Keywords: extracellular vesicles, MYC, MYCN, neuroblastoma, Warburg effect
Abstract
Amplification of the proto-oncogene MYCN is a key molecular aberration in high-risk neuroblastoma and predictive of poor outcome in this childhood malignancy. We investigated the role of MYCN in regulating the protein cargo of extracellular vesicles (EVs) secreted by tumour cells that can be internalized by recipient cells with functional consequences. Using a switchable MYCN system coupled to mass spectrometry analysis, we found that MYCN regulates distinct sets of proteins in the EVs secreted by neuroblastoma cells. EVs produced by MYCN-expressing cells or isolated from neuroblastoma patients induced the Warburg effect, proliferation and c-MYC expression in target cells. Mechanistically, we linked the cancer-promoting activity of EVs to the glycolytic kinase pyruvate kinase M2 (PKM2) that was enriched in EVs secreted by MYC-expressing neuroblastoma cells. Importantly, the glycolytic enzymes PKM2 and hexokinase II were detected in the EVs circulating in the bloodstream of neuroblastoma patients, but not in those of non-cancer children. We conclude that MYC-activated cancers might spread oncogenic signals to remote body locations through EVs.
1. Introduction
Neuroblastoma is a childhood malignancy originating from the peripheral nervous system that is still a leading cause of oncological death. About 50% of high-risk neuroblastomas are MYCN amplified and a causational link between MYCN activation in the nervous system and tumourigenesis has been firmly established [1,2]. The product of the MYCN amplicon is a transcription factor belonging to the MYC family of oncoproteins, frequently activated in neuroblastoma and other childhood malignancies [3].
In previous studies, it was shown that cancer cells expressing MYC proteins modify the tumour microenvironment via the activation or inactivation of growth factors, cytokines and immune checkpoint regulators [4–6]. The hypothesis that MYC expression could induce non-cell autonomous effects is also supported by evidence of heterogeneous (focal) amplification of MYCN in neuroblastoma tumours. Focally amplified neuroblastomas have a negative prognosis, indistinguishable from uniformly amplified cases, suggesting that MYCN could promote a cross-talk between parts of the cancer with a different amplification status [7]. We therefore hypothesized that MYCN could promote neuroblastoma growth by regulating the secretion or content of extracellular vesicles (EVs) which would modify the metabolic activity of recipient cells. EVs are formed by exosomes, of endocytic origin, and microvesicles, produced by membrane budding, that change the behaviour of recipient cells by the delivery of proteins and nucleic acids [8]. Cancer-produced EVs play a key role in the metastatic dissemination and/or pro-angiogenic activity of solid tumours [9]. While the pro-tumourigenic or pro-metastatic role of EVs has been highlighted in different types of adult cancers [10–16] it is not known whether they play a role in the context of MYCN-amplified neuroblastoma.
Using neuroblastoma as a model, we have therefore investigated the hypothesis that MYC-activated tumours could spread oncogenic signals to other body and tissue compartments via regulation of EVs protein cargo.
2. Material and methods
2.1. Clinical material
Plasma samples (10 NB patients with MYCN amplification and 10 neuroblastoma patients without MYCN amplification) were provided by the BIT-Gaslini Biobank of the IRCCS G. Gaslini Institute in Genova (Italy). The use of human biological specimens stored in the BIT-Gaslini Biobank was approved by the Ethical Committee of the Gaslini Institute. Plasma was collected by centrifuging blood at 1 200 x g for 10 min at room temperature and immediately stored at −80°C. Twenty age-matched plasma samples were obtained from leftover plasma after routine clinical analyses from outpatients. A written consent allowing the collection of samples and the use of clinical and non-genetic data for clinical research was signed by patients' guardians.
2.2. Cell culture, plasmid and siRNA transfections
The human neuroblastoma cell lines SH-EP and KELLY were purchased from the ATCC. TET21-N were kindly supplied by Prof Giovanni Perini. Neuroblastoma cells were cultured in DMEM supplemented with 10% FBS and 100 units ml−1 Penicillin/Streptomycin. TET21-N cells were routinely maintained in medium containing 0.2 mg ml−1 G-418 (11811031, GIBCO) and 0.15 mg ml−1 Hygromycin B (10453982, Invitrogen), and to switch off MYCN expression cells were cultured in the presence of 1 µg ml−1 doxycycline (D9891, Sigma-Aldrich). For EV preparation, cells were cultured in microvesicles depleted medium prepared as follows. In brief, DMEM supplemented with 20% FBS was ultra-centrifuged at 100 000 x g for 20 h. The pellet was discarded, the medium was diluted 1 : 2 with fresh DMEM and used. TET21-N cells were transfected using the transfection reagent jetPrime (114–01, Polyplus transfection) with 50 µM siRNAs targeting pyruvate kinase M2 (PKM2) (SASI_Hs01_00217689, EHU147561), Rab27a (EHU091501) or rLuc as a control (EHURLUC) purchased from Sigma-Aldrich. Forty-eight hours after transfection, the cells were collected and equal numbers were seeded on tissue culture inserts with 0.4 µm pores (83.3932.040, Sarstedt) for co-culture with SH-EP cells. The remaining cells were used for protein analysis. CD63-pEGFP C2 (62964) and pEGFP-C1-PKM2 (64698) plasmids were purchased from Addgene and transfected using the transfection reagent jetPrime.
2.3. Extracellular vesicle isolation and quantification
Cell supernatants were centrifuged at 3000 x g for 30 min and 100 000 x g for 70 min at 4°C. The pellet was resuspended in PBS and ultra-centrifuged at 100 000 x g for 70 min. The concentration of EV proteins was quantified with the bicinchoninic acid (BCA) protein assay (23225, Pierce). For EVs preparations from patients, 500 µl of frozen plasmas were processed using the Qiagen ExoEasy Maxi EVs isolation kit (76064, Qiagen). EVs were enumerated by lipophilic cationic dye staining and polychromatic flow cytometry, as described [17].
2.4. Transmission electron microscopy
EVs resuspended in PBS were fixed by mixing them in an equal volume of 4% paraformaldehyde (PFA). Glutaraldehyde was added to reach a final concentration of 1%. For negative staining, the fixed EVs were mixed with 1% sodium phosphotungstate (K-PTA), and a drop of this mixture was placed onto a carbon-coated electron microscope grid, glow discharged for 20 s. The mixture was left on the grid for 1 min and then removed almost completely with a filter paper.
2.5. Nanoparticle tracking analysis
The size distribution of EVs was quantified with a NanoSight N300 using a 640 nm laser (Malvern Instruments, Malvern). The samples were injected in the NanoSight sample cubicle and the data were analysed with the NanoSight NTA3.2 software.
2.6. PKH67 staining and Imagestream analysis
For the membrane staining of the EVs, the PKH67 green fluorescent cell linker mini kit (MINI67, Sigma-Aldrich) was used. Briefly, EVs pellets were resuspended in 200 µl diluent C; 0.8 µl of the PKH67 dye were added to 200 µl of diluent C and mixed with the resuspended EVs and incubated at room temperature for 4 min. The reaction was stopped with an equal volume (400 µl) of 1% BSA in PBS. The EVs were ultra-centrifuged at 100 000 x g at 4°C for 70 min and the pellet resuspended in PBS. The stained EVs were visualized with ImageStream flow cytometry. All samples were acquired on an ImageStreamX Mk II imaging cytometer, X60 magnification with low flow rate/high sensitivity without EDF using INSPIRE software. Channel 01 was set for bright field, channel 06 as a scattering channel and channel 02 for the required fluorescence channel (laser 488).
2.7. Proteomic analysis
The EVs preparations were resuspended in RIPA buffer (150 mM NaCl, 1 mM EDTA, 0.5% sodium deoxycolate, 0.1% SDS, Sigma) with freshly added protease and phosphatase inhibitors (Protease Inhibitor Cocktail, Sigma). After 30 min of mild sonication to extract proteins attached to the EV membranes, protein content was quantified by the Bradford method (BIO-RAD). Three aliquots of protein extracts from each condition (20 µg) were transferred onto Microcon-10 Centrifugal Filter (cut-off 10 kDa) for FASP protein digestion [18]. Proteins were reduced by 8 mM DTT and alkylated using 0.05 M iodacetamide. Each sample was then digested with trypsin overnight, after which digestions were blocked by adding formic acid to a final concentration of 0.2% (v/v). Peptides were recovered in 0.05 M AMBIC-SDS, concentrated in a speedvac and stored at −80°C until use. The digested peptides were loaded onto a Symmetry C18 (5 µm, 180 µm × 20 mm) precolumn (Waters Corp.) and subsequently separated by a 120 min reversed phase gradient at 300 nl min−1 (linear gradient, 2–85% ACN over 120 min) using a HSS T3 C18 (1.8 µm, 75 µm × 150 mm) nanoscale LC column (Waters Corp.) maintained at 40°C. The peptides were analysed by a High Definition Synapt G2-Si mass spectrometer directly coupled to the chromatographic system. Differential protein expression was evaluated with a data-independent acquisition of shotgun proteomics analysis by expression configuration mode (MSe). The mass spectrometer operated in ‘expression mode’ switching between low (4 eV) and high (15–40 eV) collision energies on the gas cell, using a scan time of 1.5 s per function over 50–2000 m/z. All spectra were acquired in ion mobility mode by applying a wave velocity for the ion separation of 900 m s−1 and a transfer wave velocity of 175 m s−1. The processing of low and elevated energy, added to the data of the reference lock mass ([Glu1]-Fibrinopeptide B Standard, Waters Corp) provided a time-aligned inventory of accurate mass-retention time components for both the low and elevated energy. Each sample was run in three technical replicates.
The analysis of differentially expressed proteins was performed according to Silva et al. [19] and Visser et al. [20]. Continuum LC-MS data from three replicates for each sample was processed for qualitative and quantitative analysis using the ProteinLynx Global Server v. 3.0.2 software (PLGS, Waters Corp.). Proteins were identified by searching the Homo Sapiens (UniProt KB/Swiss-Prot Protein reviewed) and ExoCarta databases to which sequence from Saccharomyces cerevisiae Enolase (UniProtKB/Swiss-Prot AC: P00924) was appended in order to perform the quantitative analysis. Search parameters were set as: automatic tolerance for precursor ions and for product ions, minimum three fragment ions matched per peptide, minimum three fragment ions matched per protein, minimum two peptides matched per protein, one missed cleavage, carbamydo methylation of cysteines and oxidation of methionines as fixed and variable modifications, false-positive rate of the identification algorithm under 1% and 100 fmol of the enolase set as calibration protein concentration. The expression analysis was performed considering technical replicates available for each experimental condition (i.e. untreated and treated with doxycycline) following the hypothesis that each group is an independent variable. The protein identifications were based on the detection of more than two fragment ions per peptide, more than two peptides measured per protein. The list of normalized proteins was screened according to the following criteria: protein identified in at least two out of three runs of the same sample with a fold change of regulation higher than ±20%; only modulated proteins with a p < 0.05 were considered significant.
A different EV preparation was re-suspended in lysis buffer (7 M urea, 2 M thiourea and protease inhibitors). Proteins were reduced with 10 mM DTT (Sigma-Aldrich) for 30 min at room temperature, alkylated with 40 mM iodoacetamide (Sigma-Aldrich) for 30 min in the dark and digested with trypsin (Promega) in the ratio 1 : 50 (μg trypsin : μg protein) in 50 mM ammonium bicarbonate solution at 30°C overnight. The reaction was stopped with 1% formic acid and then the sample was desalted with C18 columns (StageTips). Three biological replicates were prepared for each condition. Peptides were separated by Reprosil-Pur C18-AQ column (3 μm; Dr. Maisch GmbH, Germany) using Easy nano-LC HPLC (Proxeon, Odense, Denmark). The HPLC gradient was 0–34% B solvent (A = 0.1% formic acid; B = 90% ACN, 0.1% formic acid) in 70 min at a flow of 250 nl min−1 for a total of 93 min run. The MS analysis was performed using the LTQ-Orbitrap Velos (Thermo Scientific, Bremen, Germany). The mass range was 350–1500 m/z at a resolution of 60 000 at 400 m/z. For each MS scan, collision-induced dissociation (CID) fragmentation was performed on the 15 most intense ions in the linear ion trap. The parameters for data acquisition were activation time = 5 ms, normalized energy = 35, Q-activation = 0.25, exclusion = available with repeat count 1, exclusion duration = 30 s and intensity threshold = 5000. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE repository on https://www.ebi.ac.uk/pride/. The raw files were searched using the MaxQuant version 1.2.7.429 and the MS/MS spectra were searched using the Andromeda search engine against the Uniprot-reviewed Human Protein Database. The initial maximal allowed mass tolerance was set to 20 ppm for precursor and then set to 4.5 ppm in the main search and to 0.5 Da for fragment ions. Enzyme specificity was set to trypsin with a maximum of two missed cleavages. Carbamidomethylation of cysteine (57.02 Da) was set as a fixed modification, and oxidation of methionine (15.99 Da) and protein N-terminal acetylation (42.01 Da) were selected as variable modifications. Bioinformatics analysis was performed using the software Perseus v. 1.5.2.6 available in the MaxQuant environment and reverse and contaminant entries were excluded from further analysis. Protein FDR was calculated in the MaxQuant software and kept below 1%. Label-free quantification intensity values were considered to relatively compare the abundance of proteins present in the two conditions and regulated proteins were identified with statistical t-test with p-value < 0.05 with Benjamini–Hochberg correction.
2.8. Western blot analysis
Equal amounts of proteins were separated by SDS-PAGE electrophoresis, transferred onto nitrocellulose membranes which were blocked with 5% milk in TBS with 0.1% Tween-20 for 1 h at room temperature. The membranes were incubated over night at 4°C with primary antibodies against CD63 (1 : 200, sc-5275 Santa Cruz), eukaryotic elongation factor 2 (eEF2) (1 : 1 000, 2332, Cell Signalling), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1 : 1000, 5174S, Cell Signalling), Hexokinase I (1 : 1000, C35C4, Cell Signalling), Hexokinase II (1 : 100, C64G5, Cell Signalling), MYCN (1 : 200, sc-53993, Santa Cruz), phosphofructokinase PFKP (1 : 1 000, 8164S, Cell Signalling), PKM1/2 (1 : 1 000, 3190S, Cell Signalling), PKM2 (1 : 1 000, 4053S, Cell Signalling), rib. L10a (1 : 500, sc-100827, Santa Cruz) and β-actin (1 : 1000, 3700, Cell Signalling). After washing with TBS 0.1% Tween-20 three times for 5 min at room temperature, the membranes were incubated with horseradish peroxidase (HRP)—conjugated secondary antibodies for 1 h at room temperature (anti-mouse-HRP, 1 : 10 000, sc-2031, Santa Cruz, anti-rabbit-HRP, 1 : 10 000, sc-2313, Santa Cruz). The membranes were washed three times for 5 min at room temperature with TBS 0.1% Tween-20 and incubated with enhanced chemiluminescent substrate (32106, Pierce). The protein bands were visualized with X-ray films.
2.9. Cell counts and MTS proliferation assays
2 × 103 SH-EP cells were plated into each well of 96-well plates in the presence or absence of EVs that were replenished every day (10 µg of EV proteins per well). Cells were harvested at 24, 48 or 72 h and counted with a haemocytometer. For MTS assays, 20 µl of MTS/PMS solution (G3582, Promega) were added to each well and incubated for 4 h. Absorbance at 490 nm was measured with a spectrophotometer and expressed as optical density units.
2.10. Glycolysis assays
2 × 103 SH-EP cells were seeded in each well of 96-well plates and incubated overnight in a 5% CO2-humidified incubator at 37°C. The next morning, EVs were added to the cells (10 µg of EV proteins per well). After 24 h, the production of L-lactate was evaluated with the glycolysis cell-based assay kit (600450, Cayman Chemical). Briefly, the cell supernatants were centrifuged at 1 200 rpm for 5 min; 90 µl of assay buffer were transferred to each well of a 96-well plate plus 10 µl of cell supernatants or 10 µl of L-lactate solution used as a positive control. A 100 µl of reaction solution was added to each well and the mix was incubated for 30 min at room temperature with gentle shaking. The absorbance was measured at 490 nm using a spectrophotometer. For the SH-SY5Y cells, the consumption rate of glucose was quantified using a Glucose Uptake Colorimetric Assay Kit (Sigma-Aldrich, MAK083) according to manufacturer's instructions.
2.11. Seahorse analysis
Mitochondrial bioenergetics assays were carried out using an XFe96 analyser from Agilent Seahorse Bioscience (Santa Clara) following published protocols [21]. The XF assay medium (HCO3-free modified DMEM, Seahorse Bioscience) was supplemented with 10 mM glucose, 2 mM l-glutamine and 1 mM sodium pyruvate. 15 × 103 cells were grown in each well of Seahorse assay plates. After 4 h, at the step of Seahorse medium exchange, 100 µg ml−1 of EVs from MYCN-positive or -negative TET21-N cells or PBS were added and the cells where incubated for 20 h. The mitochondrial respiration test was performed by sequential addition of μM oligomycin, 1.5 µM FCCP and 1 µM rotenone/antimycin A to the cells. Using the XF cell Mitostress test kit (103015–100; Seahorse Bioscience), we examined the following mitochondrial parameters: basal mitochondrial respiration (basal cellular respiration minus non-mitochondrial respiration), maximal respiratory capacity (maximal uncoupled respiration minus non-mitochondrial respiration) and non-mitochondrial respiration (rotenone/antimycin A-inhibited respiration).
2.12. Immunofluorescence
The cells were cultured on coverslips or in microwells (154534, Thermo Scientific). The cells were fixed in 4% PFA for 10 min at room temperature. After washing with PBS for three times, the cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. After washing with PBS for three times, cells were blocked with 5% BSA in PBS for 30 min at 37°C. The cells were incubated with the primary antibody against p-H3 (T11) (1 : 100, ab5168, Abcam) or cMYC (1 : 800, 5605, Cell Signalling) diluted in 3% BSA in PBS for 30 min at 37°C. After washing with PBS for three times, the cells were incubated with anti-rabbit FITC secondary antibody (711-095-152, Jackson ImmunoResearch) diluted 1 : 200 in BSA 1% in PBS for 30 min at 37°C. One drop of Vectashield antifade mounting medium, containing DAPI, (H-1200, Vector Laboratories) was added to each sample before analysis. Fiji software was used for the quantification of the intensity.
2.13. Immunohistochemistry
Tumour sections were incubated in HistoChoice clearing agent (H2779, Sigma Aldrich) to remove the wax and then sequentially in 100%, 90%, 70% and 50% ethanol. In the final step, the sections were incubated in dH2O twice for 5 min. Antigens were unmasked by boiling the sections in citrate solution in a microwave oven. Endogenous peroxidase was removed with incubation in 3% H2O2 (216763, Sigma Aldrich) in distilled H2O for 20 min at RT. The sections were next incubated with mouse on mouse (MOM) mouse IgG blocking reagent (BMK-2202, Vector Laboratories) for 1 h at room temperature. After blocking, the sections were washed in PBS, incubated for 5 min in MOM diluent (BMK-2202, Vector Laboratories), the excess of diluent was removed and the sections were incubated with mouse anti-MYCN (1 : 100, OP13, Calbiochem) or anti-c-MYC (1 : 50, sc-40, Santa Cruz) primary antibodies diluted in blocking buffer 1 : 10 and incubated overnight at 4°C. The sections were washed in PBS 1X twice for 2 min and then incubated with biotinylated anti-mouse IgG reagent (BMK-2202, Vector Laboratories) for 10 min followed by incubation with Avidin, NeutrAvidin HRP conjugate (A2664, Molecular Probes) diluted 1 : 1000 in PBS for 2 h at room temperature. One drop of ImmPACT DAB chromogen (SK-4105, Vector Laboratories) was diluted in 1 ml of ImmPACT DAB diluent and applied on the sections for 2–10 min until desired colour intensity was developed. After staining with hematoxylin (H3136, Sigma Aldrich), the sections were dehydrated and mounted with a glass coverslip using one drop of DPX mountant (O6522, Sigma Aldrich).
2.14. Statistical analysis
All data are expressed as means ± s.e.m. Statistical tests used were comparison of means with two-tailed paired samples or independent samples t-test, as indicated in the figure legends; p-values less than 0.05 were considered significant. All experiments were replicated independently and the number of replicates is indicated in the figure legends. All statistical tests were performed with the IBM SPSS software.
3. Results
3.1. Extracellular vesicles secreted by neuroblastoma cells with activated MYCN are enriched in oncogenic glycolytic enzymes
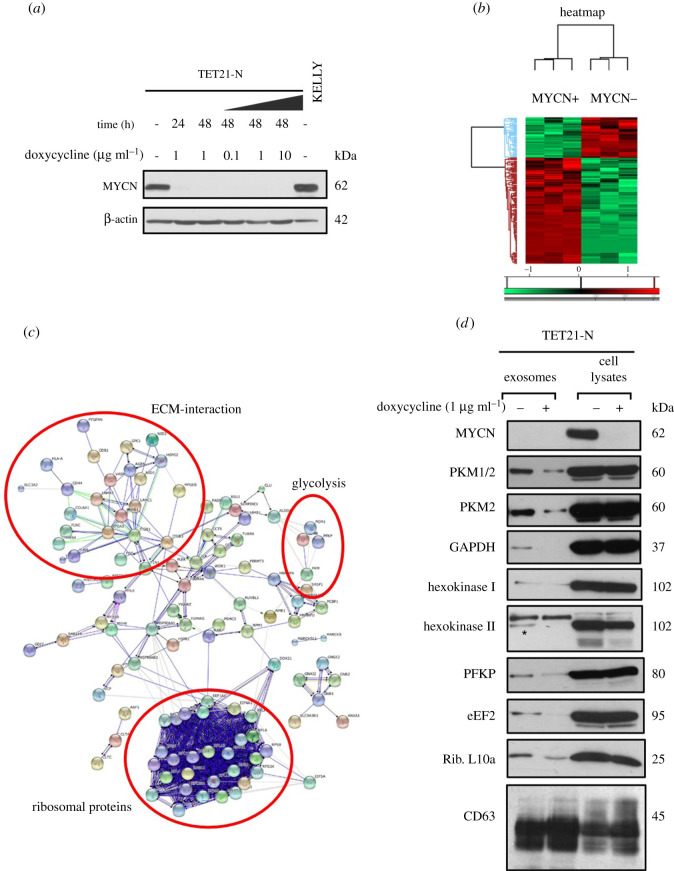
The relationship between MYCN expression and EV protein cargo was investigated using the TET21-N neuroblastoma cell line. TET21-N have been stably transfected with a MYCN, doxycycline-repressible and expression vector [22]; in the absence of doxycycline, MYCN protein is constitutively expressed (figure 1a). The supernatant of TET21-N cells, expressing or non-expressing MYCN, was collected and EVs were purified by differential ultracentrifugation. Nanoparticle tracking analysis and transmission electron microscopy confirmed the presence of exosome-like EVs with an average diameter of 100–200 nm (electronic supplementary material, figure S1A, B). The membrane integrity of the vesicles preparations was validated by staining with the membrane-specific dye PKH67 and observing the EVs with ImageStream flow cytometry (electronic supplementary material, figure S1C). The classical EV marker CD63 was used to validate the vesicles preparation (electronic supplementary material, figure S1D). Subsequently, a label-free quantitative mass spectrometry-based proteomic analysis was performed on EVs isolated from TET21-N cells expressing or non-expressing MYCN. In total, 152 EV proteins were differentially regulated between the two conditions with 111 proteins upregulated and 41 proteins downregulated upon MYCN expression (figure 1b; electronic supplementary material, Dataset S1). Pathway analysis suggests that glycolytic enzymes, ribosomal and extracellular matrix (ECM) interaction proteins were mostly enriched in the EVs secreted by the MYCN-positive cells (figure 1c). The differential expression of proteins implicated in glycolysis was validated by western blotting. PKM2, hexokinase I and hexokinase II were upregulated in the EVs secreted by TET21-N cells expressing MYCN compared to the condition in which MYCN gene expression was switched off. Other proteins linked to metabolism such as eEF2, PFKP, GAPDH and ribosomal protein L10a were also confirmed to be enriched in the EVs secreted by MYCN-positive neuroblastoma cells (figure 1d). The expression levels of vesicular proteins did not reflect those observed in the corresponding cellular lysates (figure 1d), indicating that MYCN might control their loading into EVs.
Figure 1.
MYCN expression regulates EVs protein cargo in neuroblastoma cells. (a) Western blot analysis demonstrating the conditional expression of MYCN in TET21-N cells in the presence (MYCN-off) or absence (MYCN-on) of doxycycline. Protein lysates from the MYCN-amplified cell line KELLY were used as a positive control. β-actin was used as a loading control. (b) Heat map analysis showing a drastic difference in protein expression—either upregulated (red) or downregulated (green)—in EVs secreted by TET21-N cells expressing or non-expressing MYCN. Mass spectrometry analysis was conducted in triplicate samples (biological replicates). (c) Pathway analysis of the proteins identified in EVs from MYCN-positive and -negative TET21-N cells. The protein groups enriched in the MYCN-positive condition are evidenced by the red circles. (d) Western blot analysis of EVs and total cell lysates isolated from the TET21-N cells. The EV marker CD63 was used as a loading control. An asterisk indicates the hexokinase II protein band at 105 kD, whereas the 125 kD band enriched in vesicles is of spurious nature.
3.2. Vesicles circulating in the plasma of neuroblastoma patients contain pyruvate kinase M2 and hexokinase II
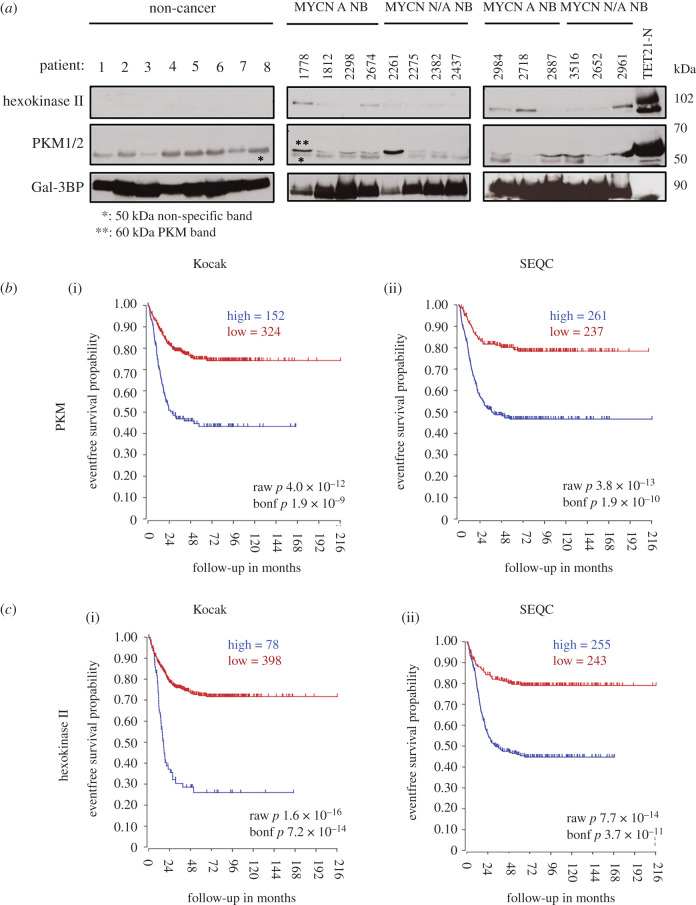
Since PKM2 and hexokinase II are glycolytic enzymes with oncogenic potential [23], detectable in the EVs of neuroblastoma cells, we wondered whether we could also detect glycolytic enzymes in the EVs circulating in the plasma of patients bearing neuroblastomas. We therefore isolated EVs from the plasmas of neuroblastoma or non-cancer patients, which were used as controls. We detected vesicles of sizes similar to those detected in the supernatant of neuroblastoma cells, which were positive to the CD63 marker (electronic supplementary material, figure S2A, B). To further validate the plasma vesicles preparations, we used an antibody against galectin-3-binding protein (Gal-3BP), which was shown to detect exosomes secreted by neuroblastoma cells [24] (electronic supplementary material, figure S2C). Remarkably, PKM2 and hexokinase II were detected in the EVs isolated from plasma samples of neuroblastoma patients, but were completely absent in the vesicles of non-cancer patients (figure 2a). The expression of the enzymes was similar in vesicles preparations from patients with MYCN-amplified or non-amplified tumours. However, MYCN non-amplified tumours often present high levels of expression of MYC (c-MYC) [25], which might explain why PKM2 and hexokinase II levels were similar in vesicles from the two groups of patients. We determined that the prominent 50 kD protein band detected in all plasma samples is of spurious nature (electronic supplementary material, figure S2D). The expression of the PKM and HK2 genes strongly correlates with poor patient prognosis, suggesting that their products might confer a growth or survival advantage to neuroblastoma cells (figure 2b,c). We analysed EVs secreted from multiple neuroblastoma cell lines to confirm that glycolytic enzymes with oncogenic potential were enriched in MYC-expressing cells. EVs or total cell lysates were prepared from MYCN-amplified (LAN5, Kelly) and non-MYCN-amplified (SH-SY5Y, SH-EP and GIMEN) neuroblastoma cell lines. The MYCN non-amplified cell lines, as expected, did not display detectable levels of MYCN protein. However, SH-SY5Y and SH-EP expressed high levels of MYC (c-MYC), whereas GIMEN cells did not express either MYC family members (electronic supplementary material, figure S3A). Consistent with the hypothesis that MYC family members regulate EV protein cargo, vesicles derived from MYC/MYCN-positive cell lines were enriched for the glycolytic enzymes hexokinase II and PKM2 in comparison to MYC-negative GIMEN cells (electronic supplementary material, figure S3B).
Figure 2.
Analysis of vesicular proteins in the plasma of neuroblastoma patients. (a) The indicated proteins were identified in the cargo of EVs isolated from the plasma of patients with (MYCN A NB) or without (MYCN N/A NB) MYCN-amplified neuroblastomas by western blot analysis. Plasmas from non-cancer patients were used as controls. Gal-3BP was used as loading control and total cell lysates from TET21-N cells were used as a positive control for PKM and hexokinase II. * indicates non-specific band; ** indicates specific band. Kaplan–Meier survival curve correlating the expression of PKM (b) and hexokinase II (c) with event-free neuroblastoma patients' survival in the Kocak (i) and SEQC (ii) datasets available in the R2 genomic visualization platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
3.3. Extracellular vesicles secreted by MYCN-positive cells induce glycolysis, increase respiration capacity, ATP production and proliferation of recipient cells
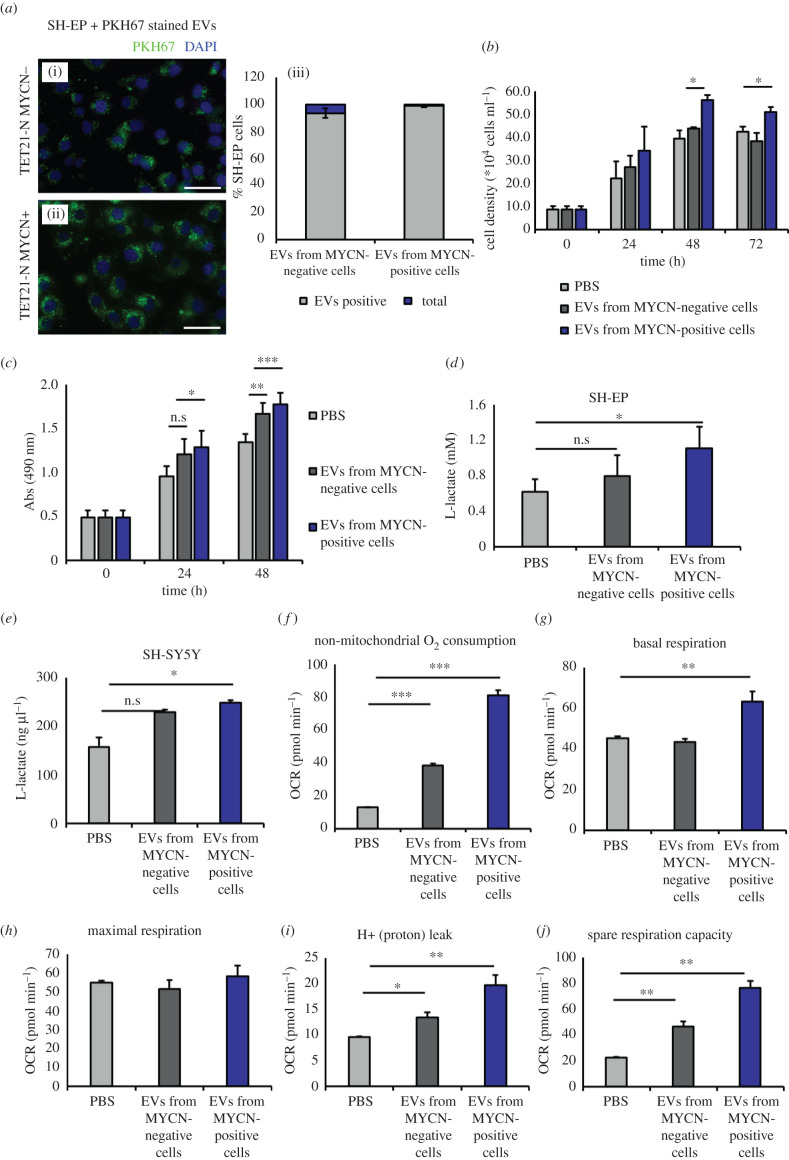
To assess the biological function of EVs in recipient cells, we used the neuroblastoma cell line SH-EP, which is stromal-type, therefore simulating the Schwann stroma surrounding the nests of highly proliferating neuroblasts in tumours. SH-EP cells were incubated with EVs purified from TET21-N neuroblastoma cells in MYCN-off or MYCN-on conditions. Vesicles were also stained with PKH67 and the ability of SH-EP cells to incorporate them was confirmed by fluorescence microscopy. The fluorescent EVs were mainly localized in the cytoplasm of recipient cells (figure 3a). To investigate whether the incorporation of EVs had biological consequences, we exposed SH-EP cells to EVs secreted by TET21-N cells in induced or un-induced conditions and measured cell proliferation and metabolic activity. Neuroblastoma cells receiving EVs produced by MYCN-activated cells were stronger inducers of cell proliferation and metabolism compared to EVs produced by MYCN non-expressing cells (figure 3b,c). Since glycolytic enzymes were particularly enriched in EVs produced by MYCN-activated cells, we also measured the glycolytic activity of recipient cells. SH-EP cells were incubated with EVs for 24 h and we observed that the secretion of L-lactate was significantly increased in the presence of EVs produced by MYCN-expressing cells (figure 3d). This effect was also observed in SH-SY5Y cells, a MYCN single-copy cell line (figure 3e). Non-mitochondrial oxygen consumption, basal respiration, maximum respiration, spare respiration capacity, proton leak and ATP production were also significantly increased in SH-SY5Y cells upon incubation with EVs secreted from MYCN-positive cells (figure 3f–j). Taken together, these experiments suggest that EVs secreted by MYCN-positive cells increase the proliferation rates and metabolic activity of stromal and MYCN single-copy neuroblastoma cells by inducing a Warburg switch.
Figure 3.
(Overleaf.) EVs secreted by MYCN-expressing cells promote glycolysis, respiration, ATP production and proliferation of recipient neuroblastoma cells. (a) Fluorescence microscopy of SH-EP cells after 24 h incubation with PKH67-labelled EVs isolated from TET21-N cells expressing or not expressing MYCN. Quantification of the percentages of cells positive to the dye in different conditions is shown on (iii). Error bars indicate mean values ± s.e.m. Scale bar 25 μm. (b) SH-EP cells were counted at 1 day intervals after being cultured in the presence or absence of EVs purified from MYCN-positive or -negative TET21-N cells (n = 3). (c) SH-EP cells were subjected to MTS assays 24 or 48 h after being cultured in the presence or absence of EVs purified from MYCN-positive or -negative TET21-N cells (n = 5). (d) L-lactate production in SH-EP (n = 4) and (e) SH-SY5Y (n = 3) neuroblastoma cells incubated for 24 h with EVs isolated from TET21-N cells with and without MYCN expression. (f–j) Seahorse analysis. MYCN single-copy cell line SH-SY5Y was used as a recipient for EVs purified from TET21-N cells with or without MYCN (n = 3). PBS was used as a control (untreated). OCR indicates oxygen consumption rate. Error bars represent mean values ± s.e.m. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
3.4. Neuroblastoma extracellular vesicles induce increased mitotic index and histone H3 (T11) phosphorylation in a pyruvate kinase M2-dependent manner
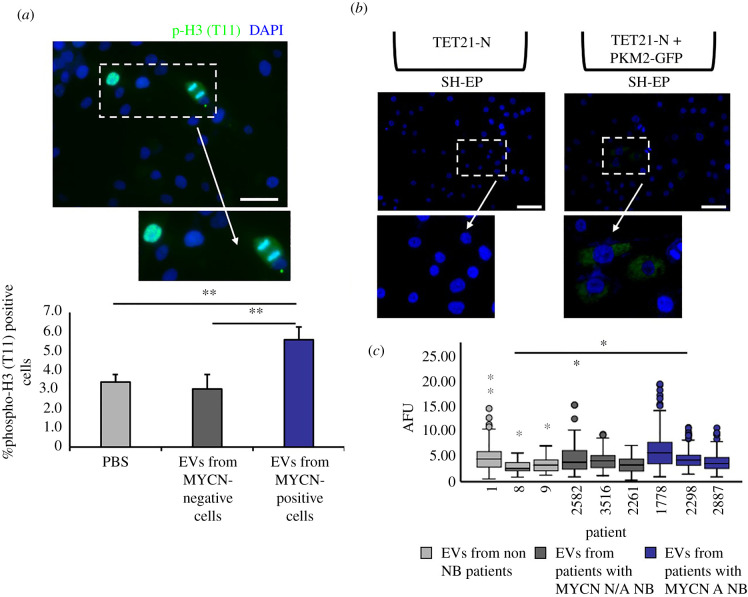
In addition to its role in glycolysis, PKM2 has been shown to cause epigenetic modifications by entering the nucleus and phosphorylating histone H3 on threonine 11. This results in the activation of signalling, transcription of target genes, such as cyclin D1 and c-MYC, and increased cell cycle progression [26]. Thus, we hypothesized that EVs enriched in PKM2 could induce phosphorylation of histone H3 in recipient cells. Indeed, histone H3 (T11) phosphorylation was increased in SH-EP cells incubated with EVs purified from MYCN positive, but not negative, cells (figure 4a). To study the role of EV PKM2 in a more physiologically relevant setting, we used a co-culture chamber system in which induced or un-induced TET21-N cells were separated by 0.4 µm pores tissue culture inserts from recipient SH-EP cells (electronic supplementary material, figure S4A). The transfer of EVs from donor to recipient cells was confirmed by transfecting TET21-N cells with a construct expressing a CD63-GFP fusion protein (electronic supplementary material, figure S4B), and observing the GFP signal in the SH-EP recipient cells after 24 h (electronic supplementary material, figure S4C). Using the same methodology, we confirmed that PKM2 is delivered to SH-EP cells co-cultured with TET21-N cells transfected with a construct expressing a PKM2-GFP fusion protein (figure 4b). To increase the clinical relevance of these results, we investigated whether EVs isolated from the plasmas of neuroblastoma patients could also induce phosphorylation of histone H3 on threonine 11, focusing on interphase cells. The intensity of histone H3 phosphorylation in interphase SH-EP cells was statistically significantly increased after incubation with EVs purified from patients with MYCN-amplified neuroblastomas, compared to patients with non-MYCN-amplified tumours or children without cancer, supporting the results obtained with the cell line systems (figure 4c).
Figure 4.
EVs produced by MYCN-expressing neuroblastoma cells induce histone H3 (T11) phosphorylation. (a) SH-EP cells were cultured for 24 h in the presence of EVs purified from the supernatants of MYCN-positive or -negative TET21-N cells and stained with an antibody against phospho-histone 3 (T11) (n = 4). Scale bar, 50 µm. (b) Fluorescence imaging of SH-EP cells after 24 h co-culture with TET21-N cells (uninduced, MYCN-on) overexpressing PKM2-GFP. Scale bar, 75 μm. (c) SH-EP cells were treated with EVs isolated from plasmas of neuroblastoma or non-cancer patients and stained with an antibody against phospho-histone 3 (T11). The intensity of the staining expressed as arbitrary fluorescence units (AFU) is displayed in the box plot and the statistical significance of the differences in intensities among groups was verified using an independent-samples t-test. Lavene's test was used to verify the assumption of equal variance. N/A NB indicates non-MYCN-amplified neuroblastoma (n = 3). Error bars represent mean values ± s.e.m. *p ≤ 0.05, **p ≤ 0.01.
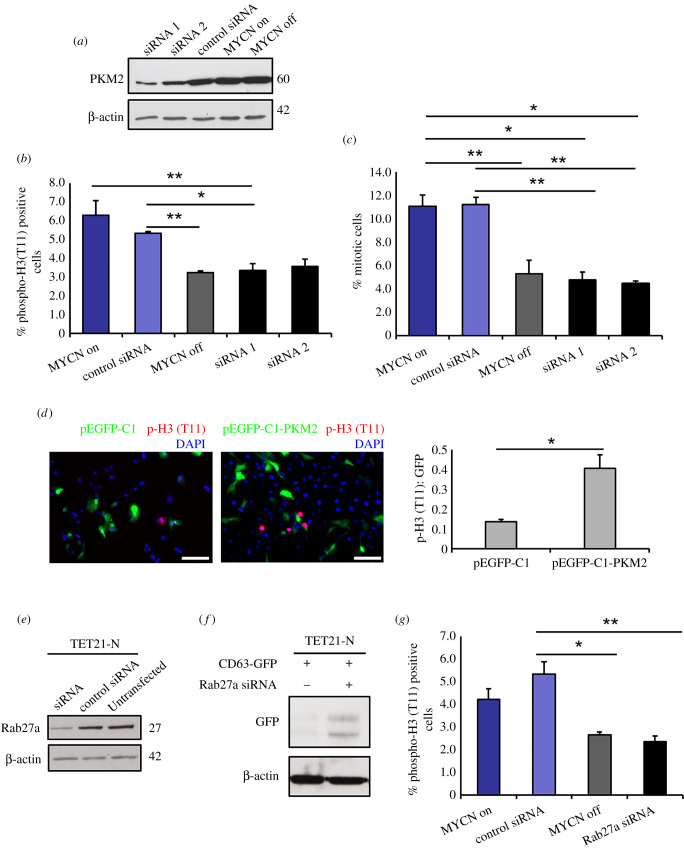
3.5. Vesicular trafficking of pyruvate kinase M2 is required for the increased levels of histone H3 phosphorylation in recipient cells following activation of MYCN in donor cells
To link the biological effects observed in recipient SH-EP cells to the transfer of vesicular PKM2, we used two independent siRNAs that caused a reduction of PKM2 expression in donor TET21 N cells (figure 5a). Histone H3 phosphorylation in SH-EP cells was significantly downregulated in the presence of the PKM2 siRNAs or when the expression of MYCN was turned off in donor cells, strongly suggesting that the transfer of PKM2 is essential to induce histone H3 phosphorylation in recipient cells (figure 5b). Since histone H3 phosphorylation is tightly linked to mitosis, we calculated the mitotic index of SH-EP cells cultured in the different conditions and verified that the mitotic index was high when MYCN was activated, but reduced to baseline levels when MYCN was switched off or PKM2 was inhibited by siRNAs in donor cells (figure 5c). To further demonstrate that the increase of histone H3 phosphorylation was caused by the transfer of PKM2 in the recipient cells, we transfected SH-EP cells with plasmids encoding GFP or PKM2-GFP, and 48 h later, we assessed phospho-histone H3 (T11). There was a significant increase of histone H3 (T11) phosphorylation in cells transfected with PKM2-GFP compared to GFP control transfection, demonstrating the direct role of the kinase in this effect (figure 5d). To ascertain whether the induction of histone H3 phosphorylation was mediated by exosomes or exosome-like EVs, we used siRNAs to downregulate Rab27a, which were previously shown to inhibit exosome secretion [27], in TET21-N cells (figure 5e). Rab27a silencing caused intracellular accumulation of the tetraspanin and EV marker CD63 fused to GFP, suggesting that exosomes were not released in the extracellular space in this setting (figure 5f). Silencing Rab27a phenocopied the loss of PKM2 expression in co-culture experiments, demonstrating that vesicular transfer of PKM2 is required to induce histone H3 phosphorylation in recipient cells (figure 5g).
Figure 5.
EVs produced by MYCN-expressing neuroblastomas induce mitotic index and histone H3 (T11) phosphorylation in a PKM2-dependent manner. (a) Western blot analysis demonstrating the downregulation of PKM2 expression by two different siRNAs. An actin antibody was used as loading control. (b) siRNA knock-down of PKM2 in TET21-N donor cells inhibits histone H3 phosphorylation in recipient cells (n = 3). (c) Mitotic index. The mitotic index is expressed as the percentage of cells in mitosis (prophase, metaphase, anaphase and telophase) over the total number of cells (n = 3). (d) SH-EP cells were transfected with pEGFP-C1 or pEGFP-C1-PKM2 and stained with an antibody against phospho-histone 3 (T11) after 48 h. The number of transfected and phospho-histone H3 (T11) positive cells was quantified in each condition (n = 3). Scale bar, 100 μm. (e) Western blot analysis demonstrating downregulation of Rab27a expression by siRNA. An actin antibody was used as loading control. (f) Western blot analysis of CD63-GFP overexpressing cells in the presence or absence of the anti-Rab27a siRNA. An actin antibody was used as loading control. (g) siRNA knock-down of Rab27a in TET21-N donor cells inhibits histone H3 phosphorylation in recipient cells (n = 3). Error bars represent mean values ± s.e.m. *p ≤ 0.05, **p ≤ 0.01.
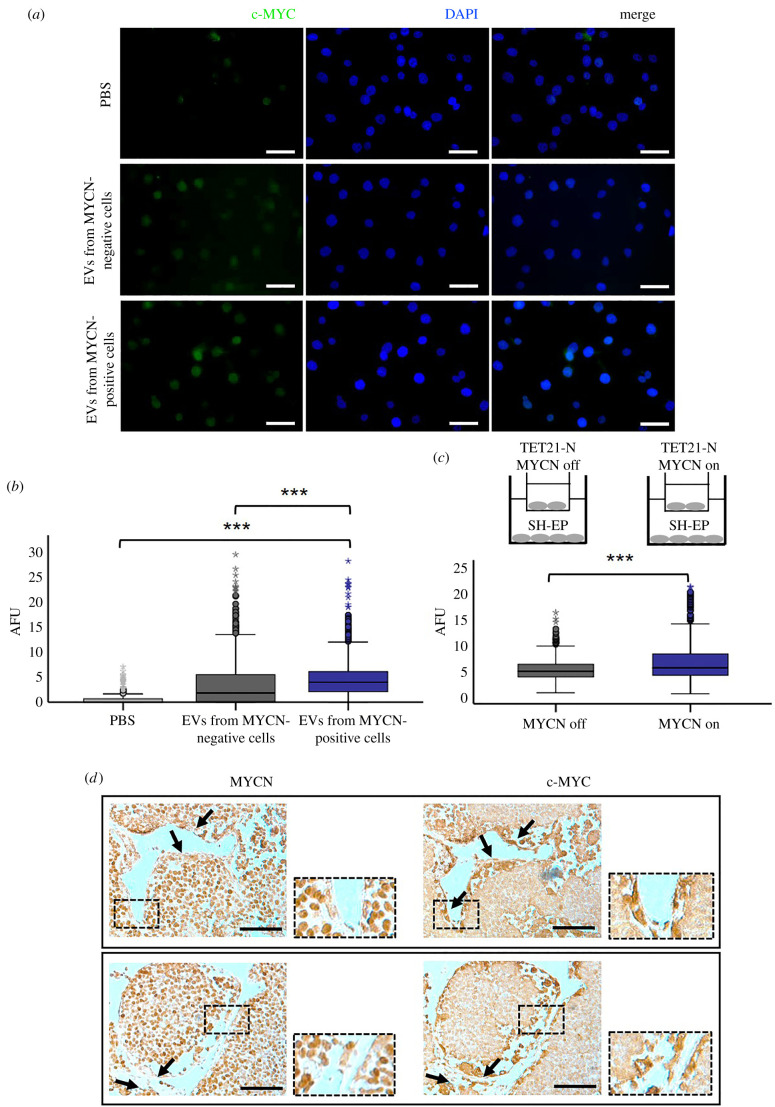
3.6. MYCN-regulated extracellular vesicles activate c-MYC expression in neuroblastoma stromal cells
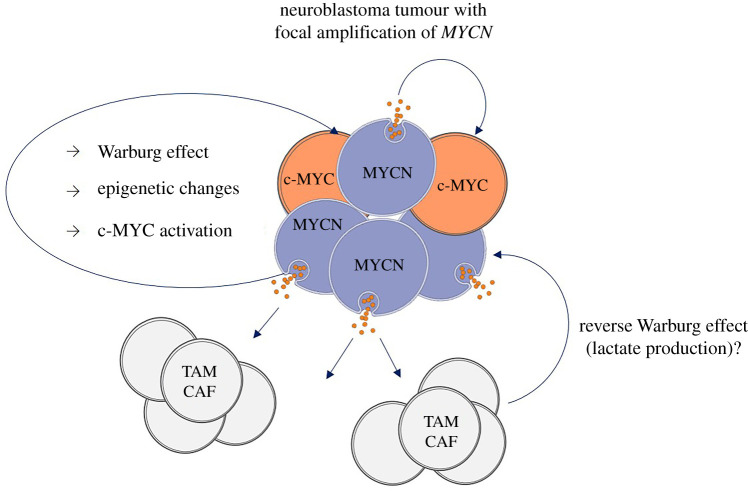
A consequence of epigenetic modification of histones induced by nuclear PKM2 is activation of c-MYC [26]. We therefore quantified c-MYC expression by immunofluorescence analysis in SH-EP cells exposed to purified EVs or co-cultured in transwell chambers with TET21-N cells in induced or un-induced conditions. EVs purified from TET21-N cells with activated MYCN induced higher expression of c-MYC than EVs purified from cells in which MYCN expression has been switched off (figure 6a,b). Similarly, c-MYC expression was significantly increased in SH-EP cells co-cultured with TET21-N cells with MYCN switched on, compared to the condition in which MYCN was switched off (figure 6c). In a syngeneic mouse model of neuroblastoma (TH-MYCN), c-MYC expression is detected in stromal cells surrounding nests of MYCN-positive neuroblastomas (figure 6d). Overall, these experiments corroborate the hypothesis that the transfer of EVs from MYCN-positive cells induces activation of c-MYC in recipient stromal cells. A cartoon illustrating how the MYCN-regulated vesicles could change the tumour microenvironment is shown in figure 7.
Figure 6.
MYCN-regulated EVs activate c-MYC expression in recipient neuroblastoma cells. (a) Immunofluorescence analysis with an antibody against c-MYC was carried out in SH-EP cells co-cultured for 24 h with EVs purified from supernatants of TET21-N cells expressing or non-expressing MYCN. PBS was used as a control. Scale bar, 50 µm. (b) Quantification of the experiment shown in (a) (n = 3). (c) Quantification of c-MYC immunofluorescence staining in SH-EP cells after 24 h co-culture with TET21-N cells expressing or non-expressing MYCN (n = 3). ***p ≤ 0.001. (d) Stromal cells surrounding MYCN-positive tumour nests express c-MYC. Neuroblastoma tumours developing in TH-MYCN mice were fixed, embedded in paraffin and serial sections stained with MYCN or c-MYC antibodies. Arrows indicate stromal cells negative for MYCN but expressing c-MYC. Scale bar, 75 µm.
Figure 7.
Cartoon illustrating a hypothetical model of neuroblastoma vesicles induced regulation of the tumour microenvironment. MYCN-amplified cells spread vesicles loaded with proteins that mediate oncogenic signals in MYCN non-amplified parts of the cancer and cells of the tumour microenvironment. TAM = tumour-associated macrophage. CAF = cancer-associated fibroblast.
4. Discussion
The role of MYC as a regulator of the tumour microenvironment, in addition to its intrinsic effects on cancer cells, is emerging. For example, c-MYC has been shown to alter immune cells metabolism and the cancer microenvironment by supporting the expression of HIF-1 and secretion of VEGF in tumour cells [28]. In neuroblastomas, MYCN amplification is conducive to reduce immune infiltration [29,30] and neuroblastoma cells have been shown to secrete macrovesicles able to stimulate the production of pro-tumourigenic signals by bone marrow stromal cells [31]. These studies prompted us to investigate the hypothesis that MYCN could exert non-cell autonomous effects by regulating EVs.
Using mass spectrometry analysis, we have identified MYCN-regulated vesicular proteins mainly clustered in three functional groups: ribosomal proteins, proteins involved in ECM interactions and glycolytic enzymes. We focused our attention on the latter group since it is well established that MYC proteins are regulators of cell metabolism and the Warburg effect. The regulation of metabolism is critical for the function of cellular processes and aggressive cancer cells are able to adapt their metabolism in a way that sustains tumour growth and metastatic dissemination. While most differentiated cells use mitochondrial oxidative phosphorylation as their main source of energy production, cancer cells shift towards aerobic glycolysis by increasing their glucose uptake and metabolizing it to lactate in aerobic conditions, a process known as the Warburg effect [32,33]. A number of enzymes implicated in the glycolytic pathway have been shown to be overexpressed in multiple cancers [34]. The M2 splice isoform of pyruvate kinase is increased in cancer with respect to the M1 variant, mainly found in normal tissues, and contributes to the metabolic adaptations required in tumourigenesis [35,36]. PKM2 has been shown to be modified by acetylation by p300, which transforms it from a cytoplasmic metabolic kinase to a nuclear protein kinase [37,38]. In glioma, PKM2 mediates phosphorylation of histone H3 on threonine 11, resulting in the transcriptional activation of c-MYC, cyclin D1 and tumour progression [26]. In addition, PKM2 regulates chromosome segregation and mitosis in cancer by interacting with the spindle checkpoint protein Bub3 [39].
MYC regulates lactate production, glutaminolysis, and increase protein translation by promoting ribosome biogenesis [40]. MYCN is also involved in metabolic reprogramming of neuroblastoma cells by enhancing glycolytic metabolism [41]. Among the glycolytic enzymes enriched in the EVs of MYCN-expressing neuroblastoma cells, we identified hexokinase II and PKM2. Notably, we detected the presence of hexokinase II and PKM2 in EVs circulating in the bloodstream of neuroblastoma patients and more frequently in those bearing MYCN-amplified tumours. The glycolytic enzymes were undetectable in the EVs of non-cancer patients, supporting the hypothesis that they are of cancer origin. Given that the expression of PKM and HK2 in neuroblastoma samples is strong predictors of negative clinical outcome, analysis of the protein content of circulating EVs could be used as a prognostic indicator and a non-invasive method for stratification of neuroblastoma patients.
MYCN-amplified cells are more sensitive to the depletion of PKM2 than MYCN non-amplified cells, suggesting oncogenic addiction [42]. This is because the PKM2 isoform is enriched in MYCN-amplified neuroblastoma cells due to the ability of the MYC transcription factors to transactivate the splicing factors PTBP1 and HNRNPA [42,43]. It is tempting to speculate that the transfer of oncogenic kinases via EVs operated by MYC mutant cells could contribute to the aggressive behaviour of the overall tumour mass by promoting the metabolic activity of cancer or stromal cells that do not carry oncogenic mutations. PKM2-enriched neuroblastoma EVs have the potential to modify epigenetically recipient cells by enhancing histone H3 phosphorylation. This would permanently fix transient oncogenic signals, perhaps explaining why MYCN-amplified tumours can revert to focally amplified, and focally amplified to non-amplified, during the evolution of the disease [7].
A limitation of this study is that we have not examined the role of nucleic acids in neuroblastoma EVs, although functional microRNAs have been previously detected in EVs secreted by neuroblastoma cell lines or immune cells infiltrating neuroblastomas [44,45]. It is not possible to exclude that the transfer of MYCN-regulated microRNAs might have contributed to some of the biological effects observed in our study. Indeed, two research groups have independently shown that microRNAs belonging to the 17–92 cluster are enriched in vesicles secreted by MYCN-amplified cells and their transfer stimulate proliferation and migration of recipient, MYCN non-amplified cells [45,46]. However, RNAi inhibition of PKM2 in MYCN-expressing donor cells completely reversed the increase in histone H3 phosphorylation in recipient MYCN single-copy cells, suggesting that this effect is regulated specifically by protein transfer. In conclusion, the results of our study suggest that MYCN-amplified neuroblastomas might promote the spreading of oncogenic kinases and other biologically active proteins to the tumour microenvironment and remote body locations.
Acknowledgements
Illary Salvatori is acknowledged for the access to the SeaHorse analyses. Giulia Catitti is kindly acknowledged for help with polychromatic flow cytometry EVs analysis.
Contributor Information
Giuseppe Palmisano, Email: palmisano.gp@usp.br.
Arturo Sala, Email: arturo.sala@brunel.ac.uk.
Data accessibility
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE repository at https://www.ebi.ac.uk/pride/.
The data are provided in the electronic supplementary material [47].
Authors' contributions
A.T. performed most of the experiments, analysed the results and wrote the manuscript. V.C.C. carried out the seahorse experiments. A.E., M.M. and G.C. provided clinical material. E.M. and O.C. helped with the characterization of neuroblastoma EVs. L.P. helped with sample preparations and M.R.L. supervised the mass spectrometry proteomic analyses. E.P. and L.C. provided neuroblastoma material from the TH-MYCN mouse model. S.B., E.C., G.S. and V.D.L. carried out the characterization of vesicles produced by neuroblastoma cell lines. G.P. carried out the mass spectrometry analysis, helped with the interpretation of the data and supervised the project. A.S. conceived and supervised the project and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Funding
The study was funded by a grant from Neuroblastoma UK and the Associazione Italiana per la Lotta al Neuroblastoma to A.S. and FAPESP SPRINT Award (grant no. 50356-4) to A.S and G.P. G.P., V.C.C. and L.P. are supported by Ricerca Finalizzata GR11-172. V.D.L. is supported by a grant from Fondazione AIRC (Italian Association for Cancer Research, ID: 20043).
References
- 1.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA. 2016. Neuroblastoma. Nat. Rev. Dis. Primers 2, 16078. ( 10.1038/nrdp.2016.78) [DOI] [PubMed] [Google Scholar]
- 2.Sala A. 2014. Editorial: targeting MYCN in pediatric cancers. Front. Oncol. 4, 330. ( 10.3389/fonc.2014.00330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz-Perez MV, Henley AB, Arsenian-Henriksson M. 2017. The MYCN protein in health and disease. Genes 8, 113. ( 10.3390/genes8040113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang J, Rychahou PG, Ishola TA, Mourot JM, Evers BM, Chung DH. 2008. N-myc is a novel regulator of PI3 K-mediated VEGF expression in neuroblastoma. Oncogene 27, 3999-4007. ( 10.1038/onc.2008.15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song L, et al. 2007. Oncogene MYCN regulates localization of NKT cells to the site of disease in neuroblastoma. J. Clin. Invest. 117, 2702-2712. ( 10.1172/JCI30751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey SC, Baylot V, Felsher DW. 2018. The MYC oncogene is a global regulator of the immune response. Blood 131, 2007-2015. ( 10.1182/blood-2017-11-742577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marrano P, Irwin MS, Thorner PS. 2017. Heterogeneity of MYCN amplification in neuroblastoma at diagnosis, treatment, relapse, and metastasis. Genes Chromosomes Cancer 56, 28-41. ( 10.1002/gcc.22398) [DOI] [PubMed] [Google Scholar]
- 8.van Niel G, D'Angelo G, Raposo G.. 2018. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213-228. ( 10.1038/nrm.2017.125) [DOI] [PubMed] [Google Scholar]
- 9.Li K, Chen Y, Li A, Tan C, Liu X. 2019. Exosomes play roles in sequential processes of tumor metastasis. Int. J. Cancer 144, 1486-1495. ( 10.1002/ijc.31774) [DOI] [PubMed] [Google Scholar]
- 10.Gopal SK, Greening DW, Hanssen EG, Zhu HJ, Simpson RJ, Mathias RA. 2016. Oncogenic epithelial cell-derived exosomes containing Rac1 and PAK2 induce angiogenesis in recipient endothelial cells. Oncotarget 7, 19 709-19 722. ( 10.18632/oncotarget.7573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Silva B, et al. 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816-826. ( 10.1038/ncb3169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josson S, et al. 2015. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 34, 2690-2699. ( 10.1038/onc.2014.212) [DOI] [PubMed] [Google Scholar]
- 13.Skog J, et al. 2008. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470-1476. ( 10.1038/ncb1800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luga V, et al. 2012. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542-1556. ( 10.1016/j.cell.2012.11.024) [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Zhang X, Zhang B, Shi H, Yuan X, Sun Y, Pan Z, Qian H, Xu W. 2016. Exosomes derived from gastric cancer cells activate NF-kappaB pathway in macrophages to promote cancer progression. Tumour Biol. 37, 12 169-12 180. ( 10.1007/s13277-016-5071-5) [DOI] [PubMed] [Google Scholar]
- 16.Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G. 2007. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 110, 2440-2448. ( 10.1182/blood-2007-03-078709) [DOI] [PubMed] [Google Scholar]
- 17.Marchisio M, et al. 2021. Flow cytometry analysis of circulating extracellular vesicle subtypes from fresh peripheral blood samples. Int. J. Mol. Sci. 22, 48. ( 10.3390/ijms22010048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Distler U, Kuharev J, Navarro P, Tenzer S. 2016. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 11, 795-812. ( 10.1038/nprot.2016.042) [DOI] [PubMed] [Google Scholar]
- 19.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. 2006. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell Proteomics 5, 144-156. ( 10.1074/mcp.M500230-MCP200) [DOI] [PubMed] [Google Scholar]
- 20.Visser NF, Scholten A, van den Heuvel RH, Heck AJ. 2007. Surface-plasmon-resonance-based chemical proteomics: efficient specific extraction and semiquantitative identification of cyclic nucleotide-binding proteins from cellular lysates by using a combination of surface plasmon resonance, sequential elution and liquid chromatography-tandem mass spectrometry. Chembiochem 8, 298-305. ( 10.1002/cbic.200600449) [DOI] [PubMed] [Google Scholar]
- 21.Wu M, et al. 2007. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 292, C125-C136. ( 10.1152/ajpcell.00247.2006) [DOI] [PubMed] [Google Scholar]
- 22.Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. 1996. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 13, 803-812. [PubMed] [Google Scholar]
- 23.Lu Z, Hunter T. 2018. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 43, 301-310. ( 10.1016/j.tibs.2018.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverman AM, Nakata R, Shimada H, Sposto R, DeClerck YA. 2012. A galectin-3-dependent pathway upregulates interleukin-6 in the microenvironment of human neuroblastoma. Cancer Res. 72, 2228-2238. ( 10.1158/0008-5472.CAN-11-2165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerman MW, et al. 2018. MYC Drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Disc. 8, 320-335. ( 10.1158/2159-8290.CD-17-0993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, et al. 2012. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685-696. ( 10.1016/j.cell.2012.07.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrowski M, et al. 2010. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19-30. ( 10.1038/ncb2000) [DOI] [PubMed] [Google Scholar]
- 28.Podar K, Anderson KC. 2010. A therapeutic role for targeting c-Myc/Hif-1-dependent signaling pathways. Cell Cycle 9, 1722-1728. ( 10.4161/cc.9.9.11358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Wu X, Basu M, Dong C, Zheng P, Liu Y, Sandler AD. 2017. MYCN amplification is associated with repressed cellular immunity in neuroblastoma: an in silico immunological analysis of TARGET Database. Front. Immunol. 8, 1473. ( 10.3389/fimmu.2017.01473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei JS, et al. 2018. Clinically relevant cytotoxic immune cell signatures and clonal expansion of T-cell receptors in high-risk MYCN-not-amplified human neuroblastoma. Clin. Cancer Res. 24, 5673-5684. ( 10.1158/1078-0432.CCR-18-0599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakata R, Shimada H, Fernandez GE, Fanter R, Fabbri M, Malvar J, Zimmermann P, DeClerck YA. 2017. Contribution of neuroblastoma-derived exosomes to the production of pro-tumorigenic signals by bone marrow mesenchymal stromal cells. J. Extracell Vesicles 6, 1332941. ( 10.1080/20013078.2017.1332941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warburg O. 1956. On the origin of cancer cells. Science 123, 309-314. ( 10.1126/science.123.3191.309) [DOI] [PubMed] [Google Scholar]
- 33.Vander HMG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029-1033. ( 10.1126/science.1160809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altenberg B, Greulich KO. 2004. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84, 1014-1020. ( 10.1016/j.ygeno.2004.08.010) [DOI] [PubMed] [Google Scholar]
- 35.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230-233. ( 10.1038/nature06734) [DOI] [PubMed] [Google Scholar]
- 36.Desai S, et al. 2014. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 5, 8202-8210. ( 10.18632/oncotarget.1159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv L, et al. 2013. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol. Cell 52, 340-352. ( 10.1016/j.molcel.2013.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amin S, Yang P, Li Z. 2019. Pyruvate kinase M2: a multifarious enzyme in non-canonical localization to promote cancer progression. Biochim. Biophys. Acta Rev. Cancer 1871, 331-341. ( 10.1016/j.bbcan.2019.02.003) [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y, et al. 2014. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol. Cell 53, 75-87. ( 10.1016/j.molcel.2013.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang CV, Le A, Gao P.. 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15, 6479-6483. ( 10.1158/1078-0432.CCR-09-0889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliynyk G, et al. 2019. MYCN-enhanced oxidative and glycolytic metabolism reveals vulnerabilities for targeting neuroblastoma. iScience 21, 188-204. ( 10.1016/j.isci.2019.10.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S, et al. 2016. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Lett. 371, 214-224. ( 10.1016/j.canlet.2015.11.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA, Bushell M, Willis AE. 2010. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene 29, 2884-2891. ( 10.1038/onc.2010.31) [DOI] [PubMed] [Google Scholar]
- 44.Neviani P, Wise PM, Murtadha M, Liu CW, Wu CH, Jong AY, Seeger RC, Fabbri M. 2019. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Res. 79, 1151-1164. ( 10.1158/0008-5472.CAN-18-0779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haug BH, Hald OH, Utnes P, Roth SA, Lokke C, Flaegstad T, Einvik C. 2015. Exosome-like extracellular vesicles from MYCN-amplified neuroblastoma cells contain oncogenic miRNAs. Anticancer Res. 35, 2521-2530. [PubMed] [Google Scholar]
- 46.Chen W, et al. 2021. MYCN-amplified neuroblastoma cell-derived exosomal miR-17–5p promotes proliferation and migration of non-MYCN amplified cells. Mol. Med. Rep. 23, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsakaneli A, et al. 2021. MYC regulates metabolism through vesicular transfer of glycolytic kinases. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Tsakaneli A, et al. 2021. MYC regulates metabolism through vesicular transfer of glycolytic kinases. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE repository at https://www.ebi.ac.uk/pride/.
The data are provided in the electronic supplementary material [47].