0.04441 | M1, L2 | Phe235, Trp236, His283 |
| DYLF | 0.888792 | 0.03488 | D1, Y2, L3, F4 | Tyr7, Phe12 |
| FPFPPTLGY | 0.888073 | 0.03811 | F1, P2, F3, P4, T6 | Phe235, Trp236, Tyr279, His283, Tyr284 |
| FGAPR | 0.876678 | 0.01649 | F1, G2, A3, R5 | Tyr7, Phe12 |
| YCTF | 0.872064 | 0.04539 | Y1, C2, T3, F4 | Phe235, Trp236, His283, Tyr284 |
| MDMP | 0.863368 | 0.08147 | D2, M3, P4 | Phe235, Trp236, Tyr279, His283 |
| FPFVPAPT | 0.836216 | 0.1015 | F1, P2, P7, T8 | Ala108*, Ser194*, Trp227*, Phe235, Trp236, His283, Tyr284, Leu285, Phe324*, Phe393, His435* |
| AMGALPM | 0.820032 | 0.1685 | A4, L5, P6, M7 | Tyr7, Phe12 |
*Hotspots.
Peptide Ranker Score of 0.8 and above is a threshold that labels the peptide as bioactive (Mooney et al., 2012).
P value for Pepsite 2 is calculated as follows: The distribution of the raw score for the peptide length is initially fitted to a Gumbel distribution. When peptide-protein match is generated, the raw score will be converted to P values based on the corresponding fitted Gumbel distribution. P value more than 0.25 is considered as insignificant or peptide did not bind to the protein (Trabuco et al., 2012).
The amino acid residues in the identified peptides varied from 2 to 9 as shown in Table 2. These outcomes are similar to those reported by (Nagaoka et al., 2001) who had previously reported the number of residues in hypocholesterolemic peptides ranges from 5 to 20. (Baba et al., 2021) and (Mudgil et al., 2019a) have also recently reported a similar number of amino acid in the identified peptides which were predicted to possess CEase and PL inhibitory properties. Moreover, peptides (GGV, IVG, and VGVL) from the Amaranth protein reported by Soares et al. (2015) demonstrated inhibition of HMG-CoA reductase enzyme, which implies a promising hypocholesterolemic effect. According to Kongo-Dia-Moukala, Nsor-Atindana, and Zhang (2011) anti- hypercholesterolemic activity of peptides has been strongly associated with the presence of hydrophobic amino acids residues e.g., alanine, phenylalanine, leucine, tyrosine and proline. Such amino acid residues were also found within the sequence of the bioactive peptides (FPFVPAPT, FPFVGP, FGAPR, FPFPPTLGY, MPFLPR) found in this study.
PepSite 2 results showed that peptides FPFPR, DFPF, MPFPR, MPFLPR, MRAF, ML, DYLF, FPFPPTLGY, FGAPR, and YCTF significantly (p < 0.05) bound to CEase as seen in Table 2. Although, none of these peptides were observed to bind on the active sites of CEase. However, FPFVPAPT was observed to be the only bioactive peptide from AB4 involved in the binding to the residue on the sites of CEase, and this could be attributed to the high number of residues interacting with the peptides. Fig. 2a shows the mass spectrum of the peptide (FPFVPAPT) which is predicted to be the most active CEase inhibiting peptide from APHs (AB-4). According to (Lin et al., 1999) the active sites of cholesterol esterase contain catalytic triad (Ser194, Asp320, and His435), together with the oxyanion hole (Gly107, Ala108, and Ala195), an alkyl chain binding site, an esteratic site, second alkyl chain biding site and/or a leaving group hydrophobic site and a leaving group hydrophilic binding site that forms a tetrahedral intermediate in the active site of CEase and are crucial for its catalytic function. Amongst the identified bioactive peptides from APHs, peptide FPFVPAPT could establish a bond with two amino acid residues present in the catalytic triad (Ser 194 and His435), and Ala108, and Phe324 which are part of an active site residue of CEase suggesting the strong inhibitory potential of this peptide against CEase. The peptides that could bind the residues of Phe324 and His435 are particularly gaining attention as CEase inhibitors, as previously reported in bioactive peptides derived from camel milk protein hydrolysates (Mudgil et al., 2019a) and camel whey protein hydrolysates (Baba et al., 2021).
Fig. 2.
Mass spectra of (a) FPFVPAPT, (b) FPFPPTLGY and (c) MPFLPR derived from hydrolysate AB-4. The window in the figure shows full analysis of the mass spectrum correlating the sequence obtained from the analysis of y-ion series and the sequence obtained from the analysis of b-ion series.
With regards to PL inhibitory peptides, results showed that APHs was able to generate higher number of peptides which were predicted to bind to higher number of residues on PL compared to peptides which could bind to CEase (Table 3). Results showed that FPFPR, DFPF, PLMF, MPFPR, FPFLPR, MPFLPR, ML, FPFPPTLGY, FGAPR, YCTF, MDMP, FPFVPAPT were predicted to significantly (p < 0.05) bind to PL. Also, all these peptides were observed to bind important amino acids of PL. Peptides generated from this study showed the superior potential of linkage to the target enzyme owing to their ability to bind up to 12 residues on the PL. For example, FGAPR, FPFVPAPT, and FPFPPTLGY could bind up to 13, 12, and 11 bound residues on the active sites of PL (i. e. Phe78, Phe216, His152, Ser153, and His264), respectively. Similar binding hot spots for lipase inhibiting peptides from camel whey protein hydrolysates have been reported by Baba et al. (2021). From the APHs (AB-4), peptide MPFLPR, FPFVGP, FPFPPTLGY, FGAPR and FPFVPAPT were predicted to bind to the highest number of hotspots on the active site of PL. An example of mass spectrum (FPFVPAPT, FPFPPTLGY and MPFLPR) is shown in Fig. 2b. Furthermore, most of the peptides also showed the capacity to bind to other bound residues (Tyr115, Leu154, Ala179, Pro181, Gly 155) that are not from the characteristic catalytic triad of PL which suggests indirect lipase inhibitory activity. Siow, Choi, and Gan (2016) also reported up to 23 AA residues in a potent lipase-inhibitory peptide CSP1 (FFRSKLLSDGAAAAKGLLPQYW) from cumin seeds, and up to 14 AA residues were reported in synthetic lipase-inhibitory peptide (RKQEEDEDEEQQRE) from soybean (Martinez-Villaluenga, Rupasinghe, Schuler, & Gonzalez de Mejia, 2010).
Table 3.
Identification of the peptides in the selected amaranth protein hydrolysate AB4 and their binding potential on PL (1ETH-a) as protein receptor.
| Peptide sequence | Peptide Ranker score1 | PepSite2 P value2 | Reactive residues in peptide | Bound residues of PL (1ETH-a) |
|---|---|---|---|---|
| FPFPR | 0.988236 | 0.002479 | F1, P2, F3, P4 | Phe78*, Tyr115, Ser153*, Leu154, Ala179, Pro181, Phe216*, His264* |
| DFPF | 0.987642 | 0.02592 | F2, P3, F4 | Phe78*, Tyr115, Ser153*, Leu154, Ala179, Pro181, Phe216*, His264* |
| PLMF | 0.979313 | 0.00124 | L2, M3, F4 | Gly77, Phe78*, Tyr115, Ser153*, Leu154, Gly155, Ala179, Pro181, Phe216*, His264* |
| MPFPR | 0.967245 | 0.002073 | M1, P2, F3, P4 | Phe78*, Tyr115, Ser153*, Leu154, Ala179, Pro181, Phe216*, His264* |
| FPFLPR | 0.96619 | 0.001776 | F1, P2, F3, L4, P5 | Gly77, Phe78*, Tyr115, Ser153*, Leu154, Gly155, Ala179, Pro181, Phe216*, His264* |
| MPFLPR | 0.93323 | 0.00285 | M1, P2, F3, L4, P5 | Gly77, Phe78*, Tyr115, His152*, Ser153*, Leu154, Gly155, Ala179, Phe216*, His264* |
| FPFVGP | 0.928911 | 0.05773 | F1, P2, F3, V4 | Gly77, Phe78*, Tyr115, His152*, Ser153*, Leu154, Gly155, Ala179, Phe216*, His264* |
| MRAF | 0.919531 | 0.05342 | M1, A3, F4 | Phe78*, Tyr115, Ser153*, Pro181, Phe216* |
| CCPDGL | 0.918875 | 0.1681 | C1, C2, P3, D4 | Lys81, Glu84, Trp253 |
| ML | 0.894564 | 0.008899 | M1, L2 | Phe78*, Tyr115, Ser153*, Phe216*, His264* |
| DYLF | 0.888792 | 0.2367 | Y2, L3, F4 | Phe78*, Try115, Ser153*, Pro181, Ile210, Phe216*, His264* |
| FPFPPTLGY | 0.888073 | 0.01714 | P2, F3, P4, P5, T6 | Gly77, Phe78*, Tyr115, His152*, Ser153*, Leu154, Gly155, Ala179, Pro181, Phe216*, His264* |
| FGAPR | 0.876678 | 0.005803 | F1, G2, A3, P4 | Gly77, Phe78*, Ile79, Tyr115, His152*, Ser153*, Leu154, Gly155, Ala179, Glu180, Pro181, Phe216*, His264* |
| YCTF | 0.872064 | 0.02824 | Y1, C2, T3, F4 | Phe78*, Tyr115, Ser153*, Leu154, Ala179, Pro181, Phe216*, His264* |
| MDMP | 0.863368 | 0.04212 | M1, M3, P4 | Phe78*, Tyr115, Ser153*, Leu154, Ala179, Phe216*, His264* |
| FPFVPAPT | 0.836216 | 0.01041 | F3, V4, P5, A6, P7, T8 | Gly77, Phe78*, Tyr115, His152*, Ser153*, Leu154, Gly155, Ala179, Glu180, Pro181, Phe216*, His264* |
| AMGALPM | 0.820032 | 0.06477 | A4, L5, P6, M7 | Phe78*, Tyr115, Ser153*, Leu154, Ala179, Pro181, Phe216*, His264* |
*Hotspots.
Peptide Ranker Score of 0.8 and above is a threshold that labels the peptide as bioactive (Mooney et al., 2012).
P value for Pepsite 2 is calculated as follows: The distribution of the raw score for the peptide length is initially fitted to a Gumbel distribution. When peptide-protein match is generated, the raw score will be converted to P values based on the corresponding fitted Gumbel distribution. P value more than 0.25 is considered as insignificant or peptide did not bind to the protein (Trabuco et al., 2012).
The presence of hydrophobic amino acid residues (leucine, glycine, proline, phenylalanine) and hydrophilic (arginine) observed in the peptides generated from APHs (AB-4) suggest the occurrence of both types of interactions between pancreatic lipase and lipase inhibiting peptides. From this study, peptides that showed the ability to bind to important amino acids of lipase were of hydrophobic amino acid supremacy i.e. leucine and proline Ngoh et al., 2017, Baba et al., 2021 observed similar results in pinto beans and camel whey protein hydrolysates, respectively. They reported that in most of the lipase inhibiting peptides, proline and leucine are identified as the interacting residues. Similarly, the existence of hydrophilic amino acid residues could help to anchor derived peptide inhibitors on PL via hydrogen bonding (Siow et al., 2016). Apart from the presence of hydrophilic amino acid residues, molecular mass, the number of negatively charged residues and isoelectric point amongst others could as well determine the inhibitory properties of bioactive peptides (Martinez-Villaluenga et al., 2010). A similar mechanism for inhibiting PL from derived peptides has been reported for pinto bean proteins by Ngoh et al. (2017). Molecular mass, isoelectric point, charge and hydrophobicity values of selected CEase and PL inhibiting peptides is shown in Table 4. All the five peptides (FPFVPAPT, MPFLPR, FPFVGP, FPFPPTLGY and FGAPR) predicted to bind to the maximum hotspots on PL and (FPFVPAPT) on CEase displayed high hydrophobicity value in the positive range (+4.34 to 9.79 Kcal * mol -1) as predicted from PepDraw website (Table 4). Novel APHs from bromelain generated at 4 h demonstrated strong prediction to possess PL inhibitory peptides by binding active site of enzyme which seems to be similar to the inhibitory mechanism of commercial lipase inhibitor. This work established that APHs could be used to alleviate the incidence of hypocholesterolemia.
Conclusion
Natural products in the present time are sought after to facilitate the development of new types of therapeutics. In this study, bioactive peptides generated from amaranth protein hydrolysates could play an important role in influencing the lipid metabolism by interacting with CEase and PL enzymes. Bromelain derived APHs at 2 h, 4 h, and 6 h led to the highest inhibition CEase and PL compared to chymotrypsin and actinase E hydrolysates. Moreover, increasing time intervals of hydrolysis in all the three enzymes decreased the IC50 values indicating that the increase in PL inhibitory activity is directly proportionate to the increase in hydrolysis time. From this study, among the different bioactive peptides identified, FPFVPAPT was predicted as a potential CEase inhibitor while MPFLPR, FPFVGP, FPFPPTLGY, FGAPR and FPFVPAPT as lipase inhibitors with binding affinity toward 5 hotspots on the active site of CEase and PL. This work established that identified peptides derived from amaranth protein hydrolysates have very fewer bound residues on the active site of CEase, in comparison to pancreatic lipase. The inhibition potential of pancreatic lipase and CEase activity established the anti-hypercholesteremic activity of bromelain-generated amaranth protein hydrolysates. The outcome of this study indicates that amaranth hydrolysates are laden with bioactive peptides, and this could lead to an innovative possibility for development of functional foods for tackling hypercholesterolemia. However, further research is required for a better understanding of the molecular mechanisms of action of the peptides and testing the purified peptides under in-vitro and in-vivo conditions.
Declaration of Competing Interest
The authors declare that they have no known competing financial or personal interest othat could have appeared to influence the work reported in this paper.
Acknowledgements
Authors are thankful to United Arab Emirates University for funding this research through a research grants (AUA-12F002) awarded to Sajid Maqsood. University Sains Malaysia RUI grant (1001/CABR/ 8011045) to Chee-Yuen Gan is also acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2021.100165.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adisakwattana S., Intrawangso J., Hemrid A., Chanathong B., Mäkynen K. Extracts of edible plants inhibit pancreatic lipase, cholesterol esterase and cholesterol micellization, and bind bile acids. Food Technology and Biotechnology. 2012;50(1):11–16. [Google Scholar]
- Al-Shamsi K.A., Mudgil P., Hassan H.M., Maqsood S. Camel milk protein hydrolysates with improved technofunctional properties and enhanced antioxidant potential in in vitro and in food model systems. Journal of Dairy Science. 2018;101(1):47–60. doi: 10.3168/jds.2017-13194. [DOI] [PubMed] [Google Scholar]
- Ashraf A., Mudgil P., Palakkott A., Iratni R., Gan C.-Y., Maqsood S., Ayoub M.A. Molecular basis of the anti-diabetic properties of camel milk through profiling of its bioactive peptides on dipeptidyl peptidase IV (DPP-IV) and insulin receptor activity. Journal of Dairy Science. 2021;104(1):61–77. doi: 10.3168/jds.2020-18627. [DOI] [PubMed] [Google Scholar]
- Awosika T.O., Aluko R.E. Inhibition of the in vitro activities of α-amylase, α-glucosidase and pancreatic lipase by yellow field pea (Pisum sativum L.) protein hydrolysates. International Journal of Food Science & Technology. 2019;54(6):2021–2034. [Google Scholar]
- Baba W.N., Mudgil P., Baby B., Vijayan R., Gan C.-Y., Maqsood S. New insights into the cholesterol esterase-and lipase-inhibiting potential of bioactive peptides from camel whey hydrolysates: Identification, characterization, and molecular interaction. Journal of Dairy Science. 2021;104(7):7393–7405. doi: 10.3168/jds.2020-19868. [DOI] [PubMed] [Google Scholar]
- Bhargava A., Shukla S., Ohri D. Genetic variability and interrelationship among various morphological and quality traits in quinoa (Chenopodium quinoa Willd.) Field Crops Research. 2007;101(1):104–116. [Google Scholar]
- Birari R.B., Bhutani K.K. Pancreatic lipase inhibitors from natural sources: Unexplored potential. Drug Discovery Today. 2007;12(19–20):879–889. doi: 10.1016/j.drudis.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Chiou, S.-Y., Lai, G.-W., Lin, L.-Y., & Lin, G. (2006). Kinetics and mechanisms of cholesterol esterase inhibition by cardiovascular drugs in vitro. Indian Journal of Biochemistry and Biophysics, 43(1), 52-55. https://doi.org/http://hdl.handle.net/123456789/3234. [PubMed]
- Fritz M., Vecchi B., Rinaldi G., Añón M.C. Amaranth seed protein hydrolysates have in vivo and in vitro antihypertensive activity. Food Chemistry. 2011;126(3):878–884. [Google Scholar]
- Frota K.M.G., Mendonça S., Saldiva P.H.N., Cruz R.J., Arêas J.A.G. Cholesterol-lowering properties of whole cowpea seed and its protein isolate in hamsters. Journal of Food Science. 2008;73(9):H235–H240. doi: 10.1111/j.1750-3841.2008.00953.x. [DOI] [PubMed] [Google Scholar]
- Gargouri Y., Julien R., Sugihara A., Verger R., Sarda L. Inhibition of pancreatic and microbial lipases by proteins. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism. 1984;795(2):326–331. doi: 10.1016/0005-2760(84)90082-1. [DOI] [PubMed] [Google Scholar]
- Gholamhoseinian, A., Shahouzehi, B., & Sharifi-Far, F. (2010). Inhibitory effect of some plant extracts on pancreatic lipase. IJP-International Journal of Pharmacology, 6(1), 18-24. https://doi.org/http://www.ansinet.org/ijp.
- Gil-Rodríguez A.M., Beresford T. Bile salt hydrolase and lipase inhibitory activity in reconstituted skim milk fermented with lactic acid bacteria. Journal of Functional Foods. 2021;77:104342. doi: 10.1016/j.jff.2020.104342. [DOI] [Google Scholar]
- Gu H., Gao J., Shen Q., Gao D., Wang Q., Tangyu M., Mao X. Dipeptidyl peptidase-IV inhibitory activity of millet protein peptides and the related mechanisms revealed by molecular docking. LWT-Food Science and Technology. 2021;138:110587. doi: 10.1016/j.lwt.2020.110587. [DOI] [Google Scholar]
- Gururaja, G., Mundkinajeddu, D., Dethe, S. M., Sangli, G. K., Abhilash, K., & Agarwal, A. (2015). Cholesterol esterase inhibitory activity of bioactives from leaves of Mangifera indica L. Pharmacognosy Research, 7(4), 355. https://doi.org/https://dx.doi.org/10.4103%2F0974-8490.159578. [DOI] [PMC free article] [PubMed]
- Jafar S., Kamal H., Mudgil P., Hassan H.M., Maqsood S. Camel whey protein hydrolysates displayed enhanced cholesteryl esterase and lipase inhibitory, anti-hypertensive and anti-haemolytic properties. LWT-Food Science and Technology. 2018;98:212–218. doi: 10.1016/j.lwt.2018.08.024. [DOI] [Google Scholar]
- Jakubczyk A., Karaś M., Złotek U., Szymanowska U. Identification of potential inhibitory peptides of enzymes involved in the metabolic syndrome obtained by simulated gastrointestinal digestion of fermented bean (Phaseolus vulgaris L.) seeds. Food Research International. 2017;100:489–496. doi: 10.1016/j.foodres.2017.07.046. [DOI] [PubMed] [Google Scholar]
- Kamal H., Mudgil P., Bhaskar B., Fisayo A.F., Gan C.-Y., Maqsood S. Amaranth proteins as potential source of bioactive peptides with enhanced inhibition of enzymatic markers linked with hypertension and diabetes. Journal of Cereal Science. 2021;101:103308. doi: 10.1016/j.jcs.2021.103308. [DOI] [Google Scholar]
- Kongo-Dia-Moukala J., Nsor-Atindana J., Zhang H. Hypocholesterolemic activity and characterization of protein hydrolysates from defatted corn protein. Asian Journal of Biochemistry. 2011;6(6):439–449. doi: 10.3923/ajb.2011.439.449. [DOI] [Google Scholar]
- Kurek M.A., Karp S., Wyrwisz J., Niu Y. Physicochemical properties of dietary fibers extracted from gluten-free sources: Quinoa (Chenopodium quinoa), amaranth (Amaranthus caudatus) and millet (Panicum miliaceum) Food Hydrocolloids. 2018;85:321–330. [Google Scholar]
- Lin G., Shieh C.-T., Tsai Y.-C., Hwang C.-I., Lu C.-P., Chen G.-H. Structure-reactivity probes for active site shapes of cholesterol esterase by carbamate inhibitors. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 1999;1431(2):500–511. doi: 10.1016/S0167-4838(99)00073-4. [DOI] [PubMed] [Google Scholar]
- Lunagariya N.A., Patel N.K., Jagtap S.C., Bhutani K.K. Inhibitors of pancreatic lipase: State of the art and clinical perspectives. EXCLI journal. 2014;13:897–921. [PMC free article] [PubMed] [Google Scholar]
- Maier S.M., Turner N.D., Lupton J.R. Serum lipids in hypercholesterolemic men and women consuming oat bran and amaranth products. Cereal Chemistry Journal. 2000;77(3):297–302. doi: 10.1094/CCHEM.2000.77.3.297. [DOI] [Google Scholar]
- Martinez-Villaluenga C., Rupasinghe S.G., Schuler M.A., Gonzalez de Mejia E. Peptides from purified soybean β-conglycinin inhibit fatty acid synthase by interaction with the thioesterase catalytic domain. The FEBS Journal. 2010;277(6):1481–1493. doi: 10.1111/j.1742-4658.2010.07577.x. [DOI] [PubMed] [Google Scholar]
- Martins J.M., Riottot M., de Abreu M.C., Viegas-Crespo A.M., Lança M.J., Almeida J.A.…Bento O.P. Cholesterol-lowering effects of dietary blue lupin (Lupinus angustifolius L.) in intact and ileorectal anastomosed pigs. Journal of Lipid Research. 2005;46(7):1539–1547. doi: 10.1194/jlr.M500129-JLR200. [DOI] [PubMed] [Google Scholar]
- Mendonça S., Saldiva P.H., Cruz R.J., Arêas J.A. Amaranth protein presents cholesterol-lowering effect. Food Chemistry. 2009;116(3):738–742. doi: 10.1016/j.foodchem.2009.03.021. [DOI] [Google Scholar]
- Montoya-Rodríguez A., De Mejía E.G., Dia V.P., Reyes-Moreno C., Milán-Carrillo J. Extrusion improved the anti-inflammatory effect of amaranth (Amaranthus hypochondriacus) hydrolysates in LPS-induced human THP-1 macrophage-like and mouse RAW 264.7 macrophages by preventing activation of NF-κB signaling. Molecular Nutrition & Food Research. 2014;58(5):1028–1041. doi: 10.1002/mnfr.201300764. [DOI] [PubMed] [Google Scholar]
- Montoya-Rodríguez A., Milán-Carrillo J., Reyes-Moreno C., De Mejía E.G. Characterization of peptides found in unprocessed and extruded amaranth (Amaranthus hypochondriacus) pepsin/pancreatin hydrolysates. International Journal of Molecular Sciences. 2015;16(4):8536–8554. doi: 10.3390/ijms16048536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney C., Haslam N.J., Pollastri G., Shields D.C., Kurgan L. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PloS One. 2012;7(10):e45012. doi: 10.1371/journal.pone.0045012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil P., Baby B., Ngoh Y.-Y., Vijayan R., Gan C.-Y., Maqsood S. Identification and molecular docking study of novel cholesterol esterase inhibitory peptides from camel milk proteins. Journal of Dairy Science. 2019;102(12):10748–10759. doi: 10.3168/jds.2019-16520. [DOI] [PubMed] [Google Scholar]
- Mudgil P., Kamal H., Yuen G.C., Maqsood S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chemistry. 2018;259:46–54. doi: 10.1016/j.foodchem.2018.03.082. [DOI] [PubMed] [Google Scholar]
- Mudgil P., Omar L.S., Kamal H., Kilari B.P., Maqsood S. Multi-functional bioactive properties of intact and enzymatically hydrolysed quinoa and amaranth proteins. LWT-Food Science and Technology. 2019;110:207–213. doi: 10.1016/j.lwt.2019.04.084. [DOI] [Google Scholar]
- Nagaoka S., Futamura Y., Miwa K., Awano T., Yamauchi K., Kanamaru Y.…Kuwata T. Identification of novel hypocholesterolemic peptides derived from bovine milk beta-lactoglobulin. Biochemical and Biophysical Research Communications. 2001;281(1):11–17. doi: 10.1006/bbrc.2001.4298. [DOI] [PubMed] [Google Scholar]
- Ngoh Y.-Y., Choi S.B., Gan C.-Y. The potential roles of Pinto bean (Phaseolus vulgaris cv. Pinto) bioactive peptides in regulating physiological functions: Protease activating, lipase inhibiting and bile acid binding activities. Journal of Functional Foods. 2017;33:67–75. doi: 10.1016/j.jff.2017.03.029. [DOI] [Google Scholar]
- Nongonierma A.B., Cadamuro C., Le Gouic A., Mudgil P., Maqsood S., FitzGerald R.J. Dipeptidyl peptidase IV (DPP-IV) inhibitory properties of a camel whey protein enriched hydrolysate preparation. Food Chemistry. 2019;279:70–79. doi: 10.1016/j.foodchem.2018.11.142. [DOI] [PubMed] [Google Scholar]
- Nongonierma A.B., FitzGerald R.J. Tryptophan-containing milk protein-derived dipeptides inhibit xanthine oxidase. Peptides. 2012;37(2):263–272. doi: 10.1016/j.peptides.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Sabbione A.C., Nardo A.E., Añón M.C., Scilingo A. Amaranth peptides with antithrombotic activity released by simulated gastrointestinal digestion. Journal of Functional Foods. 2016;20:204–214. [Google Scholar]
- Sabbione A.C., Scilingo A., Añón M.C. Potential antithrombotic activity detected in amaranth proteins and its hydrolysates. LWT-Food Science and Technology. 2015;60(1):171–177. [Google Scholar]
- Sandoval-Sicairos E.S., Milán-Noris A.K., Luna-Vital D.A., Milán-Carrillo J., Montoya-Rodríguez A. Anti-inflammatory and antioxidant effects of peptides released from germinated amaranth during in vitro simulated gastrointestinal digestion. Food Chemistry. 2021;343:128394. doi: 10.1016/j.foodchem.2020.128394. [DOI] [PubMed] [Google Scholar]
- Sarah S., Faradalila W., Salwani M., Amin I., Karsani S., Sazili A. LC–QTOF-MS identification of porcine-specific peptide in heat treated pork identifies candidate markers for meat species determination. Food Chemistry. 2016;199:157–164. doi: 10.1016/j.foodchem.2015.11.121. [DOI] [PubMed] [Google Scholar]
- Siow H.-L., Choi S.-B., Gan C.-Y. Structure–activity studies of protease activating, lipase inhibiting, bile acid binding and cholesterol-lowering effects of pre-screened cumin seed bioactive peptides. Journal of Functional Foods. 2016;27:600–611. doi: 10.1016/j.jff.2016.10.013. [DOI] [Google Scholar]
- Soares R.A.M., Mendonça S., De Castro L.Í.A., Menezes A.C.C.C.C., Arêas J.A.G. Major peptides from amaranth (Amaranthus cruentus) protein inhibit HMG-CoA reductase activity. International Journal of Molecular Sciences. 2015;16(2):4150–4160. doi: 10.3390/ijms16024150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano Santos J., Raúl R.B., Isabel G.-L., Edith P.-A., Bernardo E.-B., César A.-P.…Ruben R.-R. Dipeptidyl peptidase IV inhibitory activity of protein hydrolyzates from Amaranthus hypochondriacus L. Grain and their influence on postprandial glycemia in Streptozotocin-induced diabetic mice. African Journal of Traditional, Complementary and Alternative Medicines. 2015;12(1):90–98. [Google Scholar]
- Soriano-Santos J., Escalona-Buendía H. Angiotensin I-converting enzyme inhibitory and antioxidant activities and surfactant properties of protein hydrolysates as obtained of Amaranthus hypochondriacus L. grain. Journal of Food Science and Technology. 2015;52(4):2073–2082. doi: 10.1007/s13197-013-1223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Wang H., Ma C., Liu C., Gao C., Nie R., Tanver Rahman M.R. Hypocholesterolaemic mechanism of bitter melon aqueous extracts via inhibition of pancreatic cholesterol esterase and reduction of cholesterol micellar solubility. International Journal of Food Sciences and Nutrition. 2016;67(1):20–28. doi: 10.3109/09637486.2015.1121470. [DOI] [PubMed] [Google Scholar]
- Tovar-Pérez E.G., Lugo-Radillo A., Aguilera-Aguirre S. Amaranth grain as a potential source of biologically active peptides: A review of their identification, production, bioactivity, and characterization. Food Reviews International. 2019;35(3):221–245. [Google Scholar]
- Trabuco L.G., Lise S., Petsalaki E., Russell R.B. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Research. 2012;40(W1):W423–W427. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde-Salcedo A.J., González de Mejía E., Barba de la Rosa A.P. In: Hispanic Foods: Chemistry and Bioactive Compounds. Tunick M.H., Mejía E.G.D., editors. American Chemical Society; Washington, DC, USA: 2012. In Vitro evaluation of the antidiabetic and antiadipogenic potential of amaranth protein hydrolysates; pp. 189–198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.