Abstract
Background & Aims
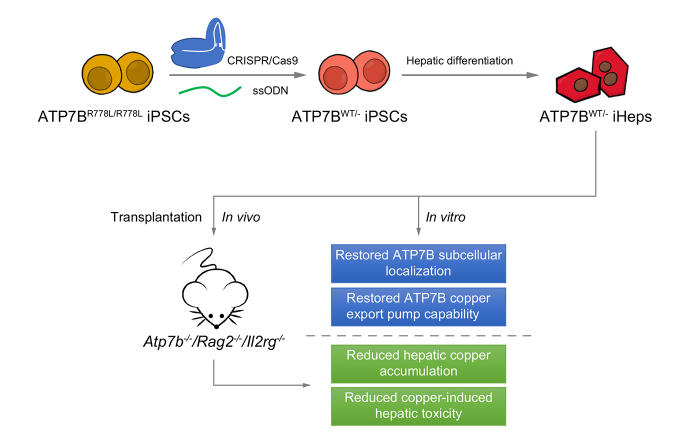
Wilson’s disease (WD) is an autosomal recessive disorder of copper metabolism caused by loss-of-function mutations in ATP7B, which encodes a copper-transporting protein. It is characterized by excessive copper deposition in tissues, predominantly in the liver and brain. We sought to investigate whether gene-corrected patient-specific induced pluripotent stem cell (iPSC)-derived hepatocytes (iHeps) could serve as an autologous cell source for cellular transplantation therapy in WD.
Methods
We first compared the in vitro phenotype and cellular function of ATP7B before and after gene correction using CRISPR/Cas9 and single-stranded oligodeoxynucleotides (ssODNs) in iHeps (derived from patients with WD) which were homozygous for the ATP7B R778L mutation (ATP7BR778L/R778L). Next, we evaluated the in vivo therapeutic potential of cellular transplantation of WD gene-corrected iHeps in an immunodeficient WD mouse model (Atp7b-/-/ Rag2-/-/ Il2rg-/-; ARG).
Results
We successfully created iPSCs with heterozygous gene correction carrying 1 allele of the wild-type ATP7B gene (ATP7BWT/-) using CRISPR/Cas9 and ssODNs. Compared with ATP7BR778L/R778L iHeps, gene-corrected ATP7BWT/- iHeps restored in vitro ATP7B subcellular localization, its subcellular trafficking in response to copper overload and its copper exportation function. Moreover, in vivo cellular transplantation of ATP7BWT/- iHeps into ARG mice via intra-splenic injection significantly attenuated the hepatic manifestations of WD. Liver function improved and liver fibrosis decreased due to reductions in hepatic copper accumulation and consequently copper-induced hepatocyte toxicity.
Conclusions
Our findings demonstrate that gene-corrected patient-specific iPSC-derived iHeps can rescue the in vitro and in vivo disease phenotypes of WD. These proof-of-principle data suggest that iHeps derived from gene-corrected WD iPSCs have potential use as an autologous ex vivo cell source for in vivo therapy of WD as well as other inherited liver disorders.
Lay summary
Gene correction restored ATP7B function in hepatocytes derived from induced pluripotent stem cells that originated from a patient with Wilson’s disease. These gene-corrected hepatocytes are potential cell sources for autologous cell therapy in patients with Wilson’s disease.
Keywords: Wilson’s disease, induced pluripotent stem cell (iPSC), iPSC-derived hepatocytes (iHeps), ATPase copper transporting beta polypeptide (ATP7B), Clustered regularly interspaced palindromic repeats (CRISPR)/Cas9, Single-stranded Oligodeoxynucleotide (ssODN), gene correction, cell therapy
Abbreviations: AFP, alpha-fetoprotein; ALB, albumin; ATP7B, ATPase copper transporting beta; EB, embryoid body; iHep(s), iPSC-derived hepatocyte(s); iPSC, induced pluripotent stem cell; RFLP, restriction fragment length polymorphism; sgRNA, single guide RNA; ssODN, single-stranded oligodeoxynucleotide; TGN, trans-Golgi network; WD, Wilson’s disease
Graphical abstract
Highlights
-
•
Correction of the ATP7B R778L mutation restored the subcellular localization of ATP7B in iHeps.
-
•
The copper exportation capability of ATP7B was restored in gene-corrected iHeps.
-
•
Gene-corrected iHeps reduced hepatic copper accumulation and copper-induced hepatic toxicity in mice with Wilson’s disease.
-
•
Gene-corrected iHeps are potential ex vivo cell sources for therapy in Wilson’s disease.
Introduction
Wilson’s disease (WD) is a monogenic autosomal recessive liver disorder caused by malfunction of ATPase copper transporting beta (ATP7B). Its incidence is approximately 1 in 30,000 live births worldwide, and around 1 in 90 healthy people carry an abnormal copy of the ATP7B gene.1 ATP7B helps deliver copper into the blood stream for use in tissues and also mediates the excretion of excess copper into bile to maintain copper homeostasis.2 Homozygous or compound heterozygous mutations in ATP7B result in defective cellular copper homeostasis and excessive copper accumulation in the liver, brain and other organs, leading to a variety of clinical manifestations.3 Left untreated, patients will develop liver failure and/or neurologic complications and die prematurely.
Liver transplantation is a potential cure for WD.4 Nonetheless its application is hampered by a scarcity of donors, immune rejection of allografts, and the adverse effects of long-term immunosuppressive therapy. It has been proposed that hepatocyte transplantation can serve as a bridging therapy or even an alternative to liver transplantation for treatment of WD.5 Prior studies have demonstrated that allogenic hepatocyte transplantation is effective in animal models of WD,6,7 and in patients with other inherited liver metabolic disorders.[8], [9], [10] Compared with liver transplantation, hepatocyte transplantation is more flexible and the surgery is much less invasive. Moreover, given the nature of WD, fewer hepatocytes can be effective in removing excess copper and ameliorating the disease. Nevertheless, immunosuppression is still needed to prevent immune rejection of the transplanted allogenic hepatocytes from heathy donors.5
Recent technological advances related to human induced pluripotent stem cells (iPSCs),11 engineered CRISPR/Cas9 nuclease-mediated genome editing,12 and pluripotent stem cell-based hepatic differentiation have enabled the generation of iPSC-derived hepatocytes (iHeps) from gene-corrected patient-specific iPSCs, which can subsequently be used for autologous cell transplantation, thereby eliminating the need for subsequent immunosuppression. Autologous iHeps derived from gene-corrected patient-specific iPSCs are a promising cell source for cell therapy. They have the potential to overcome the limitations of allogenic human hepatocytes with their limited availability and risk of immune rejection.
ATPB R778L is the most prevalent variant among Chinese patients with WD, and those with this variant predominantly present with liver disease.13 We have previously generated ATP7BR778L/R778L iPSCs from the fibroblasts of a patient with WD who was homozygous for the ATP7B R778L mutation.13 In this report, we sought to evaluate whether correction of the WD genotype could reverse the disease phenotype in vitro and to study the potential use of gene-corrected iHeps for autologous cell therapy in WD using immunocompromised WD mice. Our results provide proof-of-principle data that gene-corrected WD iPSCs can be differentiated into functional iHeps in vitro and attenuate the disease phenotype in vitro and in vivo.
Material and methods
Cell lines
We used a wild-type (ATP7BWT/WT) male human iPSC line14 and 1 male WD patient-specific iPSC line homozygous for the ATP7B R778L mutation (ATP7BR778L/R778L),13 which was previously generated in our laboratories.
Gene correction using CRISPR/Cas9 and single-stranded oligodeoxynucleotides
Single guide RNAs (sgRNAs) used in this study were designed with the online CRISPR design tool (http://crispr.mit.edu) and cloned into the PX459 plasmid (Addgene plasmid # 48139).15 The single-stranded oligodeoxynucleotide (ssODN) template for correcting ATP7B R778L was designed based on the guidelines from a previous report.16 0.15 million iPSCs were co-transfected with 0.5 μg PX459 plasmid and 0.25 μg ssODN, and targeted single cell-derived clones selected using a restriction fragment length polymorphism (RFLP) assay and confirmed by Sanger sequencing. Detailed methodology can be found in the supplementary methods.
Differentiation of hepatocytes from pluripotent stem cells
Our previously modified 3-step protocol was used for differentiation of iHeps from iPSCs.14 Details can also be found in the supplementary methods.
Transplantation of hepatocytes into immunodeficient Wilson’s disease mice
Sixteen-week-old immunodeficient WD mice (Atp7b-/-/ Rag2-/-/ Il2rg-/-, ARG mice) were used for iHep transplantation. One million iHeps per mouse were transplanted using the method described previously.14,17
For further details regarding the materials used, please refer to the CTAT table and supplementary information.
Results
Correction of ATP7B R778L in WD iPSCs using CRISPR/Cas9 and single-stranded oligodeoxynucleotides
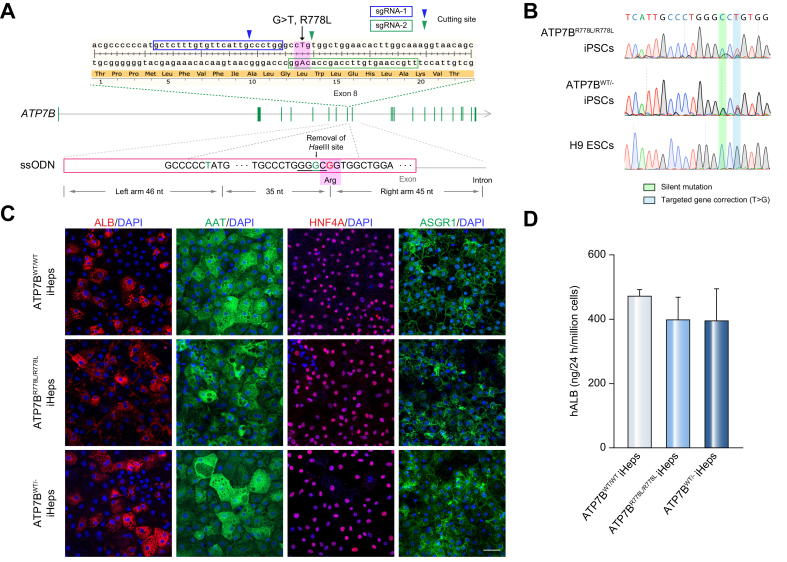
Since any extra DNA fragment left in the genome, even in the intron or 3’ downstream elements, may interrupt target gene expression (e.g., the loxP site),18 we used ssODNs combined with CRISPR/Cas9 to correct the mutation in a footprint-free manner. Two sgRNAs were designed to target the genome adjacent to the R778L site (Fig. 1A), and the single-strand annealing assay was performed to assess CRISPR/Cas9 cutting efficiency (Fig. S1A). sgRNA-2 was used in subsequent experiments since it showed a relatively higher cutting activity. To avoid second cutting after the homologous recombination, a C>G silent mutation was introduced into the PAM domain of the ssODNs (Fig. 1A). This silent mutation also removed the HaeIII cutting site, useful for subsequent screening of clones with homologous recombination, by RFLP assay. Target clones with homologous recombination were expected to have an ∼300 bp band resistant to HaeIII cutting.
Fig. 1.
Correction of ATP7B R778L in ATP7BR778L/R778L iPSCs and generation of iHeps from iPSCs.
(A) Design of sgRNAs and ssODNs for correction of ATP7B R778L variant (G>T). Arrow heads indicate the cutting sites of sgRNA-1 (blue) or sgRNA-2 (green). Pink shaded areas indicate the R778L variant (in genome) or the corrected sequence (in ssODN). “G” highlighted in green in ssODN indicates the C>G silent mutation site. The pink rectangle in ssODNs indicates the exon region of ATP7B. (B) Sanger sequencing confirmed the correction of R778L mutant (T>G) in ATP7BR778L/R778L iPSCs. (C) Immunofluorescence staining showed that iHeps highly express hepatic specific markers (ALB, AAT, HNF4A and ASGR1). Scale bar represents 50 μm. (D) ELISA result showed the secreted hALB level in the supernatant of indicated iHeps at day 17 to 18 of differentiation (n = 7). Error bars indicate SEM. hALB, human albumin; iHep(s), iPSC-derived hepatocyte(s); iPSC, induced pluripotent stem cell; sgRNA, single guide RNA; ssODN, single-stranded oligodeoxynucleotide;
We have a well-established workflow for ssODN-mediated genome editing (Fig. S1B). First, ssODNs and plasmids encoding sgRNA-2 and Cas9 were co-transfected into ATP7BR778L/R778L iPSCs by electroporation, then target cells were enriched by puromycin treatment. Using an RFLP assay, a small proportion of cells with homologous recombination of ssODNs was observed (Fig. S1C). To further screen out target cells, single cell-derived colonies were isolated and screened for using an RFLP assay. Positive colonies were maintained for subsequent Sanger sequencing (Fig. S1D). A targeted corrected clone (ATP7BWT/- iPSC) with heterozygous knock-in of ssODNs – 1 corrected allele and the other non-corrected allele with indels (Fig. 1B and Fig. S1E) – was selected for further analysis. Given that WD carriers with 1 allele of wild-type ATP7B remain healthy, achieving a heterozygous correction should be sufficient to restore ATP7B functions.
We observed that pluripotency markers (OCT4, NANOG, SSEA-4 and TRA-1-60) were highly expressed in ATP7BWT/- iPSCs as well as the parent ATP7BR778L/R778L iPSCs (Fig. S2A), indicating that ATP7BWT/- iPSCs maintained their pluripotent characteristics after genome editing. In addition, karyotype analysis revealed that ATP7BWT/- iPSCs maintained a normal 46, XY karyotype, the same as their parent line (Fig. S2B). To further confirm their differentiation capacity, embryoid body (EB)-mediated in vitro differentiation was performed. Both ATP7BR778L/R778L and ATP7BWT/- iPSCs formed EBs during suspension culturing in EB medium (Fig. S2C) and underwent spontaneous differentiation into cells positive for alpha-fetoprotein (AFP: endoderm marker), smooth muscle actin (mesoderm marker) and microtubule-associated protein 2 (ectoderm marker) (Fig. S2D). Therefore, ATP7BWT/- iPSCs, as well as the parent ATP7BR778L/R778L iPSCs, maintained the potential for differentiation into all 3 germ layers.
Potential off-target effects of CRISPR/Cas9 are a safety concern. To determine the possible off-target events introduced by CRISPR/Cas9 cleavage in the genome of engineered iPSCs, the top 10 potential off-target loci selected by the sgRNA Design server15 and the COSMID server19 were analyzed. Off-target events were evaluated by Sanger sequencing of the selected regions amplified by high-fidelity PCR that monitors potential small indels generated from non-homologous end joining of CRISPR/Cas9 cleavages. Our results showed that none of the candidate regions had detectable indels from non-homologous end joining in ATP7BWT/- iPSCs (Table S1).
In conclusion, genome editing using CRISPR/Cas9 and ssODNs neither altered the pluripotent properties nor gave rise to any obvious off-target events in our modified iPSCs.
In vitro differentiation of hepatocytes from pluripotent stem cells
We previously established a 3-step hepatic differentiation protocol based on several former studies.14,20 iPSCs were induced to definitive endoderm cells (day 3), hepatoblasts (day 10) and hepatocytes (day 17) in a stepwise manner (Fig. S3A). After 17 days of differentiation, iHeps from ATP7BWT/WT iPSCs, ATP7BR778L/R778L iPSCs and ATP7BWT/- iPSCs highly expressed hepatocyte markers including albumin (ALB), AAT (alpha 1 antitrypsin), HNF4A (hepatocyte nuclear 4 alpha), and ASGR1 (asialoglycoprotein receptor 1) (Fig. 1C). These iHeps could synthesize and secrete substantial human ALB into the supernatant (Fig. 1D), and also possessed other essential hepatic functions such as glycogen and lipid storage (Fig. S3B). To conclude, iHeps derived from ATP7BWT/WT iPSCs, ATP7BR778L/R778L iPSCs and ATP7BWT/- iPSCs had essential features of functional hepatocytes.
Correction of ATP7B R778L restores ATP7B subcellular localization and its trafficking in response to copper overload
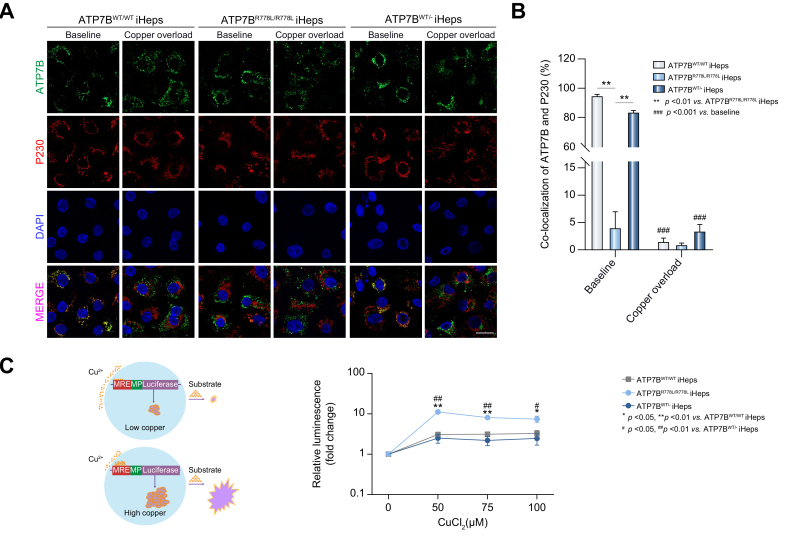
Next, differentiated iHeps were used as the cell model to determine whether correction of the genotype could rescue the disease phenotype. At relatively low copper levels, ATP7B protein resides on the membrane of the trans-Golgi network (TGN), where it delivers copper to copper-dependent proteins such as ceruloplasmin.2 We used immunofluorescence analysis to reveal the relative intracellular location of ATP7B protein in the TGN. Under normal culture conditions (baseline), the distribution of ATP7B protein was highly co-localized with P230 (a marker of the TGN membrane) in ATP7BWT/WT iHeps (Fig. 2A & B). In ATP7BR778L/R778L iHeps, ATP7B and P230 were extensively mis-co-localized, indicating that most of the ATP7B mutant protein has failed to reach the TGN – its functional site (Fig. 2A & B). Notably, in the ATP7BWT/- iHeps, ATP7B and P230 were again highly co-localized, indicating that the subcellular localization of ATP7B was restored after gene correction (Fig. 2A & B). In response to intracellular copper elevation, ATP7B traffics from the TGN membrane towards the post-Golgi compartments where it sequesters excess copper and promotes copper excretion.21 ATP7B trafficking is essential for intercellular copper regulation.21 To further explore the trafficking of ATP7B protein in response to copper overload, iHeps were exposed to CuCl2 for 2 hours. As expected, in both ATP7BWT/WT iHeps and ATP7BWT/- iHeps, the ATP7B protein travelled away from the TGN (Fig. 2A) and the co-localization rate significantly decreased relative to normal culture conditions (Fig. 2B).
Fig. 2.
ATP7BWT/R778L gene-corrected iHeps display normal ATP7B subcellular localization and function.
(A) Representative confocal images show different co-localization patterns of ATP7B and P230 in the indicated iHeps in normal culture conditions (baseline) or after 200 μM CuCl2 treatment for 2 hours (copper overload). Scale bar represents 20 μm. (B) Percentage of ATP7B and P230 co-localization in the indicated iHeps with/without copper treatment (n = 5). Error bars indicate SEM; ∗∗∗p <0.001, p value was obtained using two-way ANOVA. (C) Left panel: Schematic view of the copper-responsive element luciferase reporter assay. Right panel: Line chart shows luminescence induction fold-change of the indicated iHeps after 0 μM, 50 μM, 75 μM and 100 μM CuCl2 treatment for 24 hours (n = 4). Error bars indicate SEM, p values are indicated on the figure and were obtained using two-way ANOVA. iHep(s), iPSC-derived hepatocyte(s); iPSC, induced pluripotent stem cell; MP, minimal promoter. MRE, metal-responsive element; WT, wild-type.
Our results thus demonstrate that correction of the ATP7B R778L mutation, even in a heterozygous manner, restores ATP7B protein localization on the TGN and ATP7B trafficking.
ATP7B correction rescues copper exportation capability in WD iHeps
To further determine whether the copper exportation function was recovered following gene correction, an MRE-driven luciferase reporter assay was performed (Fig. 2C left panel). iHeps transfected with MRE-driven luciferase were treated with different doses of CuCl2 for 24 hours, and the relative intracellular copper level was measured by reading the induced luminescence signals. Interestingly, ATP7BR778L/R778L iHeps showed significantly increased luminescence signals after copper treatment in comparison with ATP7BWT/WT iHeps or ATP7BWT/R778L iHeps (Fig. 2C right panel), indicating a much higher intracellular copper level in ATP7BR778L/R778L iHeps than in ATP7BWT/WT and ATP7BWT/- iHeps. These results confirmed that ATP7BR778L/R778L iHeps were less capable of exporting excess copper from the cell, and that the copper exporting deficiency could be rescued after gene correction in ATP7BWT/- iHeps.
Transplantation of corrected iHeps alleviated liver injury and liver fibrosis in WD mice
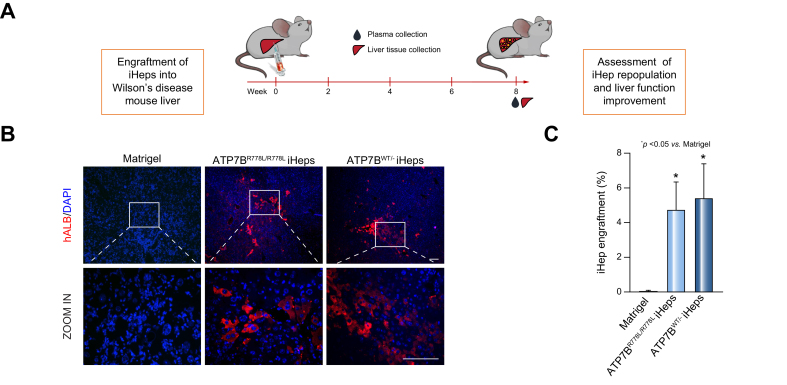
To determine whether gene-corrected iHeps possess the capacity to rescue the disease phenotype in vivo, iHeps were transplanted into the liver of immunocompromised WD mice (Atp7b-/- / Rag2-/- / Il2rg-/-, ARG) via intra-splenic injection. Intra-splenic injection of Matrigel was also performed as a sham-operation group. At 8 weeks post engraftment, animals were sacrificed, then plasma and tissue samples were collected for further assessments (Fig. 3A). The engrafted iHeps were visualized by immunofluorescence staining for human specific marker – human ALB. As shown in Fig. 3B and Fig. S6A, positive cells were detected in the liver of mice transplanted with ATP7BR778L/R778L and ATP7BWT/- iHeps, indicating that iHeps could integrate into the mouse liver and survive in vivo for at least 8 weeks. The engraftment rates of ATP7BR778L/R778L iHeps and ATP7BWT/- iHeps were similar at around 5% (Fig. 3C).
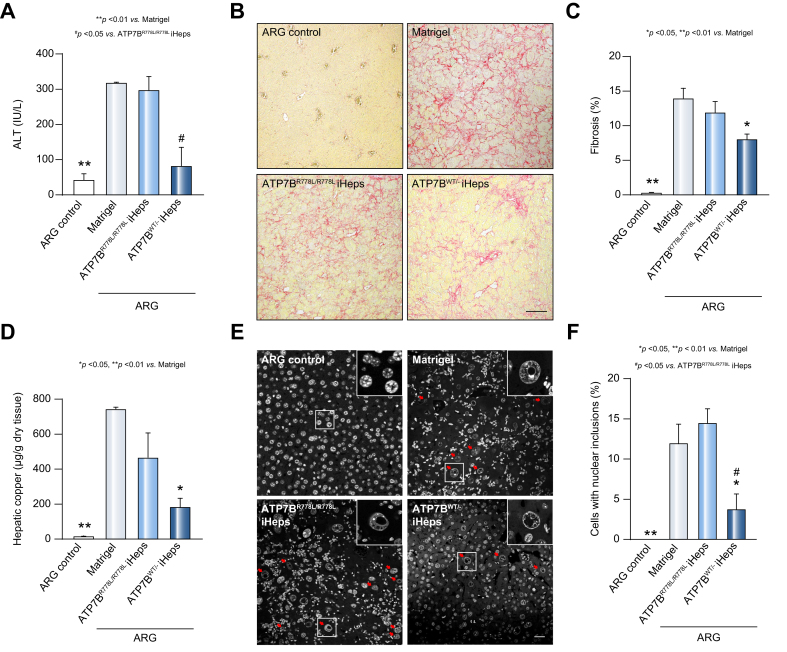
Fig. 3.
Transplantation of iHeps into Atp7b-/-/ Rag2-/-/ Il2rg-/- Wilson’s disease mouse liver.
(A) Schematic of the engraftment of iHeps into the liver of ARG mice (Atp7b-/-/ Rag2-/-/ Il2rg-/-). (B) Immunofluorescence staining for hALB shows iHeps incorporated into Wilson’s disease mouse livers at 8 weeks post engraftment. Rectangles indicate the zoom-in areas. Scale bar represents 100 μm. (C) Percentage of human albumin positive cells in mouse livers transplanted with the indicated iHeps, indicating similar engraftment efficiencies between the ATP7BR778L/R778L and ATP7BWT/R778L iHeps group. Error bars indicate SEM (n = 6 in Matrigel group, n = 5 in ATP7BR778L/R778L iHep group, n = 4 in ATP7BWT/R778L iHep group, and n represents animal number; 2 liver sections and 10 random fields (200x magnification) from each section were calculated for each animal). P values were obtained using one-way ANOVA adjusted with Dunnett’s multiple comparison; error bars indicate SEM. hALB, human albumin; iHep(s), iPSC-derived hepatocyte(s); iPSC, induced pluripotent stem cell; WT, wild-type.
Next, we evaluated the therapeutic effect of engrafted iHeps in ARG mice. The Atp7b+/- / Rag2-/- / Il2rg-/- mice were used as healthy controls (ARG control), as they were WD-free (Atp7b+/-) and had an identical genetic background to ARG mice. Plasma alanine transaminase level was first measured to generally assess the severity of liver injury and revealed a significant reduction in the ATP7BWT/- iHeps group compared with the sham-operation group (Matrigel) and ATP7BR778L/R778L iHeps group, indicating that liver injury was alleviated after ATP7BWT/- iHeps transplantation (Fig. 4A). Apart from ARG control mice, all ARG WD mice had large areas of liver fibrosis, as determined by Picrosirius red staining (Fig. 4B & Fig. S6B). Although there was no significant difference between the ATP7BR778L/R778L iHeps and sham-operation group, the ATP7BWT/- iHeps group had significantly reduced liver fibrosis (Fig. 4C). Consistent with the severity of liver injury, mice in the Matrigel and ATP7BR778L/R778L iHep groups showed significantly increased macrophage infiltration compared with ARG control mice (Fig. S7A & B). Although we could not ignore the possibility that these macrophages were contributing to copper clearance, considering that macrophages were present in the 3 experimental groups and less enriched in the less affected transplanted group, we could conclude that the major effects came from the transplanted gene-corrected iHeps.
Fig. 4.
Transplantation of ATP7BWT/R778L iHeps attenuates liver injury and reduces hepatic copper content in Wilson’s disease mice.
(A) Plasma ALT level in ARG control mice (Atp7b+/-/ Rag2-/-/ Il2rg-/-) or ARG mice (Atp7b-/-/ Rag2-/-/ Il2rg-/-) engrafted with the indicated iHeps at 8 weeks post transplantation. (n = 3 in ARG control mice group or ARG mice transplanted with ATP7BWT/R778L iHeps group, n = 5 in ARG mice transplanted with Matrigel or ATP7BR778L/R778L iHeps groups; n represents animal number). (B) Representative liver sections with Picrosirius red staining show the status of liver fibrosis in each group of animals. Scale bar represents 100 μm. (C) Percentage of fibrotic areas calculated according to Picrosirius red staining. (n = 3 in ARG control mice group or ARG mice transplanted with ATP7BWT/R778L iHeps group, n = 5 in ARG mice transplanted with Matrigel or ATP7BR778L/R778L iHeps groups, and n represents animal number; 5 different liver sections and 3 random fields (100x magnification) per section were calculated for each animal). (D) Hepatic copper content of the indicated mouse livers, measured by inductively coupled plasma mass spectrometry. (n = 3 in ARG mice transplanted with Matrigel or ATP7BWT/R778L iHeps groups, n = 5 in ARG control mice or ARG mice transplanted with ATP7BWT/R778L iHep group, and n represents animal number). (E) Representative images of nuclei staining with DAPI show hepatocyte nucleic structures of liver sections in the indicated mouse group, and cells with nuclear inclusions are indicated with red arrows. Scale bar represents 25 μm. (F) Percentage of cells with nuclear inclusions in each mouse group calculated according to DAPI staining. (n = 3 in ARG control mice group or ARG mice transplanted with ATP7BWT/R778L iHeps group, n = 5 in ARG mice transplanted with Matrigel or ATP7BR778L/R778L iHeps groups, and n represents animal number; 2 different sections and 10 random fields (200x magnification) from each section were calculated for each animal). P values were obtained using one-way ANOVA adjusted with Dunnett’s multiple comparison; error bars indicate SEM. ALT, alanine aminotransferase; iHep(s), iPSC-derived hepatocyte(s); iPSC, induced pluripotent stem cell; WT, wild-type.
Transplantation of gene-corrected iHeps reduced hepatic copper accumulation and copper-induced hepatotoxicity
Since excessive hepatic copper accumulation is the primary cause of WD,2 it is important to determine whether engraftment of iHeps to restore ATP7B function can remove excessive copper from the liver. Rhodanine staining for copper in liver sections revealed that a reduced number of cells with copper deposition was observed in mice engrafted with ATP7BWT/- iHeps compared with the sham-operation and WD groups (Fig. S5A,B). To further quantify the hepatic copper content, a whole right lobe from each mouse liver was collected and copper content measured by ICP-MS. Mice engrafted with ATP7BWT/- iHeps showed significantly reduced liver copper content compared with the sham-operation group (Fig. 4D). Mice engrafted with WD iHeps also showed a decreasing trend but were not significantly different to the sham-operation group. These data suggest that transplantation of gene-corrected iHeps facilitated copper removal and reduced hepatic copper accumulation. Transplantation of WD iHeps also tended to decrease hepatic copper levels, possibly due to the residual function of the R778L mutant protein.
Nuclear inclusion is considered one of the most frequent ultrastructural changes in hepatocytes of patients with WD,2,22 and the formation of nuclear inclusions is correlated with copper-induced oxidative stress that causes hepatocyte toxicity.23 As shown in Fig. 4E, no obvious nuclear inclusions were found in any liver section from the ARG control mice, indicating that nuclear inclusions are not normally generated in the healthy liver. As expected, many nuclear inclusions were observed in ARG mice following a sham-operation or ATP7BR778L/R778L iHep transplantation. On the contrary, ARG mice transplanted with ATP7BWT/- iHeps had significantly fewer nuclear inclusions (Fig. 4F). This indicates that transplantation of ATP7BWT/- iHeps decreased copper-induced hepatocyte toxicity in the ARG mouse liver.
Taken together, gene-corrected ATP7BWT/- iHeps restored ATP7B functions In vitro and in vivo. Transplantation of ATP7B competent ATP7BWT/- iHeps decreased copper accumulation in WD mouse livers and alleviated copper-induced hepatocyte toxicity and liver fibrosis.
Discussion
In this study, we corrected the ATP7B R778L mutant in WD iPSCs with CRISPR/Cas9 and ssODNs in a footprint-free manner, and hence restored the subcellular location of ATP7B and its function in copper regulation. To the best of our knowledge, this is the first report of a corrected endogenous ATP7B mutation in iPSCs derived from patients with WD. We further demonstrated that transplantation of gene-corrected iHeps alleviated liver injury and hepatic copper accumulation in a mouse model of WD disease. In this respect, our footprint-free gene-corrected iHeps hold great potential for the development of autologous cell therapies for WD as well as other inherited liver diseases.
Previous studies have shown that overexpression of R778L mutant protein in several human cell lines[24], [25], [26] leads to misfolding of the ATP7B protein, and consequent failure to localize to its functional site – the Golgi apparatus. In line with previous findings, our results demonstrated that ATP7BR778L/R778L iHeps, with the same genetic background as the patient with WD, had extensively mis-colocalized ATP7B protein and a significantly decreased ability to export copper (Fig. 2). Our results demonstrated that patient-specific iHeps recapitulated the disease phenotype and could serve as a reliable cell model for studying disease mechanisms and testing novel therapies for WD.
The CRISPR/Cas9 system is a powerful technique for genome editing. In combination with homologous recombination templates, patient-specific iPSC lines can be genetically modified.[27], [28], [29], [30] ssODN is one of the widely used homologous recombination templates and has been shown to genetically modify the target gene efficiently in a scar-free manner. In our study, we corrected the ATP7B R778L mutant with CRISPR/Cas9 and ssODNs. Since there were no selection markers employed in ssODNs, the targeting rate was relatively low (around 1%), while the benefits of not using selective markers are significant. There is no need to remove the selection markers, saving time and effort in subsequent experiments. More importantly, it is a “footprint-free” genetic modification as it avoids introducing foreign DNA fragments into the host genome. Given that integration of foreign DNA sequences has the risk of disturbing some functional elements and adversely affecting the phenotype of the cells, the “footprint-free” gene editing approach makes the gene-corrected cells potentially safer for clinical use.
After gene correction with CRISPR/Cas9 and ssODNs, the subcellular location and copper regulatory function of the ATP7B protein were restored in the gene-corrected ATP7BWT/- iHeps. Since ATP7BWT/- iPSCs were a heterozygous line knocked-in with ssODNs, this indicates that correction of one allele of ATP7B was sufficient to restore the protein function to a wild-type level. Since obtaining a heterozygous knock-in iPSC line is easier than that of homozygous knock-in, our study shows evidence of an efficient approach to functionally recover ATP7B by correcting only one allele of ATP7B and provides insight for future gene and cell therapies.
In line with our in vitro findings, our animal experiments showed that transplantation of ATP7BWT/WT iHeps and gene-corrected ATP7BWT/- iHeps alleviated hepatic fibrosis and reduced hepatic copper accumulation in mouse models of WD, showing that these iPSC-derived iHeps offer promising therapeutic potential for cell replacement therapy in WD. The iHep engraftment efficiency, ∼5% in our WD mouse model, is not high, but comparable with previous studies from other groups14,31,32 (ranging from 2%–17%). In the setting of WD, other studies have also shown that disease correction can be achieved even with modest numbers of healthy transplanted wild-type hepatocytes.33 The reason for this is that a limited number of functional hepatocytes is sufficient to clear excess copper from blood, which reinforces the potential therapeutic interest of transplanted gene-corrected iPSC-derived iHeps for treating WD. Hence, although the overall engraftment efficiency was not high, the small proportion of engrafted cells helped slow the progression of liver pathology induced by progressive copper accumulation.
Increasing the in vivo repopulation efficiency is a challenge for stem cell-derived hepatocytes. The iHep engraftment efficiency, in this study as well as many other similar studies,14,32,34,35 was much lower than that achieved by engrafting primary human hepatocytes.36,37 The low engraftment rate of iHeps is likely attributed to their relative immaturity. The 3-stage hepatic differentiation protocol14 employed in this study is based on a classic method used in several studies,[38], [39], [40] but these iHeps derived from iPSCs were relatively immature, as indicated by continuous expression of fetal liver markers and low expression of some adult liver markers.39,41 Similarly, we observed a high expression of AFP, a fetal liver marker, in all groups of iHeps after 17 days of differentiation (Fig. S4), indicating that the iHeps we obtained remained the property of fetal livers and were not functionally competent. Nonetheless our results showed that a small number of transplanted cells was still beneficial in alleviating the disordered liver copper metabolism. This implies that in the future we might be able to cure WD if a relatively high repopulation efficiency of functional iHeps can be achieved.
Other than hepatic manifestations, neurological impairment is also commonly seen in WD disease pathogenesis. In this study, we focused only on hepatic presentation because the neurological impairments are more difficult to study due to the rarity of neurological symptoms in WD animal models.[42], [43], [44] Interestingly, it has been reported that transplantation of liver cells in WD mouse livers led to reduced copper levels in the liver and extrahepatic tissues, but not in the brain.43 This implies that liver cell transplantation may not be capable of clearing copper inside the brain, or that the transplanted cells require more time to clear accumulated copper before a reversal in brain copper levels is evident. Therefore, for patients with mainly neurological impairment, the therapeutic effects of iHep therapy may be limited.
In conclusion, our study provides proof-of-principle that correction of the genotype of WD iHeps can reverse the localization and function of the ATP7B protein, and these gene-corrected iHeps showed promising therapeutic effects in a mouse model of WD. This technique offers a potential unlimited cell source for autologous cell replacement therapy for WD as well as other monogenic hepatic disorders.
Financial support
This work was supported by the Shenzhen-Hong Kong Technology Cooperation Funding Scheme [GHP/130/18/SZ (Hong Kong); SGLH20180627143202102 (Shenzhen)], the Guangdong-Hong Kong Technology Cooperation Funding Scheme [GHP/046/17GD (Hong Kong); 2017B050506007 (Guangdong)], the National Natural Science Foundation of China (81873521) and the Innovative Team Program of the Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (2018GZR110103001 to M.A.E.).
Authors’ contributions
HF.T, M.A.E and J.Y designed the study. R.W and J.Y designed and performed most of the experiments. CW.C and KM.N assisted in designing and performing the experiments. CW.C, WI.H, N.L, and Y.H assisted in most parts of the in vivo experiments. LY.W bred the mice and assisted in most of the surgical operations. X.H, J.F, B.Y, Y.L, L.J assisted in the gene correction. WH.L, KW.A, WL,T, YL.T and KM.N supplied experimental materials and resources. R.W, J.Y, HF.T, M.A.E analyzed the data and wrote the manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author, [HF.T and M.A.E], upon reasonable request.
Conflict of interest
The authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100389.
Contributor Information
Miguel A. Esteban, Email: miguel@gibh.ac.cn.
Hung-Fat Tse, Email: hftse@hku.hk.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Huster D. Wilson disease. Best Pract Res Clin Gastroenterol. 2010;24:531–539. doi: 10.1016/j.bpg.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Ala A., Walker A.P., Ashkan K., Dooley J.S., Schilsky M.L. Wilson's disease. Lancet. 2007;369:397–408. doi: 10.1016/S0140-6736(07)60196-2. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes J., Saudubray J.-M., Van den Berghe G., Walter J.H. Springer Science & Business Media; 2006. Inborn metabolic diseases: diagnosis and treatment. [Google Scholar]
- 4.Schilsky M.L. Liver transplantation for Wilson's disease. Ann N Y Acad Sci. 2014;1315:45–49. doi: 10.1111/nyas.12454. [DOI] [PubMed] [Google Scholar]
- 5.Filippi C., Dhawan A. Current status of human hepatocyte transplantation and its potential for Wilson's disease. Ann N Y Acad Sci. 2014;1315:50–55. doi: 10.1111/nyas.12386. [DOI] [PubMed] [Google Scholar]
- 6.Sauer V., Siaj R., Stoppeler S., Bahde R., Spiegel H.U., Kohler G., et al. Repeated transplantation of hepatocytes prevents fulminant hepatitis in a rat model of Wilson's disease. Liver Transpl. 2012;18:248–259. doi: 10.1002/lt.22466. [DOI] [PubMed] [Google Scholar]
- 7.Irani A.N., Malhi H., Slehria S., Gorla G.R., Volenberg I., Schilsky M.L., et al. Correction of liver disease following transplantation of normal rat hepatocytes into Long-Evans Cinnamon rats modeling Wilson's disease. Mol Ther. 2001;3:302–309. doi: 10.1006/mthe.2001.0271. [DOI] [PubMed] [Google Scholar]
- 8.Soltys K.A., Setoyama K., Tafaleng E.N., Soto Gutierrez A., Fong J., Fukumitsu K., et al. Host conditioning and rejection monitoring in hepatocyte transplantation in humans. J Hepatol. 2017;66:987–1000. doi: 10.1016/j.jhep.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorns C., Nowak G., Nemeth A., Zemack H., Mork L.M., Johansson H., et al. De novo donor-specific HLA antibody formation in two patients with Crigler-najjar syndrome type I following human hepatocyte transplantation with partial hepatectomy preconditioning. Am J Transpl. 2016;16:1021–1030. doi: 10.1111/ajt.13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox I.J., Chowdhury J.R., Kaufman S.S., Goertzen T.C., Chowdhury N.R., Warkentin P.I., et al. Treatment of the Crigler-Najjar syndrome type I with hepatocyte transplantation. N Engl J Med. 1998;338:1422–1426. doi: 10.1056/NEJM199805143382004. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S., Chen S., Li W., Guo X., Zhao P., Xu J., et al. Rescue of ATP7B function in hepatocyte-like cells from Wilson's disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20:3176–3187. doi: 10.1093/hmg/ddr223. [DOI] [PubMed] [Google Scholar]
- 14.Yang J., Wang Y., Zhou T., Wong L.Y., Tian X.Y., Hong X., et al. Generation of human liver chimeric mice with hepatocytes from familial hypercholesterolemia induced pluripotent stem cells. Stem Cell Rep. 2017;8:605–618. doi: 10.1016/j.stemcr.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F., Pruett-Miller S.M., Huang Y., Gjoka M., Duda K., Taunton J., et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Wong L.Y., Tian X.Y., Wei R., Lai W.H., Au K.W., et al. A familial hypercholesterolemia human liver chimeric mouse model using induced pluripotent stem cell-derived hepatocytes. J Vis Exp. 2018 doi: 10.3791/57556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease–based gene targeting and the piggyBac transposon. Nat Protoc. 2013;8:2061. doi: 10.1038/nprot.2013.126. [DOI] [PubMed] [Google Scholar]
- 19.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polishchuk R.S., Polishchuk E.V. From and to the Golgi - defining the Wilson disease protein road map. FEBS Lett. 2019;593:2341–2350. doi: 10.1002/1873-3468.13575. [DOI] [PubMed] [Google Scholar]
- 22.Gerosa C., Fanni D., Congiu T., Piras M., Cau F., Moi M., et al. Liver pathology in Wilson's disease: from copper overload to cirrhosis. J Inorg Biochem. 2019;193:106–111. doi: 10.1016/j.jinorgbio.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Thakur P., Lamoke F., Chaffin J.M., Bartoli M., Lee J.R., Duncan M.B. Dysplastic hepatocytes develop nuclear inclusions in a mouse model of viral hepatitis. PloS one. 2014;9 doi: 10.1371/journal.pone.0099872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Berghe P.V., Stapelbroek J.M., Krieger E., de Bie P., van de Graaf S.F., de Groot R.E., et al. Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin. Hepatology. 2009;50:1783–1795. doi: 10.1002/hep.23209. [DOI] [PubMed] [Google Scholar]
- 25.Zhu M., Dong Y., Ni W., Wu Z.Y. Defective roles of ATP7B missense mutations in cellular copper tolerance and copper excretion. Mol Cell Neurosci. 2015;67:31–36. doi: 10.1016/j.mcn.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Forbes J.R., Cox D.W. Copper-dependent trafficking of Wilson disease mutant ATP7B proteins. Hum Mol Genet. 2000;9:1927–1935. doi: 10.1093/hmg/9.13.1927. [DOI] [PubMed] [Google Scholar]
- 27.Xu X., Tay Y., Sim B., Yoon S.-I., Huang Y., Ooi J., et al. Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in Huntington disease patient-derived induced pluripotent stem cells. Stem Cell Rep. 2017;8:619–633. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 29.Burnight E.R., Gupta M., Wiley L.A., Anfinson K.R., Tran A., Triboulet R., et al. Using CRISPR-Cas9 to generate gene-corrected autologous iPSCs for the treatment of inherited retinal degeneration. Mol Ther. 2017;25:1999–2013. doi: 10.1016/j.ymthe.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei R., Yang J., Tse H.-F. Seamless gene correction of LDLR in familial hypercholesterolemia induced pluripotent stem cells mediated by CRISPR/Cas9 and PiggyBac. Am Heart Assoc. 2017 [Google Scholar]
- 31.Liu H., Kim Y., Sharkis S., Marchionni L., Jang Y.-Y. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Trans Med. 2011;3 doi: 10.1126/scitranslmed.3002376. 82ra39-82ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y., Li Y., Wang X., Zhang W., Sauer V., Chang C.-J., et al. Amelioration of hyperbilirubinemia in gunn rats after transplantation of human induced pluripotent stem cell-derived hepatocytes. Stem Cell Rep. 2015;5:22–30. doi: 10.1016/j.stemcr.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S. Cell therapy to remove excess copper in Wilson's disease. Ann N Y Acad Sci. 2014;1315:70–80. doi: 10.1111/nyas.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Sun D., Liang Z., Wang J., Zhong X., Lyu Y., et al. Generation of human hepatocytes from extended pluripotent stem cells. Cell Res. 2020;30:810–813. doi: 10.1038/s41422-020-0293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H., Kim Y., Sharkis S., Marchionni L., Jang Y.Y. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Trans Med. 2011;3:82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azuma H., Paulk N., Ranade A., Dorrell C., Al-Dhalimy M., Ellis E., et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bissig K.D., Le T.T., Woods N.B., Verma I.M. Repopulation of adult and neonatal mice with human hepatocytes: a chimeric animal model. Proc Natl Acad Sci USA. 2007;104:20507–20511. doi: 10.1073/pnas.0710528105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hay D.C., Zhao D., Fletcher J., Hewitt Z.A., McLean D., Urruticoechea-Uriguen A., et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem cells. 2008;26:894–902. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- 39.Rashid S.T., Corbineau S., Hannan N., Marciniak S.J., Miranda E., Alexander G., et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajiwara Correction for Kajiwara et al., Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci. 2012;109 doi: 10.1073/pnas.1209979109. 14716-14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baxter M., Withey S., Harrison S., Segeritz C.P., Zhang F., Atkinson-Dell R., et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J Hepatol. 2015;62:581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutsenko S. Atp7b−/− mice as a model for studies of Wilson's disease. Biochem Soc Trans. 2008;36:1233–1238. doi: 10.1042/BST0361233. [DOI] [PubMed] [Google Scholar]
- 43.Allen K.J., Cheah D.M., Wright P.F., Gazeas S., Pettigrew-Buck N.E., Deal Y.H., et al. Liver cell transplantation leads to repopulation and functional correction in a mouse model of Wilson's disease. J Gastroenterol Hepatol. 2004;19:1283–1290. doi: 10.1111/j.1440-1746.2004.03451.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee B.H., Kim J.H., Kim J.-M., Heo S.H., Kang M., Kim G.-H., et al. The early molecular processes underlying the neurological manifestations of an animal model of Wilson's disease†. Metallomics. 2013;5:532–540. doi: 10.1039/c3mt20243g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, [HF.T and M.A.E], upon reasonable request.