Abstract
Objective
In many cases, genetic testing labs provide their test reports as portable document format files or scanned images, which limits the availability of the contained information to advanced informatics solutions, such as automated clinical decision support systems. One of the promising standards that aims to address this limitation is Health Level Seven International (HL7) Fast Healthcare Interoperability Resources Clinical Genomics Implementation Guide-Release 1 (FHIR CG IG STU1). This study aims to identify various data content of some genetic lab test reports and map them to FHIR CG IG specification to assess its coverage and to provide some suggestions for standard development and implementation.
Materials and Methods
We analyzed sample reports of 4 genetic tests and relevant professional reporting guidelines to identify their key data elements (KDEs) that were then mapped to FHIR CG IG.
Results
We identified 36 common KDEs among the analyzed genetic test reports, in addition to other unique KDEs for each genetic test. Relevant suggestions were made to guide the standard implementation and development.
Discussion and Conclusion
The FHIR CG IG covers the majority of the identified KDEs. However, we suggested some FHIR extensions that might better represent some KDEs. These extensions may be relevant to FHIR implementations or future FHIR updates.
The FHIR CG IG is an excellent step toward the interoperability of genetic lab test reports. However, it is a work-in-progress that needs informative and continuous input from the clinical genetics’ community, specifically professional organizations, systems implementers, and genetic knowledgebase providers.
Keywords: biomedical informatics, FHIR, genetic test reporting, interoperability, genetic results
INTRODUCTION
Genetic test reports are the main communication channel between clinical testing laboratories, clinicians, and genetic counselors. These reports usually contain descriptions of identified genetic variants, testing methods, interpretations, and recommended actions.1 However, these reports are usually provided as portable document formats or scanned images, limiting their computational availability for both primary and secondary uses of their information content. Nevertheless, emerging interoperability standards offer an opportunity to support a standards-based exchange, processing, and using clinical genetic information included in genetic lab test reports.2
Health Level Seven International (HL7) provides a list of standards that can support computable and standards-based interoperability between genetic testing labs and hospitals.3 One standard is the Fast Healthcare Interoperability Resources Clinical Genomics Implementation Guide-Release 1 (FHIR CG IG STU1) released in November 2019.4 FHIR CG IG is a promising standard representing genetic testing information at 2 levels: the laboratory observation level and the diagnostic report level. The diagnostic report may include one or more genetic observations, such as single nucleotide variants, cytogenetic abnormalities, and large copy number variations. The diagnostic report also tries to structure the interpretation sections and provides a suggested standards-based coding approach. The FHIR CG IG includes numerous and interconnected data elements that may appear different to those familiar with the usual organization of conventional genetic test reports. FHIR CG IG intends to serve a broad set of genetic test types. Therefore, moving this standard from the specification phase to the implementation phase requires matching the information content of the result report of a specific test type to the corresponding data elements of FHIR CG IG. A previous publication describes a similar effort that was made for whole-exome sequencing test reports.5 However, many other test types need to be considered.
The purpose of this study was to identify key data elements (KDEs) in the result reports of 4 types of genetic tests and map them to FHIR CG IG resources and their attributes. To avoid missing any recommended KDEs, we also analyzed relevant professional reporting guidelines provided by organizations such as the American College of Medical Genetics and Genomics (ACMG), the Association for Molecular Pathology (AMP), the International System for Human Cytogenomic Nomenclature (ISCN), and the European Society of Human Genetics (ESHG).
BACKGROUND ON FAST HEALTHCARE INTEROPERABILITY RESOURCES
Fast Healthcare Interoperability Resources (FHIR) is an HL7 standard that specifies how to represent clinical information and knowledge, and how to query, retrieve, and send this information.6 The foundation of FHIR is the Resource that represents an entity or a concept, such as Patient, or a clinical Condition. Throughout this paper, FHIR Resource names are italicized.
FHIR Resources are connected to each other to “build a web of information about healthcare” using a referencing approach, where each Resource can reference other relevant Resources.7 For example, the Condition resource references Patient and Practitioner who are involved in asserting this clinical condition. This referencing approach ensures granularity while facilitating the integration of various Resource types.
Each Resource is modeled to include a predefined set of attributes that describe the corresponding entity or concept. For example, the Patient resource contains the following attributes: name, date of birth, patient identifiers, contact information, and a patient’s photo. Each attribute has a specific data type and cardinality.8 For example, the Patient date of birth has the “date” data type and can either be absent or mentioned only one time. The FHIR data type “CodeableConcept” allows for representing attributes in one or more codes in addition to its descriptive text.9 The code may be bound to a standard terminology system, such as LOINC and SNOMED-CT.10,11 This approach of describing attributes using specific standard terminologies is called “terminology binding.”
Resources can be represented using various formats, such as JavaScript Object Notation (JSON) and Extensible Markup Language (XML), and can be searched, retrieved, and sent using representational state transfer (RESTful) application program interfaces.8,12 This adoption of widely used web technologies facilitates the development and implementation of numerous informatics tools, such as clinical decision support systems (CDSs). In addition, Resources can be customized to fit specific use cases. This customization process is called “profiling” or “to profile,” while the profiled Resource is called “profile.”13 For example, GenomicsReport is a profile of the more general FHIR DiagnosticReport resource, customized to better represent the genomic lab test reporting.14
FHIR Resources, terminology bindings, specified communication approaches, and FHIR profiles are paramount for the standards-based interoperability of biomedical data among various stakeholders, such as hospitals and testing laboratories.
FHIR Genomics includes resources, profiles, extensions, and implementation guides around clinical genomics—from family history pedigrees to molecular sequencing.15 FHIR has evolved over the years and has been leveraged for apps that integrate with Electronic Health Records (EHR’s), labs, and/or Genomic Archiving Communication System (GACS) for precision medicine use cases.16 FHIR CG IG STU1 is part of FHIR Genomics and will be the focus of this paper.
MATERIALS AND METHODS
We used a qualitative approach to identify KDEs within genetic lab test reports and professional reporting guidelines and aligned them to FHIR CG IG (STU1).4 We also sought KDEs that are not currently represented in the FHIR implementation that could be considered for inclusion by HL7 in future FHIR CG IG updates. This study was submitted to the Institutional Review Boards of the University of Utah and Intermountain Healthcare (IHC) and deemed exempted on January 17, 2019, and April 1, 2019, respectively. We describe the steps we followed in the subsequent sections.
Sample reports and KDEs
We retrieved tallies of genetic test types that were ordered by IHC17 and performed by ARUP Laboratories18 from December 2017 to November 2018, then we identified the most frequently ordered test types . AK retrieved their corresponding sample reports from the ARUP web portal18 and conducted a preliminary analysis of these report samples to identify the corresponding testing method and the included report sections, such as patient information, lab information, genetic result, and interpretation.
The project’s principal investigator (SMH) was consulted to select 4 test types to be included in this study based on the preliminary analysis and in his capacity as the chief medical informatics officer of IHC. The selection criteria include:
Frequently ordered genetic tests, to impact large patient population.
Test method that is common among other tests, to support future analyses and interoperability projects.
SMH provided additional deidentified sample reports for the selected test types, and AK analyzed them for KDEs, while SMH reviewed the results of the analysis. We followed a saturation sampling approach, where we obtained 5 sample reports for each test type, and if we identified additional KDEs in each set of reports, we could analyze additional samples obtained from IHC until saturation occurred.
Mappings
For each of the 4 test types selected, the corresponding professional reporting guidelines were identified, and the recommended information content was mapped to the initial set of identified KDEs. If a new KDE was identified, it was flagged as not present in the sample reports and included in the whole set of KDEs per each type of test report.
AK mapped the complete set of KDEs to the corresponding elements of the FHIR CG IG (STU1).4 We made some suggestions to better represent some KDEs as extensions (that may alternatively be done as additionally constrained coded elements, in some cases) to be considered in future updates of the implementation guide. The study team reviewed the results to validate the identified KDEs and their corresponding mappings. The study team includes experts in genomic medicine, biomedical informatics terminologies and communication standards, laboratory information systems, and health information exchange. Suggestions were collected and required edits were made, such as merging, deleting, and adding some KDEs and their corresponding mappings.
We did not consider mapping reporting guidelines that were not related to data requirements and structure, such as recommended formatting, clarity of the report, conciseness, sections, and pagination. However, if the reporting guideline clearly states the need to provide a narrative description for a coded KDE, we considered it an additional KDE. For example, we considered the ISCN nomenclature of a cytogenetic change and its corresponding narrative description as 2 KDEs, although they may be modeled as one element from the FHIR perspective.
RESULTS
We retrieved the results of 187 tests between December 2017 and November 2018. Twenty-two test types were ordered over 49 times throughout this period. The top 22 test type reports were subjected to the preliminary analysis for the testing methods, report sections, and broad information categories, such as patient, result, and interpretation.
Test types and relevant professional reporting guidelines
We had selected 4 test types for the study, and their corresponding professional reporting guidelines were identified and analyzed, as mentioned in Table 1.
Table 1.
Selected genetic test types and their corresponding reporting guidelines
| Test name | Reporting guidelines |
|---|---|
| Chromosome Analysis Bone Marrow (CHR-BM)19 | “Section E6.1–6.4 of the ACMG technical standards and guidelines: chromosome studies of neoplastic blood and bone marrow–acquired chromosomal abnormalities”20 |
| Methylenetetrahydrofolate Reductase (MTHFR) 2 Variants21 | “Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology”22 |
| Cytogenomic SNP Microarray (CMA-SNP)23 |
|
| Chromosome FISH Interphase (CHR-FISHI)31 |
|
Mappings
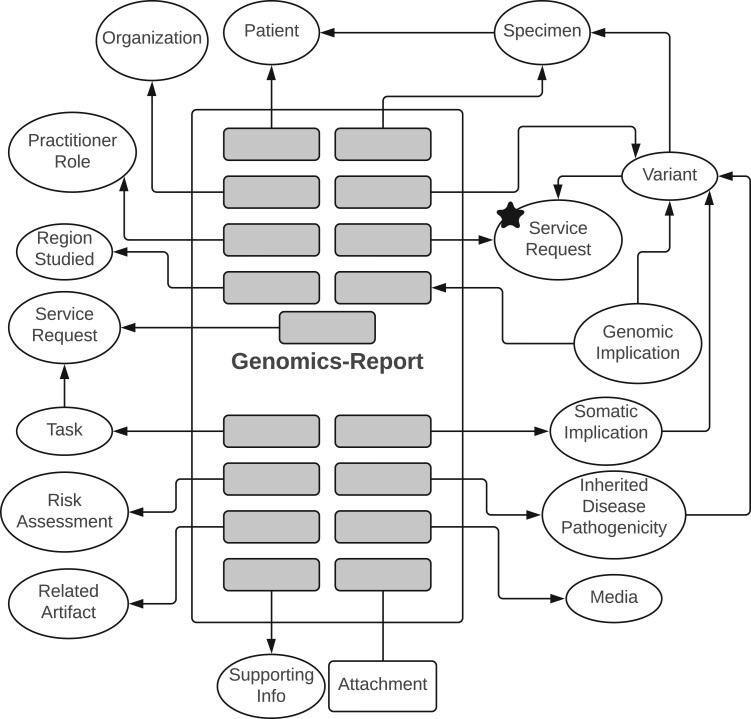
We retrieved and analyzed at least 7 sample reports per test type. Two were available from the ARUP web portal, and the rest were deidentified and retrieved from IHC. KDEs were relatively uniform within each set of sample reports. The sample reports were very similar in their information content (ie, KDEs). Therefore, saturation was reached in the first round of sampling and review, and no additional reports were needed. Table 2 describes the identified information categories and their corresponding FHIR resources and profiles. Some of the identified profiles are abstract profiles, that is, nonimplementable profiles that represent common data elements shared by conformant implementable profiles.4 A KDE may be included in more than one resource or profile according to the use case and the agreement between the lab and the hospital. For example, the patient’s KDEs and their corresponding Patient’s attributes are part of the Specimen, Variant, and GenomicsReport profiles. Figure 1 depicts an illustrative subset of the most informative relations between the identified FHIR Resources (simplified for clarity). The ServiceRequest (marked with a star in Figure 1) would be the first resource to be created in an FHIR-based communication between the hospital and the lab. Table 3 includes sample mappings of the “chromosome analysis bone marrow” lab test report.
Table 2.
Data categories and relevant FHIR resources
| Data category | Corresponding FHIR Resources | Examples of key data elements |
|---|---|---|
| Patient | Patient | Name, identifiers, date of birth |
| Physician | Practitioner, PractitionerRole | Name, qualification, contact information |
| Test interpreter | Practitioner, PractitionerRole | Name, qualification, contact information |
| Hospital | Organization | Name, identifiers, contact information |
| Laboratory | Organization | Name, identifiers, contact information |
| Testing method | Variant, §Genomic Implication, RegionStudied | How is DNA obtained and analyzed, test limitation, testing probe that was used |
| Specimen | Specimen | Type, body location, collection date |
| Genetic finding description | Variant, RegionStudied, *Genotype, *Haplotype, *§GenomicFinding, *SequencePhase | Gene name, variant nomenclature, cytogenetic location |
| Result interpretation | GenomicsReport, RiskAssessment, §Genomic Implication, §SomaticImplication, InheritedDiseasePathogenicity, *§MedicationImplication | Disease or condition association, penetrance, inheritance, risk |
| Recommendation | GenomicsReport (RecommendedAction), Task, ServiceRequest, *MedicationRequest, *RequestGroup, CarePlan, *DeviceRequest, *NutritionOrder, *SupplyRequest | Follow-up test, family testing, consultation |
| Image or attachment | Media, Attachment | Karyotype (slide image), patient questionnaire, report as portable document format |
| Note or disclaimer | RelatedArtifact, SupportingInfo, Variant(note) | Disclosure of recessive carrier status, submission to public databases, informed consent requirement |
| Educational material | RelatedArtifact, SupportingInfo | Support or advocacy group, related organization, educational web page |
| Test order | ServiceRequest, *Questionnaire | Date, requisition number, reason for referral |
| Report | GenomicsReport | Accession number, date, status (eg, preliminary, or final) |
Notes: “*” indicates that the corresponding FHIR Resource was not used in mapping but may be used by other test types. “§” indicates that the corresponding FHIR Profile is an abstract profile.
FHIR: Fast Healthcare Interoperability Resources.
Figure 1.
Genomic lab test report as a composition of multiple FHIR Resources. The star indicates the first resource that may need to be established in an FHIR-based communication. There are 2 ServiceRequest resources: one represents the test order that initiated the conducted genetic test (denoted by star), while the other represents future services that are recommended based on the test results. More details are in Table 2. Arrows directions represent referencing directions between depicted FHIR resources and profiles. “Attachment” is an FHIR datatype. FHIR: Fast Healthcare Interoperability Resources.
Table 3.
Sample mappings of Chromosome Analysis Bone Marrow lab test report
| Data element | ACMG [reference] | FHIR GenomicsReport | FHIR Variant | Extension needed | Comment on FHIR mapping |
|---|---|---|---|---|---|
|
Yes | Yes (as a reference to ServiceRequest resource that represents the test order) | Yes | Yes | The current mapping perfectly accommodates the required information. However, an extension could be added to identify the indication for the test within the GenomicsReport or the Variant level. Because, the ServiceRequest resource may not be available for referencing, and extra effort from the lab or the EHR may be needed in order to be developed. |
| Test performed | Not mentioned | Yes | Yes | No | This is a codableConcept that can accommodate multiple codes and descriptions, either locally defined or according to standards terminologies. |
| Number of cells karyotyped | Not mentioned | Yes | Not available | Yes | A dedicated extension to describe the number of cells counts, analyzed, and karyotyped is needed. This could be a slice with the procedure code as the “Discriminator.” |
| ISCN band level | Not mentioned | Yes | Yes | No | This can be described by the “method” element as one of the codings of this codableConcept. |
| Banding method | Not mentioned | Yes | Yes | No | |
| A note about performing a related test | Yes | Yes | Yes | No |
|
| Electronic signature of the approving physician | Not mentioned | Yes | Not applicable | Yes |
|
Table 4 includes numbers of KDEs per test type, KDEs not identified in sample reports but recommended by reporting guidelines, and suggested FHIR extensions. It also includes the total number of common KDEs among all tests. These common KDEs include data about patient, test, test report, ordering physician, referring hospital, method of testing, the reason for referral, attachments, various notes, and disclaimers. The detailed mappings are provided in the Supplemental Material.
Table 4.
Tallies of KDEs per test type and common KDEs
| Test type | Total KDEs | KDEs not in sample reports*, n (%) | Suggested FHIR extensions |
|---|---|---|---|
| Chromosome analysis bone marrow | 45 | 5 (11) | 11 |
| Methylenetetrahydrofolate Reductase (MTHFR) 2 variants | 63 | 18 (29) | 21 |
| Cytogenomic SNP microarray | 83 | 24 (29) | 31 |
| Chromosome FISH interphase | 83 | 41(49) | 33 |
| Common KDEs (eg, patient, hospital, ordering physician, specimen, and test report) | 36 | 5 (14) | 10 |
KDEs not in sample reports should not imply noncompliance with reporting guidelines. See “Discussion” section for more detailed explanation.
Suggested extensions
A total of 46 unique extensions among all test reports are suggested to better represent the identified KDEs. Table 5 lists these extensions and categorizes them into 9 groups. More details about the proposed extensions are provided in the Supplemental Material (Extensions Sheet).
Table 5.
Suggested extensions and their corresponding groups
| Extension groups | Extensions |
|---|---|
| Testing methods | An additional method of testing; investigational nature of the testing; Probe(s) used; Resolution of the cytogenetic analysis; Gene amplification |
| Test limitations and sensitivity | Limitations; Analytical Sensitivity and Specificity; Clinical Sensitivity |
| Inheritance | Inheritance pattern; Carrier status; Recessive nature of the condition |
| Cell source and tallies | Cell source; Number of cells counted; Number of cells analyzed; Number of cells karyotyped; Total number of abnormal cells; Percentage of abnormal cells |
| Genetic findings | Penetrance; Incidental (secondary) finding; Historical variant nomenclature; variants refractory to detection by this method; In silico analysis summary; Variants of uncertain significance; DNA region name; level of mosaicism; Variant implication; Constitutional versus clonal abnormality; Chromosome modal number; Genetic predisposition concerning the associated disease/condition; Sex chromosome complement; The total number of genes in the detected copy number variation interval |
| Assessment of recurrence | Future pregnancy diagnosis; Assessment of recurrence |
| Notes and disclaimers | Disclaimer; Compliance Statement [Web address]; Note-Informed Consent requirement; A note that the test is not diagnostic and needs confirmation; Note about the disclosure of recessive carrier status; Submission to public databases note |
| Patient educational materials and resources | Patient and family education materials; Clinical trials; Research groups; Related organizations and patient advocacy and support groups |
| Reason for referral | Reason for referral and Indication for testing |
| Miscellaneous | A brief description of the lab; Age; Electronic Signature of the approving physician |
DISCUSSION
FHIR CG IG can convey most of the KDEs we identified from the sample test reports and their corresponding professional reporting guidelines. Some of the identified KDEs may be better considered as additional FHIR extensions or additional constrained codes by the systems implementing FHIR specification for these specific reports. The suggested extensions may be considered for inclusion in future releases of the FHIR CG IG. Therefore, we encourage similar effort to describe more clinically actionable details for genomic use cases, such as risk assessments and service requests.
The current FHIR CG IG can describe almost all of the genetic variations identified within the sample reports. Some KDEs can be represented directly by the FHIR CG IG GenomicsReport profile, while other KDEs are better represented by the Variant, or Genomic Implication-derived profiles. Some KDEs may be represented by both profiles, such as patient identifiers, and the performing lab. In these cases, the mapping considered both profiles for the corresponding FHIR data elements. This is not redundant as each resource is modeled according to the broadest set of use cases it may be used for and the required attributes that may support these use cases. Almost all KDEs can be represented as narrative data elements. However, this limits their computability, such as use by CDSs and machine learning research tools.
KDEs that were not mentioned by the corresponding reporting guideline may be an important information that are naturally expected as part of the test report or covered by another reporting guideline that we did not review, such as test name, and number of cells karyotyped. KDEs that were not identified in the sample reports, yet recommended by professional reporting guidelines, do not imply that the sample reports do not follow the reporting guidelines. They may not fit such specific test type but may fit other tests that employ the same testing methods. Similarly, the suggested FHIR extensions do not imply that the current FHIR implementation guide does not cover this information. However, these suggestions may help labs better implement FHIR CG IG for these specific use cases and their agreement with their client hospitals and clinics.
Although the suggested extensions may play direct roles in patients care (eg, carrier status, recurrence, and inheritance pattern), they also may serve other purposes including patient education, quality control, and scientific research. For example, patient and family educational materials can be identified and automatically communicated to the patients or their families. Testing methods, cell sources, and tallies extensions may support quality assurance purposes. In addition, extensions such as variants of uncertain significance and variants refractory to detection could support various research purposes and evidence generation. This information if conveyed in a structured approach can make the development of relevant medical application more efficient for both clinical and nonclinical purposes.
It is essential to note that the genetic lab test report is not only a description of genetic variants and additional interpretation sections. The reports are also the means of communication between the testing lab, the clinician, and the healthcare system. The optimal interpretation of a genetic result may need additional information from the clinician besides the standard information that accompanies the test requisition, such as patient medical history.33 Although these information types are not part of the actual lab test report, FHIR considers them part of the clinical context from which the interpretation would be concluded. Where a resource that describes a lab test request will be referenced by another resource that describes the lab observation made, and in turn, both of them will be referenced by the FHIR diagnostic report. These linked resources allow efficient traversing of the available information as needed for either primary or secondary use of data.
The report may also contain disease associations, risk assessments, recommendations, and other clinically actionable details. These types of information elements can be represented using the general FHIR artifacts, such as Questionnaire, RiskAssessment, and ServiceRequest resources.34 FHIR CG IG already makes use of some of these resource types, but it has not customized (ie, profiled) them to fit genomic medicine use cases. Profiling efforts need continuous input from the clinical genetics community and relevant professional organizations to ensure their feasibility, such as ACMG, AMP, College of American Pathologists (CAP), ISCN, and ESHG.
In addition, the reports usually contain substantial genetic information that is retrieved from genetic knowledge bases, such as disease associations, reference sequences, genetic locations, and recommended genetic tests. Therefore, genetic knowledge bases and professional organizations can play a significant role in genetic data interoperability by directly providing their content and guidelines as FHIR resources. These resources can then be readily used and referenced by FHIR Variant and GenomicsReport profiles. FHIR is already considering these kinds of knowledge representations independent of a specific patient, such as Evidence and ActivityDefinition resources.35–37
Systematic identification of relevant KDEs across multiple healthcare providers and testing labs would facilitate the development and implementation of supporting interoperability standards. Professional and standard development organizations such as ACMG and HL7 encourage the participation of subject matter experts and relevant stakeholders to identify reporting guidelines and frequently used data elements to be standardized. However, healthcare providers and testing labs can support these organizations by sharing enough samples of their testing reports and clear information about how they communicate and use these reports, for example, using specific standards, or as free text. Researchers, subject matter experts, and stakeholder representatives would be able to systematically analyze these reports and information to identify currently available KDEs, desired information content, and their optimal representations. Moreover, machine learning and artificial intelligence methods may be used to support this identification process. This corpus of data would provide real use cases and statistics of KDEs across multiple organizations and domains.
More involvement from the professional community will make this effort even more valuable to genetic lab testing use cases and to genomic medicine as a whole. They may incorporate these types of informatic standards within their reporting guidelines following the method we described in this paper but in a larger scale. We also encourage the genetic community to support FHIR workgroups in building and customizing these resource types for clinical genetic use cases. Investment in this kind of effort will benefit the efficiency and outcomes of both clinical practice and genomic medicine research, which are the main components of a learning health system for genomic medicine.
Strengths and limitations
This study is a fundamental analysis of optimal development and implementation of FHIR CG IG for specific use cases. We analyzed frequently ordered test types ordered by a large healthcare system (IHC) and produced by a large reference lab (ARUP); and provided detailed mappings for KDEs in those reports. Therefore, the results provide further guidance beyond the FHIR CG IG for the implementation of a standards-based health information exchange.
This study has several limitations. First, we did not study other promising test types that involve next-generation sequencing because these test types were not frequently ordered by IHC. This probably represents the timeframe of the study, as next-generation sequencing was just beginning to emerge in clinical practice in 2017–2018. Other investigators have begun to study these reports5 and this will be an important focus for our future research. Second, although we followed a saturation sampling approach, we may have missed some KDEs, and prospective analysis with a larger sample size may identify other KDEs. Third, we reviewed only a limited set of guidelines, although these are the most frequently referenced guidelines by referral testing laboratories in the United States. However, other guidelines may be present and were not considered by our study. Fourth, a more comprehensive assessment of the landscape is necessary to inform the development of an international standard.
During the publishing process of this manuscript, FHIR CG IG STU2 was subject to balloting (Ballot 1). We encourage healthcare providers and system vendors to consider the current version of the standard for implementation and interpret our results in light of it.
We tried to provide a computable representation for each of the identified KDEs. However, some stakeholders may find the text representation of KDEs to be sufficient. The KDEs, mapping decisions, and suggestions are based on the study team’s point of view. While the study team had representation of a number of stakeholder perspectives: geneticists, information modelers, and clinicians, others may have different points of view. We view this report as a starting point for discussions that can provide a more in-depth analysis to inform development of the standard. We are confident that informed and transparent discussions on KDEs and best modeling and standardization approaches by all stakeholders are paramount to achieving an optimal health information exchange.
Future work
We look forward to studying additional genetic testing report types to support their FHIR implementations, especially methods utilizing next-generation sequencing technologies. We also aim to study how FHIR can support more genomic medicine use cases besides genetic test reporting.
CONCLUSION
We identified KDEs in the results report of 4 frequently ordered genetic tests and their relevant professional reporting guidelines and mapped them to the current FHIR CG IG data elements. The FHIR CG IG covers the majority of the identified KDEs. However, we provided some suggested FHIR extensions that might better represent some KDEs in structured and standards-based formats. These extensions may be relevant to FHIR implementations or future FHIR updates.
The FHIR CG IG is an excellent step toward the interoperability of genetic lab test reports. However, it is a work-in-progress that needs informative and continuous input from the clinical genetics community—specifically, professional organizations, systems implementers, and genetic knowledgebase providers. This collaboration would make the genetic lab test reports more interoperable, actionable, and accessible at different levels of the learning health system.
AUTHOR CONTRIBUTIONS
AK and SMH obtained the sample reports. AK analyzed the reports and extracted KDEs. SMH reviewed the identified KDEs. AK mapped the identified KDEs to FHIR CG IG (STU1) and made some suggestions for its future development and implementation. AK, CCM, JHG, MSW, GDF, BRJ, SBB, GA, and SMH reviewed the mapping and the suggestions. AK and SMH wrote the first draft of the manuscript, and CCM, JHG, MSW, GDF, BRJ, SBB, and GA revised it critically. All authors approved the final version for submission and agree to be accountable for all aspects of the work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article are available in the article and in its online supplementary material. However, the Institutional Review Board (IRB) restricts the sharing of patients’ reports. Therefore, those reports are not available. We provided links to similar sample reports provided publicly by ARUP Laboratories.
Supplementary Material
REFERENCES
- 1.Technical Standards and Guidelines. https://www.acmg.net/ACMG/Medical-Genetics-Practice-Resources/Technical_Standards_and_Guidelines.aspx Accessed September 30, 2020.
- 2.ACMG Board of Directors. Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genet Med 2017; 19: 721–2. [DOI] [PubMed] [Google Scholar]
- 3.Clinical Genomics—Products | HL7 International. http://www.hl7.org/Special/committees/clingenomics/products.cfm Accessed October 1, 2020.
- 4.HL7 Standards Product Brief—HL7 FHIR® Implementation Guide: Clinical Genomics, Release 1 | HL7 International. http://www.hl7.org/implement/standards/product_brief.cfm?product_id=508 Accessed September 30, 2020.
- 5. Swaminathan R, Huang Y, Astbury C, et al. Clinical exome sequencing reports: current informatics practice and future opportunities. J Am Med Inform Assoc 2017; 24 (6): 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Summary—FHIR v4.0.1. https://www.hl7.org/fhir/summary.html Accessed January 18, 2020.
- 7.References—FHIR v4.0.1. https://www.hl7.org/fhir/references.html Accessed October 4, 2020.
- 8.Resource Formats—FHIR v4.0.1. https://www.hl7.org/fhir/formats.html Accessed October 4, 2020.
- 9.Datatypes—FHIR v4.0.1. https://hl7.org/fhir/datatypes.html#CodeableConcept Accessed October 1, 2020.
- 10.LOINC Home Page. https://loinc.org/ Accessed March 20, 2020.
- 11.SNOMED Home Page. https://www.snomed.org/ Accessed March 20, 2020.
- 12.Http—FHIR v4.0.1. https://www.hl7.org/fhir/http.html Accessed October 16, 2020.
- 13.Profiling—FHIR v4.0.1. https://www.hl7.org/fhir/profiling.html Accessed October 4, 2020.
- 14.Genomics Reporting Implementation Guide. http://hl7.org/fhir/uv/genomics-reporting/STU1/genomics-report.html Accessed October 4, 2020.
- 15. Alterovitz G, Warner J, Zhang P, et al. SMART on FHIR genomics: facilitating standardized clinico-genomic apps. J Am Med Inform Assoc 2015; 22 (6): 1173–8. [DOI] [PubMed] [Google Scholar]
- 16. Alterovitz G, Heale B, Jones J, et al. FHIR genomics: enabling standardization for precision medicine use cases. NPJ Genomic Med 2020; 5: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Intermountain Healthcare Home Page. https://intermountainhealthcare.org/ Accessed October 1, 2020.
- 18.ARUP Laboratories Home Page. https://www.aruplab.com/ Accessed September 30, 2020.
- 19.Chromosome Analysis, Bone Marrow | ARUP Lab Test Directory. https://ltd.aruplab.com/Tests/Pub/2002292 Accessed September 15, 2020.
- 20. Mikhail FM, Heerema NA, Rao KW, et al. Section E6.1–6.4 of the ACMG technical standards and guidelines: chromosome studies of neoplastic blood and bone marrow–acquired chromosomal abnormalities. Genet Med 2016; 18 (6): 635–42. [DOI] [PubMed] [Google Scholar]
- 21.Methylenetetrahydrofolate Reductase (MTHFR) 2 Variants | ARUP Lab Test Directory. https://ltd.aruplab.com/Tests/Pub/0055655 Accessed September 15, 2020.
- 22. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17 (5): 405–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cytogenomic SNP Microarray | ARUP Lab Test Directory. https://ltd.aruplab.com/Tests/Pub/2003414 Accessed September 15, 2020.
- 24. Kearney HM, Thorland EC, Brown KK, et al. ; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011; 13 (7): 680–5. [DOI] [PubMed] [Google Scholar]
- 25. Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med 2020; 22 (2): 245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rehder CW, David KL, Hirsch B, et al. American College of Medical Genetics and Genomics: standards and guidelines for documenting suspected consanguinity as an incidental finding of genomic testing. Genet Med 2013; 15 (2): 150–2. [DOI] [PubMed] [Google Scholar]
- 27. Kearney HM, South ST, Wolff DJ, et al. ; Working Group of the American College of Medical Genetics. American College of Medical Genetics recommendations for the design and performance expectations for clinical genomic copy number microarrays intended for use in the postnatal setting for detection of constitutional abnormalities. Genet Med 2011; 13 (7): 676–9. [DOI] [PubMed] [Google Scholar]
- 28. Silva M, de Leeuw N, Mann K, et al. European guidelines for constitutional cytogenomic analysis. Eur J Hum Genet 2019; 27 (1): 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Claustres M, Kožich V, Dequeker E, et al. ; European Society of Human Genetics. Recommendations for reporting results of diagnostic genetic testing (biochemical, cytogenetic and molecular genetic). Eur J Hum Genet 2014; 22 (2): 160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30., , . Stevens-Kroef M, Simons A, Rack K, Hastings RJ. Cytogenetic Nomenclature and Reporting. In: Wan T, ed. Cancer Cytogenetics. Methods in Molecular Biology, vol. 1541. New York, NY: Humana Press; 2017. 10.1007/978-1-4939-6703-2_24 [DOI] [PubMed] [Google Scholar]
- 31.Chromosome FISH, Interphase | ARUP Lab Test Directory. https://ltd.aruplab.com/Tests/Pub/2002298 Accessed September 15, 2020.
- 32. Mascarello JT, Hirsch B, Kearney HM, et al. ; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. Section E9 of the American College of Medical Genetics technical standards and guidelines: fluorescence in situ hybridization. Genet Med 2011; 13 (7): 667–75. [DOI] [PubMed] [Google Scholar]
- 33. Grebe TA, Khushf G, Chen M, et al. ; ACMG Social, Ethical and Legal Issues Committee. The interface of genomic information with the electronic health record: a points to consider statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2020; 22 (9): 1431–6. [DOI] [PubMed] [Google Scholar]
- 34.Resource Index—FHIR v4.0.1. https://hl7.org/fhir/resourcelist.html Accessed October 1, 2020.
- 35.Representing Knowledge Artifacts—FHIR v4.0.1. https://www.hl7.org/fhir/clinicalreasoning-knowledge-artifact-representation.html Accessed October 1, 2020.
- 36.Evidence Resource—FHIR v4.0.1. https://hl7.org/fhir/evidence.html Accessed October 1, 2020.
- 37.Activity Definition Resource—FHIR v4.0.1. http://www.hl7.org/fhir/activitydefinition.html Accessed October 16, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. However, the Institutional Review Board (IRB) restricts the sharing of patients’ reports. Therefore, those reports are not available. We provided links to similar sample reports provided publicly by ARUP Laboratories.