Abstract
Objective
Neural network deidentification studies have focused on individual datasets. These studies assume the availability of a sufficient amount of human-annotated data to train models that can generalize to corresponding test data. In real-world situations, however, researchers often have limited or no in-house training data. Existing systems and external data can help jump-start deidentification on in-house data; however, the most efficient way of utilizing existing systems and external data is unclear. This article investigates the transferability of a state-of-the-art neural clinical deidentification system, NeuroNER, across a variety of datasets, when it is modified architecturally for domain generalization and when it is trained strategically for domain transfer.
Materials and Methods
We conducted a comparative study of the transferability of NeuroNER using 4 clinical note corpora with multiple note types from 2 institutions. We modified NeuroNER architecturally to integrate 2 types of domain generalization approaches. We evaluated each architecture using 3 training strategies. We measured transferability from external sources; transferability across note types; the contribution of external source data when in-domain training data are available; and transferability across institutions.
Results and Conclusions
Transferability from a single external source gave inconsistent results. Using additional external sources consistently yielded an F1-score of approximately 80%. Fine-tuning emerged as a dominant transfer strategy, with or without domain generalization. We also found that external sources were useful even in cases where in-domain training data were available. Transferability across institutions differed by note type and annotation label but resulted in improved performance.
Keywords: deidentification, generalizability, transferability, domain generalization
INTRODUCTION
Within biomedical informatics, deidentification refers to the task of removing protected health information (PHI) from clinical text. It aims to unlink health information from patients so that health information can be used without potential infringement of patient privacy. Due to the high cost and limited practicality of manual deidentification,1 automated deidentification systems have been developed. Early efforts at automated deidentification focused on rule-based methods,2,3 using dictionaries of names and common patterns to identify PHI. Subsequent approaches used supervised machine learning methods, treating deidentification as a sequence labeling problem. These approaches included Conditional Random Fields (CRF),4 Support Vector Machines,5 and Hidden Markov Models.6,7 The current state-of-the-art deidentification systems are neural network based8–11 and have demonstrated promising performance in recent natural language processing (NLP) shared tasks.12,13 When training and test sets are from the same distribution, these systems detect PHI with high accuracy. However, they fail to generalize to corpora that differ in their distribution of PHI.14
A handful of studies have tried to mitigate this problem with domain generalization (Domain adaptation, domain generalization, and transfer learning are closely related concepts. Although there are slight differences in the definitions of these learning concepts, for simplicity, we refer to them interchangeably in this paper.) . Lee et al.15 combined rule-based methods with dictionaries and regular expressions alongside 2 CRF-based taggers which both incorporated data from the i2b2/UTHealth-2014 dataset using EasyAdapt,16 a domain adaptation technique. Lee et al.17 examined the generalizability of feature-based machine learning deidentification classifiers. They evaluated several domain generalization techniques including instance pruning, instance weighting, and feature augmentation16 and concluded that feature augmentation was the most effective. Lee et al.18 evaluated transfer learning methods on an architecture similar to NeuroNER.8 They fine-tuned the trained source dataset to the target dataset by measuring which model parameters affected performance the most. The authors noted that transfer learning with only 16% of available in-domain training data achieved comparable performance as in-domain learning with 34% of the training data.
Our deidentification methods focus on finding and removing tokens that correspond to PHI categories defined by the Health Insurance Portability and Accountability Act (HIPAA) Safe Harbor (https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html) (see Supplementary AppendixTable SA.1) and mark the PHI type of each token. We add “Doctor” and “Hospital” categories to the HIPAA PHI, since their removal may improve the privacy protection.5
Table 1.
Overview of the deidentification datasets
| i2b2-2006 | i2b2/UTHealth-2014 | CEGS-NGRID-2016 | UW | |
|---|---|---|---|---|
| Institution | Partners HealthCare | Partners HealthCare | Partners HealthCare | UW Medical Center and Harborview Medical Center |
| Record types | Discharge notes | Diabetic longitudinal notes | Psychiatric intake notes | Admit, Discharge, Emergency department, Nursing, Pain management, Progress, Psychiatry, Radiology, Social work, and Surgery notes |
| Total no. of records (training/test) | 889 | 1,304 | 1,000 | 1,000 |
| (669/220) | (790/514) | (600/400) | (800/200) | |
| Total no. of tokens | 586,186 | 1,066,224 | 2,305,206 | 1,900,402 |
| Average no. of tokens per record | 659 | 818 | 2,305 | 1,900 |
| Total no. of PHIa | 19,498 | 25,260 | 21,121 | 44,701 |
| Average no. of PHI per record | 22 | 19 | 21 | 44 |
| Average no. of token per PHI token | 30 | 42 | 110 | 42 |
The distribution of the PHI types covered in our datasets is presented in Supplementary Appendix Table SB.2.
PHI: protected health information.
In this article, we evaluate the transferability of a state-of-the-art neural network deidentification system, NeuroNER, with and without architectural modifications for domain generalization, and under various training strategies, for identifying narrative text tokens corresponding to PHI. NeuroNER (http://neuroner.com/) is available online and readily lends itself to architectural modifications. Our architectural modifications include joint learning of domain information with deidentification (referred henceforth as Joint-Domain Learning [JDL]) and adoption of the state-of-the-art domain generalization approach of Common-Specific Decomposition (CSD).19 We apply 3 training strategies, sequential, fine-tuning, and concurrent, with NeuroNER, JDL, and CSD, as appropriate. We experiment with datasets that represent different distributions of note types and/or come from different institutions. Different note types represent the clinical sublanguage of their corresponding medical specializations.20 As a result, note types often differ in their linguistic representation, and in their use of PHI. Hence, each dataset can be viewed as being from a separate domain. We define source as the dataset to learn from and target as the dataset of application. We define an external source as a source that is drawn from a different domain than the target. In-domain data refer to data that come from the same domain as the target. Given our neural network architectures, training strategies, and domains, we investigate the following research questions.
(Q1) Can models trained on single domain datasets transfer well?
Individual data sets provide a start to deidentification. However, different individual datasets may transfer differently on new target data. We evaluate each of our datasets for transferability to other domains.
(Q2) Can models become more generalizable by adding more external sources?
With in-domain training, larger training data are generally beneficial in making models more generalizable.18 However, it is unclear whether using sources from multiple different domains improves generalizability, and by how much, for deidentification. Moreover, different training strategies can be applied when multiple sources are available. For example, one can train a model sequentially using external sources one by one in multi-iteration, or combine sources and train them concurrently in one iteration.
(Q3) Are domain generalization algorithms helpful for the transferability of deidentification systems?
Domain generalization methods attempt to generate common knowledge from external sources with the goal of benefiting from this knowledge when deidentifying as-yet unseen targets. In this paper, we integrate 2 representative domain generalization methods into NeuroNER architecture.
(Q4) Are external sources useful when sufficient in-domain training data are available?
Neural network deidentification systems using in-domain training data have been demonstrated to perform well in many cases. It is less clear, however, if external sources can be helpful even in cases where sufficient in-domain training data are available. On one hand, differences from external sources may introduce noise to training parameters and deteriorate overall performance. On the other hand, data from different domains may increase the general understanding of the task and may be useful to reduce the amount of required in-domain training data and/or improve performance.
(Q5) Can models transfer across institutions?
Transferability across institutions can be different from that within the same institution. For example, although different institutions may use different note authoring conventions, structures, and styles,21 models trained on a certain note type source may transfer well to the same note type target in another institution because they at least share the clinical sublanguage of their corresponding medical specializations. Transferability may also be affected by the nature of the PHI contained in the data. Often, the available external deidentification data contain surrogate PHI, that is, realistic placeholder PHI that obfuscate the originals. Target data may in contrast contain authentic PHI. We investigated transferability across institutions using data from 2 different institutions. One of our institutions provides surrogate deidentification data whereas the second one contains authentic PHI.
Our work builds on previous findings, adding the following contributions. First, we integrate domain generalization into the architecture of our state-of-the-art deidentification system, NeuroNER. Second, we compare NeuroNER with and without domain generalization in multiple experiments using a variety of dataset combinations and 3 training strategies. Third, we identify the best-performing architecture and training strategy combination and evaluate them on external institutions’ data when one of the datasets contains surrogate while the other contains authentic PHI.
DATASETS
We used 4 datasets for our experiments: i2b2-2006, i2b2/UTHealth (University of Texas Health Science Center)-2014, CEGS-NGRID-2016 datasets, and the University of Washington (UW) deidentification dataset. The first 3 datasets come from multiple clinical contexts and time periods within Partners HealthCare: i2b2-2006 consists of discharge summaries,22 i2b2/UTHealth-2014 consists of a mixed set of note types including discharge summaries, progress reports, and correspondences between medical professionals as well as communication with patients,12,23 and CEGS-NGRID-2016 consists of 1000 psychiatric intake records.13 The Partners HealthCare data contain surrogate PHI that have been generated to be realistic in their context (The details of the surrogate generation process are described in Stubbs et al.12). The UW dataset comes from the UW Medical Center and Harborview Medical Center and includes 10 note types (These include admit, discharge, emergency department, nursing, pain management, progress, psychiatry, radiology, social work, and surgery notes.) from 2018 to 2019. The UW dataset contains authentic PHI.
In order for the transferability between domains to be possible, datasets must be consistently labeled. We reviewed the annotation guidelines for each dataset and made adjustments in order to ensure label consistency. We refer to Supplementary Appendix SB.1 for details.
An overview of the deidentification datasets after the adjustment is shown in Table 1.
MATERIALS AND METHODS
Baseline deidentification approach—NeuroNER
To measure the transferability of neural network deidentification models, we used our NeuroNER,8 a representative, publicly available, state-of-the-art bidirectional Long Short-Term Memory (biLSTM) with CRF system which utilizes character and word embeddings24 as features. Each character in an input sentence is assigned to a unique character embedding, which is then passed through a character-level biLSTM layer. The output from that layer is concatenated with pretrained word embeddings to create character-enhanced token embeddings which are fed into a token-level biLSTM layer. The system performs multilabel prediction of PHI by delivering token-level biLSTM output into a fully connected layer and produces probabilities for individual PHI labels for each token in the sentence. The final PHI label for each token is determined by a CRF that maximizes the likelihood of the PHI label sequence for a sentence.
Domain generalization
We incorporated 2 domain generalization algorithms into NeuroNER.
Common-Specific Decomposition
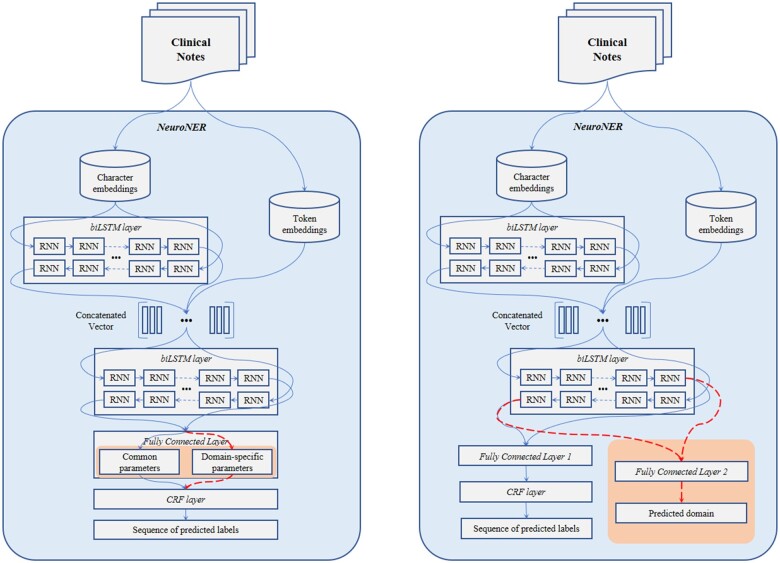
Decomposition is one of the major themes in domain generalization.19,25–27 In general decomposition training, parameters in a neural network layer are considered as the sum of their common parameters (ie, general parameters across multidomains) and domain-specific parameters (ie, specifically related to one domain). Piratla et al.19 suggest an efficient variation of this technique using common-specific low-rank decomposition. Assuming there are common features that are consistent across domains, they decompose the last layer of a neural network system into common parameters and domain-specific parameters. The domain-specific parameters are calculated as being low rank and orthogonal to the common parameters. The losses from both the common and domain-specific parameters are backpropagated during training. When making predictions, however, domain-specific parameters are dropped, and only the common parameters are used. In this way, the effect of domain differences is minimized. The overview of the CSD implementation on NeuroNER is shown in the left side of Figure 1 where CSD was integrated into the penultimate layer of the original NeuroNER.
Figure 1.
Overview of Common-Specific Decomposition (left) and Join-Domain Learning (right). Structures for domain generalization are shaded. The dotted lines are used only in training.
Joint-Domain Learning
JDL builds on the principles of joint learning.28 It treats deidentification and domain prediction as 2 tasks that can be learned jointly and that can inform each other. In JDL, a separate fully connected layer enables NeuroNER to jointly learn both a sequence of labels and a domain per each sentence. As shown in the right side of Figure 1, the Fully Connected Layer 2 in the shaded area is used only to learn and predict the domain of the input sentence, while the Fully Connected Layer 1 is used for label prediction. During training, losses from the label prediction and domain prediction layers are combined and backpropagated to update parameters in earlier layers. The 2 losses can be combined either equally or disproportionately. We experimented with various ratios and found the best performance with 85% of loss from label prediction and 15% of loss from domain prediction. For testing, the model predicts based on Fully Connected Layer 1 followed by CRF and does not use the Fully Connected Layer 2.
EXPERIMENTS
Experimental setting
Our experiments evaluated 3 architectures, NeuroNER, JDL, and CSD, using different training strategies. We defined 3 training strategies, sequential, fine-tuning, and concurrent as follows.
Sequential
This approach starts with training using a single external dataset to create pretrained weights which can be further refined on the external datasets in an iterative fashion. We experimented with different sequential ordering of domains; however, only the highest performing sequence of each combination was used for the follow-up experiments. Note that sequential training starts with learning from a single external dataset and can only be used for training NeuroNER. JDL and CSD by default require 2 or more domains to learn from and are therefore excluded from sequential training experiments.
Fine-tuning
A special form of sequential training where the in-domain training data are the final sequentially trained dataset.
Concurrent
Concurrent training combines multiple external sources and trains in a single iteration.
Experiments were conducted to respond to the previously presented research questions:
To assess whether models trained on single domain datasets transfer well (Q1), we train on individual i2b2-2006, i2b2/UTHealth-2014, and CEGS-NGRID-2016 datasets and evaluate against each of their respective test sets (eg, training and testing on i2b2-2006) as well as against each other’s test sets (eg, training i2b2-2006 and testing on i2b2/UTHealth-2014) using NeuroNER to establish baselines.
To check whether models become more generalizable by adding more external sources (Q2) and whether domain generalization architectures are helpful for transferability (Q3), we train with each of the 3 architectures with different training strategies using only external sources (eg, training a mix of i2b2-2006 and i2b2/UTHealth-2014, and testing on CEGS-NGRID-2016).
To evaluate whether external sources are useful when sufficient in-domain training data are available (Q4), we repeat the experiments above but include in-domain training data (eg, training a mix of i2b2-2006, i2b2/UTHealth-2014, CEGS-NGRID-2016, and testing on CEGS-NGRID-2016).
To measure transferability across institutions (Q5), we apply the best-performing architecture and training strategy combination across institutions, using one institution as the source and the other as the target.
Training
We tune our hyperparameters on the entire Partners HealthCare training set (Note that it would also have been valid to tune the parameters to individual datasets. However, our experiments showed that the above parameters worked well for individual data sets as well with very few exceptions where dataset-specific parameters would have affected performance. For the sake of simplicity, we chose to adopt the above parameters for all experiments.). The resulting hyperparameters were as follows: character embeddings dimension 25; token embeddings dimension 100; hidden layer dimension for LSTM over character 25; hidden layer dimension for LSTM over concatenation of (character LSTM output, token embeddings) 100; dropout 0.5; SGD optimizer; learning rate 0.005; maximum number of epochs 100; and patience 10.
Evaluation
Among various evaluation metrics for deidentification,12 we used PHI (entity-based) F1-scoring which is common in named entity recognition evaluation. In our case, PHI represent entities, with F1 calculated as 2 × (precision × recall)/(precision + recall), where precision is defined as true positives over the sum of true positives and false positives, and recall is defined as true positives over the sum of true positives and false negatives. The entity-based F1-score requires exact PHI spans to be found and counts partial predictions as false. For example, the entity “Jane Doe” is composed of 2 tokens, both of which would have to be marked as “Patient” PHI in order to count as correct. Partial labels that include only “Jane” or “Doe” are considered incorrect.
RESULTS
Table 2 presents the performance of NeuroNER on the individual i2b2 datasets (in response to Q1). Using in-domain training data, the system achieved F1-scores of 96.9, 94.0, and 92.5 on the i2b2-2006, i2b2/UTHealth-2014, and CEGS-NGRID-2016 test sets, respectively. The F1-scores of the models trained on a single external source were notably poorer, ranging from 58.2 to 82.3. The model trained on the i2b2-2006 dataset was the least generalizable and achieved F1-scores of 64.4 and 58.2 on the i2b2/UTHealth-2014 and CEGS-NGRID-2016 test data. The model trained using i2b2/UTHealth-2014 dataset was the most generalizable, achieving F1-scores of 75.2 and 82.3 on the i2b2-2006 and CEGS-NGRID-2016 test sets. This may be because i2b2/UTHealth-2014 dataset contains various note types whereas i2b2-2006 and CEGS-NGRID-2016 datasets consist of a single note type.
Table 2.
Baseline results
| Architecture | Source | Target |
||
|---|---|---|---|---|
| i2b2-2006 | i2b2/UTHealth-2014 | CEGS-NGRID-2016 | ||
| NeuroNER | 2006 | 97.8/96.1/96.9 | 73.9/57.0/64.4 | 68.6/50.5/58.2 |
| 2014 | 77.4/73.2/75.2 | 94.3/93.8/94.0 | 84.0/80.1/82.3 | |
| 2016 | 68.1/60.9/64.3 | 75.4/71.6/73.5 | 93.4/91.6/92.5 | |
Note: Performance when trained on individual datasets (%, P/R/F1). Bold indicates in-domain evaluation.
To check whether models become more generalizable by adding more external sources (Q2) and whether domain generalization is helpful for transferability (Q3), we experimented using 2 external sources with the NeuroNER, CSD, and JDL architectures. Results are shown in Table 3. Compared to Table 1, the results were largely consistent for all targets and higher overall, with NeuroNER using a concurrent training strategy achieving the best results. CSD and JDL performed higher on the i2b2-2006 dataset but generally performed worse than NeuroNER for all training strategies. JDL performed higher than CSD in all cases.
Table 3.
Performance of trained models without in-domain data (%, P/R/F1)
| Architecture | Training | Source | Target |
||
|---|---|---|---|---|---|
| Strategy | i2b2-2006 | i2b2/UTHealth-2014 | CEGS-NGRID-2016 | ||
| NeuroNER | Sequential | 2014→2016 | 77.4/73.2/75.2 | – | – |
| 2016→2014 | 78.2/79.4/78.8 | – | – | ||
| 2006→2016 | – | 80.2/77.4/78.8 | – | ||
| 2016→2006 | – | 75.4/71.6/73.5 | – | ||
| 2006→2014 | – | – | 85.5/82.3/83.9 | ||
| 2014→2006 | – | – | 84.0/80.1/82.3 | ||
| Concurrent | 2014 + 2016 | 77.6/81.5/79.5 | – | ||
| 2006 + 2016 | – | 80.1/77.1/78.6 | – | ||
| 2006 + 2014 | – | 82.7/80.8/81.7 | |||
| CSD | Concurrent | 2014 + 2016 | 82.8/78.6/80.6 | – | – |
| 2006 + 2016 | – | 82.1/69.2/75.1 | – | ||
| 2006 + 2014 | – | 81.4/65.5/72.6 | |||
| JDL | Concurrent | 2014 + 2016 | 81.8/81.7/81.7 | – | – |
| 2006 + 2016 | – | 80.6/71.8/76.0 | – | ||
| 2006 + 2014 | – | – | 79.5/68.8/73.8 | ||
Notes: Bold indicates the best performance for each target test set. → indicates sequential learning. + indicates concurrent learning. Results are reported only on the target test set. That is, i2b2-2006→i2b2/UTHealth-2014 sequentially trains on i2b2-2006 training set, continues training on the i2b2/UTHealth-2014 training set, and applies the resulting model to CEGS-NGRID-2016 test set.
CSD: Common-Specific Decomposition; JDL: Joint-Domain Learning.
To evaluate whether external sources are useful when sufficient in-domain training data are available (Q4), we repeated the experiments above but included in-domain training data for fine-tuning. Results are shown in Table 4. Fine-tuning experiments with concurrent training and NeuroNER architecture showed improvement over results in Table 2 using only in-domain training data, and a significantly higher performance than Table 3 external source-only experiments. As in Table 3, F1-scores for fine-tuned CSD and JDL architectures performed at or slightly higher than NeuroNER fine-tuned on the i2b2-2006 test dataset, but worse on the i2b2/UTHealth-2014 and CEGS-NGRID-2016 datasets. As in Table 3, JDL performed higher than CSD in all cases.
Table 4.
Performance of trained models with in-domain datasets (%, P/R/F1)
| Architecture | Training | Source | Target |
||
|---|---|---|---|---|---|
| Strategy | i2b2-2006 | i2b2/UTHealth-2014 | CEGS-NGRID-2016 | ||
| NeuroNER | Fine-tuning from Sequential | 2016→2014→2006 | 97.8/96.5/97.2 | – | – |
| 2006→2016→2014 | – | 95.8/94.9/95.3 | – | ||
| 2006→2014→2016 | – | – | 93.8/92.0/92.9 | ||
| Fine-tuning from Concurrent | 2014 + 2016→2006 | 97.9/96.8/97.3 | – | – | |
| 2006 + 2016→2014 | – | 95.9/95.2/95.5 | – | ||
| 2006 + 2014→2016 | – | – | 93.9/92.6/93.2 | ||
| Concurrent | 2006 + 2014 + 2016 | 96.6/97.2/96.8 | 95.6/94.9/95.3 | 94.1/92.0/93.0 | |
| CSD | Fine-tuning from Concurrent | 2014 + 2016→2006 | 98.1/96.4/97.2 | – | – |
| 2006 + 2016→2014 | – | 94.9/91.0/92.9 | – | ||
| 2006 + 2014→2016 | – | – | 92.1/86.3/89.1 | ||
| Concurrent | 2006 + 2014 + 2016 | 96.3/95.9/96.1 | 94.7/91.4/93.0 | 92.6/84.1/88.1 | |
| JDL | Fine-tuning from Concurrent | 2014 + 2016→2006 | 98.0/97.0/97.5 | – | – |
| 2006 + 2016→2014 | – | 95.6/91.2/93.4 | – | ||
| 2006 + 2014→2016 | – | – | 90.9/87.6/89.2 | ||
| Concurrent | 2006 + 2014 + 2016 | 96.5/95.6/96.1 | 95.0/91.2/93.1 | 91.3/85.3/88.2 | |
Notes: Bold indicates the best performance for each target test set by F1-score. → indicates sequential learning. + indicates concurrent learning. Results are only reported on the target test set.
CSD: Common-Specific Decomposition; JDL: Joint-Domain Learning.
Finally, we explored transferability across institutions (Q5) by first training a model using the training data of the combined Partners HealthCare datasets and applying the model to UW test data. We then reversed the direction on these experiments and applied the model from UW to Partners HealthCare data. Results are shown in Table 5. As previously mentioned, Partners HealthCare data contain surrogate PHI that are generated to be realistic placeholders while the UW data contain authentic PHI.
Table 5.
Transferability across institutions (%, P/R/F1)
| Architecture | Training | Source | Target | Performance |
|---|---|---|---|---|
| Strategy | ||||
| NeuroNER | In-domain | UW | UW | 90.5/91.8/91.1 |
| External source | 2006 + 2014 + 2016 | UW | 77.7/79.2/78.5 | |
| Concurrent | 2006 + 2014 + 2016+UW | UW | 90.1/91.9/91.0 | |
| Fine-tuning | 2006 + 2014 + 2016→UW | UW | 91.1/92.3/91.7 | |
| In-domain | 2006 + 2014 + 2016 | 2006 + 2014 + 2016 | 91.1/86.3/88.6 | |
| External source | UW | 2006 + 2014 + 2016 | 85.3/65.2/73.9 | |
| Concurrent | UW + 2006 + 2014 + 2016 | 2006 + 2014 + 2016 | 92.2/85.9/89.0 | |
| Fine-tuning | UW→2006 + 2014 + 2016 | 2006 + 2014 + 2016 | 92.1/86.7/89.3 |
Note: Best performance by F1-score is in bold.
F1-performance of in-domain UW training was 91.1. We achieved the best performance by fine-tuning a pretrained model, which improved performance to 91.7 (+0.6) on the UW test set. The improvement was statistically significant (p ≤ .05). (We calculated the statistical significance of differences using approximate randomization29 with 10,000 shuffles.) In the reverse direction, the combined Partners HealthCare datasets achieved the best performance also using fine-tuning with an F1-score of 89.3 versus a concurrent training score of 89.0. The difference was also statistically significant.
The sensitivity for the transfer across institutions varied per note type and label. To demonstrate performance differences across note types and labels, more granular examination of results can be found in Supplementary Appendix SC.
DISCUSSION
Overall, in response to whether models trained on single domain datasets can transfer well (Q1), we found that models trained on single domain datasets showed low and inconsistent performance when applied to external target datasets. Among the Partners HealthCare datasets, only the i2b2/UTHealth-2014 dataset includes multiple note types and achieved the best performance among external training sets on i2b2-2006 and CEGS-NGRID-2016 test sets in our baseline experiments using NeuroNER. This suggests that single domain datasets with a greater variety of note types can be expected to generalize better than those with only a single note type.
In terms of whether combining data from multiple source domains improves generalizability to target domains (Q2), as shown in Table 3, we found this to be true but only under certain conditions. When using sequential training, we found the order of the training sequence to be critical. When trained in certain sequences, sequential training of NeuroNER yielded +3.6, +5.3, and +1.6 F1-score improvements over the baseline results on all Partners HealthCare datasets. When trained in the opposite order from the best-scoring sequences, however, the models failed to improve at all and performance deteriorated as training progressed. We suspect the similarity of the external and in-domain corpora play a role in the success of transfer, but defining similarity is beyond the scope of this article and is left for future work. This suggests that careful examination of the relatedness between domains is necessary when choosing the sequence of model training.
This lesson was also true for domain generalization algorithms (Q3). These algorithms generated transferable models. They even provided a stronger model for transfer to the i2b2-2006 dataset, but generally performed worse than concurrent and best-performing sequential training of NeuroNER. Domain generalization approaches benefited from fine-tuning, as did other concurrent training strategies. Perhaps unsurprisingly, the domain generalization architectures were most effective when domain differences among sources were not significant. When the i2b2-2006 dataset (which was the least generalizable in the baseline) was used for training, domain generalization approaches did not generalize as well. However, it should be noted that domain generalization approaches thrive when given larger number of training domains. For example, CSD used data from 25 to 76 domains when it achieved the state-of-the-art performance.19 Therefore, domain generalization performance may be different if more sources from different note types were available. In all cases of our experiments, JDL performed higher than CSD.
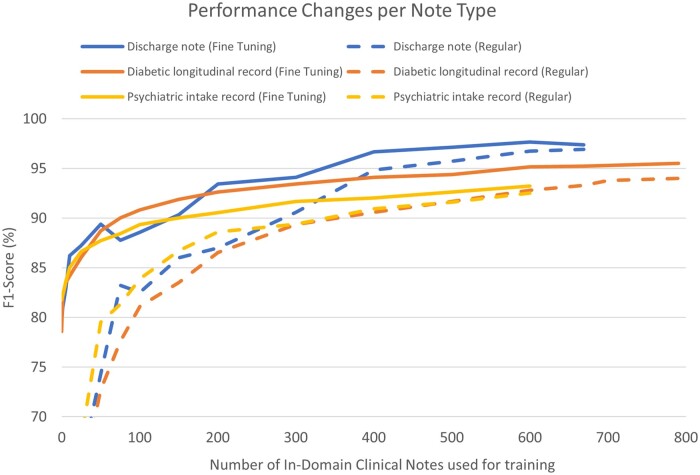
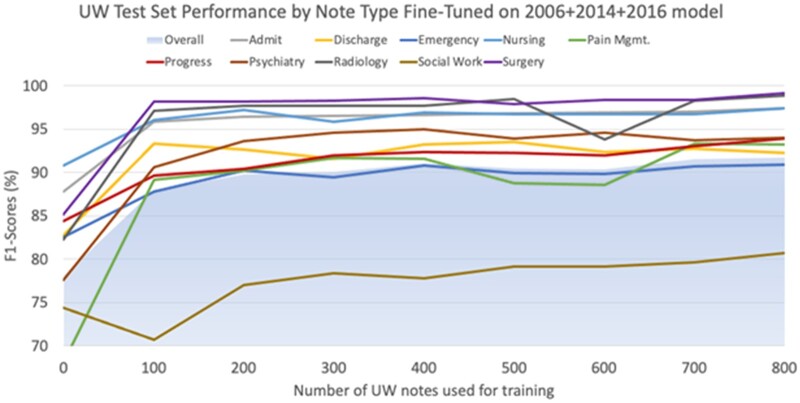
We also observed that even in cases when sufficient in-domain data are available, a model pretrained using external sources provides advantages (Q4). To test performance gains from external data, in the presence of in-domain data, we used 2 of the Partners HealthCare corpora as external sources while using the third one as in-domain fine-tuning data and tested performance on the corresponding in-domain test data. We increased the amount of available in-domain fine-tuning data at each iteration; we compared each iteration to the baseline of training on the same amount of in-domain training data and testing on the corresponding in-domain test data. In Figure 2, the dotted lines represent the baseline, and the solid lines represent the model fine-tuned from external data. The solid lines not only ascend faster than the dotted lines but also ultimately reach a higher level of performance. Repeating the same experiment on UW data, Figure 3 shows performance changes on the UW test set when fine-tuned from combined Partners HealthCare data using increasing UW training notes in increments of 100. UW note types with comparatively less PHI, such as Surgery, Radiology, and Nursing (see Supplementary AppendixTable SB.3) and Admit notes (which we hypothesize are structurally similar to Partners HealthCare notes) perform reasonably well using only 100 notes (10 of each note type), exceeding F1-scores of 95%. Other UW note types which have greater amounts of PHI or tend to be structurally more different than Partners HealthCare notes perform worse using 100 notes, but generally exceed an F1-score of 90% using 200 or so notes. The exception to this is Social Work notes, which exceeds an F1-score of 80% only when using 800 total training notes.
Figure 2.
Learning curve per in-domain training size using i2b2-2006, i2b2/UTHealth-2014, and CEGS-NGRID-2016 datasets (%, F1).
Figure 3.
Learning curve per in-domain UW training size using fine-tuning on a 2006 + 2014 + 2016 pretrained model. Each iteration incrementally increased the number of UW notes for training, with each note type representing 10% of training data (eg, for 100 total UW training notes, 10 from each note type were randomly chosen). Performance is measured by F1-scores using the same 200 UW test notes at each stage. UW: University of Washington.
Based on these findings, if the aim is to achieve an F1-score of 90%, a researcher must annotate about 300 in-domain notes regardless of note type if no external data are present. However, using a pretrained model, our findings suggest that only 50–150 in-domain annotated notes would be needed, depending on the note type. These results are largely consistent with the findings of previous studies.17,18
Regarding transferability across institutions (Q5), we found that models were able to transfer, but as noted earlier sensitivity varied by note type and label. While we expected the i2b2-2006 and CEGS-NGRID-2016 results to show the best performance on UW notes of the same type (discharge and psychiatry), Supplementary Appendix SC shows the i2b2/UTHealth-2014 dataset generalized well (and often better) for both note types, suggesting that certain corpora of mixed note types (such as i2b2/UTHealth-2014) may generalize better than those of single note types.
In addition, we found that domain transfer performance varied significantly by PHI type (see results in Supplementary Appendix SC). In general, “Date” types are the easiest to be transferred, as in most cases, dates are transcribed in only a handful of number formats and with a relatively limited vocabulary compared to other PHI. The second most transferable PHI type was “Doctor,” which we assume to be due to the common usage of honorifics and degrees such as “Dr.” and “M.D.,” or indicators such as “dictated by” and “signed by,” which are used relatively consistently across institutions. Other PHI types, particularly “ID,” showed generally poor performance in transferability. “Age” types, as limited to only cases with patients over 90 years and thus rarely present in the training data, proved to predictably show the poorest performance.
CONCLUSION
In this study, we investigated the transferability of neural network clinical deidentification methods from the practical perspective of researchers having a limitation on preparing enough in-domain annotated data. We found deidentification model transfers from a single external source to be limited. However, a more generalizable model can be produced using 2 or more external sources, which consistently achieved an F1-score of approximately 80%. We also found that domain generalization techniques generated transferable models but could also benefit from fine-tuning. In addition, external sources were useful even in cases where in-domain annotated data exist by reducing the required amount of in-domain annotation data and/or further improving model performance. The models could be transferred across institutions; however, the sensitivity for the transfer differed between note types and labels. When transferring from another institution, data of mixed note types performed higher than single note type data. External sources from different institutions were still useful to further improve performance in cases where in-domain training data were available. We expect that these findings will be useful in allowing deidentification models to be more easily leveraged across institutions.
FUNDING
This work was supported in part by the National Library of Medicine under Award Number R15LM013209 and by the National Center for Advancing Translational Sciences of National Institutes of Health under Award Number UL1TR002319.
AUTHOR CONTRIBUTIONS
KL and NJD contributed equally to this work. They incorporated domain generalization algorithms into NeuroNER, evaluated the systems’ performance, created figures, and wrote the paper. BM, MY, and ÖU formulated the original problem, provided direction and guidance, and gave helpful feedback on the paper.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The Partners HealthCare data underlying this article are available in the Department of Biomedical Informatics (DBMI) at Harvard Medical School as n2c2 (National NLP Clinical Challenges), at https://portal.dbmi.hms.harvard.edu/. The UW data cannot be shared publicly due to the privacy of patients.
Supplementary Material
ACKNOWLEDGMENTS
Experiments on the UW data were run on computational resources generously provided by the UW Department of Radiology. The authors gratefully acknowledge the work of David Wayne (UW) in the annotation of clinical notes.
REFERENCES
- 1. Dorr DA, Phillips WF, Phansalkar S, Sims SA, Hurdle JF.. Assessing the difficulty and time cost of de-identification in clinical narratives. Methods Inf Med 2006; 45 (3): 246–52. [PubMed] [Google Scholar]
- 2. Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. AMIA Annu Symp Proc 1996; 333–7. [PMC free article] [PubMed] [Google Scholar]
- 3. Meystre SM, Friedlin FJ, South BR, et al. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol 2010; 10: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gardner J, Xiong L.. HIDE: an integrated system for health information DE-identification. Proc IEEE Symp Comput Med Syst 2008; 254–9. doi: 10.1109/CBMS.2008.129. [Google Scholar]
- 5. Uzuner Ö, Sibanda TC, Luo Y, Szolovits P.. A de-identifier for medical discharge summaries. Artif Intell Med 2008; 42 (1): 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen T, Cullen RM, Godwin M.. Hidden Markov model using Dirichlet process for de-identification. J Biomed Inform 2015; 58 (Suppl): S60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wellner B, Huyck M, Mardis S, et al. Rapidly retargetable approaches to de-identification in medical records. J Am Med Inform Assoc 2007; 14 (5): 564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dernoncourt F, Lee JY, Uzuner O, Szolovits P.. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 24 (3): 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khin K, Burckhardt P, Padman R. A deep learning architecture for de-identification of patient notes: implementation and evaluation. 2018; 1–15. http://arxiv.org/abs/1810.01570 Accessed May 26, 2021.
- 10. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. 2018. http://arxiv.org/abs/1802.05365 Accessed May 26, 2021.
- 11. Liu Z, Tang Z, Wang X, Chen Q.. De-identification of clinical notes via recurrent neural network and conditional random field. J Biomed Inform 2017; 75 (1): S34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stubbs A, Kotfila C, Uzuner Ö.. Automated systems for the de-identification of longitudinal clinical narratives: overview of 2014 i2b2/UTHealth shared task Track 1. J Biomed Inform 2015; 58 Suppl (2015): S11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stubbs A, Filannino M, Uzuner Ö.. De-identification of psychiatric intake records: overview of CEGS N-GRID shared tasks Track 1. J Biomed Inform 2017; 75 (2017): S4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trienes J, Trieschnigg D, Seifert C, Hiemstra D.. Comparing rule-based, feature-based and deep neural methods for de-identification of Dutch medical records. CEUR Workshop Proc 2020; 2551 (2020): 3–11. [Google Scholar]
- 15. Lee H-J, Wu Y, Zhang Y, Xu J, Xu H, Roberts K.. A hybrid approach to automatic de-identification of psychiatric notes. J Biomed Inform 2017; 75 (2017): S19–27. doi: 10.1016/j.jbi.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daumé H. III Frustratingly easy domain adaptation. In: ACL 2007—Proceedings of the 45th Annual Meeting of the Association for Computational Linguistics 2007: 256–63. http://arxiv.org/abs/0907.1815 Accessed May 26, 2021. [Google Scholar]
- 17. Lee H-J, Zhang Y, Roberts K, Xu H.. Leveraging existing corpora for de-identification of psychiatric notes using domain adaptation. AMIA Annu Symp Proc 2018; 2017: 1070–9. [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JY, Dernoncourt F, Szolovits P.. Transfer learning for named-entity recognition with neural networks. In: Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018). European Language Resources Association (ELRA); 2017; 4470–3. https://aclanthology.org/L18-1708 Accessed May 26, 2021. [Google Scholar]
- 19. Piratla V, Netrapalli P, Sarawagi S. Efficient domain generalization via common-specific low-rank decomposition. 2020. http://arxiv.org/abs/2003.12815 Accessed May 26, 2021.
- 20. Weng W-H, Wagholikar KB, McCray AT, et al. Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Med Inform Decis Mak 2017; 17 (1): 155–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferraro JP, Ye Y, Gesteland PH, et al. The effects of natural language processing on cross-institutional portability of influenza case detection for disease surveillance. Appl Clin Inform 2017; 8 (2): 560–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uzuner O, Luo Y, Szolovits P.. 2007. Evaluating the state of the art in automatic de-identification. J Am Med Inform Assoc 2007; 14 (5): 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar V, Stubbs A, Shaw S, et al. Creation of a new longitudinal corpus of clinical narratives. J Biomed Inform 2015; 58 (Suppl): S6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pennington J, Socher R, Manning C.. Glove: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Association for Computational Linguistics; 2014; 1532–43. doi: 10.3115/v1/D14-1162. http://aclweb.org/anthology/D14-1162 Accessed May 26, 2021. [Google Scholar]
- 25. Li D, Yang Y, Song YZ, Hospedales TM.. 2018. Learning to generalize: meta-learning for domain generalization. 32nd AAAI Conf Artif Intell AAAI 2018 2018; 3490–7. https://www.aaai.org/ocs/index.php/AAAI/AAAI18/paper/viewPaper/16067 Accessed May 26, 2021. [Google Scholar]
- 26. Muandet K, Balduzzi D, Schölkopf B.. 2013. Domain generalization via invariant feature representation. 30th Int Conf Mach Learn ICML 2013 2013; 28 (PART 1): 10–8. http://proceedings.mlr.press/v28/muandet13.html Accessed May 26, 2021. [Google Scholar]
- 27. Shankar S, Piratla V, Chakrabarti S, Chaudhuri S, Jyothi P, Sarawagi S.. Generalizing across domains via cross-gradient training. In: Int Conf Learn Represent 2018 2018: 1–12. http://arxiv.org/abs/1804.10745 Accessed May 26, 2021. [Google Scholar]
- 28. Devlin J, Chang M-W, Lee K, Toutanova K.. BERT: pre-training of deep bidirectional transformers for language understanding. MLM 2018. http://arxiv.org/abs/1810.04805 Accessed May 26, 2021. [Google Scholar]
- 29. Noreen EW. Computer-Intensive Methods for Testing Hypotheses. New York, NY: Wiley; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Partners HealthCare data underlying this article are available in the Department of Biomedical Informatics (DBMI) at Harvard Medical School as n2c2 (National NLP Clinical Challenges), at https://portal.dbmi.hms.harvard.edu/. The UW data cannot be shared publicly due to the privacy of patients.