Abstract
Background
Cranial radiation therapy is essential in treating many pediatric cancers, especially brain tumors; however, its use comes with the risk of developing second malignancies. Cranial radiation-induced gliomas (RIGs) are aggressive high-grade tumors with a dismal prognosis, for which no standard therapy exists. A definitive molecular signature for RIGs has not yet been established. We sought to address this gap by performing a systematic review and meta-analysis of the molecular features of cranial RIGs.
Methods
A systematic review of the literature was performed according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines. Articles and case reports that described molecular analyses of cranial radiation-induced high-grade gliomas were identified and evaluated, and data extracted for collation.
Results
Of 1727 records identified, 31 were eligible, containing 102 unique RIGs with molecular data. The most frequent genetic alterations in RIGs included PDGFRA or TP53 mutations, PDGFRA or CDK4 amplifications, and CDKN2A deletion, along with 1q gain, 1p loss and 13q loss. Of note, mutations in ACVR1, EGFR, H3F3A, HIST1H3B, HIST1H3C, IDH2, SMARCB1 or the TERT promoter were not observed. A comparative analysis revealed that RIGs are molecularly distinct from most other astrocytomas and gliomas and instead align most closely with the pedGBM_RTK1 subgroup of pediatric glioblastoma.
Conclusions
This comprehensive analysis highlights the major molecular features of RIGs, demonstrates their molecular distinction from many other astrocytomas and gliomas, and reveals potential genetic drivers and therapeutic targets for this currently fatal disease.
Keywords: Radiation-induced glioma, molecular, pediatric, radiation, cancer
Key Points.
A comprehensive meta-analysis of the molecular features of radiation-induced glioma.
Radiation-induced gliomas are genetically distinct from most other brain tumors.
Radiation-induced gliomas share many genetic features with pedGBM_RTK1 pediatric glioblastoma.
Importance of Study.
Investigations into the genetic features of radiation-induced gliomas (RIGs) are few and are limited by the small number of cases in each study. Consequently, a definitive molecular signature for RIGs has not yet been established, highlighting a gap in the knowledge of potentially actionable drivers of this aggressive disease. To our knowledge, this is the most comprehensive systematic review and meta-analysis of the genetic features of cranial RIGs. This study identified recurrent molecular alterations in these tumors and demonstrated that they are molecularly distinct from many other brain tumor types that they are commonly diagnosed and treated as. Despite most RIGs being diagnosed during adulthood, we identified that RIGs share the largest genetic overlap with the pedGBM_RTK1 subtype of pediatric glioblastoma, which may have implications for future clinical management. These findings reveal that molecular classification of RIGs may complement existing tools for pathological diagnosis of these tumors in the future.
Cranial radiation therapy is a key treatment modality for many pediatric cancers, particularly brain tumors where radiotherapy is routinely delivered to the brain or entire craniospinal axis. Cranial or craniospinal irradiation is associated with a myriad of significant long-term complications1,2 including the development of second malignant neoplasms.3–5 The cumulative risk of developing a brain tumor following cranial radiation therapy ranges from 0.5% to 2.7% at 15 years.6 A recent study of 1294 medulloblastoma patients treated with radiation therapy between 1973 and 2014 reported that these patients developed second central nervous system (CNS) tumors at 40 times the rate expected in the general population.4 Furthermore, several large cohort studies have demonstrated a direct correlation between the cumulative dose of radiation received and the risk of subsequent CNS tumor development.5,7,8 This is particularly pertinent for young children, as those under 5 years of age are more susceptible to the development of radiation-associated gliomas compared with children receiving radiotherapy at a later age.5
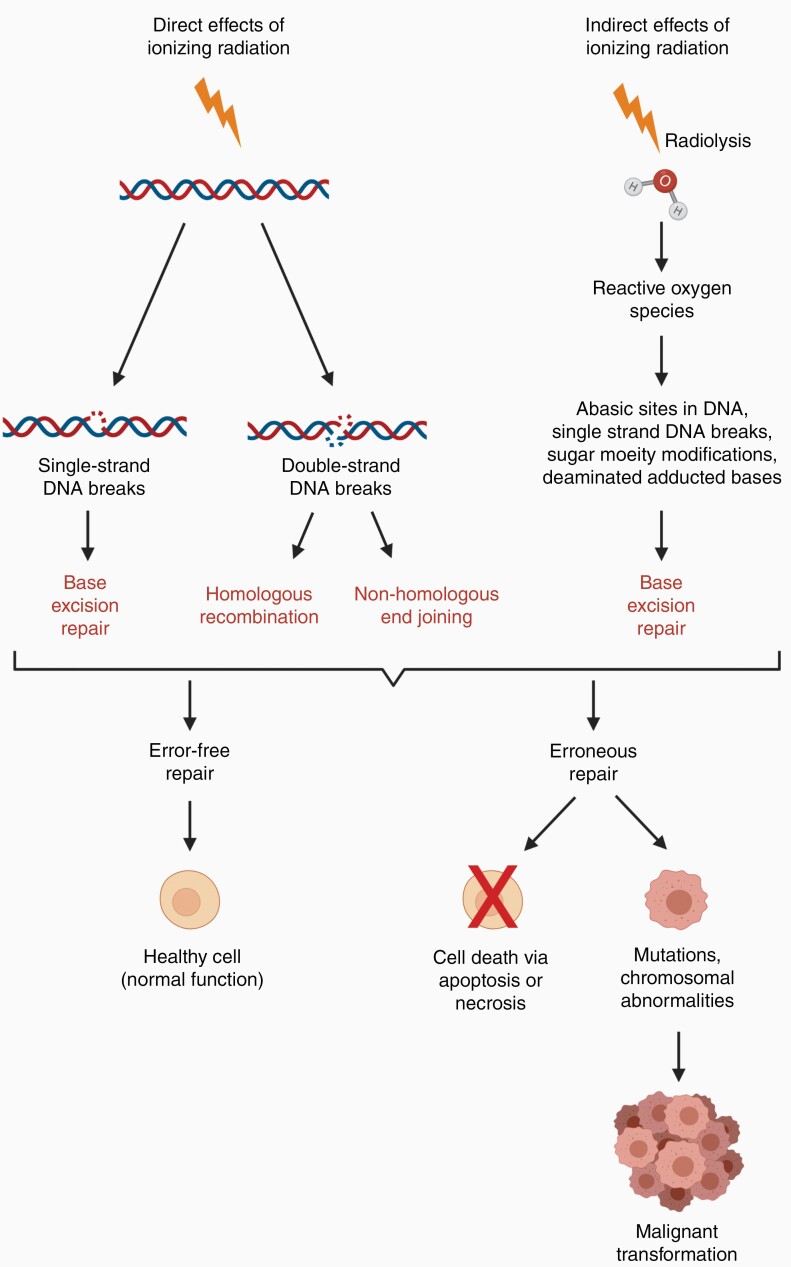
Ionizing radiation directly damages DNA by inducing both single- and double-strand breaks, with the latter being the most deleterious.9 Indirect DNA damage can also occur via radiolysis of water molecules which produces reactive oxygen species, in turn causing single-strand breaks and other alterations to DNA10,11 (Figure 1). Imperfect repair of this damage can result in point mutations, gene fusions, large-scale deletions or translocations, all with the potential to activate oncogenes or inactivate tumor-suppressor genes. These changes are often associated with ongoing genomic instability and thus an increased risk of developing cancer.9,11,12 In the case of radiation-induced second malignancies, genomic instability is thought to persist for multiple generations of cells over many years prior to oncogenic transformation, resulting in a significant latency period between the exposure event and the development of radiation-induced cancer.12
Figure 1.
Mechanisms of DNA damage caused by ionizing radiation. Ionizing radiation damages DNA both directly and indirectly. Depending on the type of damage caused, cells will attempt to repair DNA lesions by base excision repair, homologous recombination or non-homologous end-joining. Successful repair results in a healthy cell with normal function whereas unsuccessful repair may result in cell death or the accumulation of mutations or chromosomal abnormalities, potentially leading to malignant transformation.
A seminal study by Cahan et al13 established a widely accepted set of criteria that define radiation-induced malignancies. These are: (1) the tumor must arise within the irradiated field, (2) a sufficient latency period must have passed between the time of irradiation and the development of the second tumor (measured in years), (3) the second tumor must be histologically distinct from the primary tumor, and (4) the patient must have no genetic history of cancer predisposition (eg, Li-Fraumeni Syndrome or Neurofibromatosis).
The most common radiation-induced CNS neoplasms following radiation treatment for childhood cancer are gliomas and meningiomas.5,8,14 The survival rates for patients that develop glioma following cranial radiation treatment are far poorer compared with those that develop meningioma, with a 5-year relative survival rate of just 4% for radiation-induced gliomas (RIGs) compared with 77%–84% for radiation-induced meningiomas.6,15 For this meta-analysis, we have focused on the more aggressive cranial RIGs.
A comprehensive epidemiological meta-analysis of patients diagnosed with RIG16 showed that the most frequent primary tumors were hematological malignancies (35%), followed by medulloblastoma (13%) and pituitary adenoma (12%). The median overall survival following RIG diagnosis was just 11 months, highlighting the aggressive nature of these cancers. The median overall latency of disease onset is approximately 9 years between radiotherapy for the primary lesion and diagnosis of the RIG.5,17 Over 50% of RIGs are diagnosed during adulthood,16 and arise more frequently in patients who received radiotherapy early in life.4,5,17
Several reviews have comprehensively detailed the epidemiological and clinical aspects of RIGs.5,14,16,17 In contrast, few investigations have described the molecular features of these tumors. Those that do exist are limited by the small number of cases available for analysis, with 1 study reporting that RIGs are genetically similar to pilocytic astrocytoma (PA),18 while another study suggests they are analogous to primary adult glioblastoma (GBM).19 Recently DNA methylation analysis has enhanced traditional diagnostic methods to significantly improve the accuracy of brain tumor classification.20 Despite this, a definitive molecular signature for RIGs has not yet been established, highlighting a gap in the knowledge of potentially actionable drivers of this extremely aggressive disease. Indeed, there are currently no clear diagnostic criteria for RIGs and, as a consequence, no consistent or optimal treatment regimen has been defined. With this in mind, we have performed a systematic review and meta-analysis of the genetic features of cranial RIGs reported in the literature to identify recurrent changes that may be characteristic of this disease.
Methods
Systematic review and meta-analysis
A systematic search of the literature (from database inception up to April 7, 2021) was performed using PubMed according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 guidelines.21 The terms used in the search were “radiation-induced” or “radiation-associated” or “treatment-induced” or “treatment-associated” each in combination with “glioma” or “astrocytoma” or “glioblastoma” or “ependymoma.” Titles and abstracts of articles and case reports written in English were screened for cases of radiation-induced high-grade gliomas (HGGs, WHO grade III and IV) located in the human brain. Records describing cases with a genetic predisposition to cancer development or where the second glioma may have been a relapse of the initial tumor were excluded. The full-text versions of these articles were obtained in their entirety and examined for analysis of molecular features for inclusion in this study. The reference lists of eligible articles were also examined for further records not obtained through the database search. Only peer-reviewed articles were eligible for inclusion, with unpublished data excluded.
All individual cases were examined for the satisfaction of Cahan’s criteria,13 detailed in Table 1. The molecular data available from all eligible RIG cases in the literature were examined and compiled. All data were independently studied by 3 reviewers (J.P.W., M.H., and R.E.), who assessed the eligibility of each case and extracted data from the publications. Cases without information on family history of genetic cancer predisposition or matched germline DNA were included where available clinical evidence (latency, location of RIG, and/or distinction from primary malignancy) was suggestive of the second tumor being a RIG. Details on individual case exclusions and a brief description of analysis techniques employed by the primary sources are detailed in Supplementary Figure 1.
Table 1.
Satisfaction of Cahan’s Criteria13 for Radiation-induced Glioma Cases with Molecular Data Examined in This Study
| Reference | Year | Genetic Predisposition | Latency Postradiation Treatment (Years) | RIG Occurred in an Irradiated Field | Distinct From Primary Malignancy (Primary Tumor Diagnosis) |
|---|---|---|---|---|---|
| Tada22 | 1997 | No family history of genetic predisposition, no germline p53 mutation (other germline predispositions not tested for). | 10 | Yes | Yes (suprasellar germ cell tumor) |
| Matsumura23 | 1998 | No family history of genetic predisposition. Germline DNA not tested. | 8 | Yes | Yes (subependymal giant cell astrocytoma) |
| Brat24 | 1999 | No family history provided. Germline DNA not tested. | 5-23 | Yes | Yes (Hodgkin’s disease, pituitary adenoma, rhabdomyosarcoma, craniopharyngioma, ALL, pineal tumor, ependymoma, lymphoblastic lymphoma) |
| Yang25 | 2005 | No family history of genetic predisposition, and no TP53 mutation in the primary MB (TP53 mutation present in the RIG). Germline DNA not tested. | 10 | Yes | Yes (MB) |
| Berman26 | 2007 | Satisfaction of Cahan’s criteria stated. | 9 | Yes | Yes (arteriovenous malformation) |
| Donson18 | 2007 | No family history provided. Germline DNA not tested. | 3–15 | Yes | Yes (Burkitt’s lymphoma, MB, pilocytic astrocytoma, ALL, ependymoma) |
| Romeike27 | 2007 | No family history of genetic predisposition. Germline DNA not tested. | 7–14 | Yes | Yes (MB, ALL) |
| Gessi28 | 2008 | No family history of genetic predisposition, and no TP53 mutation in the primary MB or germline DNA (TP53 mutation present in the RIG). | 8 | Yes | Yes (MB) |
| Salvati29 | 2008 | Satisfaction of Cahan’s criteria stated for all cases. No family history of genetic predisposition. Germline DNA not tested. | 6–26 | Yes | Yes (MB, cavernous angioma, tinea capitis, cutaneous hemangioma, scalp hemangioma, ALL) |
| Sasayama30 | 2008 | No family history of genetic predisposition, no germline p53 mutation (other germline predispositions not tested for). | 28 | Yes | Yes (MB) |
| Garcia-Navarro31 | 2009 | No family history provided. Germline DNA not tested. | 8 | Yes | Yes (pineal germinoma) |
| Kamide32 | 2010 | Satisfaction of Cahan’s criteria stated. No family history provided. Germline DNA not tested. | 29 | Yes | Yes (MB) |
| Paugh33 | 2010 | No family history provided. Germline DNA not tested. | Not described | Yes | Yes (ALL, germinoma, ependymoma, MB) |
| Ohba34 | 2011 | No family history of genetic predisposition. Germline DNA not tested. | 4 | Yes | Yes (meningothelial meningioma) |
| Khoo35 | 2012 | Satisfaction of Cahan’s criteria stated. | 30 | Yes | Yes (diffuse astrocytoma) |
| Mascelli36 | 2012 | No family history provided. Germline DNA only analyzed for IDH1/2 mutation. | 7 | Yes | Yes (sellar/suprasellar craniopharyngioma) |
| Ahmed37 | 2014 | No family history provided. Germline DNA not tested. | 10 | Yes | Yes (ALL) |
| Nakao38 | 2017 | Satisfaction of Cahan’s criteria stated for all cases. No family history of genetic predisposition. Germline DNA not tested. | 22–29 | Yes | Yes (MB, craniopharyngioma, primitive neuroectodermal tumor, pituitary adenoma) |
| Ng39 | 2017 | No family history of Neurofibromatosis 1 or 2. Germline DNA not tested. | 6 | Yes | Yes (vestibular schwannoma) |
| Gits19 | 2018 | Germline DNA tested for 2/3 cases (case #6 without germline DNA was reanalyzed with matched germline DNA available in Whitehouse et al40) | 4–12 | Yes | Yes (MB) |
| Izycka-Swieszewska41 | 2018 | No family history provided. Germline DNA not tested. | 3–6.5 | Yes | Yes (ALL) |
| Kajitani42 | 2018 | No family history of genetic predisposition. Germline DNA not tested. | 5–10 | Yes | Yes (ALL) |
| Phi43 | 2018 | Germline DNA only available for 4/5 patients. | 4.3–10 years post primary diagnosis | Yes | Yes (MB) |
| Porter44 | 2018 | No family history provided. Germline DNA not tested. | 19 | Yes | Yes (MB) |
| Wang45 | 2018 | No family history of genetic predisposition. Germline DNA not tested. | 8 | Yes | Yes (MB) |
| Lopez46 | 2019 | Satisfaction of Cahan’s criteria stated for all cases. No family history of genetic predisposition. Germline DNA tested for 6/12 cases. | 4–41 | Yes | Yes (MB, intracranial germinoma, leukemia, Hodgkin’s lymphoma, craniopharyngioma, pineocytoma) |
| Mucha-Malecka47 | 2019 | Satisfaction of Cahan’s criteria stated. Germline DNA not tested. | 12 | Yes | Yes (MB) |
| Biswas48 | 2020 | No family history of genetic predisposition. One case of unilateral breast cancer in paternal grandmother. | 5 | Yes | Yes (ALL) |
| Smith49 | 2020 | No family history provided. Germline DNA not tested. | Not described | Yes | Yes (MB) |
| Whitehouse40 | 2020 | Satisfaction of Cahan’s criteria stated. No family history of genetic predisposition. Germline DNA tested. | 11 | Yes | Yes (MB) |
| Woo50 | 2021 | Satisfaction of Cahan’s criteria stated. | 6–23 | Yes | Yes (nasopharyngeal carcinoma, primary intracranial germinoma) |
ALL, acute lymphoblastic leukemia; GBM, glioblastoma; MB, medulloblastoma; RIG, radiation-induced glioma.
t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis
Raw IDAT files from both reference set (https://academic.oup.com/neuro-oncology/article/23/1/34/5948536?login=true) and reported patient-derived xenograft (PDX) sample and from both array types (450k or EPIC) were loaded into the R environment (version 4.0.1) using the minfi package (version 1.21.4). CpG site probes present on both arrays have been selected; sample signal intensities have been normalized, filtered, and log2-transformed (limma package version 3.30.11). Unsupervised t-SNE analysis was performed on the top 10 000 most variable (row variance) CpG probes applying the Rtsne package (version 0.15), with the following parameters: pca = F, max_iter = 2500, theta = 0, perplexity = 35.
Statistical Analysis
Statistical analyses were performed using the GraphPad Prism software, version 8. Comparison of means for latency, anatomical location, and recurrent genetic alterations was performed using a 1-way ANOVA with a nonparametric Kruskal–Wallis test.
Results
The Profile of Recurrent Genetic Alterations in RIGs Is Distinct From Most Other Astrocytomas or Gliomas
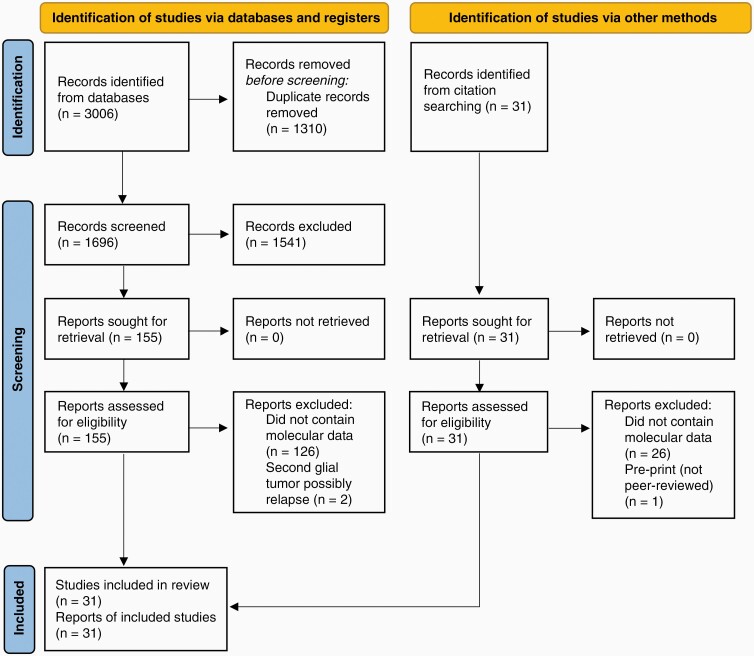
Database searching identified 3006 records. After duplicates were removed, 1696 records were screened, from which 155 full-text articles were obtained. A further 31 records were identified from searching the reference lists of eligible articles and full-text versions of these obtained. A total of 152 reports were excluded as they did not contain molecular analysis, 2 reports were excluded as the RIG described may have been a relapse of the initial tumor,51,52 and 1 report was excluded as it had not been peer-reviewed.53 A final total of 31 reports were included, describing 102 unique cases of high-grade cranial RIGs with molecular data (Figure 2).
Figure 2.
Flow chart of literature searching strategy detailing the report selection procedure as outlined in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2020 guidelines.21 A total of 31 reports were eligible for inclusion in this systematic review.
To identify recurrent themes and ensure confidence in interpretations, only genetic alterations that were tested in at least 10 unique RIGs are reported here (summarized in Table 2), with all remaining analyzed data available in Supplementary Table 1. Herein, the frequency of each genetic alteration in RIGs is expressed both as a percentage (%) and as a fraction of the total number of cases tested for that alteration (denoted as n/n).
Table 2.
Summary of molecular alterations tested in at least ten unique radiation-induced glioma cases
| DNA Alterations | RNA Expression | Protein Expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Function | Gene | Amplified | Deleted | Mutation/Fusion/Promoter Methylation (MGMTonly) | Low | High | Low | High | Total Unique cases | References |
| Receptor | ACVR1 | 0/9 | 0/9 | 0/12 | NR | NR | NR | NR | 12 | 19,46 |
| EGFR | 4/31 | 0/10 | 0/14 | 1/5 | 0/5 | 14/22 | 4/22 | 48 | 18, 24–28, 33, 40, 41, 46, 48, 49 | |
| PDGFRA | 10/21 | 0/18 | 7/16 | 0/5 | 5/5 | 2/7 | 3/7 | 32 | 18 a , 19, 28, 33, 40, 43, 46 | |
| Signal transduction | BRAF | 0/9 | 0/9 | 3/19 | NR | NR | NR | NR | 19 | 19, 38b, 42 c , 46 |
| NF-1 | 0/18 | 0/18 | 2/19 | 1/1 | 0/1 | NR | NR | 19 | 33,40,46 | |
| PIK3CA | 0/9 | 0/9 | 3/12 | NR | NR | NR | NR | 12 | 19,40,46 | |
| PTEN | 0/9 | 4/25 | 1/22 | 0/1 | 0/1 | 0/4 | 4/4 | 32 | 18, 19, 24, 28, 40, 41, 43, 46 | |
| Cellular metabolism | IDH1 | 0/9 | 0/9 | 1/47 | 0/1 | 1/1 | NR | NR | 47 | 34 d, 36, 38d,e, 39d, 40, 41d, 42, 43d, 46, 47d, 50 |
| IDH2 | 0/9 | 0/9 | 0/16 | NR | NR | NR | NR | 16 | 36, 38e, 42, 46 | |
| Cell cycle regulation | CDK4 | 4/10 | 0/10 | 0/10 | NR | NR | NR | NR | 10 | 46,49 |
| CDKN2A | 0/10 | 13/28 | 0/10 | NR | NR | 1/3 | 2/3 | 31 | 19, 24, 33, 40, 42, 46, 49 | |
| Transcriptional regulation/ chromatin modification |
ATRX | 0/10 | 0/10 | 3/10 | NR | NR | 0/12 | 12/12 | 22 | 38, 39, 41, 42, 45, 46, 48 |
| H3F3A | 0/9 | 0/9 | 0/21 | NR | NR | NR | NR | 21 | 19, 38 f , 39f, 40, 42g, 46 | |
| HIST1H3B | 0/9 | 0/9 | 0/12 | NR | NR | NR | NR | 12 | 19,46 | |
| MYCN | 1/10 | 0/10 | 0/10 | NR | NR | NR | NR | 10 | 46,49 | |
| SMARCB1 | 0/10 | 0/10 | 0/10 | NR | NR | NR | NR | 10 | 46,49 | |
| TERT promoter | 0/9 | 0/9 | 0/11 | NR | NR | NR | NR | 11 | 38,46 | |
| TP53 | 0/13 | 2/14 | 14/30 | 0/1 | 0/1 | 20/42 | 12/42 | 61 | 18, 19, 22–28, 30, 31, 34, 37, 38, 40–43, 45, 46, 48, 49 | |
| DNA repair | MGMT | NR | NR | 8/29 promoter methylated | NR | NR | 4/10 | 6/10 | 33 | 28,29,32,35,39,41,44,45,50 |
Data expressed as the total number of tumors positive for the described alteration as a fraction of the total number of tumors tested for that alteration (n/n). Genes are grouped based on function.
NR, not reported.
aPooled microarray data excluded from analysis as individual tumor data unavailable.
bMutational status determined by BRAF V600E direct sequencing.
cMutational status determined by BRAF V600E IHC.
dMutational status determined by IDH1 mutant-specific antibody via IHC.
eMutational status determined by direct sequencing.
fMutational status determined by H3K27M IHC.
gMutational status determined by K27M, G34R, G34V mutation detection by direct sequencing.
PDGFRA was the most frequently altered gene in cranial RIGs, with amplification of this gene observed in 48% of tumors (10/21), and 44% (7/16) of cases harboring PDGFRA mutations. Another frequently altered gene was TP53, with 47% (14/30) of cases harboring mutations and 14% (2/14) demonstrating TP53 deletion. Deletion of CDKN2A (often also including CDKN2B) was reported in 46% (13/28) of tumors, and CDK4 amplification was also frequent (40%; 4/10).
Three out of 10 RIGs with sequence data demonstrated mutations in ATRX. In contrast, ATRX protein was detected via immunohistochemistry in 100% of samples tested (12/12 cases). Loss of ATRX protein strongly correlates with ATRX mutation by immunohistochemistry.54,55 Although certain missense ATRX mutations may result in positive staining in a small percentage of cases,55,56 immunohistochemistry for ATRX is routinely used histopathologically as a surrogate for ATRX mutation in the absence of genetic data. The immunohistochemistry results compiled here are suggestive of a very low mutation rate in this gene in cranial RIGs, in contrast to the sequencing results. To reduce the risk of bias, we have included results from both techniques in this analysis, resulting in an overall ATRX mutation rate of 14% (3/22).
Molecular alterations in other oncogenes or tumor-suppressor genes known to be associated with human glioma have also been reported for cranial RIGs including PTEN deletion (16%; 4/25 of cases tested) or mutation (5%; 1/22), amplification of EGFR (13%; 4/31) or MYCN (10%; 1/10), and mutations in PIK3CA (25%; 3/12), NF1 (11%; 2/19), BRAF (5%; 1/19) or IDH1 (2%; 1/47). An intragenic deletion in BRAF (5%; 1/19) and a GTF2I-BRAF fusion (5%; 1/19) were also reported.46 The MGMT promoter was methylated in 28% (8/29) of cases tested for this alteration. Correspondingly, MGMT protein expression was low or undetectable in 40% (4/10) of samples tested by immunohistochemical techniques. Of note, none of the cranial RIGs examined harbored mutations in other genes frequently altered in HGG or other cancers, including ACVR1 (0/12), EGFR (0/14), H3F3A (0/21), HIST1H3B (0/12), IDH2 (0/16), SMARCB1 (0/10) or the TERT promoter (0/11).
RIGs Demonstrate Unique Recurrent Chromosomal Alterations
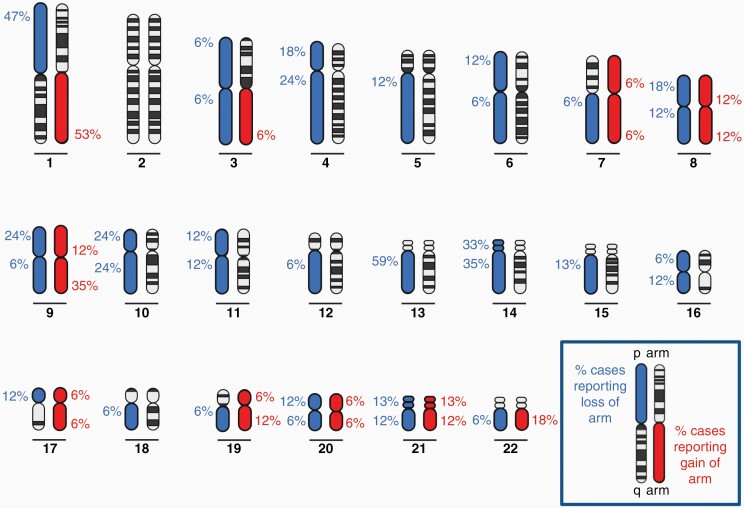
The karyotypes of HGGs are often highly complex, with increasing complexity associated with increasing tumor grade.57–59 A small cytogenetic study of 3 RIGs reported extremely complex karyotypes18; however, few studies have extensively examined broad chromosomal gains or losses in cranial RIGs. Paugh et al33 performed the largest analysis to date examining 10 RIG samples, to which we have added seven more cases here34,40,43,49 (Figure 3 and Supplementary Tables 2 and 3). Overall, the most frequent copy number changes observed in RIGs were loss of 13q (observed in 59% of samples reported), gain of 1q (53%), and loss of 1p (47%). This was followed by gain of 9q (35%), loss of 14q (35%) and loss of 14p (33%). Of these changes, it has been reported that gains of chromosome 1q and 9q and losses of 1p and 13q are significantly more frequent in RIGs compared with other pediatric and adult HGG.33
Figure 3.
Frequency of gains and losses of autosomal chromosomal arms observed in cranial radiation-induced gliomas. Blue indicates loss and red indicates gain of chromosomal arms, with percentages of cases reporting gain or loss for each arm shown.33,34,40,43,49
Anatomical Distribution of Recurrent Genetic Alterations and Latency Intervals of RIGs
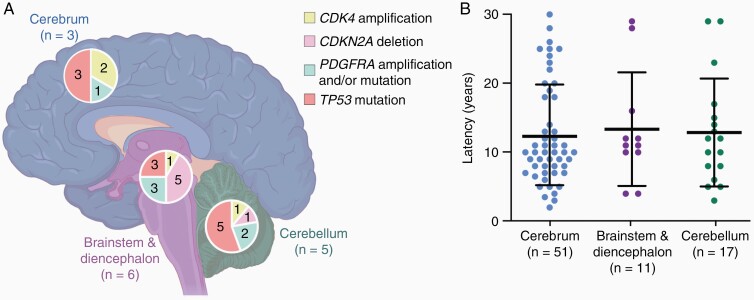
Previously, glioma-associated genetic alterations have correlated closely with tumor type, age of onset and location of disease.60 To determine whether this was also the case in RIG, we examined whether there was a correlation between RIG location and genetic features. The most frequent recurrent genetic alterations found in RIGs (CDK4 amplification, CDKN2A deletion, PDGFRA amplification and/or mutation, or TP53 mutation) were mapped to 3 brain regions: the cerebrum, cerebellum, or brainstem and diencephalon. Deletion of CDKN2A was most commonly observed in RIGs located in the brainstem and diencephalon region, while TP53 mutations were most often found in cerebellar RIGs (Figure 4A), with many tumors harboring more than 1 genetic alteration (Supplementary Table 4). However, the anatomical location of RIGs harboring these recurrent genetic alterations was only reported for a small number of cases (n=14) which significantly limits the strength of this correlation and highlights the need for further research in this area.
Figure 4.
Relationship of recurrent radiation-induced glioma (RIG) mutations and disease latency with anatomical brain location. (A) RIGs with the indicated genetic alterations were grouped according to the locations shown (cerebrum, blue; brainstem and diencephalon, purple; or cerebellum, green). The number of tumors harboring each alteration (see key) are shown within each segment of the pie chart for each region, with the total number of tumors for each location (n) shown. Some tumors harbored multiple genetic alterations; data shown represent all reported genetic alterations for that location, and do not imply mutual exclusivity of genetic alterations within tumors.19,22,25,28,40,46 (B) Scatter plot of latency intervals for RIGs arising in the three defined brain regions.18,19,22,23,25–32,34–38,40–42,45–48,50 Bars show mean ± SD.
We also investigated whether the most frequently observed genetic alterations correlated with time to RIG onset, although the latency interval of RIGs harboring these was only reported for a total of 18 unique cases. Using this limited data set we observed no correlation between disease latency and genetic alterations (Supplementary Table 4) however, further data are required to confidently assess this relationship.
Given these limitations within the existing studies, we examined if there was any correlation between RIG location and disease latency interval using all the cases in this series where location information was available (n = 79 cases). We found no correlation between anatomical location and the time between radiation treatment and RIG development (Figure 4B; Supplementary Table 5). Again, the small number of cases in this series must be taken into consideration when drawing conclusions from these data.
DNA Methylation Analysis of RIGs and Associated PDX models
The assessment of global DNA methylation is rapidly becoming a widely used method to classify CNS tumors. The majority of RIG cases described in the literature were published before the DNA methylation array was fully appreciated as a useful classification tool for CNS tumor diagnosis.20 Indeed, only 2 recent studies performed methylation array on RIG cases,40,49 which limits our ability to analyze these data in great detail. Smith et al49 performed methylation clustering analysis using an online classification tool (www.molecularneuropathology.org, version 11b4) and reported that the RIG and the PDX derived from that tumor were most similar to the GBM, IDH-wild type, subclass midline (GBM, MID) methylation class. Of note, within the reference cohort,20 there are no clearly defined RIG reference samples and a RIG-specific methylation profile has not been established. The GBM, MID methylation subclass describes tumors that are located in the midline and have a median age at diagnosis of 13 years. These tumors typically lack H3K27M mutations and frequently demonstrate amplification of PDGFRA, CDKN2A/B loss, and mutations in FGFR120.
Of the only other published RIG where methylation data are available, similar DNA methylation analysis was performed using the same web platform (www.molecularneuropathology.org, version 11b4); however, this tumor failed to cluster with any of the defined subclasses, as previously described.40 Using DNA isolated from a PDX derived from this RIG, we repeated methylation-based clustering analysis using a more recently reported reference cohort of well-characterized astrocytic gliomas from both adults and children.61 Analysis by t-SNE revealed that this RIG-derived PDX clustered with the pedGBM_RTK1 methylation subclass (Supplementary Figure 2). Tumors classified as pedGBM_RTK1 are characterized by frequent PDGFRA amplification, TP53 mutation, and homozygous CDKN2A deletion and lack mutations in the TERT promoter or EGFR amplification.62 Of note, this subclass is not described in the online methylation classifier used by Smith et al49. Upon comparison, there is substantial overlap between the GBM, MID and pedGBM_RTK1 methylation subclasses (David Jones, personal communication, July 26, 2021). Consequently, the next version of the molecular neuropathology classifier (version 12) will no longer include a GBM, MID subclass, but will instead include subclasses of pedGBM_RTK1 (David Jones, personal communication, July 26, 2021). Given this, it is likely that the RIG described in Smith et al49 may correspond to a pedGBM_RTK1 subclass in this new version of the classifier.
Discussion
Historically, RIGs have been clinically diagnosed as diffuse intrinsic pontine glioma (DIPG),19,40 anaplastic astrocytoma (AA),24,30,31,36–38,41,46,47,63 or GBM22–25,28,29,34,38,39,41,42,45,46,48,50 based primarily on histological and/or radiographical characteristics. In very rare cases, the diagnosis of anaplastic ependymoma has also been reported.35 RIGs have even been mistaken for recurrent medulloblastoma43,49 or radiation necrosis.63 By compiling molecular data from 31 publications describing RIG we strove to identify the most common genetic alterations in human cranial RIG. We found that the most frequent alterations in RIGs occurred in genes involved in cellular growth (PDGFRA), cell cycle regulation (CDKN2A, CDK4), DNA repair, and induction of apoptosis (TP53), with approximately half of the tumors tested for these changes demonstrating alterations in one or more of these genes.
In this study, we aimed to determine whether RIGs are molecularly similar to other brain tumor types or instead form their own distinct subgroup. As mentioned, RIGs have often been diagnosed as AA.24,30,36,38,41,46,47,63 AAs are histologically WHO grade III diffuse HGGs with invasive and aggressive tendencies and arise most often in adults.64 The most common WHO-defined variant is “Anaplastic astrocytoma, IDH-mutant”.65 These tumors are characterized by IDH1/2 mutation, along with mutations in TP53 and ATRX,66–68 with an associated loss of ATRX protein.69,70 Additionally, mutations in NOTCH pathway genes, and less frequently PIK3CA, PIK3R1 and DSG3 have been reported in AAs.68 While we observed a high frequency of TP53 mutations in RIGs and a small number of cases harbored mutations in ATRX, ATRX protein expression was detected in 100% of RIG cases by immunohistochemistry and mutations in NOTCH and IDH1/2 were rare (Table 2; Supplementary Table 1), suggesting low concordance between the genetic profiles of these 2 tumor types.
A previous analysis of 5 RIGs suggested that their gene expression profile resembled PAs, with a 39% overlap in highly expressed genes reported between these 2 tumor types.18 Of those genes, only PDGFRA was reliably overexpressed in RIGs from our analysis, with limited information available on expression levels of the other genes reported. A lack of IDH1/2 mutations was the only other genetic similarity observed between these tumor types.71 A hallmark feature of PAs is mitogen-activated protein kinase pathway dysregulation.72 While alterations in BRAF were observed in a small number of RIG cases (16%; 3/19 tumors), the characteristic KIAA1549:BRAF fusion and other less frequent genetic alterations reported in PAs, such as FGFR1 mutations or NTRK2 fusions,73–75 were not found (Supplementary Table 1). Our analysis suggests there are few genetic similarities between PA and RIGs. Indeed, Donson et al18 suggested that the similar RNA expression profiles they observed may have been due to these tumors sharing a common precursor cell, particularly given the considerable differences in tumor grade and patient outcome between PA and RIG.
Due to disease location, radiological features and their aggressive nature, RIGs have also been clinically diagnosed as DIPG or brainstem glioma.19,40 DIPGs are rapidly growing, diffuse gliomas arising in the brainstem, most commonly observed in children.76 Nearly 80% of pediatric DIPG harbor a lysine 27 to methionine (K27M) mutation in histone H377–79, which led to the WHO reclassifying this tumor type as “diffuse midline glioma (DMG), H3 K27M-mutant” in 201665,80. We specifically compared the recurrent genetic alterations reported for RIG with those reported for DIPG/DMG. Several genetic alterations were shared between DIPG/DMG and RIG, including a high frequency of TP53 mutations and PDGFRA amplification, moderate frequency of CDK4 amplification,65 and lower frequencies of IDH1/2, NF1, and PIK3CA mutations, and PTEN deletion/mutation.33,78,81–83 However, in contrast, all RIGs lacked mutations in ACVR1 or PPM1D, and few exhibited MYC or MYCN amplification (Table 2; Supplementary Table 1) previously described in DIPG/DMG. Most notably, the hallmark H3 K27M mutation that is now pathognomonic for DMG was not reported in any RIG within the cases examined here.58,65,78,81,82 Furthermore, nearly half of the RIGs analyzed showed homozygous deletion of CDKN2A, which is almost never observed in DIPG/DMG, and 16% of RIGs harbored BRAF mutations that are absent in DIPG/DMG.58,81,82 Our analysis demonstrates that while some key genetic alterations are shared between these tumor types, significant differences (most notably the lack of hallmark mutations) exist, rendering RIGs molecularly distinct from DIPG/DMG.
It has been proposed that pediatric RIGs share molecular similarities with adult primary GBMs.19 GBMs are highly aggressive WHO Grade IV diffuse gliomas that can arise as a primary tumor (primary GBM) or result from progression of a lower-grade II/III glioma (secondary GBM)84 and are most commonly wild type for IDH.65 While our analysis showed some similarities between RIGs and adult primary GBMs, including a high frequency of PDGFRA amplification/mutation, TP53 mutations, CDKN2A deletion, and absence of IDH1/2 mutations,85–87 a number of key differences were evident. Most notably, several hallmark alterations of adult primary GBM, such as PTEN mutation/deletion, EGFR mutation/amplification and TERT promoter mutation,85–87 were either absent or only rarely observed in the cranial RIGs assessed in our analysis.
A large-scale study of mostly adult GBM samples identified 4 genetic subtypes of GBM: proneural (characterized by alterations in IDH1, PDGFRA and TP53), neural (no defining genetic alterations but overexpress neural markers), classical (often harbor EGFR amplifications/mutations and CDKN2A homozygous deletion) and mesenchymal (commonly demonstrate NF1 mutations, some with concurrent PTEN mutations).88 Sturm et al89 later built on this work using combined pediatric and adult samples and defined a total of 6 epigenetic and biologic subgroups of GBM. Of these, the RTK I subgroup can be considered most closely aligned to RIGs from our analysis. This subgroup is generally typified as having wild-type IDH1, PDGFRA amplification, and CDKN2A deletion, with around half of the samples also harboring alterations in TP53. Mutations in H3F3A are also absent from this methylation subclass.89
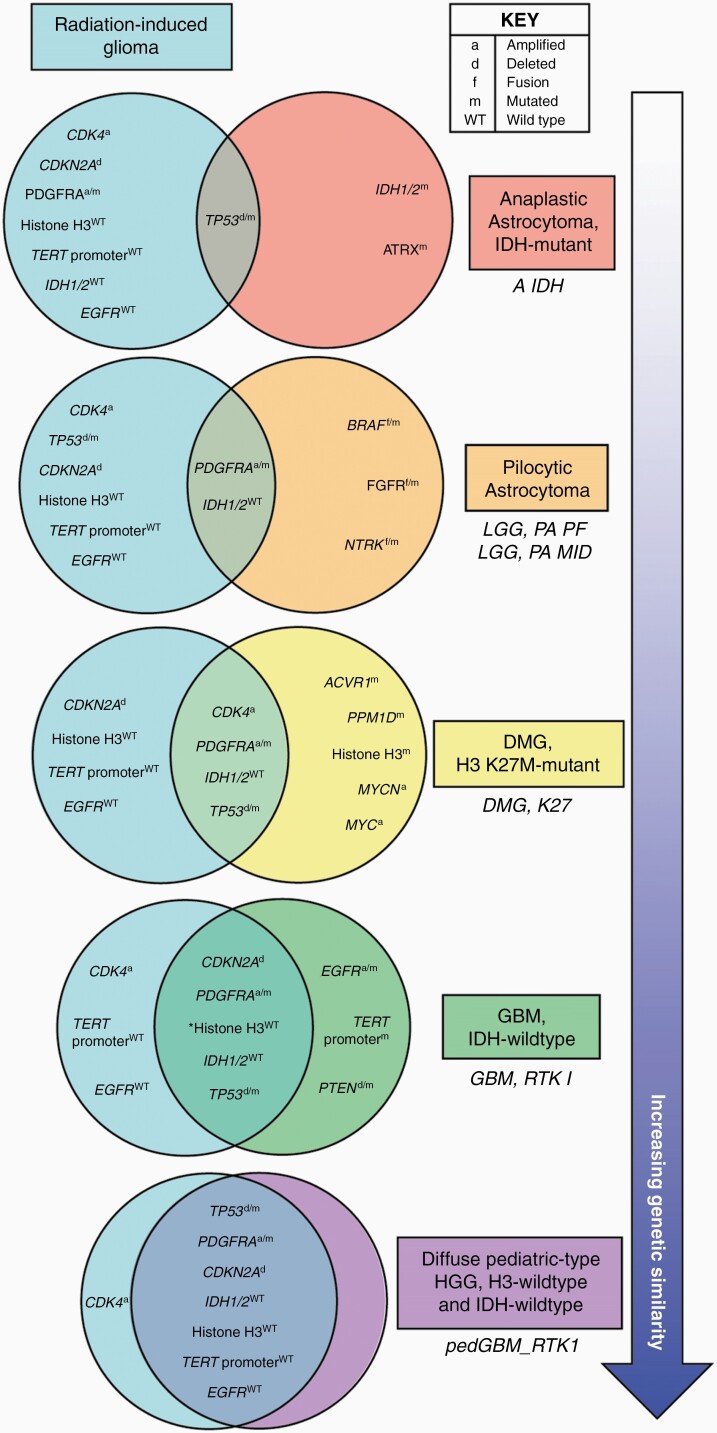
From our comparisons thus far, it is evident that the genetic features of RIG are most closely aligned to the WHO diagnostic classification GBM IDH-wild type, and more specifically the RTK I methylation subclass. The tumors that fell into this methylation subclass described by Sturm et al89 occurred in both adult and pediatric populations, while the “GBM, IDH wild type, subclass RTK I” described in Capper et al20 was solely made up of adult patients. In contrast, Korshunov et al62 specifically investigated histone H3-/IDH1-wild-type pediatric GBM. This integrated analysis identified 3 distinct molecular subgroups of pediatric GBM designated pedGBM_MYCN (demonstrating a high frequency of MYCN amplification), pedGBM_RTK1 (enriched for PDGFRA amplification), and pedGBM_RTK2 (characterized by EGFR amplification). Similar to the GBM RTK I methylation subclass,89 the pedGBM_RTK1 subpopulation has frequent PDGFRA amplification and TP53 mutation, as well as a considerable proportion of tumors harboring homozygous CDKN2A deletion. Additionally, pedGBM_RTK1 tumors do not exhibit mutations in the TERT promoter or amplifications of EGFR and have a comparable frequency of MYCN amplification and PTEN loss to RIGs examined in our analysis. Thus, the features of this subgroup closely correlate with recurrent genetic alterations of the RIGs compiled here, and our data show that a RIG PDX model (TK-RIG915) clusters with the pedGBM_RTK1 subgroup by methylation profiling. Preliminary data from others support this correlation, with 26 out of 36 gliomas that developed after therapy clustering with the pedGBM_RTK1 subgroup by methylation array.90 In summary, despite most RIGs being diagnosed in adulthood,16 our comparisons of cranial RIGs with multiple other types of glioma revealed that these tumors most closely resemble the pedGBM_RTK1 molecular subgroup of pediatric gliomas (depicted in Figure 5).
Figure 5.
Radiation-induced gliomas (RIGs) are genetically distinct from most other astrocytoma and glioma brain tumor types and share the highest number of common genetic alterations with the pedGBM_RTK1 methylation subclass of pediatric glioblastoma. Major recurrent genetic features of RIGs were defined as occurring in more than 40% or cases and are compared here with hallmark genetic features of the indicated WHO-defined CNS tumors65 (colored boxes ordered by increasing similarity to RIG): IDH-mutant anaplastic astrocytoma,65–68,91–93 pilocytic astrocytoma,18,65,71,73–75,91,94 diffuse midline glioma (DMG), H3 K27M-mutant,33,58,65,77–79,81–83 IDH-wild type glioblastoma (GBM).65,85–89 The closest corresponding methylation subclass20,62 is indicated beneath each WHO-defined tumor type in italics. Comparison of recurrent genetic features of RIGs with the pedGBM_RTK1 methylation subclass of pediatric GBM62 is also shown. Being pediatric-specific, this methylation class does not align with a specific entity in the 2016 WHO diagnostic guidelines65 therefore we have used a new entity proposed in the 2021 edition (diffuse pediatric-type high-grade glioma (HGG), H3-wild type and IDH-wild type).95 Common astrocytoma/glioma genetic alterations that were only observed in a small proportion of RIGs (e.g. ATRX mutation (14%), BRAF mutation (10%), BRAF fusion (5%), EGFR amplification (13%), IDH1 mutation (2%), MYCN amplification (10%), PTEN deletion (16%)) were considered infrequent and not deemed to be a defining characteristic of RIG. Asterisk indicates a genetic feature associated only with the GBM, RTK I methylation subclass and has not been described as a feature of GBM, IDH-wild type. Abbreviations for the methylation classes are as previously defined20,62: A IDH—IDH glioma, subclass astrocytoma; LGG, PA PF—low-grade glioma, subclass posterior fossa pilocytic astrocytoma; LGG, PA MID—low-grade glioma, subclass midline pilocytic astrocytoma; DMG K27—diffuse midline glioma H3 K27M mutant; GBM, RTK I—glioblastoma, IDH wild type, subclass RTK I; pedGBM_RTK1—pediatric glioblastoma enriched for PDGFRA amplification.
Limitations
This analysis used data reported from a limited number of published cases, rather than from a comprehensive large-scale genome-wide study using primary tumor tissue. As a result, our study was restricted to the genetic alterations reported in the original sources, with not all alterations tested across all samples, resulting in loss of power through missing information. Additionally, the limited number of samples precluded any meaningful correlative analysis of RIG location or latency and the underlying genetic features of these tumors. The correlation of recurrent RIG genetic alterations with those observed in the methylation subclass pedGBM_RTK1, and the positive association of the TK-RIG915 PDX with this methylation class, raises the question of whether other RIGs would cluster similarly using DNA methylation-based techniques. Furthermore, preliminary data by Lucas et al90 corroborate our assessment, where they report that RIG DNA methylation profiles are similar to pedGBM_RTK1 methylation subgroup tumors. Additional research using a larger number of samples is essential to determine whether RIGs truly are genetically similar to pediatric GBM or if they represent a unique tumor subclass.
For the majority of cases, information on germline mutations predisposing to cancer and/or a family history of cancer was available. Where this information was not explicitly stated, cases were included based on the clinical evidence available; however, these cases are acknowledged as a limitation of this analysis as germline predisposition cannot be confidently excluded in these instances. Although Cahan’s criteria state that cases with germline alterations predisposing to cancer should not be classified as RIGs, a more recent analysis reported that a high proportion of patients that developed glioma following therapy had frequent pathogenic germline alterations in DNA repair genes, including BARD1, BRCA1, BRCA2, ATR, and PMS196. These data suggest that these patients may be at higher risk of RIG development, and as such may require increased surveillance following radiation therapy.
Finally, there is some controversy regarding radiation-induced tumors and the role of chemotherapy in contributing to their development. The combination of radiation and chemotherapy has been reported to have a synergistic effect on the development of treatment-induced gliomas4,97 or result in a shorter latency period.16 In contrast, others report statistically significant increases in the risk of glioma development with increasing radiation dose, but no further increased risk with the addition of chemotherapy.5,8 Given these inconsistent findings in the literature, we cannot rule out the possibility that chemotherapy may have contributed to the effects of radiation in the initiation of the glioma cases we report here, despite satisfaction of Cahan’s criteria in most cases. For this review, we have chosen to retain the more widely-used term “radiation-induced glioma”, but acknowledge that broader terms such as “radiation-associated” 98 or “treatment-induced” 90 may prove to be more accurate should chemotherapy be shown to be a contributing factor in the development of these tumors in the future.
Future directions
Currently, a disease-specific treatment approach for RIG remains undefined and patients are treated in accordance with the broad treatment regimens assigned to the tumor type histologically and/or radiologically classified at the time of diagnosis (eg, AA, DIPG/DMG, GBM). However, our analysis reveals that the mutational and gene expression profiles of RIGs are distinct from most other astrocytomas and gliomas, aligning most closely with the pedGBM_RTK1 subgroup of pediatric GBM.
Given the paucity of molecular data currently available for RIGs, there is a clear need for research that further defines the molecular characteristics of RIG. This can only be achieved by increasing the number of samples comprehensively analyzed at the genetic, epigenetic and transcriptional level in a patient-specific manner, an opinion supported by other research groups.90,99 This large-scale analysis will help the field conclusively determine if RIGs are molecularly distinct from, or should be considered synonymous with, the pedGBM_RTK1 tumors. Proposed plans to distinguish between pediatric and adult diffuse HGG in the next edition of the WHO classification of CNS tumors may assist further with a more accurate diagnosis of these tumors.95
The findings of this meta-analysis may have implications for future clinical management of this disease, particularly given that most RIGs are diagnosed in adulthood. Additionally, molecular classification of RIGs may complement existing tools for pathological diagnosis of these tumors in the future. Preclinical models such as genetically engineered mouse models of RIG susceptibility100 and PDX models of RIG40 can not only aid in our understanding of the biological pathways relevant to the development of these tumors but are also an essential tool in bridging the gap between potential therapeutic approaches and rational clinical trial design. Currently, these models are rare, highlighting the need to focus future efforts on their development to increase the number of relevant models available to the research community.
Given the rarity of these tumors, accurate characterization of RIGs moving forward will require a global collaborative effort from both the research and clinical communities to collectively advance the knowledge of the field. Indeed, the development of a global RIG registry would facilitate the centralized collection of relevant clinical, pathological and molecular data defining these rare tumors. Identifying the molecular features of these tumors will help us to understand the mechanisms that drive RIG initiation and progression, as well as potentially uncover therapeutic targets for this currently incurable disease.
Supplementary Material
Acknowledgments
Funding
R.E. has support on a Brainchild Fellowship from the Pirate Ship Foundation. N.G.G. is funded by the Stan Perron Chair of Paediatric Haematology and Oncology. This study was conducted with support from the Pirate Ship Foundation.
Conflict of Interest statement. All authors state they have no conflicts of interest to declare.
Authorship Statement Conceptualization: J.P.W. Data curation and analysis: J.P.W., M.H., A.F., M.K., R.E. Data interpretation and visualization: J.P.W., M.H., A.F., M.K., R.E., N.G.G. Funding acquisition: R.E., N.G.G. Methodology: J.P.W. Writing – original draft: J.P.W. Writing – critical review and editing: J.P.W., M.H., A.F., M.K., R.E., N.G.G.
References
- 1. Bernier V, Klein O. Late effects of craniospinal irradiation for medulloblastomas in paediatric patients. Neurochirurgie. 2021;67(1):83–86. [DOI] [PubMed] [Google Scholar]
- 2. Fossati P, Ricardi U, Orecchia R. Pediatric medulloblastoma: toxicity of current treatment and potential role of protontherapy. Cancer Treat Rev. 2009;35(1):79–96. [DOI] [PubMed] [Google Scholar]
- 3. Braunstein S, Nakamura JL. Radiotherapy-induced malignancies: review of clinical features, pathobiology, and evolving approaches for mitigating risk. Front Oncol. 2013;3:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nantavithya C, Paulino AC, Liao K, et al. . Observed-to-expected incidence ratios of second malignant neoplasms after radiation therapy for medulloblastoma: a surveillance, epidemiology, and end results analysis. Cancer. 2021;127(13):2368–2375. [DOI] [PubMed] [Google Scholar]
- 5. Neglia JP, Robison LL, Stovall M, et al. . New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. J Natl Cancer Inst. 2006;98(21):1528–1537. [DOI] [PubMed] [Google Scholar]
- 6. Paulino AC, Mai WY, Chintagumpala M, Taher A, Teh BS. Radiation-induced malignant gliomas: is there a role for reirradiation? Int J Radiat Oncol Biol Phys. 2008;71(5):1381–1387. [DOI] [PubMed] [Google Scholar]
- 7. Taylor AJ, Little MP, Winter DL, et al. . Population-based risks of CNS tumors in survivors of childhood cancer: the British Childhood Cancer Survivor Study. J Clin Oncol. 2010;28(36):5287–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walter AW, Hancock ML, Pui CH, et al. . Secondary brain tumors in children treated for acute lymphoblastic leukemia at St Jude Children’s Research Hospital. J Clin Oncol. 1998;16(12):3761–3767. [DOI] [PubMed] [Google Scholar]
- 9. Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin Oncol (R Coll Radiol). 2013;25(10):578–585. [DOI] [PubMed] [Google Scholar]
- 10. Borrego-Soto G, Ortiz-López R, Rojas-Martínez A. Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet Mol Biol. 2015;38(4):420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu J, Bi K, Yang R, Li H, Nikitaki Z, Chang L. Role of DNA damage and repair in radiation cancer therapy: a current update and a look to the future. Int J Radiat Biol. 2020;96(11):1329–1338. [DOI] [PubMed] [Google Scholar]
- 12. Suzuki K, Ojima M, Kodama S, Watanabe M. Radiation-induced DNA damage and delayed induced genomic instability. Oncogene. 2003;22(45):6988–6993. [DOI] [PubMed] [Google Scholar]
- 13. Cahan WG, Woodard HQ. Sarcoma arising in irradiated bone; report of 11 cases. Cancer. 1948;1(1):3–29. [DOI] [PubMed] [Google Scholar]
- 14. Pettorini BL, Park YS, Caldarelli M, Massimi L, Tamburrini G, Di Rocco C. Radiation-induced brain tumours after central nervous system irradiation in childhood: a review. Childs Nerv Syst. 2008;24(7):793–805. [DOI] [PubMed] [Google Scholar]
- 15. Yamanaka R, Hayano A, Kanayama T. Radiation-induced meningiomas: an exhaustive review of the literature. World Neurosurg. 2017;97:635–644 e638. [DOI] [PubMed] [Google Scholar]
- 16. Yamanaka R, Hayano A, Kanayama T. Radiation-induced gliomas: a comprehensive review and meta-analysis. Neurosurg Rev. 2018;41(3):719–731. [DOI] [PubMed] [Google Scholar]
- 17. Elsamadicy AA, Babu R, Kirkpatrick JP, Adamson DC. Radiation-induced malignant gliomas: a current review. World Neurosurg. 2015;83(4):530–542. [DOI] [PubMed] [Google Scholar]
- 18. Donson AM, Erwin NS, Kleinschmidt-DeMasters BK, Madden JR, Addo-Yobo SO, Foreman NK. Unique molecular characteristics of radiation-induced glioblastoma. J Neuropathol Exp Neurol. 2007;66(8):740–749. [DOI] [PubMed] [Google Scholar]
- 19. Gits HC, Anderson M, Stallard S, et al. . Medulloblastoma therapy generates risk of a poorly-prognostic H3 wild-type subgroup of diffuse intrinsic pontine glioma: a report from the International DIPG Registry. Acta Neuropathol Commun. 2018;6(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Capper D, Jones DTW, Sill M, et al. . DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Page MJ, Moher D, Bossuyt PM, et al. . PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. 2021;372:n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tada M, Sawamura Y, Abe H, Iggo R. Homozygous p53 gene mutation in a radiation-induced glioblastoma 10 years after treatment for an intracranial germ cell tumor: case report. Neurosurgery. 1997;40(2):393–396. [DOI] [PubMed] [Google Scholar]
- 23. Matsumura H, Takimoto H, Shimada N, Hirata M, Ohnishi T, Hayakawa T. Glioblastoma following radiotherapy in a patient with tuberous sclerosis. Neurol Med Chir (Tokyo). 1998;38(5):287–291. [DOI] [PubMed] [Google Scholar]
- 24. Brat DJ, James CD, Jedlicka AE, et al. . Molecular genetic alterations in radiation-induced astrocytomas. Am J Pathol. 1999;154(5):1431–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang SY, Wang KC, Cho BK, et al. . Radiation-induced cerebellar glioblastoma at the site of a treated medulloblastoma: case report. J Neurosurg. 2005;102(4 Suppl):417–422. [DOI] [PubMed] [Google Scholar]
- 26. Berman EL, Eade TN, Brown D, et al. . Radiation-induced tumor after stereotactic radiosurgery for an arteriovenous malformation: case report. Neurosurgery. 2007;61(5):E1099; discussion E1099. [DOI] [PubMed] [Google Scholar]
- 27. Romeike BF, Kim YJ, Steudel WI, Graf N. Diffuse high-grade gliomas as second malignant neoplasms after radio-chemotherapy for pediatric malignancies. Childs Nerv Syst. 2007;23(2):185–193. [DOI] [PubMed] [Google Scholar]
- 28. Gessi M, Maderna E, Guzzetti S, et al. . Radiation-induced glioblastoma in a medulloblastoma patient: a case report with molecular features. Neuropathology. 2008;28(6):633–639. [DOI] [PubMed] [Google Scholar]
- 29. Salvati M, D’Elia A, Melone GA, et al. . Radio-induced gliomas: 20-year experience and critical review of the pathology. J Neurooncol. 2008;89(2):169–177. [DOI] [PubMed] [Google Scholar]
- 30. Sasayama T, Nishihara M, Tanaka K, et al. . Two metachronous tumors induced by radiation therapy: case report and review of the literature. J Neurooncol. 2008;88(3):315–320. [DOI] [PubMed] [Google Scholar]
- 31. García-Navarro V, Tena-Suck ML, Celis MA, Vega R, Rembao D, Salinas C. Anaplastic astrocytoma post radiotherapy of pineal germinoma. Arq Neuropsiquiatr. 2009;67(3A):707–709. [DOI] [PubMed] [Google Scholar]
- 32. Kamide T, Nakada M, Hayashi Y, et al. . Radiation-induced cerebellar high-grade glioma accompanied by meningioma and cavernoma 29 years after the treatment of medulloblastoma: a case report. J Neurooncol. 2010;100(2):299–303. [DOI] [PubMed] [Google Scholar]
- 33. Paugh BS, Qu C, Jones C, et al. . Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohba S, Shimizu K, Shibao S, et al. . A glioblastoma arising from the attached region where a meningioma had been totally removed. Neuropathology. 2011;31(6):606–611. [DOI] [PubMed] [Google Scholar]
- 35. Khoo HM, Kishima H, Kinoshita M, et al. . Radiation-induced anaplastic ependymoma with a remarkable clinical response to temozolomide: a case report. Br J Neurosurg. 2013;27(2):259–261. [DOI] [PubMed] [Google Scholar]
- 36. Mascelli S, Raso A, Biassoni R, et al. . Analysis of NADP+-dependent isocitrate dehydrogenase-1/2 gene mutations in pediatric brain tumors: report of a secondary anaplastic astrocytoma carrying the IDH1 mutation. J Neurooncol. 2012;109(3):477–484. [DOI] [PubMed] [Google Scholar]
- 37. Ahmed I, Krishnamurthy S, Kakkar A, Julka PK, Rath GK. Primary anaplastic astrocytoma of the brain after prophylactic cranial irradiation in a case of acute lymphoblastic leukemia: Case report and review of the literature. Indian J Med Paediatr Oncol. 2014;35(1):86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakao T, Sasagawa Y, Nobusawa S, et al. . Radiation-induced gliomas: a report of four cases and analysis of molecular biomarkers. Brain Tumor Pathol. 2017;34(4):149–154. [DOI] [PubMed] [Google Scholar]
- 39. Ng I, Tan CL, Yeo TT, Vellayappan B. Rapidly fatal radiation-induced Glioblastoma. Cureus. 2017;9(6):e1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Whitehouse JP, Howlett M, Hii H, et al. . A Novel Orthotopic Patient-Derived Xenograft Model of Radiation-Induced Glioma Following Medulloblastoma. Cancers (Basel). 2020;12(10):2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Izycka-Swieszewska E, Bien E, Stefanowicz J, et al. . Malignant Gliomas as second Neoplasms in pediatric cancer survivors: neuropathological study. Biomed Res Int. 2018;2018:4596812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kajitani T, Kanamori M, Saito R, et al. . Three case reports of radiation-induced glioblastoma after complete remission of acute lymphoblastic leukemia. Brain Tumor Pathol. 2018;35(2):114–122. [DOI] [PubMed] [Google Scholar]
- 43. Phi JH, Park AK, Lee S, et al. . Genomic analysis reveals secondary glioblastoma after radiotherapy in a subset of recurrent medulloblastomas. Acta Neuropathol. 2018;135(6):939–953. [DOI] [PubMed] [Google Scholar]
- 44. Porter AB, Sio TT, Nelson KD, Raghunathan A, Bendok BR, Mrugala MM. Disseminated High-grade Glioma in a Long-term Survivor of Medulloblastoma: Implications and Management of Radiation-induced Malignancies. Neurologist. 2018;23(6):191–193. [DOI] [PubMed] [Google Scholar]
- 45. Wang Y, Song S, Su X, et al. . Radiation-induced glioblastoma with rhabdoid characteristics following treatment for medulloblastoma: A case report and review of the literature. Mol Clin Oncol. 2018;9(4):415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. López GY, Van Ziffle J, Onodera C, et al. . The genetic landscape of gliomas arising after therapeutic radiation. Acta Neuropathol. 2019;137(1):139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mucha-Małecka A, Wojton-Dziewonska D, Adamek D, Urbanek K, Małecki K. Radiation-induced anaplastic astrocytoma following treatment of medulloblastoma. Folia Neuropathol. 2019;57(1):80–86. [DOI] [PubMed] [Google Scholar]
- 48. Biswas A, Kashyap L, Bakhshi S. Radiation-Associated Glioblastoma after Prophylactic Cranial Irradiation in a Patient of ALL: Review of Literature and Report of a Rare Case. Pediatr Neurosurg. 2020;55(6):409–417. [DOI] [PubMed] [Google Scholar]
- 49. Smith KS, Xu K, Mercer KS, et al. . Patient-derived orthotopic xenografts of pediatric brain tumors: a St. Jude resource. Acta Neuropathol. 2020;140(2):209–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woo PYM, Lee JWY, Lam SW, et al. . Radiotherapy-induced glioblastoma: distinct differences in overall survival, tumor location, pMGMT methylation and primary tumor epidemiology in Hong Kong Chinese patients. Br J Neurosurg. 2021:1–8. doi: 10.1080/02688697.2021.1881445 [DOI] [PubMed] [Google Scholar]
- 51. Madden JR, Addo-Yobo SO, Donson AM, et al. . Radiation-induced glioblastoma multiforme in children treated for medulloblastoma with characteristics of both medulloblastoma and glioblastoma multiforme. J Pediatr Hematol Oncol. 2010;32(7):e272–e278. [DOI] [PubMed] [Google Scholar]
- 52. Martin SE, Brat DJ, Vance GH, et al. . Glioblastoma occurring at the site of a previous medulloblastoma following a 5-year remission period. Neuropathology. 2012;32(5):543–550. [DOI] [PubMed] [Google Scholar]
- 53. DeSisto J, Lucas JT, Xu K, et al. . Comprehensive molecular characterization of pediatric treatment-induced high-grade glioma: a distinct entity despite disparate etiologies with defining molecular characteristics and potential therapeutic targets. bioRxiv, doi: 10.1101/809772, October 25, 2019, preprint: not peer reviewed. [DOI] [Google Scholar]
- 54. Ikemura M, Shibahara J, Mukasa A, et al. . Utility of ATRX immunohistochemistry in diagnosis of adult diffuse gliomas. Histopathology. 2016;69(2):260–267. [DOI] [PubMed] [Google Scholar]
- 55. Liu XY, Gerges N, Korshunov A, et al. . Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124(5):615–625. [DOI] [PubMed] [Google Scholar]
- 56. Tanboon J, Williams EA, Louis DN. The Diagnostic use of immunohistochemical surrogates for signature molecular genetic alterations in gliomas. J Neuropathol Exp Neurol. 2016;75(1):4–18. [DOI] [PubMed] [Google Scholar]
- 57. Kim KE, Kim KU, Kim DC, Park JI, Han JY. Cytogenetic characterizations of central nervous system tumors: the first comprehensive report from a single institution in Korea. J Korean Med Sci. 2009;24(3):453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mackay A, Burford A, Carvalho D, et al. . Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537 e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Magnani I, Guerneri S, Pollo B, et al. . Increasing complexity of the karyotype in 50 human gliomas. Progressive evolution and de novo occurrence of cytogenetic alterations. Cancer Genet Cytogenet. 1994;75(2):77–89. [DOI] [PubMed] [Google Scholar]
- 60. Sturm D, Pfister SM, Jones DTW. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. 2017;35(21):2370–2377. [DOI] [PubMed] [Google Scholar]
- 61. Sievers P, Sill M, Schrimpf D, et al. . A subset of pediatric-type thalamic gliomas share a distinct DNA methylation profile, H3K27me3 loss and frequent alteration of EGFR. Neuro-Oncology. 2020;23(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Korshunov A, Schrimpf D, Ryzhova M, et al. . H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol. 2017;134(3):507–516. [DOI] [PubMed] [Google Scholar]
- 63. Kato A, Nagashima G. A case of multiple radiation-induced gliomas 24 years after radiation therapy against pituitary adenoma. Clin Case Rep. 2016;4(4):356–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smoll NR, Hamilton B. Incidence and relative survival of anaplastic astrocytomas. Neuro Oncol. 2014;16(10):1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 66. Arcella A, Limanaqi F, Ferese R, et al. . Dissecting molecular features of Gliomas: genetic loci and validated biomarkers. Int J Mol Sci. 2020;21(2):685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jiao Y, Killela PJ, Reitman ZJ, et al. . Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Killela PJ, Pirozzi CJ, Reitman ZJ, et al. . The genetic landscape of anaplastic astrocytoma. Oncotarget. 2014;5(6):1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Takano S, Ishikawa E, Sakamoto N, et al. . Immunohistochemistry on IDH 1/2, ATRX, p53 and Ki-67 substitute molecular genetic testing and predict patient prognosis in grade III adult diffuse gliomas. Brain Tumor Pathol. 2016;33(2):107–116. [DOI] [PubMed] [Google Scholar]
- 70. Wiestler B, Capper D, Holland-Letz T, et al. . ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126(3):443–451. [DOI] [PubMed] [Google Scholar]
- 71. Korshunov A, Meyer J, Capper D, et al. . Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118(3):401–405. [DOI] [PubMed] [Google Scholar]
- 72. Jones DT, Gronych J, Lichter P, Witt O, Pfister SM. MAPK pathway activation in pilocytic astrocytoma. Cell Mol Life Sci. 2012;69(11):1799–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Forshew T, Tatevossian RG, Lawson AR, et al. . Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218(2):172–181. [DOI] [PubMed] [Google Scholar]
- 74. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project . Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jones DT, Kocialkowski S, Liu L, et al. . Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vanan MI, Eisenstat DD. DIPG in Children - what can we learn from the past? Front Oncol. 2015;5:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. . K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu G, Broniscer A, McEachron TA, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project . Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Louis DN, Giannini C, Capper D, et al. . cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639–642. [DOI] [PubMed] [Google Scholar]
- 81. Buczkowicz P, Hawkins C. Pathology, molecular genetics, and epigenetics of diffuse intrinsic pontine glioma. Front Oncol. 2015;5:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wu G, Diaz AK, Paugh BS, et al. . The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zarghooni M, Bartels U, Lee E, et al. . Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28(8):1337–1344. [DOI] [PubMed] [Google Scholar]
- 84. Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. [DOI] [PubMed] [Google Scholar]
- 85. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sturm D, Witt H, Hovestadt V, et al. . Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 90. Lucas J, DeSisto J, Xu K, et al. . HGG-57. Whole-genome sequencing, methylation analysis, and single-cell RNA-seq define unique characteristics of pediatric treatment-induced high-grade glioma and suggest oncogenic mechanisms. Neuro-Oncology. 2020;22(Suppl 3):iii354–iii354. [Google Scholar]
- 91. Batista R, Cruvinel-Carloni A, Vinagre J, et al. . The prognostic impact of TERT promoter mutations in glioblastomas is modified by the rs2853669 single nucleotide polymorphism. Int J Cancer. 2016;139(2):414–423. [DOI] [PubMed] [Google Scholar]
- 92. Mosaab A, El-Ayadi M, Khorshed EN, et al. . Histone H3K27M mutation overrides histological grading in pediatric gliomas. Sci Rep. 2020;10(1):8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Smith JS, Tachibana I, Passe SM, et al. . PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93(16):1246–1256. [DOI] [PubMed] [Google Scholar]
- 94. Carvalho PO, Uno M, Oba-Shinjo SM, et al. . Activation of EGFR signaling from pilocytic astrocytomas to glioblastomas. Int J Biol Markers. 2014;29(2):e120–e128. [DOI] [PubMed] [Google Scholar]
- 95. Louis DN, Perry A, Wesseling P, et al. . The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lucas JT, DeSisto J, Wu G, et al. . Comprehensive molecular characterization of pediatric treatment-induced glioblastoma: germline DNA repair defects as a potential etiology. Journal of Clinical Oncology. 2018;36(15 Suppl):10573–10573. [Google Scholar]
- 97. Relling MV, Rubnitz JE, Rivera GK, et al. . High incidence of secondary brain tumours after radiotherapy and antimetabolites. Lancet. 1999;354(9172):34–39. [DOI] [PubMed] [Google Scholar]
- 98. Loeffler JS, Niemierko A, Chapman PH. Second tumors after radiosurgery: tip of the iceberg or a bump in the road? Neurosurgery. 2003;52(6):1436–1440; discussion 1440-1432. [DOI] [PubMed] [Google Scholar]
- 99. Váňová K, Vícha A, Krsková L, et al. . HGG-31. Unique biological characteristics of radiation-induced gliomas. Neuro-Oncology. 2020;22(Suppl 3):iii349–iii349. [Google Scholar]
- 100. Todorova PK, Fletcher-Sananikone E, Mukherjee B, et al. . Radiation-Induced DNA Damage Cooperates with Heterozygosity of TP53 and PTEN to Generate High-Grade Gliomas. Cancer Res. 2019;79(14):3749–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.