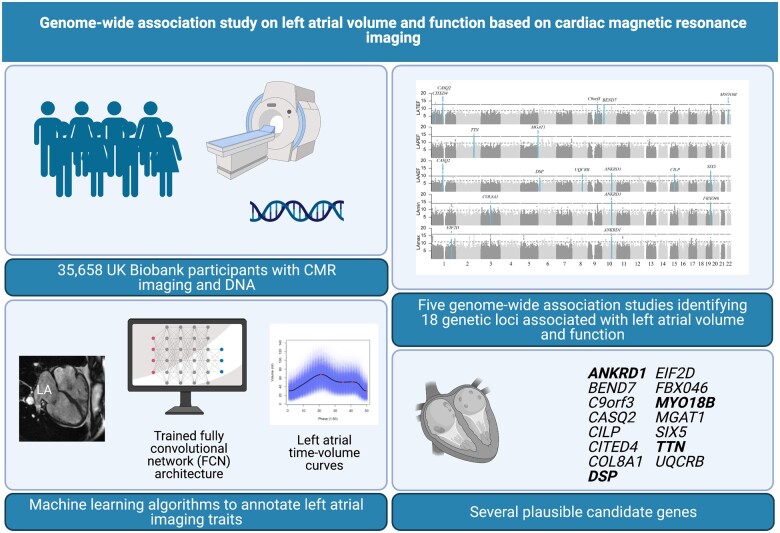
Abstract
Aims
Left atrial (LA) volume and function impose significant impact on cardiovascular pathogenesis if compromised. We aimed at investigating the genetic architecture of LA volume and function using cardiac magnetic resonance imaging data.
Methods and results
We used the UK Biobank, which is a large prospective population study with available phenotypic and genetic data. On a subset of 35 658 European individuals, we performed genome-wide association studies on five volumetric and functional LA variables, generated using a machine learning algorithm. In total, we identified 18 novel genetic loci, mapped to genes with known roles in cardiomyopathy (e.g. MYO18B, TTN, DSP, ANKRD1) and arrhythmia (e.g. TTN, CASQ2, MYO18B, C9orf3). We observed high genetic correlation between LA volume and function and stroke, which was most pronounced for LA passive emptying fraction (rg = 0.40, P = 4 × 10−6). To investigate whether the genetic risk of atrial fibrillation (AF) is associated with LA traits that precede overt AF, we produced a polygenetic risk score for AF. We found that polygenetic risk for AF is associated with increased LA volume and decreased LA function in participants without AF [LAmax 0.25 (mL/m2)/standard deviation (SD), 95% confidence interval (CI) (0.15; 0.36), P = 5.13 × 10−6; LAmin 0.21 (mL/m2)/SD, 95% CI (0.15; 0.28), P = 1.86 × 10−10; LA active emptying fraction −0.35%/SD, 95% CI (−0.43; −0.26), P = 3.14 × 10−14].
Conclusion
We report on 18 genetic loci associated with LA volume and function and show evidence for several plausible candidate genes important for LA structure.
Keywords: Cardiac magnetic resonance, UK Biobank, GWAS, Left atrium
Graphical Abstract
See page 4533 for the editorial comment for this article ‘Genetics, atrial cardiomyopathy, and stroke: enough components for a sufficient cause?’, by S. Camen and R.B. Schnabel,https://doi.org/10.1093/eurheartj/ehab523.
Translational perspective
Atrial cardiomyopathy has been suggested to play an important role in the development of atrial fibrillation (AF). Using machine-learning algorithms, we segmented and annotated the left atrium (LA) in cardiac magnetic resonance data from 35 658 individuals. We identified 18 novel genetic loci associated with LA volume and function, many near well-established cardiomyopathy and arrhythmia genes. We observed high genetic correlation between LA volume and function and stroke. We found that the genetic risk of AF influences LA structure prior to diagnosis of AF, suggesting an intrinsic relationship.
Introduction
The left atrium (LA) serves primarily as a reservoir chamber of blood entering the heart from the pulmonary veins and is important for enhancing ventricular filling. Changes in left atrial (LA) volume and function have been associated with impaired cardiac performance,1 arrhythmogenesis,2 risk of stroke,3 and mortality.4 Left atrial assessment is part of the standard echocardiographic examination5 and changes in the LA are traditionally considered secondary to other cardiovascular diseases. However, the pathology behind changes in LA volume and function and the association with atrial fibrillation (AF), heart failure (HF), and risk of stroke is poorly characterized.6 , 7 , 8 An increasing interest in LA pathology and atrial tissue prompted the need for defining atrial cardiomyopathy.9 In a recent expert consensus document, written by internationally recognized experts, atrial cardiomyopathy was defined as structural, architectural, contractile, or electrophysiological changes affecting the atria with the potential to produce clinically relevant manifestations.6
Cardiac magnetic resonance (CMR) imaging is at present the gold standard for assessing cardiac volumes, with a superior accuracy and precision compared with other imaging modalities such as echocardiography.10 The clinical use of CMR is currently expanding due to lowered costs and increased accessibility, but the widespread use of the CMR modality is still limited.
Genetics play a major role in ventricular cardiomyopathies and could also play a role in several of the pathophysiological mechanisms behind atrial cardiomyopathy,6 but large-scale genetic studies of LA volume and function are lacking. Prior genome-wide association studies (GWAS) on cardiac chamber volumes and function have been limited to echocardiographic measurements11 or CMR imaging-derived left ventricular (LV) phenotypes.12
UK Biobank (UKBB) provides extensive phenotypic and genotypic data on more than ∼500 000 individuals from the British general population.13 At the time of this study, a subset of UKBB participants had CMR imaging data available, which enabled the coupling of genetic data with these state-of-the-art imaging variables. Using a validated machine learning algorithm to annotate the LA, we performed GWAS on five LA volume and function traits.
Methods
UK Biobank
UK Biobank is a large, population-based, prospective cohort study with ∼500 000 participants aged 40–69 years at the time of recruitment (2006–10). The biobank holds a variety of health-related information on participants, including health records, imaging data, lifestyle indicators, cognitive function, biomarkers from blood and urine, imputed genome-wide genotypes, information on population structure, relatedness, and genotype-level quality. A detailed description of data collection and study protocol has been provided previously by Bycroft et al.13
All participants gave written informed consent and the study complies with the Declaration of Helsinki.
Derivation of cardiac magnetic resonance measurements
The UKBB CMR protocol has previously been described in detail.14 In brief, CMR scans were performed using a 1.5 Tesla scanner (MAGNETOM Aera, Syngo Platform VD13A, Siemens Healthcare, Erlangen, Germany) using an 18-channel body coil. Long-axis cines and a complete short axis stack of balanced steady-state free precession cines were acquired at one slice per breath hold.
Cardiac magnetic resonance DICOM images on >43 000 individuals were converted into NifTI format. In an automated image analysis, the NifTI images were quality controlled and followed by annotation of cardiac imaging traits from the segmentations, using a trained fully convolutional network architecture. The method is described in detail by Bai et al.,15 see URLs for source code and model availability. The reported mean Dice metric for LA segmentation of the model was 0.93 for LA (2-ch) and 0.95 for LA (4-ch).15 The following traits were derived as described below: indexed LA maximum (LAmax) and LA minimum (LAmin) volumes, LA active emptying fraction (LAAEF), LA passive emptying fraction (LAPEF), and LA total emptying fraction (LATEF).
Annotations of cardiac atrial imaging traits were defined by the identification of the global maximum LA volume in a phase, followed by a local minimum (LA mid-diastolic volume; LAmdv), a subsequent local maximum (LA volume before atrial contraction; LAbac) and lastly a global minimum volume (Supplementary material online, Figure S1). Cardiac magnetic resonance images were excluded if any of these points could not be identified. Left atrial volumes were indexed by body surface area, calculated on each participant using Du Bois formula,16 denoted LAmax and LAmin (mL/m2). Left atrial passive emptying fraction was calculated as (LAmax – LAmdv)/LAmax * 100, LAAEF as (LAbac – LAmin)/LAbac * 100 and LATEF as (LAmax – LAmin)/LAmax * 100. See Supplementary material online, Methods for details on CMR quality control. A subset of images was manually segmented and the reliability between automatic and manual segmentation was evaluated (Supplementary material online, Methods).
Study population
From the individuals with available CMR images, we identified an ethnically homogeneous subpopulation using principal component (PC) analyses. UK Biobank provides quality control sheets with information such as relatedness and PCs. We removed individuals with sex mismatch, heterozygosity rate outliers, missing genotypes or excess relatives.
We then calculated the mean and standard deviations (SD) of PC 1–5 given by individuals classified ‘white British ancestry’ in UKBB data field 22006. We included individuals with a sum total of less than 6 SDs in PC 1–5 who also self-reported ‘white’ ancestry (see Supplementary material online, Methods and Figure S6). To minimize confounding by pre-existing conditions, participants with previous myocardial infarction, diagnosis of HF or cardiomyopathy as well as body mass index (BMI) <16 or >40 kg/m2 were excluded from analyses (see Supplementary material online, Methods for phenotype definitions).
Genetic analysis
UK Biobank version 3 is imputed with the haplotype reference consortium (HRC) reference panel and a merged UK10K and 1000 Genomes reference panel and has ∼90 million imputed variants.
We extracted a subset of UKBB imputed autosomal genotypes from the selected study population. Info scores and minor allele frequencies (MAF) were calculated using QCTOOL on the subset of genotypes. Variants with a MAF <0.5% or info score <0.8 were excluded, leaving 10.6 million quality-controlled variants available for analysis. New PCs were generated with fastPCA v.2.017 (see Supplementary material online, Methods for further details).
The GWASs were performed on autosomal chromosomes assuming an additive genetic model based on genotype dosages with BOLT-LMM v.2,18 which employs a linear mixed model algorithm. For the model single nucleotide polymorphisms (SNPs) used to assess random effect in BOLT, we had 581 883 hard called genotypes with a MAF > 0.5%, call rate > 95% and linkage disequilibrium (LD) < 0.9. Single nucleotide polymorphisms residing in the major histocompatibility (MHC) complex (chr6:28–35 Mb) or the chromosome 8 inversion (chr8:7–13 Mb) were removed from the analyses. Outcomes were rank-based inverse transformed residuals obtained by regressing LA traits on the covariates sex, age, PC 1–10, genotyping array (UKBB and UK BiLEVE array) and assessment centre (UKBB data field 54, instance 2).
To test the reproducibility and robustness of our results, we conducted three sensitivity analyses: excluding individuals with AF diagnosis, excluding individuals diagnosed with valvular heart disease and adjusting for systolic blood pressure (SBP; see Supplementary material online, Methods for definition of covariates). Systolic blood pressure was corrected for antihypertensive treatment by adding 10 mmHg to the observed blood pressure.19
Variance component analysis
Single nucleotide polymorphism heritability () and genetic correlation (rg) were estimated with the BOLT-REML algorithm for variance component analysis.20 As input, we used the model SNPs described in the genetic analysis above. Sex, age, genotyping array, assessment centre, and PC 1–10 were used as covariates in the analysis. The variance explained by each sentinel SNP was calculated with effect allele frequency (f) and beta (B) estimates, in SD−1, with the formula R2 = B2(1-f)2f.21 For each trait separately the total variance explained by the sentinel SNPs was calculated by summing the variance explained by each sentinel.
Correlation between left atrial and left ventricular traits
We compared the phenotypic correlation between LA traits using Pearson’s correlation. Furthermore, we evaluated the correlation between LA traits and measurements of LV ejection fraction (LVEF), LV end-diastolic volume (LVEDV), and LV end-systolic volume (LVESV). Left ventricular measurements were derived simultaneously to the annotation of the LA, as described by Bai et al.15
Genetic correlation of left atrial traits with other traits
Using summary statistics, we applied LD score regression (LDSC software) to estimate the genetic correlation between LA traits and 10 traits selected based on availability and relevance for cardiovascular disease [AF, type 2 diabetes (T2D), systolic and diastolic blood pressure, any stroke, any ischaemic stroke, cardioembolic stroke (CES), HF, overall health rating, BMI; Supplementary material online, Data S2, stroke definitions from the MEGASTROKE Consortium].22 We excluded the MHC region and SNPs with MAF <1%. Only phenotypes with a mean χ2 > 1.02 were considered. Pre-computed LD scores for HapMap3 SNPs were used in the regression (see URLs).
Biological and functional annotation
Loci were formed around sentinel SNPs. Sentinel SNPs were chosen by the lowest P-value. Variants that were nominally significant (P < 0.05) and within 1000 kb and a r 2 > 0.1 of the sentinel SNPs were assigned to a locus. The locus region was defined by the SNPs assigned to a locus. To identify independently associated variants within a locus, we performed conditional and joint analyses using GCTA-cojo v.1.91 with default settings.23
Co-localization
We investigated co-localization with expression quantitative trait loci (eQTL) using coloc.24 We tested all genes with a significant cis-eQTL association within 250 kb of the sentinel SNP in each locus. Pre-computed cis-eQTLs from LV and atrial appendage tissue were obtained from GTEx v.8 (see URLs). We used the coloc.abf function with default settings and tested variants intersecting both GTEx and GWAS datasets. We reported on eQTLs for genes with high evidence of co-localization (posterior probability >0.75).
Transcriptome-wide analyses
We used MetaXcan v0.625 to investigate mediating effects of gene expression levels on LA traits. Using summary statistics and eQTL data (GTEx v.7 atrial appendage and LV heart tissue, see URLs) we performed gene-based association analyses of predicted gene expression and phenotype risk. We tested 5369 genes with available expression data in the LV and 5982 in the atrial appendage. A P < 9.3 × 10−6 was considered significant in the LV and a P < 8.4 × 10−6 was considered significant in atrial appendage tissue. See Supplementary material online, Methods for further details.
FINEMAP
In an effort to identify plausible causal variants, we employed FINEMAP v1.426 at each of the 18 identified loci. All variants within 500 kb of a sentinel SNP were used to model each locus. Variants with a log10 Bayes factor (log10BF) >2 were functionally annotated using snpEFF v.4.327 and dbNSFP v2.9.28
Conditional analysis
Mediating effects on loci by AF or HF were analysed by performing conditional analyses with multi-trait conditional and joint analysis (mtCojo) from GCTA v1.92.29 The mtCojo method is able to condition phenotype y on phenotype x using GWAS summary statistics. These estimates are free from collider bias, which otherwise can occur when adjusting for heritable covariates.30 For estimation of LD structure, a reference sample set of 12 000 individuals without CMR imaging and free from AF or HF was randomly selected from UKBB. Left atrial traits summary statistics were adjusted on GWAS summary statistics of AF31 and HF.32
Polygenetic risk score for atrial fibrillation
We derived a polygenetic risk score (PRS) for AF using stacked clumping and thresholding.33 A penalized regression was used for stacking the clumping and threshold predictions. Therefore, the penalized regression was fitted on a training set and then predicted onto the CMR cohort. Unrelated UKBB participants with European ancestry and without available CMR (n = 332 673) were used for developing the PRS (the training set). Input beta estimates and P-values came from summary statistics from a GWAS on AF not containing the UKBB cohort.34 We used SNPs with MAF >1% and a grid of varying thresholds for P-values, base size for clumping window and LD (r2) to derive intermediate PRSs (Supplementary material online, Table S1). The intermediate PRSs were stacked into one PRS, weighted by a penalized logistic regression. The polygenetic risk for AF was then calculated for each individual with available CMR and free of AF included in this study. Additionally, we employed a two-sample 2-stage least square (2SLS) regression in order to explore the impact of LA traits on AF, HF, and stroke (Supplementary material online, Methods).
Mendelian randomization analyses
We used Mendelian randomization to investigate causal effects between the five exposures AF,31 HF,32 SBP, coronary artery disease (CAD), and T2D and LA traits. We employed the inverse-variance weighted method35 as primary model, and Mendelian randomization-Egger regression, weighted median and mode-based estimates as sensitivity analyses. All analyses were performed using the R package TwoSampleMR.36 The MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) method was used to assess whether any detectable effect was mediated through outliers. We selected SNPs that were genome-wide significant (P < 5 × 10−8), and removed SNPs in LD using default settings (r 2 < 0.001) in order to obtain independent variants. A P < 0.05/5 traits = 0.01 was considered statistically significant. See Supplementary material online, Methods for references to MR tools.
Associations between left ventricular loci and left atrial traits
To assess the role of LV loci in LA traits, we queried the genetic variants associated with LV by Aung et al.12 in our summary statistics. To correct for multiple testing, variants with a P < 0.05/(11 loci * 5 LA traits) = 0.0009 were considered significantly associated with LA traits.
Results
The overall study design is illustrated in Figure 1. Baseline characteristics of the study population are shown in Table 1 and summary of CMR traits in Table 2. A comprehensive description of the selection of study population and covariates is available in Supplementary material online, Methods.
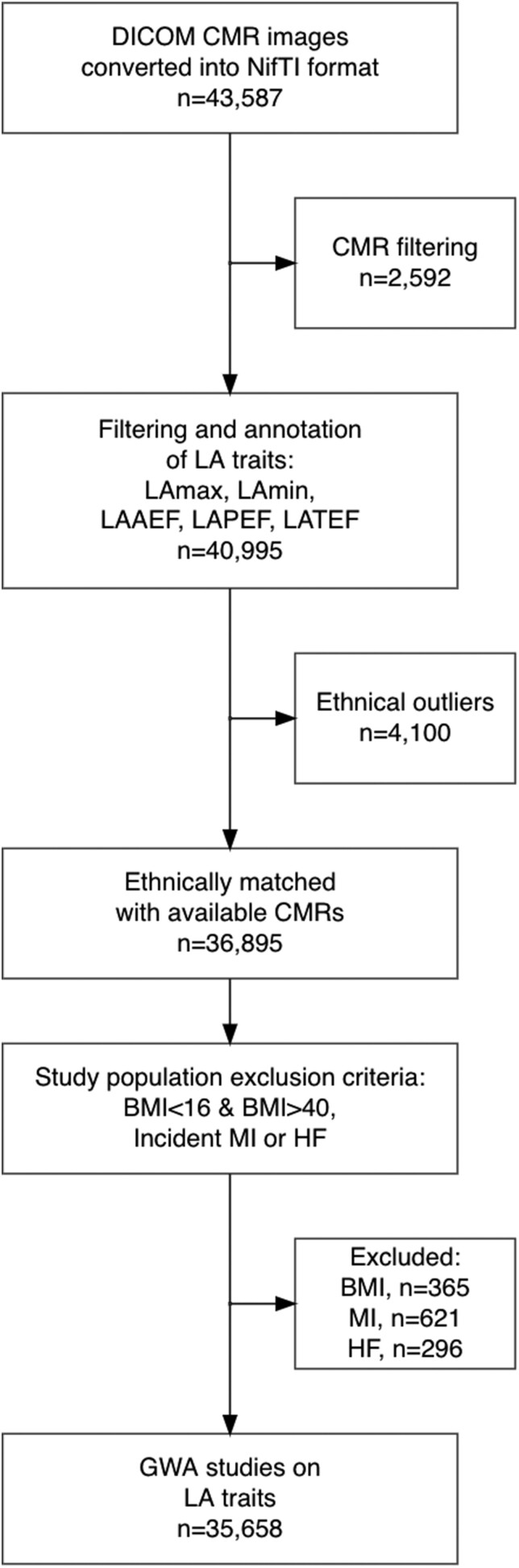
Figure 1.
Sample inclusion flowchart. Please note that some individuals may fulfil multiple exclusion criteria and can be listed multiple times in the exclusion boxes, e.g. having both extreme BMI and MI. BMI, body mass index; CMR, cardiac magnetic resonance; GWAS, genome-wide association study; HF, heart failure; LA, left atrial; LAAEF, left atrial active emptying fraction; LAPEF, left atrial passive emptying fraction; LATEF, left atrial total emptying fraction; LAV, left atrial volume; MAF, minor allele frequency; MI, myocardial infarction.
Table 1.
Baseline characteristics
| N | 35 658 |
| Age, years, median (IQR) | 64 (58–70) |
| Male sex, n (%) | 16 979 (48) |
| Height, cm, mean (SD) | 169 (9) |
| Weight, kg, mean (SD) | 76 (10) |
| BMI, kg/m2, mean (SD) | 26 (4) |
| Body surface area, m2, mean (SD) | 1.86 (0.2) |
| Systolic/diastolic blood pressure, mmHg, mean (SD) | 130 (18)/77 (10) |
| Pulse rate, b.p.m., mean (SD) | 67 (11) |
| Stroke, n (%) | 341 (1) |
| AF, n (%) | 713 (2) |
| T2DM, n (%) | 1070 (3) |
| CAD, n (%) | 552 (1.5) |
| HT, n (%) | 9272 (26) |
| Valvular heart disease, n (%) | 102 (0.3) |
AF, atrial fibrillation; BMI, body mass index; CAD, coronary artery disease; HT, hypertension; IQR, interquartile range; SD, standard deviation; T2DM, type 2 diabetes mellitus.
Table 2.
Cardiac magnetic resonance variables
| LAmax | 38.9 (10.5) |
| LAmin | 15.5 (6.3) |
| LAmdv | 25.9 (8.4) |
| LAbac | 28.9 (8.6) |
| LAAEF | 47.5 (8.7) |
| LAPEF | 33.9 (7.8) |
| LATEF | 61.1 (7.9) |
| LVEF | 59.7 (5.8) |
| LVEDV | 148.0 (33.0) |
| LVESV | 60.0 (18.3) |
All data are presented as mean (SD). Volume measures for LA are indexed by BSA.
bac, before atrial contraction; BSA, body surface area; CMR, cardiac magnetic resonance; LA, left atrium; LATEF, left atrial active emptying fraction; LAPEF, left atrial passive emptying fraction; LATEF, left atrial total emptying fraction; LVEF, left ventricular ejection fraction; LVEDV, left ventricular end-diastolic volume; LVESV, left ventricular end-systolic volume; mdv, mid-diastolic volume.
A total of 35 658 individuals were included. The median age was 64 years (IQR 58–70), average BMI was 26 (SD = 4) and 52% were women. Biplane LAmax had a mean of 39 mL/m2 (SD = 11 mL/m2), biplane LAmin had a mean of 16 mL/m2 (SD = 6 mL/m2), and LATEF 61% (SD = 8%), which are comparable to previous studies on reference values on the UKBB cohort37 (Supplementary material online, Figure S2). The reproducibility of the included images with automatic and manual measurements was high (intraclass correlation coefficient [ICC]lamax = 0.93, ICClamin = 0.88, n = 50; Supplementary material online, Methods).
Identification of loci associated with left atrial volume and function
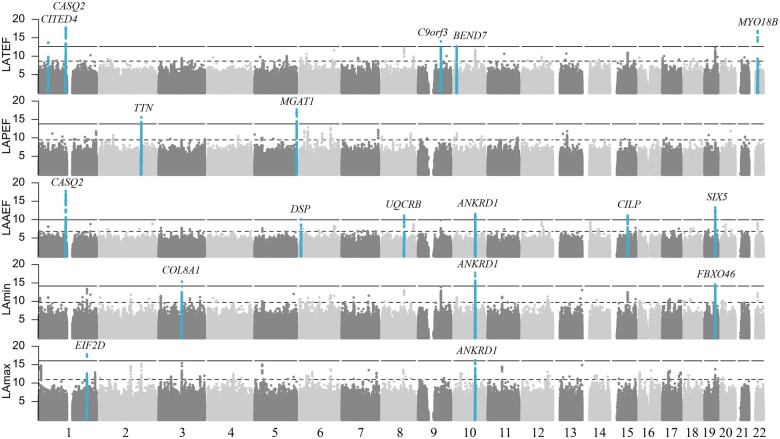
We identified 18 novel loci associated with LA volume and function (LAmax n = 2; LAmin n = 3; LAAEF n = 6; LAPEF n = 2; LATEF n = 5, Figure 2, Supplementary material online, Data S3). Of these, 15 constituted unique loci. Several loci were associated with multiple LA traits, e.g. rs9664170 in proximity of ANKRD1 reached genome-wide significance in LAmax, LAmin, and LAAEF (MAF = 41%, LAmax/LAmin/LAAEF P = 4.3 × 10−8/6.8 × 10−10/3.4 × 10−9). There was little evidence of population stratification or cryptic relatedness (Supplementary material online, Figures S6 and S7 and Table S2). Regional plots for each locus are shown in Supplementary material online, Figure S8. We conducted three sensitivity analyses, to test the robustness and reproducibility of our results. The main results did not materially change, when we excluded individuals with AF, valvular heart disease or when adjusting for blood pressure (Supplementary material online, Data S4). Moreover, nominally significant variants (P < 0.05) in the reported loci correlated with beta estimates obtained in the sensitivity models (min r > 0.98).
Figure 2.
Genome-wide associations. Manhattan plots with the −log10(P-value) plotted on the y-axis and chromosomal position on the x-axis. Each locus is labelled with name of nearest gene to sentinel SNP. All sentinel SNPs are listed in Supplementary material online, Data S3. LA, left atrial; LAAEF, left atrial active emptying fraction; LAPEF, left atrial passive emptying fraction; LATEF, left atrial total emptying fraction; SNP, single nucleotide polymorphism.
Variance explained by loci was 0.29/0.18/0.63/0.21/0.50% for LAmax/LAmin/LAAEF/LAPEF/LATEF, respectively (Supplementary material online, Data S5).
Heritability and genetic correlation of left atrial traits
Heritability estimates were = 0.24/0.22/0.17/0.18/0.16 for LAmax/LAmin/LAAEF/LAPEF/LATEF, respectively (Supplementary material online, Figure S9 and Data S6). We observed a high genetic correlation between LAmax and LAmin (rg = 0.94), as well as LAAEF and LATEF (rg = 0.92). Left atrial emptying fractions had a negative genetic correlation with LA volumes, e.g. LATEF with LAmax/min (rg = −0.59/−0.84). Left atrial traits had a lower genetic correlation with LV traits (LVEDV, LVESV, LVEF). The highest absolute genetic correlation with LV traits was observed between LAmax and LVEDV (rg = 0.36). Left ventricular end-diastolic volume also carried the largest heritable component ( = 0.39).
Characterization of left atrial associated loci
We examined co-localization of loci and eQTLs by conducting a Bayesian co-localization analysis.24 Using GTEx v.8 tissue samples from atrial appendage (n = 372) and LV (n = 386), we found evidence of co-localization (posterior probability > 0.75) between loci and eQTL for 10 genes (Supplementary material online, Data S7). Left atrial volume associated loci co-localized with ANKRD1. Left atrial function associated loci co-localized with C9orf3 (also known as AOPEP), CASQ2, RP11-10A14.4, VANGL1, and RP11-236B18.2. Loci associated with both LA function and volume co-localized with BHMG1, DMWD, PCGF5, and SYMPK. Interestingly, the genes BHMG1, DMWD, and SYMPK reside in the same gene dense on chromosome 19. We also investigated the effects of gene expression levels on LA traits in a transcriptome-wide analysis (TWAS) using MetaXcan. This revealed 20 associations between the predicted gene expression and LA volume/function (Supplementary material online, Methods, Figures S10 and S11, and Data S8–S17). Interestingly, the expression of C9orf3 was associated with increased LA function and decreased LA volume in atrial appendage tissue samples (LAmin/LAAEF/LATEF, P = 9.1 × 10−8/4.7 × 10−6/3.0 × 10−7).
In a further effort to identify genes that might be causally linked to the associated loci, we used FINEMAP26 to identify credibly causal alleles. We identified seven missense variants in genes associated with LA function (Supplementary material online, Data S18), e.g. missense variants associated with LAAEF in the genes CASQ2, CILP, DSP, and SIX5, and a missense variant in TTN associated with LAPEF. Two missense variants associated with LATEF were identified in CASQ2 and MYO18B. Functional characterization and evidence linking genes to loci are summarized in Table 3.
Table 3.
Summary of functional characteristics in loci
| Chr: Pos | Sentinel rsid | Phenotype | Nearest gene | eQTL co-localization | Fine mapped missense variants (gene, missense rsID, CADD score) |
|---|---|---|---|---|---|
| 1:41348272 | rs12026695 | LATEF | CITED4 | ||
| 1:116310967 | rs4074536 | LAAEF | CASQ2 | CASQ2, VANGL1 | CASQ2, rs4074536, 0.027 |
| 1:116310967 | rs4074536 | LATEF | CASQ2 | CASQ2, VANGL1 | CASQ2, rs4074536, 0.027 |
| 1:206796179 | rs4845119 | LAmax | EIF2D | ||
| 2:179381323 | rs6735077 | LAPEF | TTN | TTN, rs9808377, 8.518 | |
| 3:99156961 | rs114825396 | LAmin | COL8A1 | ||
| 5:180209454 | rs655437 | LAPEF | MGAT1 | ||
| 6:7573455 | rs17143007 | LAAEF | DSP | DSP, rs17604693, 26.7 | |
| 8:97223162 | rs35216833 | LAAEF | UQCRB | RP11-10A14.4 | |
| 9:97477793 | rs4744370 | LATEF | C9orf3 | C9orf3 | |
| 10:13474369 | rs113306707 | LATEF | BEND7 | ||
| 10:92692047 | rs9664170 | LAmax | ANKRD1 | PCGF5 | |
| 10:92692047 | rs9664170 | LAmin | ANKRD1 | PCGF5, ANKRD1 | |
| 10:92692047 | rs9664170 | LAAEF | ANKRD1 | PCGF5, RP11-236B18.2 | |
| 15:65493852 | rs2073708 | LAAEF | CILP | CILP, rs2073711, 17.71 | |
| 19:46213416 | rs62111731 | LAmin | FBXO46 | BHMG1, SYMPK, DMWD | |
| 19:46269076 | rs2014576 | LAAEF | SIX5 | BHMG1, SYMPK, DMWD | SIX5, rs2014576, 17.36 |
| 22:26159289 | rs133885 | LATEF | MYO18B | MYO18B, rs133885, 16.91 |
Genetic correlation with cardiovascular traits
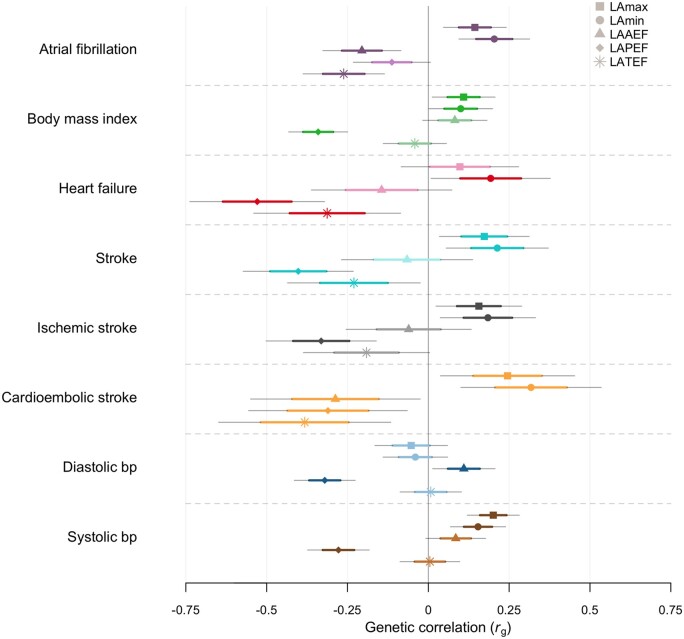
Using LD score regression, we investigated the genetic correlation of LA volume and function with selected cardiovascular diseases (Figure 3 for nominal significant associations, Supplementary material online, Data S19 for all associations). Atrial fibrillation, CES, and HF had higher genetic correlation estimates with LAmin than LAmax (LAmin-AF rg = 0.20, P = 3 × 10−4; LAmin-CES rg = 0.32, P = 4 × 10−3; LAmin-HF rg = 0.19, P = 4 × 10−2). For LA functional traits, we observed the highest genetic correlation in LAPEF with all-cause stroke (AS) and HF (rg = 0.40, P = 4 × 10−6; rg = 0.53, P = 6 × 10−7), whereas the genetic correlation between LAPEF and AF did not reach nominal significance. Left atrial passive emptying fraction had higher genetic correlation estimates than other LA traits with overall health rating, DBP and SBP (rg = −0.33, P = 4 × 10−12; rg = −0.32, P = 3 × 10−11; rg = −0.28, P = 1 × 10−8). Among LA functional traits, LATEF had the highest genetic correlation with AF and CES (rg = 0.26, P = 5 x 10−5; rg = −0.38, P = 5 × 10−3).
Figure 3.
Genetic correlation between left atrial traits and cardiovascular phenotypes using LDSC. Sample sizes of external genome-wide association studies are shown in Supplementary material online, Data S2. Phenotypes with negative log10(P) are displayed on the y-axis. X-axis shows genetic correlation (rg). Point estimates shown as dots with thick lines indicate mean and standard error (SE) and thin lines 95% confidence intervals (1.96xSE). Nominal significance (P < 0.05) is denoted with bright colour and non-significant correlations with faded colour. Standard errors and P-values were derived using block jackknife resampling.
Structural effects of polygenetic risk for atrial fibrillation
To identify whether genetic risk of AF yields LA structural changes without AF diagnosis, we produced a PRS for AF. We calculated the estimated polygenetic risk for AF in each included individual with available CMR and without AF (n = 34 945, the test set), given our stated definition of AF (Supplementary material online, Methods). Fitting a linear model adjusted for age and sex, we found that the AF PRS was associated with decreased LAAEF [−0.35%/SD, 95% confidence interval (CI) (−0.43; −0.26), P = 3.14 × 10−14] and LATEF [−0.27 SD, 95% CI (−0.35; −0.19), P = 4.57 × 10−11], but not with LAPEF (P = 0.54). Atrial fibrillation PRS was also associated with increased LAmax and LAmin [LAmax 0.25 (mL/m2)/SD, 95% CI (0.15; 0.36), P = 5.13 × 10−6; LAmin 0.21 (mL/m2)/SD, 95% CI (0.15; 0.28), P = 1.86 × 10−10]. Furthermore, 2SLS regression of sentinel SNP allele scores indicated that LA function has an impact on AF risk [LAAEF/SD OR = 0.69, 95% CI (0.57; 0.84), P = 2 × 10−4; LAPEF/SD OR = 0.43, 95% CI (0.31; 0.61), P = 1.0 × 10−6; LATEF/SD OR = 0.41, 95% CI (0.33; 0.51), P = 1.0 × 10−15; Supplementary material online, Data S20]. For LAAEF and LATEF, we also identified a nominal significant modification on the effect of AF-PRS for individuals with incident AF (LAAEF P = 0.019, LATEF P = 0.036; Supplementary material online, Methods and Data S21).
Conditional analyses and Mendelian randomization
To estimate whether the associations between genetics and LA traits were mediated by AF or HF, we performed a conditional analysis using mtCOJO.29 All identified sentinel SNPs with their respective LA trait were conditioned on either AF or HF, using publicly available summary statistics. We did not observe any mediating effect on beta estimates greater than the estimated standard error, which suggests that SNP effect sizes are independent from AF and HF (Supplementary material online, Figure S12 and Data S22).
Next, we estimated the causal effects of AF, HF, SBP, CAD, and T2D on LA traits using MR.
We found supporting evidence for a casual effect of SBP on all LA traits (Supplementary material online, Data S23). Systolic blood pressure increases LA volume and decreases LA function (beta LAmax/LAmin/LAAEF/LAPEF/LATEF = 0.23/0.21/−0.01/−0.18/−0.11 SD/SD−1, P < 0.01). The effect of SBP was larger on LAPEF than on LAAEF, which is in line with other findings on LAPEF. We also found robust support for a casual effect of AF, which show congruent causal effects on LA volume, LAAEF, and LATEF (Supplementary material online, Data S23). Atrial fibrillation was similarly associated with an increase in LAmin/max, (beta LAmax and LAmin = 0.07 and 0.1 SD, respectively, P < 0.0.1) and decrease in LAAEF and LATEF (beta LAAEF and LATEF = −0.11 and 0.10 SD, respectively, P < 0.01), but had no statistically significant effect on LAPEF.
Role of left ventricular loci in left atrial traits
Three LV loci previously associated with LV mass, LVEDV/LVESV/LVEF, and LV mass to end-diastolic volume ratio, respectively, were significantly associated with LAPEF (rs2255167, P = 1.5 × 10−6, TTN; rs2042995, P = 3.2 × 10−7, TTN; rs146170154, P = 2.7 × 10−6, CDKN1A; Supplementary material online, Data S24).
Discussion
This is, to our knowledge, the first GWAS to date investigating the genetic background of LA traits derived from CMR. We identified 18 previously unreported loci associated with LA volume and function and demonstrated a genetic link between LA traits and incident AF (Graphical abstract). Furthermore, we established a set of credible causal genes, of which many have a previously established relation to AF or cardiomyopathy. We evaluated the casual relationship between AF and LA traits with MR analyses and found supporting evidence of AF being causal for increased LA volume and decreased active and total emptying function. We show that polygenetic risk for AF is associated with increased LA volume and decreased LA function, in participants without prior diagnosis of AF. This suggests that structural and functional dysfunction may appear before clinically diagnosed AF.
Genome-wide association study on left atrial volume and function based on cardiac magnetic resonance imaging. Imaging data from 35 658 individuals were exposed to machine learning algorithms to annotate five left atrial traits. A total of 18 novel genetic loci were associated with left atrial volume and function. Of these, 15 constituted unique loci. Genes marked with bold have previously been associated with cardiomyopathy.
Many of the genes identified in this study have previously been linked to cardiomyopathy or muscular dystrophy. Using fine-mapping, seven plausibly causal missense variants associated with LA function in the genes DSP, SIX5, MYO18B, CILP, TTN, and CASQ2. SIX5 resides in a gene dense region on chromosome 19, with neighbouring genes such as DMKP and DMWD. This region is known to cause myotonic dystrophy type 1 through expansion of CTG repeats.38 MYO18B has recently been associated with AF.31 Zebrafish mutants with loss of function of the myo18b gene show compromised sarcomere assembly and atrial enlargement,39 a hallmark of atrial cardiomyopathies. Similar findings have been reported in zebrafish with titin-truncating variants.40 CILP, encoding cartilage intermediate layer protein 1, has been shown as a marker for fibrosis, which can lead to HF.41 Mutations in DSP, encoding the protein desmoplakin, are known causes of several types of cardiomyopathy.42 , 43 The TTN gene encodes the protein titin, which is responsible for the passive elasticity of the cardiomyocytes and has been associated with dilated cardiomyopathy and AF.40 , 44 , 45rs9664170, in proximity to ANKRD1 encoding the cardiac ankyrin repeat protein, was associated with LAAEF and LAmax/min and showed evidence of co-localization with eQTLs affecting expression of ANKRD1 and RP11-236B18.2. RP11-236B18.2 is a long non-coding RNA (lncRNA), residing adjacent to ANKRD1. Long non-coding RNA has been shown to regulate gene expression.46 Cardiac ankyrin repeat protein functions as a transcription co-factor and is a member of the titin-N2A mechanosensory complex that translocate to the cell nucleus in response to stretch, and has previously been associated with dilated cardiomyopathy.47 Cardiac ankyrin repeat protein expression is up-regulated in HF, where it has been shown to suppress the expression of sarcomere proteins.47 The SNP rs4074536 was associated with LAAEF and LATEF and showed evidence of co-localization with CASQ2 eQTLs, also supported by TWAS. CASQ2, encoding the gene calsequestrin 2, is an important gene in calcium handling and cardiac contraction, which is functionally in line with its association with LAAEF. CASQ2 has previously been associated with AF48 and catecholaminergic polymorphic ventricular tachycardia.49 Although some of the identified loci have previously been associated with AF and HF, adjusting our analyses for AF or HF genetics with conditional analyses did not show any mediating effects.
Due to a growing interest in atrial cardiomyopathy, a number of studies focusing on LA volume and function have previously been conducted. These studies indicate that LA traits are independent predictors of a number of clinical outcomes such as stroke, HF, and AF.50 In this study, we estimate that these LA traits have a SNP heritability of ∼16–26%. Left atrial volume is more heritable than LA function. Left atrial volume traits are highly genetically correlated but LAAEF and LAPEF are different genetic entities based on genetic correlation.
Recent GWAS on AF have implicated numerous genes associated with cardiomyocyte function.31 Moreover, a growing body of evidence suggests that rare variants in structural genes cause AF.40 , 51 Previous reports have shown that decreased LAAEF is associated with an increased risk of AF.52 To evaluate the influence of predisposition to AF on the volumetric and functional properties of the atria, we produced an AF-PRS. We found that our AF-PRS was associated with lower LAAEF in individuals without known AF. This suggests that the increased risk of AF in part is mediated by predisposition to alterations in atrial function that precede the development of AF. In fact, LAAEF was associated with genes previously associated with cardiomyopathy. Whether the same genes expressed in the atria give rise to an atrial cardiomyopathy that precedes overt AF remains to be established.
By separating the passive and active emptying function in our genetic analyses of the LA, we discern that these traits have vastly different genetic bases. We observed a higher genetic correlation between LAAEF, AF, and CES compared with LAPEF. With regards to LAPEF, we found no significant association between the genetically predictive risk of AF and LAPEF. However, we found larger genetic correlations for LAPEF with traditional cardiovascular risk factors and diseases (e.g. BMI, all-cause stroke, blood pressure, and HF) compared with LAAEF. This could be of importance since the mechanisms giving rise to stroke may differ and the link between the atria and CES needs further refinement. When looking at the overlap between LV and LA genetics, we found LV loci close to the TTN gene associated significantly with LAPEF. It is possible that this overlap could be mediated by the gene TTN, which plays an important role in ventricular cardiomyocyte sarcomere elasticity. Impaired ventricular elasticity could in theory influence the passive emptying of the atria.
Traditionally, LAmax is used to assess LA volume. However, LAmin has previously been found to have a stronger predictive value of AF, as well as subclinical AF, than LAmax . 2 , 52 In line with these reports, our genetic analyses show that LAmin has a larger genetic overlap with AF and CES than LAmax. Using MR analyses, we measured that AF had a larger effect on LAmin than LAmax, altogether reiterating the role of LAmin as important when evaluating cardiac disease.
Our findings should be interpreted within the context of their limitations. To avoid population stratification bias, we excluded persons with non-European ancestry from analyses. Our findings are therefore limited to European ancestry only and cannot be directly extrapolated to other populations. However, they do inform us of fundamental important biological processes in the LA. Annotation of LA traits is based on machine learning algorithms with the resulting strengths and weaknesses. To evaluate the accuracy of the deep learning model, we performed manual segmentation on a subset of the dataset which validated the performance of the model. Although our manual segmentation was highly correlated with the automated segmentation, an overestimation was present for the automated segmentation compared with the manual. The overestimation was systematic and should therefore not influence the genetic results. Bai et al.15 have previously validated the algorithm on UKBB CMR at a larger scale with a Dice metric of 0.93 for LA (2-ch) and 0.95 LA (4-ch). Also, they note that the automated method is trained by multiple observers which may make it less susceptible to biases by learning a consensus estimate, since manual inter-observer variability is to be expected. However, the LA is challenging to measure and even after quality control filtering, errors might remain that dilute the genetic signal. In the future, refinements addressing these issues might be able to enhance the genetic signal of these traits. Although the majority of our findings are plausible candidate genes with well-described roles in cardiac pathophysiology, future studies are needed to cement their role in LA volume and function.
In conclusion, we report on 18 genetic loci associated with LA function and volume. We show evidence for several plausible candidate genes important for atrial structure. Many of the genes associated with LA function have previously been linked with cardiomyopathy. The results found in this study suggest that compromised LA function may share genetic underpinning with cardiomyopathy. Furthermore, we show that the genetic risk of AF influences LA structure prior to diagnosis of AF, suggesting an intrinsic relationship.
URLs
FCN model: https://github.com/baiwenjia/ukbb_cardiac
Precomputed LD scores etc.: https://data.broadinstitute.org/alkesgroup/LDSCORE/
LDSC: https://github.com/bulik/ldsc
GTEx v.8: https://www.gtexportal.org/
MetaXcan transcriptome models: http://predictdb.org/
Supplementary material
Supplementary material is available at European Heart Journal online.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank resource under application number 43247. The data used for the analyses described in this manuscript were obtained from: https://www.gtexportal.org/ the GTEx Portal on 2020-04-20.
Funding
This work was supported by the John and Birthe Meyer Foundation, the Research Foundation of the Heart Centre, Rigshospitalet, the Research Council at Rigshospitalet, The Hallas-Møller Emerging Investigator Novo Nordisk (NNF17OC0031204), and the Arvid Nilsson Foundation. G.A. has received grant from the Novo Nordisk Foundation, BRIDGE—Translational Excellence Programme, Ref #NNF18SA0034956. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html (see Supplementary material online for list of authors).
Conflict of interest: none declared.
Data availability
Summary statistics are available for download at https://zenodo.org/record/5074929. Data generated in this study have been returned to the UKBB. The UKBB will make these data available to all researchers who apply by standard access procedures. Please see the UKBB’s website for access procedures (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access).
Contributor Information
Gustav Ahlberg, Laboratory for Molecular Cardiology, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Henrik Harpestrengs Vej 4C, 2100 Copenhagen, Denmark; Department of Biomedical Sciences, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
Laura Andreasen, Laboratory for Molecular Cardiology, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Henrik Harpestrengs Vej 4C, 2100 Copenhagen, Denmark; Department of Biomedical Sciences, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
Jonas Ghouse, Laboratory for Molecular Cardiology, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Henrik Harpestrengs Vej 4C, 2100 Copenhagen, Denmark; Department of Biomedical Sciences, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
Litten Bertelsen, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Inge Lehmanns Vej 7, 2100 Copenhagen, Denmark.
Henning Bundgaard, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Inge Lehmanns Vej 7, 2100 Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
Stig Haunsø, Laboratory for Molecular Cardiology, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Henrik Harpestrengs Vej 4C, 2100 Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
Jesper H Svendsen, Laboratory for Molecular Cardiology, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Henrik Harpestrengs Vej 4C, 2100 Copenhagen, Denmark; Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Inge Lehmanns Vej 7, 2100 Copenhagen, Denmark; Department of Clinical Medicine, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
Morten S Olesen, Laboratory for Molecular Cardiology, Department of Cardiology, Heart Centre, Rigshospitalet, University Hospital of Copenhagen, Henrik Harpestrengs Vej 4C, 2100 Copenhagen, Denmark; Department of Biomedical Sciences, University of Copenhagen, Blegdamsvej 3, 2200 Copenhagen, Denmark.
This work was carried out at the Department of Biomedical Sciences, University of Copenhagen, Copenhagen, Denmark.
References
- 1. Kaminski M, Steel K, Jerosch-Herold M, Khin M, Tsang S, Hauser T, Kwong RY. Strong cardiovascular prognostic implication of quantitative left atrial contractile function assessed by cardiac magnetic resonance imaging in patients with chronic hypertension. J Cardiovasc Magn Reson 2011;13:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Olsen FJ, Møgelvang R, Jensen GB, Jensen JS, Biering-Sørensen T. Relationship between left atrial functional measures and incident atrial fibrillation in the general population: the Copenhagen City Heart Study. JACC Cardiovasc Imaging 2019;12:981–989. [DOI] [PubMed] [Google Scholar]
- 3. Habibi M, Zareian M, Venkatesh BA, Samiei S, Imai M, Wu C, Launer LJ, Shea S, Gottesman RF, Heckbert SR, Bluemke DA, Lima JAC. Left atrial mechanical function and incident ischemic cerebrovascular events independent of AF: insights from the MESA study. JACC Cardiovasc Imaging 2019;12:2417—2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gupta S, Matulevicius SA, Ayers CR, Berry JD, Patel PC, Markham DW, Levine BD, Chin KM, Lemos J. D, Peshock RM, Drazner MH. Left atrial structure and function and clinical outcomes in the general population. Eur Heart J 2013;34:278—285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, Lancellotti P, Muraru D, Picard MH, Rietzschel ER, Rudski L, Spencer KT, Tsang W, Voigt JU. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur Heart J Cardiovasc Imaging 2015;16:233—270. [DOI] [PubMed] [Google Scholar]
- 6. Goette A, Kalman JM, Aguinaga L, Akar J, Cabrera JA, Chen SA, Chugh SS, Corradi D, D'Avila A, Dobrev D, Fenelon G, Gonzalez M, Hatem SN, Helm R, Hindricks G, Ho SY, Hoit B, Jalife J, Kim Y-H, Lip GYH, Ma C-S, Marcus GM, Murray K, Nogami A, Sanders P, Uribe W, Van Wagoner DR, Nattel S; Document Reviewers. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: definition, characterization, and clinical implication. Europace 2016;18:1455—1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stefanadis CDernellis J, Toutouzas P. A clinical appraisal of left atrial function. Eur Heart J 2001;22:22—36. [DOI] [PubMed] [Google Scholar]
- 8. Gal PMarrouche NF. Magnetic resonance imaging of atrial fibrosis: redefining atrial fibrillation to a syndrome. Eur Heart J 2017;38:14—19. [DOI] [PubMed] [Google Scholar]
- 9. Kottkamp H. Human atrial fibrillation substrate: towards a specific fibrotic atrial cardiomyopathy. Eur Heart J 2013;34:2731—2738. [DOI] [PubMed] [Google Scholar]
- 10. Grothues F, Smith GC, Moon JCC, Bellenger NG, Collins P, Klein HU, Pennell DJ. Comparison of interstudy reproducibility of cardiovascular magnetic resonance with two-dimensional echocardiography in normal subjects and in patients with heart failure or left ventricular hypertrophy. Am J Cardiol 2002;90:29—34. [DOI] [PubMed] [Google Scholar]
- 11. Vasan RS, Larson MG, Aragam J, Wang TJ, Mitchell GF, Kathiresan S, Newton-Cheh C, Vita JA, Keyes MJ, O'Donnell CJ, Levy D, Benjamin EJ. Genome-wide association of echocardiographic dimensions, brachial artery endothelial function and treadmill exercise responses in the Framingham Heart Study. BMC Med Genet 2007;8:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aung N, Vargas JD, Yang C, Cabrera CP, Warren HR, Fung K, Tzanis E, Barnes MR, Rotter JI, Taylor KD, Manichaikul AW, Lima JAC, Bluemke DA, Piechnik SK, Neubauer S, Munroe PB, Petersen SE. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation 2019;140:1318—1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203—209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petersen SE, Matthews PM, Francis JM, Robson MD, Zemrak F, Boubertakh R, Young AA, Hudson S, Weale P, Garratt S, Collins R, Piechnik S, Neubauer S. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson 2016;18:8—7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bai W, Sinclair M, Tarroni G, Oktay O, Rajchl M, Vaillant G, Lee AM, Aung N, Lukaschuk E, Sanghvi MM, Zemrak F, Fung K, Paiva JM, Carapella V, Kim YJ, Suzuki H, Kainz B, Matthews PM, Petersen SE, Piechnik SK, Neubauer S, Glocker B, Rueckert D. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 2018;20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Du Bois D, Du Bois EF. A formula to estimate the approximate surface area if height and weight be known. 1916. Nutrition 1989;5:303—311; discussion 312–313. [PubMed] [Google Scholar]
- 17. Abraham G, Qiu Y, Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics 2017;33:2776—2778. [DOI] [PubMed] [Google Scholar]
- 18. Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet 2018;50:906—908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med 2005;24:2911—2935. [DOI] [PubMed] [Google Scholar]
- 20. Loh P-R, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ, de Candia TR, Lee SH, Wray NR, Kendler KS, O'Donovan MC, Neale BM, Patterson N, Price AL; Schizophrenia Working Group of Psychiatric Genomics Consortium. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance components analysis. Nat Genet 2015;47:1385—1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 2016;35:1880—1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291—295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369—375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, Stahl EA, Huckins LM, Nicolae DL, Cox NJ, Im HK; GTEx Consortium. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018;9:1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benner C, Spencer CCA, Havulinna AS, Salomaa V, Ripatti S, Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 2016;32:1493—1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012;6:80—92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 2013;34:E2393—E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, Robinson MR, McGrath JJ, Visscher PM, Wray NR, Yang J. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 2018;9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet 2015;96:329—339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, Surakka I, Mathis MR, Yamazaki M, Crawford RD, Gabrielsen ME, Skogholt AH, Holmen OL, Lin M, Wolford BN, Dey R, Dalen H, Sulem P, Chung JH, Backman JD, Arnar DO, Thorsteinsdottir U, Baras A, O'Dushlaine C, Holst AG, Wen X, Hornsby W, Dewey FE, Boehnke M, Kheterpal S, Mukherjee B, Lee S, Kang HM, Holm H, Kitzman J, Shavit JA, Jalife J, Brummett CM, Teslovich TM, Carey DJ, Gudbjartsson DF, Stefansson K, Abecasis GR, Hveem K, Willer CJ. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 2018;50:1234—1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, Hedman ÅK, Wilk JB, Morley MP, Chaffin MD, Helgadottir A, Verweij N, Dehghan A, Almgren P, Andersson C, Aragam KG, Ärnlöv J, Backman JD, Biggs ML, Bloom HL, Brandimarto J, Brown MR, Buckbinder L, Carey DJ, Chasman DI, Chen X, Chen X, Chung J, Chutkow W, Cook JP, Delgado GE, Denaxas S, Doney AS, Dörr M, Dudley SC, Dunn ME, Engström G, Esko T, Felix SB, Finan C, Ford I, Ghanbari M, Ghasemi S, Giedraitis V, Giulianini F, Gottdiener JS, Gross S, Guðbjartsson DF, Gutmann R, Haggerty CM, van der Harst P, Hyde CL, Ingelsson E, Jukema JW, Kavousi M, Khaw K-T, Kleber ME, Køber L, Koekemoer A, Langenberg C, Lind L, Lindgren CM, London B, Lotta LA, Lovering RC, Luan J, Magnusson P, Mahajan A, Margulies KB, März W, Melander O, Mordi IR, Morgan T, Morris AD, Morris AP, Morrison AC, Nagle MW, Nelson CP, Niessner A, Niiranen T, O'Donoghue ML, Owens AT, Palmer CNA, Parry HM, Perola M, Portilla-Fernandez E, Psaty BM, Rice KM, Ridker PM, Romaine SPR, Rotter JI, Salo P, Salomaa V, van Setten J, Shalaby AA, Smelser DT, Smith NL, Stender S, Stott DJ, Svensson P, Tammesoo M-L, Taylor KD, Teder-Laving M, Teumer A, Thorgeirsson G, Thorsteinsdottir U, Torp-Pedersen C, Trompet S, Tyl B, Uitterlinden AG, Veluchamy A, Völker U, Voors AA, Wang X, Wareham NJ, Waterworth D, Weeke PE, Weiss R, Wiggins KL, Xing H, Yerges-Armstrong LM, Yu B, Zannad F, Zhao JH, Hemingway H, Samani NJ, McMurray JJV, Yang J, Visscher PM, Newton-Cheh C, Malarstig A, Holm H, Lubitz SA, Sattar N, Holmes MV, Cappola TP, Asselbergs FW, Hingorani AD, Kuchenbaecker K, Ellinor PT, Lang CC, Stefansson K, Smith JG, Vasan RS, Swerdlow DI, Lumbers RT; Regeneron Genetics Center. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Privé F, Vilhjálmsson BJ, Aschard H, Blum MGB. Making the most of clumping and thresholding for polygenic scores. Am J Hum Genet 2019;105:1213—1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, Lin H, Arking DE, Smith AV, Albert CM, Chaffin M, Tucker NR, Li M, Klarin D, Bihlmeyer NA, Low S-K, Weeke PE, Müller-Nurasyid M, Smith JG, Brody JA, Niemeijer MN, Dörr M, Trompet S, Huffman J, Gustafsson S, Schurmann C, Kleber ME, Lyytikäinen L-P, Seppälä I, Malik R, Horimoto ARVR, Perez M, Sinisalo J, Aeschbacher S, Thériault S, Yao J, Radmanesh F, Weiss S, Teumer A, Choi SH, Weng L-C, Clauss S, Deo R, Rader DJ, Shah SH, Sun A, Hopewell JC, Debette S, Chauhan G, Yang Q, Worrall BB, Paré G, Kamatani Y, Hagemeijer YP, Verweij N, Siland JE, Kubo M, Smith JD, Van Wagoner DR, Bis JC, Perz S, Psaty BM, Ridker PM, Magnani JW, Harris TB, Launer LJ, Shoemaker MB, Padmanabhan S, Haessler J, Bartz TM, Waldenberger M, Lichtner P, Arendt M, Krieger JE, Kähönen M, Risch L, Mansur AJ, Peters A, Smith BH, Lind L, Scott SA, Lu Y, Bottinger EB, Hernesniemi J, Lindgren CM, Wong JA, Huang J, Eskola M, Morris AP, Ford I, Reiner AP, Delgado G, Chen LY, Chen Y-DI, Sandhu RK, Li M, Boerwinkle E, Eisele L, Lannfelt L, Rost N, Anderson CD, Taylor KD, Campbell A, Magnusson PK, Porteous D, Hocking LJ, Vlachopoulou E, Pedersen NL, Nikus K, Orho-Melander M, Hamsten A, Heeringa J, Denny JC, Kriebel J, Darbar D, Newton-Cheh C, Shaffer C, Macfarlane PW, Heilmann-Heimbach S, Almgren P, Huang PL, Sotoodehnia N, Soliman EZ, Uitterlinden AG, Hofman A, Franco OH, Völker U, Jöckel K-H, Sinner MF, Lin HJ, Guo X, Dichgans M, Ingelsson E, Kooperberg C, Melander O, Loos RJF, Laurikka J, Conen D, Rosand J, van der Harst P, Lokki M-L, Kathiresan S, Pereira A, Jukema JW, Hayward C, Rotter JI, März W, Lehtimäki T, Stricker BH, Chung MK, Felix SB, Gudnason V, Alonso A, Roden DM, Kääb S, Chasman DI, Heckbert SR, Benjamin EJ, Tanaka T, Lunetta KL, Lubitz SA, Ellinor PT; AFGen Consortium; METASTROKE Consortium of the ISGC, Neurology Working Group of the CHARGE Consortium. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet 2017;49:946—952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658—665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petersen SE, Aung N, Sanghvi MM, Zemrak F, Fung K, Paiva JM, Francis JM, Khanji MY, Lukaschuk E, Lee AM, Carapella V, Kim YJ, Leeson P, Piechnik SK, Neubauer S. Reference ranges for cardiac structure and function using cardiovascular magnetic resonance (CMR) in Caucasians from the UK Biobank population cohort. J Cardiovasc Magn Reson 2017;19:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thornton CA. Myotonic dystrophy. Neurol Clin 2014;32:705—719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gurung R, Ono Y, Baxendale S, Lee SLC, Moore S, Calvert M, Ingham PW. A zebrafish model for a human myopathy associated with mutation of the unconventional myosin MYO18B. Genetics 2017;205:725—735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahlberg G, Refsgaard L, Lundegaard PR, Andreasen L, Ranthe MF, Linscheid N, Nielsen JB, Melbye M, Haunsø S, Sajadieh A, Camp L, Olesen S-P, Rasmussen S, Lundby A, Ellinor PT, Holst AG, Svendsen JH, Olesen MS. Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat Commun 2018;9:4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Nieuwenhoven FA, Munts C, op’t Veld RC, González A, Díez J, Heymans S, Schroen B, van Bilsen M. Cartilage intermediate layer protein 1 (CILP1): a novel mediator of cardiac extracellular matrix remodelling. Sci Rep 2017;7:16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Norgett EE, Hatsell SJ, Carvajal-Huerta L, Cabezas JC, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP. Recessive mutation in desmoplakin disrupts desmoplakin–intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet 2000;9:2761—2766. [DOI] [PubMed] [Google Scholar]
- 43. Norman M, Simpson M, Mogensen J, Shaw A, Hughes S, Syrris P, Sen-Chowdhry S, Rowland E, Crosby A, McKenna WJ. Novel mutation in desmoplakin causes arrhythmogenic left ventricular cardiomyopathy. Circulation 2005;112:636—642. [DOI] [PubMed] [Google Scholar]
- 44. Herman DS, Lam L, Taylor MRG, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJR, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619—628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi SH, Weng L-C, Roselli C, Lin H, Haggerty CM, Shoemaker MB, Barnard J, Arking DE, Chasman DI, Albert CM, Chaffin M, Tucker NR, Smith JD, Gupta N, Gabriel S, Margolin L, Shea MA, Shaffer CM, Yoneda ZT, Boerwinkle E, Smith NL, Silverman EK, Redline S, Vasan RS, Burchard EG, Gogarten SM, Laurie C, Blackwell TW, Abecasis G, Carey DJ, Fornwalt BK, Smelser DT, Baras A, Dewey FE, Jaquish CE, Papanicolaou GJ, Sotoodehnia N, Wagoner DRV, Psaty BM, Kathiresan S, Darbar D, Alonso A, Heckbert SR, Chung MK, Roden DM, Benjamin EJ, Murray MF, Lunetta KL, Lubitz SA, Ellinor PT; For the DiscovEHR study and the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. Association between titin loss-of-function variants and early-onset atrial fibrillation. JAMA 2018;320:2354—2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, Shao W, Lu Z, Li H, Na J, Tang F, Wang J, Zhang YE, Shen X. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 2016;18:637—652. [DOI] [PubMed] [Google Scholar]
- 47. Moulik M, Vatta M, Witt SH, Arola AM, Murphy RT, McKenna WJ, Boriek AM, Oka K, Labeit S, Bowles NE, Arimura T, Kimura A, Towbin JA. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol 2009;54:325—333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dörr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Völker U, Völzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JCM, Kao WHL, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjögren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BHC, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kääb S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 2012;44:670—675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roston TM, Yuchi Z, Kannankeril PJ, Hathaway J, Vinocur JM, Etheridge SP, Potts JE, Maginot KR, Salerno JC, Cohen MI, Hamilton RM, Pflaumer A, Mohammed S, Kimlicka L, Kanter RJ, LaPage MJ, Collins KK, Gebauer RA, Temple JD, Batra AS, Erickson C, Miszczak-Knecht M, Kubuš P, Bar-Cohen Y, Kantoch M, Thomas VC, Hessling G, Anderson C, Young M-L, Choi SHJ, Cabrera Ortega M, Lau YR, Johnsrude CL, Fournier A, Van Petegem F, Sanatani S. The clinical and genetic spectrum of catecholaminergic polymorphic ventricular tachycardia: findings from an international multicentre registry. Europace 2018;20:541—547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gottdiener JS, Kitzman DW, Aurigemma GP, Arnold AM, Manolio TA. Left atrial volume, geometry, and function in systolic and diastolic heart failure of persons ≥65 years of age (The Cardiovascular Health Study). Am J Cardiol 2006;97:83—89. [DOI] [PubMed] [Google Scholar]
- 51. Orr N, Arnaout R, Gula LJ, Spears DA, Leong-Sit P, Li Q, Tarhuni W, Reischauer S, Chauhan VS, Borkovich M, Uppal S, Adler A, Coughlin SR, Stainier DYR, Gollob MH. A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat Commun 2016;7:11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bertelsen L, Diederichsen SZ, Haugan KJ, Brandes A, Graff C, Krieger D, Kronborg C, Køber L, Højberg S, Vejlstrup N, Svendsen JH. Left atrial volume and function assessed by cardiac magnetic resonance imaging are markers of subclinical atrial fibrillation as detected by continuous monitoring. Europace 2020;22:724—731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics are available for download at https://zenodo.org/record/5074929. Data generated in this study have been returned to the UKBB. The UKBB will make these data available to all researchers who apply by standard access procedures. Please see the UKBB’s website for access procedures (https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access).