Abstract
Introduction
In cases of lung tumors that occur after treatment for malignancies in other organs, the tumor may represent either a primary lung cancer or a solitary pulmonary metastasis from the other tumor. Because some lung tumors are difficult to differentiate on the basis of imaging and pathologic findings, treatment selection is often difficult. In this study, we attempted to make a genomic diagnosis of primary and metastatic lung tumors by analyzing tumor samples using next-generation sequencing and evaluated the efficacy and validity of the genomic diagnosis.
Methods
A total of 24 patients with a solitary lung nodule and a history of other malignancies were enrolled in this study. Tumor cells were selected from tissue samples using laser capture microdissection. DNA was extracted from those cells and subjected to targeted deep sequencing of 53 genes.
Results
The driver mutation profiles of the primary lung tumors were discordant from those of the primary tumors in other sites, whereas the mutation profiles of pulmonary metastases and previous malignancies were concordant. In all 24 patients, we could diagnose either primary lung cancer (six patients) or lung metastases (18 patients) on the basis of whether gene mutation profiles were concordant or discordant. In 12 patients (50.0%), discrepancies were observed between the genomic and clinical or histopathologic diagnoses.
Conclusions
In patients with a solitary lung lesion and a history of cancer, tumor-specific mutations can serve as clonal markers, affording a more accurate understanding of the pathological condition and thus possibly improving both treatment selection and patient outcome.
Keywords: Lung cancer, Metastasis, Sequencing, Mutation, Genomic diagnosis
Introduction
In recent years, the number of patients with lung cancer has been increasing both in Japan and worldwide.1,2 Furthermore, with improvements in surgical outcomes and the advent of an aging society, an increasing number of patients who have previously been treated for malignancies at other primary sites develop lung tumors.3,4 When lung tumors arise in patients who have previously been treated for malignancies at other primary sites, the tumors may represent either primary lung cancer or a solitary pulmonary metastasis, and the appropriate treatment varies depending on pathological condition. For example, a common treatment for solitary pulmonary metastasis after surgery for esophageal cancer consists of partial resection of the lung followed by chemotherapy for esophageal cancer. In contrast, primary lung cancer occurring after surgery for esophageal cancer is treated with lobectomy, and postoperative chemotherapy is unnecessary in principle.
If individual tumors are histologically inconsistent in terms of histologic feature or cellular atypism, a diagnosis of multiple primary cancers is highly likely. Nevertheless, although differentiation between primary lung tumors and solitary metastases has been discussed in the field of lung surgery for many years, there are no specific radiologic, clinical, or histologic features that can be universally used to accurately distinguish pulmonary metastases from primary lung cancers. Usually, diagnosticians (e.g., clinicians, radiologists, and pathologists) evaluate imaging findings, clinical courses, and pathologic findings and ultimately diagnose, on the basis of their subjective views, whether a patient is likely to have a primary or metastatic tumor. Nevertheless, different diagnosticians often reach different diagnoses. In such cases, subsequent treatment is based on an uncertain diagnosis instead of a definitive diagnosis. Consequently, the diagnosis does not agree with the subsequent clinical course, and there are presumably a considerable number of cases that are misdiagnosed and mistreated.5
We previously developed and reported a molecular pathologic method to differentiate primary and metastatic tumors in patients with multicentric lung cancers.6,7 In this previous study, we reported that the gene mutation profile of a tumor is a potential clonal marker specific to each tumor and that evaluation of differences in these profiles allows differentiation between primary and metastatic tumors, provision of appropriate treatment on the basis of pathological condition, and prediction of outcomes.6,7 In the present study, we evaluated whether our previous findings could be used to differentiate primary lung cancers and solitary pulmonary metastases from malignancies at other sites. We attempted to make a genomic diagnosis of primary or metastatic lung tumors by analyzing tumor samples using next-generation sequencing (NGS) and evaluated the efficacy and validity of the genomic diagnosis. The goals of this study were to develop a new diagnostic method that is much more accurate than the conventional pathologic diagnostic methods and present basic data for clinical application.
Materials and Methods
Patients and Sample Preparation
This study included 24 patients with a history of treated malignancies in organs other than the lung (e.g., colorectal cancer, gastric cancer, head and neck cancer, and cervical cancer) who subsequently developed lung tumors and underwent lung tumor resection at our hospital between April 2014 and April 2021. Written informed consent was obtained from all patients for genetic research studies, which were performed according to the protocols approved by the Institutional Review Board in Yamanashi Central Hospital. Histologic typing was performed according to the WHO classification (fifth edition), and clinical staging was performed according to the International Union Against Cancer TNM classification (eighth edition).8
A serial section from formalin-fixed, paraffin-embedded (FFPE) tissue was stained with hematoxylin and eosin and subsequently microdissected using an ArcturusXT laser capture microdissection system (Thermo Fisher Scientific, Tokyo, Japan). DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Tokyo, Japan). FFPE DNA quality was verified using primers for the ribonuclease P locus. Peripheral blood was drawn from each patient immediately before surgery. The buffy coat was isolated by centrifugation, and DNA was extracted from these cells using the QIAamp DNA Blood Mini Kit (Qiagen).
Targeted Deep Sequencing and Data Analysis
A panel that covered the coding regions of 53 cancer-related genes (Supplementary Table 1) was designed in-house to perform targeted sequencing. Ion AmpliSeq designer software (Thermo Fisher Scientific) was used for the primer composition, as previously reported.10, 11, 9 The Ion AmpliSeq Library kit (Thermo Fisher Scientific) was used to prepare sequencing libraries. The library samples were barcoded using the Ion Xpress Barcode Adapters kit (Thermo Fisher Scientific), purified using Agencourt AMPure XP reagent (Beckman Coulter, Tokyo, Japan), and subsequently quantified using the Ion Library Quantitation Kit (Thermo Fisher Scientific). The libraries were templated using the Ion PI Template OT2 200 Kit v3 (Thermo Fisher Scientific). Sequencing was performed using the Ion Proton (Ion Torrent) and the Ion PI Sequencing 200 Kit v3.
The sequence data were processed on standard Ion Torrent Suite Software. Raw signal data were evaluated using Torrent Suite version 4.0. The pipeline consisted of signaling processing, base calling, quality score assignment, read alignment to the human genome 19 reference, quality control of mapping, and coverage analysis. After data analysis, the annotation of single-nucleotide variants and insertions and deletions was performed using the Ion Reporter Server System (Thermo Fisher Scientific). Blood cell DNA extracted from peripheral blood was used as the normal control to detect variants (tumor-normal pair analysis). Sequencing data were visually analyzed using the Integrative Genomics Viewer.
Results
Patient Characteristics
The 24 patients recruited in this study (age range: 45–83 y; mean age = 68.3 ± 10.1 y) included 17 men and seven women (Table 1). The patients had previously undergone surgery for the following cancers: colorectal cancer in 13 patients, cervical cancer in one patient, urothelial cancer in one patient, pancreatic cancer in one patient, gastric cancer in three patients, esophageal cancer in two patients, choriocarcinoma in one patient, pharyngeal cancer in two patients, and gingival cancer in one patient. Regarding the timing of tumor development, tumors developed synchronously and metachronously in nine and 15 patients, respectively (Table 1).
Table 1.
Patients’ Characteristics
| Case No. | Sex | Age (y) | Other Malignancy | Interval Between Detection of the Tumors |
|---|---|---|---|---|
| 1 | F | 64 | Gingival ca. | 1 y 8 mo |
| 2 | M | 79 | Pharyngeal ca. | 4 y 3 mo |
| 3 | M | 57 | Choriocarcinoma | Synchronous |
| 4 | M | 70 | Pharyngeal ca./esophageal ca. | 13 y 0 mo/3 y 3 mo |
| 5 | M | 65 | Esophageal ca. | 1 y 2 mo |
| 6 | M | 65 | Gastric ca. | 2 y 6 mo |
| 7 | M | 78 | Gastric ca. | Synchronous |
| 8 | M | 66 | Gastric ca. | Synchronous |
| 9 | M | 73 | Pancreatic ca. | 2 y 4 mo |
| 10 | F | 75 | Colorectal ca. | Synchronous |
| 11 | F | 75 | Colorectal ca. | Synchronous |
| 12 | M | 77 | Colorectal ca. | 1 y 0 mo |
| 13 | M | 45 | Colorectal ca. | 1 y 0 mo |
| 14 | M | 49 | Colorectal ca. | 5 y 0 mo |
| 15 | F | 72 | Colorectal ca. | Synchronous |
| 16 | M | 72 | Colorectal ca. | Synchronous |
| 17 | M | 72 | Colorectal ca. | Synchronous |
| 18 | M | 60 | Colorectal ca. | Synchronous |
| 19 | M | 83 | Colorectal ca. | 1 y 6 mo |
| 20 | M | 83 | Colorectal ca. | 1 y 6 mo |
| 21 | F | 69 | Colorectal ca. | 6 mo |
| 22 | F | 54 | Colorectal ca. | 6 mo |
| 23 | F | 64 | Cervical ca. | 2 y 9 mo |
| 24 | M | 70 | Urothelial ca. | 2 y 5 mo |
ca., cancer; F, female; M, male.
Somatic Mutations Identified by Targeted Sequencing
Targeted sequencing was performed for 24 surgically resected tumors obtained from 24 patients; blood samples were used as normal controls. The mean coverage depths were 1044-fold for cancer samples (range: 226- to 5048-fold) and 1326-fold for blood cell samples (range: 214- to 4030-fold). Sequence analyses detected 102 somatic mutations with an allele fraction greater than or equal to 1% from 53 cancer lesions (1–12 mutations per tumor) (Supplementary Table 2).
In six patients, the gene, amino acid substitution, and nucleotide changes caused by these somatic mutations within individual tumors were discordant (Fig. 1 and Supplementary Table 2), that is, there were no shared or overlapping mutations among the individual tumors detected in these patients. This finding revealed that the lung tumors in these cases were independently developing primary cancers (Fig. 1). Nevertheless, in 18 patients, the gene mutation profiles of the lung tumors and the original tumors at another primary site were concordant, suggesting the presence of intrapulmonary metastasis (Fig. 2). Importantly, in these cases, mutation variance was entirely consistent at the nucleotide level among the tumors (Supplementary Table 2).
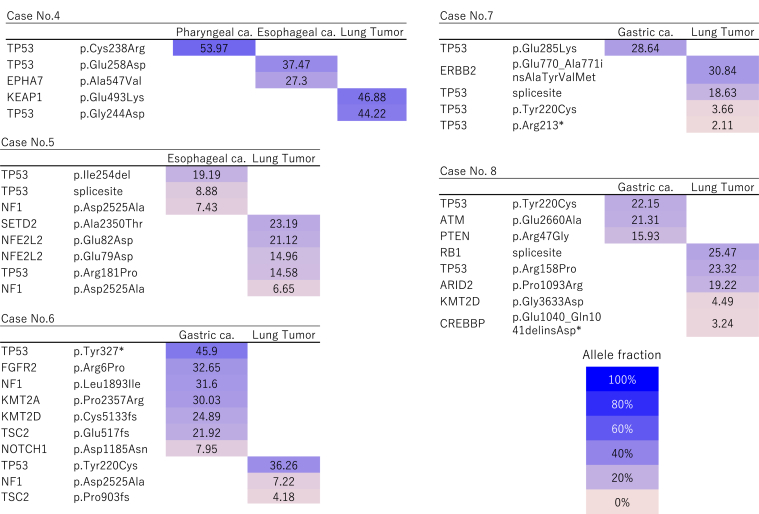
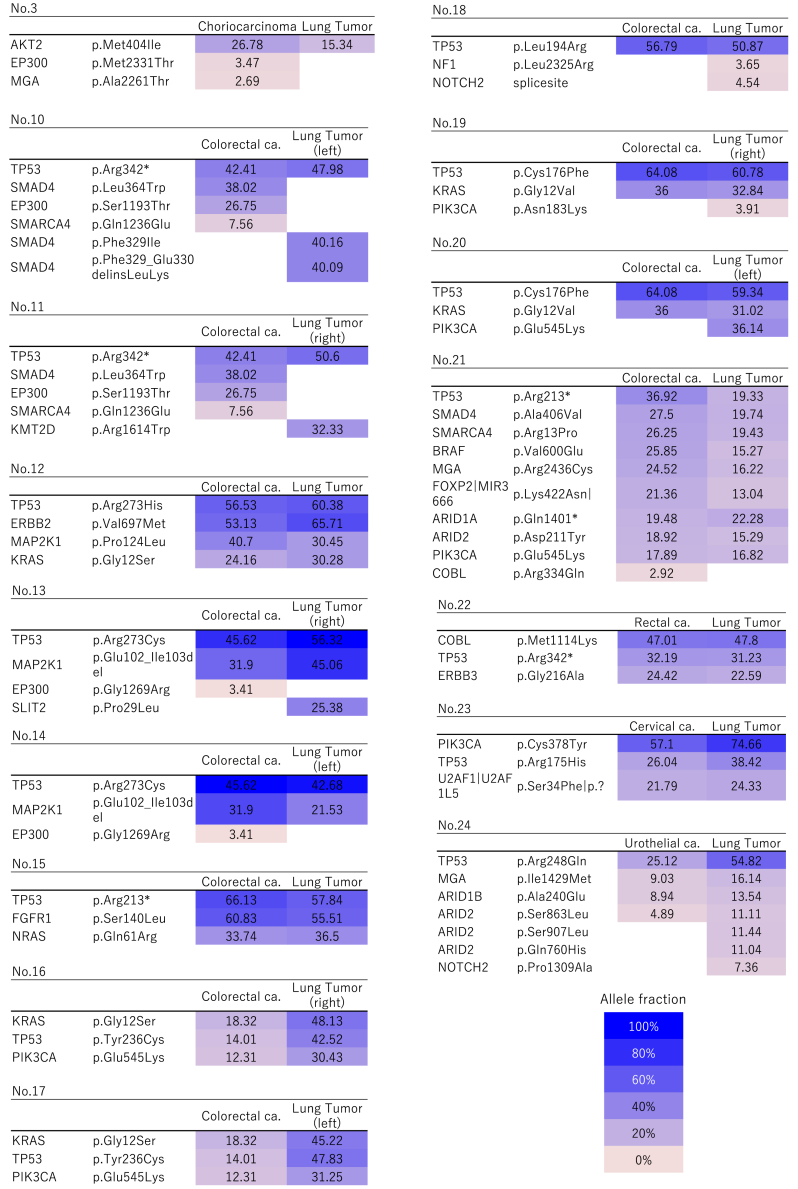
Figure 1.
Heatmap of gene mutations in patients with double or triple primary lung cancers. These maps visualize the gene mutations present in each cancer. Two or three tumors in each patient were characterized by different mutation profiles, and all patients were diagnosed with having multicentric primary cancers. The remaining case (case 2 in Tables 1 and 2) not illustrated in this figure is described in detail in the Case Presentations section and in Figure 4. ca., cancer.
Figure 2.
Heatmap of gene mutations in patients with solitary lung metastases. The mutation profiles were concordant between the individual tumors in each case, and the tumors were identified as intrapulmonary metastases. The remaining cases (cases 1 and 9 in Tables 1 and 2) not illustrated in this figure are described in detail in the Case Presentations section and in Figures 3 and 5. ca., cancer.
Case Presentations
Three representative cases are presented subsequently in detail.
Case I (Case 1 in Tables 1 and 2)
Table 2.
Clinical, Pathologic, and Genomic Diagnosis of the Lung Tumors
| Case No. | Clinical Diagnosis | Pathologic Diagnosis | Genomic Diagnosis |
|---|---|---|---|
| 1 | Lung cancer | Lung cancer | Meta,b |
| 2 | Met | Met | Lung cancera,b |
| 3 | Met | Met | Met |
| 4 | Lung cancer | Lung cancer | Lung cancer |
| 5 | Lung cancer | Met | Lung cancera,b |
| 6 | Lung cancer | Lung cancer | Lung cancer |
| 7 | Lung cancer | Lung cancer | Lung cancer |
| 8 | Lung cancer | Lung cancer | Lung cancer |
| 9 | Lung cancer | Lung cancer | Meta,b |
| 10 | Lung cancer | Met | Metb |
| 11 | Met | Lung cancer | Meta,b |
| 12 | Met | Met | Met |
| 13 | Met | Met | Met |
| 14 | Lung cancer | Met | Metb |
| 15 | Lung cancer | Met | Metb |
| 16 | Met | Met | Met |
| 17 | Met | Met | Met |
| 18 | Lung cancer | Met | Metb |
| 19 | Met | Met | Met |
| 20 | Met | Met | Met |
| 21 | Lung cancer | Met | Metb |
| 22 | Lung cancer | Met | Metb |
| 23 | Lung cancer | Met | Metb |
| 24 | Met | Met | Met |
Note: Cases in which the clinicopathologic and genomic diagnoses or the pathologic and genomic diagnoses were discordant are indicated by b and a, respectively.
Met, metastasis.
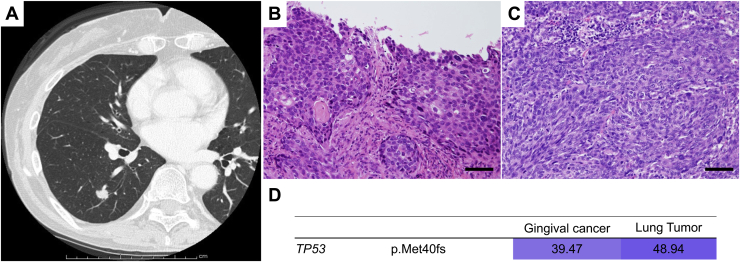
A 64-year-old woman received chemoradiation therapy for gingival cancer. After 1 year and 8 months, she developed a nodule in the right lower lobe and underwent right lower lobectomy. Morphologically, the lung tumor had an irregular surface; therefore, it was diagnosed as primary lung cancer (Fig. 3A). Pathologically, both lesions were identified as squamous cell carcinoma, although the lung tumor was histologically more poorly differentiated (Fig. 3B and C). Therefore, the tumor was pathologically diagnosed as primary lung cancer. Nevertheless, genomically, the mutation profiles between these two tumors were completely concordant, suggesting the lung lesion was a metastasis of the gingival cancer (Fig. 3D). At the patient’s request, she was placed on follow-up without any postoperative adjuvant chemotherapy. The patient has remained alive 2 years postoperatively without recurrence.
Figure 3.
Radiologic, histopathologic, and genomic findings in case I. (A) One pulmonary nodule appeared in the right lower lobe 2 years after treatment for gingival cancer. (B, C) Histologically, both the (B) gingival cancer and (C) lung nodule were squamous cell carcinoma. Each scale bar indicates 50 μm. (D) The heatmap revealed that the same mutation profiles were shared between the two tumors.
Case II (Case 2 in Tables 1 and 2)
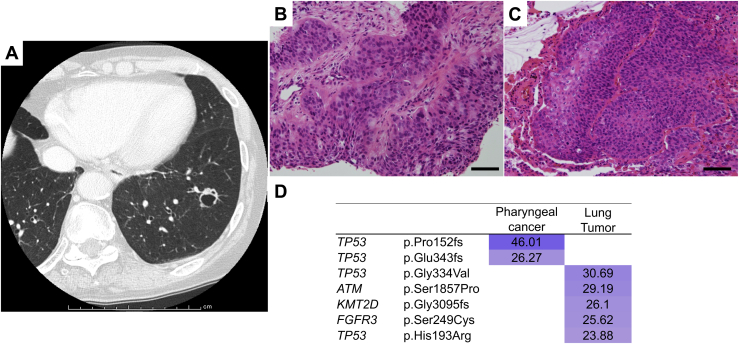
A 79-year-old man presented with a 1.2-cm–sized cavitary nodule in the left lower lobe 4 years after undergoing radiation therapy for pharyngeal cancer (Fig. 4A). Morphologically, the lung nodule seemed to be metastatic. After 4 months of follow-up, there was no increase in the number of lung lesions, suggesting indolent oligometastasis, for which left lower lobectomy was performed. The tumor was a squamous cell carcinoma, as was the patient’s pharyngeal cancer (Fig. 4B and C), which led to a diagnosis of pulmonary metastasis. Nevertheless, genomically, the mutation profiles of the two tumors were discordant, suggesting the lung lesion was a second primary cancer (Fig. 4D). The patient has remained alive without recurrence for 2 years after the surgery.
Figure 4.
Radiologic, histopathologic, and genomic findings of case II. (A) A cavitary nodule appeared in the left lower lobe 4 years after radiotherapy for pharyngeal cancer. (B, C) Histopathology of the (B) pharyngeal cancer and (C) pulmonary nodule. Each scale bar indicates 50 μm. (D) Heatmap of the gene mutations in the two lung tumors. The mutation profiles of the two tumors were completely different.
Case III (Case 9 in Tables 1 and 2)
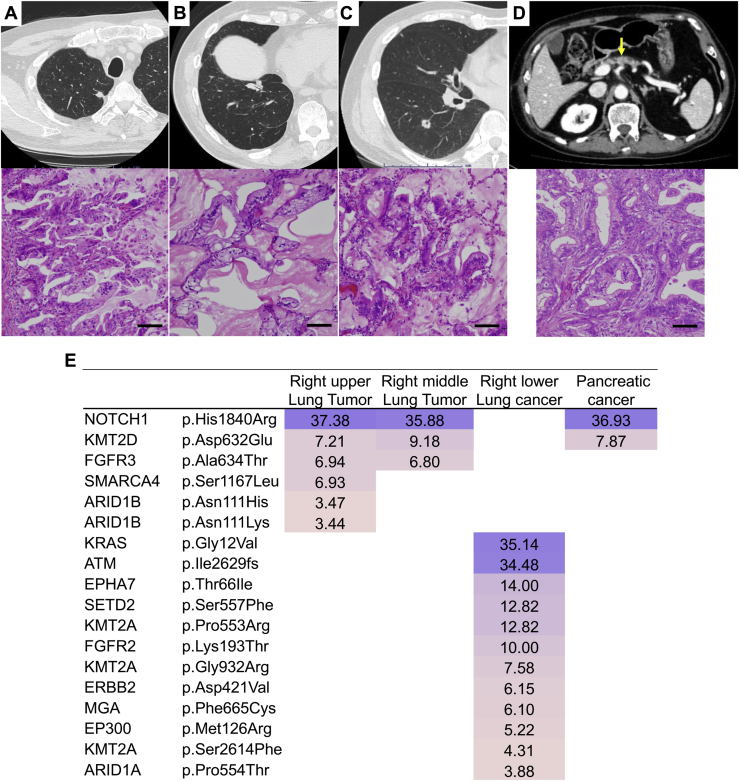
A 73-year-old man presented with tumors measuring 1.1 cm and 2.0 cm in the right upper and middle lobes, respectively, for which partial resections of the lung were performed (Fig. 5A and B). The patient had undergone right lower lobectomy for lung cancer 3 years previously and pancreaticoduodenectomy for pancreatic cancer 2 years previously (Fig. 5C and D). As these three lung tumors exhibited similar histopathology of invasive mucinous adenocarcinoma, the tumors located in the right upper and middle lobes were assumed to be pulmonary metastases from the lung cancer in the right lower lobe (Fig. 5A–C). Nevertheless, the genomic mutation profiling revealed that the pulmonary metastases had arisen not from the lung cancer but from the pancreatic cancer (Fig. 5E). The patient received postoperative chemotherapy for pancreatic cancer and has remained alive without recurrence for 3 years after the last surgery.
Figure 5.
Radiologic, histopathologic, and genomic findings of case III. (A, B) Two pulmonary nodules appeared in the right upper and middle lobes after right lower lobectomy. A, a nodule in the right upper lobe; B, a nodule in the middle lobe. Each scale bar indicates 100 μm. (C, D) Computed tomography and histopathology of the (C) previous lung cancer and (D) the pancreatic cancer. The arrow indicates the pancreatic cancer. Each scale bar indicates 100 μm. (E) Heatmap of the gene mutations of the three lung tumors and the pancreatic cancer. The dominant mutations identified in the right upper and middle lobe tumors were homologous to those detected in the pancreatic cancer.
Study on the Discrepancies Between the ClinicalorHistopathologic Diagnoses and the Genomic Diagnosis
Table 2 reveals the discrepancies among the clinical, pathologic, and genomic diagnoses of the primary or metastatic lesions in all 24 patients. The clinical diagnoses were comprehensively determined by the cancer board (comprising thoracic surgeons, pulmonologists, pathologists, and radiologists) based mainly on imaging findings and clinical courses. The pathologic diagnoses were subjectively determined on the basis of postoperative pathologic findings, especially differences in tissue form and cellular atypia detected by pathologists. The genomic diagnoses were determined on the basis of digital and statistical analyses of similarities in the mutation profiles of the individual tumors. The genomic diagnosis and clinical or histopathologic diagnoses were discordant in 12 patients (50.0%), whereas the genomic diagnosis and histopathologic diagnosis were discordant in five patients (20.8%).
In case 5, previous esophageal carcinoma and lung tumor exhibited almost the same histological findings of poorly differentiated squamous cell carcinoma, leading to the pathologic diagnosis of metastatic disease, whereas it turned out primary lung cancer by genomic analysis. Likewise, in case 11, although positive thyroid transcription factor-1 staining in lung tumor favored the diagnosis of primary lung cancer, genomic analysis revealed it to be metastatic.
Among the nine patients with synchronous tumors, primary and metastatic tumors were eventually diagnosed on the basis of genomic diagnosis in two and seven patients, respectively. Among the 15 patients with metachronous tumors, primary and metastatic tumors were diagnosed in four and 11 patients, respectively, in the same manner. Therefore, there was no association between synchronous or metachronous appearance of the lung tumor and the primary or metastatic status (p = 0.81, chi-square test).
Discussion
Solitary lung nodules that appear in patients who have previously been treated for malignancies in other organs may represent either pulmonary metastasis from the previous malignancy or a primary lung cancer. Nevertheless, in the clinical setting, it is often difficult to differentiate between primary and metastatic lesions on the basis of imaging and pathologic findings. Such cases render treatment selection challenging. In this study, we analyzed tumor samples using NGS to diagnose whether lung tumors were primary or metastatic. We reported that cancer-specific mutations could serve as clonal markers, enabling discrimination of primary and metastatic lung tumors.
Because advances in personalized treatment have improved cancer treatment options, differentiation between primary and metastatic tumors in patients with multiple lung cancers has become increasingly important, and there have been only sporadic reports on new methods to differentiate these tumors.12,13 Although most of these reports present methods to estimate clonality on the basis of immunostaining results, other pathologic findings, clinical data, or imaging findings, no accurate diagnostic method applicable to all patients has been found. Detterbeck et al.14 reviewed reports using clinical and pathologic diagnostic criteria to differentiate primary and metastatic tumors in patients with multiple lung cancers in 2016. They concluded that there are no definitive diagnostic criteria and that no definitive diagnosis can be made even if the similarity between two lesions is evaluated by conventional methods.15 Although a few studies focusing on gene mutations have attempted differentiation, these studies focused only on typical mutations of several (one to five) genes, such as TP53 and EGFR, and used estimation methods for clonality.16, 17, 18, 19, 20 Nevertheless, because of intratumoral heterogeneity, different cells in the same lung tumor harbor various gene mutations.21 In addition, the possibility of chance co-occurrence of the same mutation in multiple lesions cannot be excluded completely; therefore, cases that could be differentiated by the methods used were limited. Hence, their significance as a diagnostic method was questioned, and they were not clinically applied.
We previously developed and reported a comprehensive analysis of gene mutations for diagnosing primary and metastatic tumors in patients with multiple lung cancers.6,7 In this method, gene mutations in each tumor are comprehensively analyzed. First, trunk mutations are identified. Then, the profiles of these trunk mutations are compared between tumors to determine whether the clonality is concordant or discordant. Because genetic mutation underlies the pathological condition of cancer, the diagnostic criteria on the basis of our method may be theoretical and essential, unlike the conventional criteria, which are stochastic and suggestive. Furthermore, the assay is performed only using NGS. If the analysis results are displayed using heatmaps, a diagnosis can be made instinctively and easily (Figs. 1 and 2). When we applied our method to differentiate solitary pulmonary metastasis from malignancies in other organs and primary lung cancer in this study, our method yielded a clear genomic diagnosis in all patients. In other words, no equivocal diagnosis was obtained in any of the patients. Moreover, the genomic diagnoses made in this study differed from the pathologic diagnoses in five of the 24 patients (20.8%). Our novel approach may help resolve the dilemma of misdiagnosis in the clinical setting. We anticipate that our method will be broadly used as a standard diagnostic approach in daily clinical practice in the near future.
Consistency in multiple mutations, with complete concordance in the position and patterns of base-pair substitutions or insertions and deletions, cannot occur as a coincidental phenomenon. Although discordance in the genomic profiles of two tumors was noted in mutations with low allele fractions, this can be explained by tumor heterogeneity.21, 22, 23 Cancers are composed of populations of cells with distinct molecular and phenotypic features, a phenomenon described as intratumoral heterogeneity.21, 22, 23 This may afford tumor adaptation, cancer progression and metastasis, and therapeutic failure through negative selection.21,24 Conversely, a driver mutation triggers clonal expansion and is retained ubiquitously within the tumors of the same clone.24, 25, 26 Therefore, clonally dominant mutations are important clonal markers and primary and metastatic tumors can be differentiated by determining whether such ubiquitous driver mutations are concordant.
Although it is easy to diagnose multicentric primary cancers with distinct histologic findings, it is often difficult to determine whether a lesion is a primary tumor or a metastasis when it shares similar histologic findings with the first primary cancer. Particularly in cases of multiple tumors classified as squamous cell carcinoma (such as cases I and II), differentiation on the basis of histopathology alone is impractical. Nevertheless, even when morphologic and immunohistologic features are nonhomogeneous among different parts of the tumors, the driver mutation is ubiquitously retained within the tumors of the same clone.24,25,27 Therefore, distinction of clonality on the basis of mutational profiles may be more specific and definitive than histologic distinction.
Meanwhile, in the clinical settings, it is unnecessary to perform NGS in all cases from the viewpoint of medical economy; NGS can be practically eliminated by the pathologist's confident diagnosis of primary lung cancer or metastatic lung tumor. In contrast, with the pathologist's relatively equivocal judgment not leading to a definitive diagnosis, NGS confirmation such as our method contributes a final judgment. Furthermore, in such cases, NGS should be performed at an early stage after surgery if the indications and regimen for postoperative drug therapy differ depending on the determination of primary/metastasis. It is also very important to confirm the primary site by NGS in cases of metastases or recurrent foci appearing in patients in postoperative follow-up, because, in metastasis or recurrent cancer cases, determining from which cancer the metastatic or recurrent tumor originates allows applications of personalized medicine on the basis of genotypes and phenotypes of each tumor, thereby contributing to improved prognosis.
This study had some limitations. First, the patient cohort was relatively small owing to the single-institution design. Second, patient survival could not be analyzed as no patient developed recurrence. Generally, patients with oligometastasis are indicated for surgery and can be completely cured in some cases. Nevertheless, their postoperative prognosis is supposed to be poorer than that of patients with primary lung cancer.28 Therefore, postoperative treatments and surveillance strategies need to be tailored on the basis of the diagnosis and predicted outcome. If a large number of patients had been enrolled in this study, we could have more closely examined clinical adjustment on the basis of different prognoses and the tumor pathological characteristics. A larger study with more patients needs to be performed to validate our findings. Nevertheless, because the major aim of this preliminary analysis was the identification of clonal markers in multicentric cancers to determine whether this method should be prioritized for clinical development, even a modestly sized sample can provide useful insights.
In conclusion, we developed a genomic diagnostic method with a focus on gene mutations. This method allows physicians to make a more objective and scientific diagnosis on the basis of DNA sequence data. This method is novel and revolutionary because it allows accurate differentiation between primary and metastatic tumors through statistical processing of DNA data, even in cases where pathologic distinction is impossible or equivocal. Clinical application of this study may allow physicians to prescribe appropriate treatment on the basis of an accurate diagnosis, thereby fundamentally improving patient outcomes. Furthermore, we expect that this diagnostic method will contribute to the establishment of new therapeutic systems for pathologic conditions that may be pulmonary metastasis from malignancies in other organs.
CRediT Authorship Contribution Statement
Rumi Higuchi: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Writing—original draft.
Taichiro Goto: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Writing—review and editing.
Takahiro Nakagomi, Kenji Amemiya: Data curation, Investigation.
Yosuke Hirotsu: Conceptualization, Methodology, Data curation, Investigation, Resources, Writing—original draft.
Toshio Oyama: Data curation.
Hitoshi Mochizuki: Data curation, Methodology, Formal analysis.
Masao Omata: Conceptualization, Methodology, Formal analysis, Resources, Supervision.
Acknowledgments
This study was supported by a grant-in-aid for Genome Research Project from Yamanashi Prefecture (Drs. Hirotsu and Omata) and by the Japan Society for the Promotion of Science KAKENHI grant number JP21K08894 (Dr. Goto). The funder had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. The authors greatly appreciate Yoshihiro Miyashita, Yumiko Kakizaki, and Toshiharu Tsutsui for helpful scientific discussions.
Footnotes
Drs. Higuchi and Goto contributed equally to this work.
Disclosure: The authors declare no conflict of interest.
Cite this article as: Higuchi R, Goto T, Nakagomi T, et al. Discrimination between primary lung cancer and lung metastases by genomic profiling. JTO Clin Res Rep. 2021;2:100255.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2021.100255.
Supplementary Data
References
- 1.Bong C.Y., Smithers B.M., Chua T.C. Pulmonary metastasectomy in the era of targeted therapy and immunotherapy. J Thorac Dis. 2021;13:2618–2627. doi: 10.21037/jtd.2020.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiono S. The role of pulmonary metastasectomy for pulmonary metastasis from head and neck cancer. J Thorac Dis. 2021;13:2643–2648. doi: 10.21037/jtd.2020.04.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seitlinger J., Prieto M., Siat J., Renaud S. Pulmonary metastasectomy in renal cell carcinoma: a mainstay of multidisciplinary treatment. J Thorac Dis. 2021;13:2636–2642. doi: 10.21037/jtd-2019-pm-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerull W.D., Puri V., Kozower B.D. The epidemiology and biology of pulmonary metastases. J Thorac Dis. 2021;13:2585–2589. doi: 10.21037/jtd.2020.04.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichinose J., Shinozaki-Ushiku A., Takai D., Fukayama M., Nakajima J. Differential diagnosis between primary lung squamous cell carcinoma and pulmonary metastasis of head and neck squamous cell carcinoma. Expert Rev Anticancer Ther. 2016;16:403–410. doi: 10.1586/14737140.2016.1147352. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi R., Nakagomi T., Goto T., et al. Identification of clonality through genomic profile analysis in multiple lung cancers. J Clin Med. 2020;9:573. doi: 10.3390/jcm9020573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto T., Hirotsu Y., Mochizuki H., et al. Mutational analysis of multiple lung cancers: discrimination between primary and metastatic lung cancers by genomic profile. Oncotarget. 2017;8:31133–31143. doi: 10.18632/oncotarget.16096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chansky K., Detterbeck F.C., Nicholson A.G., et al. The IASLC Lung Cancer Staging Project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2017;12:1109–1121. doi: 10.1016/j.jtho.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Goto T., Hirotsu Y., Amemiya K., et al. Distribution of circulating tumor DNA in lung cancer: analysis of the primary lung and bone marrow along with the pulmonary venous and peripheral blood. Oncotarget. 2017;8:59268–59281. doi: 10.18632/oncotarget.19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goto T., Hirotsu Y., Oyama T., Amemiya K., Omata M. Analysis of tumor-derived DNA in plasma and bone marrow fluid in lung cancer patients. Med Oncol. 2016;33:29. doi: 10.1007/s12032-016-0744-x. [DOI] [PubMed] [Google Scholar]
- 11.Nakagomi T., Goto T., Hirotsu Y., et al. Genomic characteristics of invasive mucinous adenocarcinomas of the lung and potential therapeutic targets of B7-H3. Cancers (Basel) 2018;10:478. doi: 10.3390/cancers10120478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C., Huang X., Peng M., Liu W., Yu F., Wang X. Multiple primary lung cancer: a rising challenge. J Thorac Dis. 2019;11(suppl 4):S523–S536. doi: 10.21037/jtd.2019.01.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue X., Liu Y., Pan L., et al. Diagnosis of multiple primary lung cancer: a systematic review. J Int Med Res. 2013;41:1779–1787. doi: 10.1177/0300060513504707. [DOI] [PubMed] [Google Scholar]
- 14.Detterbeck F.C., Nicholson A.G., Franklin W.A., et al. The IASLC Lung Cancer Staging Project: summary of proposals for revisions of the classification of lung cancers with multiple pulmonary sites of involvement in the forthcoming eighth edition of the TNM classification. J Thorac Oncol. 2016;11:639–650. doi: 10.1016/j.jtho.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Detterbeck F.C., Franklin W.A., Nicholson A.G., et al. The IASLC Lung Cancer Staging Project: background data and proposed criteria to distinguish separate primary lung cancers from metastatic foci in patients with two lung tumors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:651–665. doi: 10.1016/j.jtho.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 16.van Oijen M.G., Leppers Vd Straat F.G., Tilanus M.G., Slootweg P.J. The origins of multiple squamous cell carcinomas in the aerodigestive tract. Cancer. 2000;88:884–893. doi: 10.1002/(sici)1097-0142(20000215)88:4<884::aid-cncr20>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 17.van der Sijp J.R., van Meerbeeck J.P., Maat A.P., et al. Determination of the molecular relationship between multiple tumors within one patient is of clinical importance. J Clin Oncol. 2002;20:1105–1114. doi: 10.1200/JCO.2002.20.4.1105. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu S., Yatabe Y., Koshikawa T., et al. High frequency of clonally related tumors in cases of multiple synchronous lung cancers as revealed by molecular diagnosis. Clin Cancer Res. 2000;6:3994–3999. [PubMed] [Google Scholar]
- 19.Ono K., Sugio K., Uramoto H., et al. Discrimination of multiple primary lung cancers from intrapulmonary metastasis based on the expression of four cancer-related proteins. Cancer. 2009;115:3489–3500. doi: 10.1002/cncr.24382. [DOI] [PubMed] [Google Scholar]
- 20.Takamochi K., Oh S., Matsuoka J., Suzuki K. Clonality status of multifocal lung adenocarcinomas based on the mutation patterns of EGFR and K-ras. Lung Cancer. 2012;75:313–320. doi: 10.1016/j.lungcan.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Goto T., Hirotsu Y., Amemiya K., Mochizuki H., Omata M. Understanding intratumor heterogeneity and evolution in NSCLC and potential new therapeutic approach. Cancers (Basel) 2018;10:212. doi: 10.3390/cancers10070212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap T.A., Gerlinger M., Futreal P.A., Pusztai L., Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 23.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto T., Hirotsu Y., Mochizuki H., et al. Stepwise addition of genetic changes correlated with histological change from “well-differentiated” to “sarcomatoid” phenotypes: a case report. BMC Cancer. 2017;17:65. doi: 10.1186/s12885-017-3059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yatabe Y., Matsuo K., Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Fujimoto J., Zhang J., et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Bruin E.C., McGranahan N., Mitter R., et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otake S., Goto T. Stereotactic radiotherapy for oligometastasis. Cancers (Basel) 2019;11:133. doi: 10.3390/cancers11020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.