Abstract
Skeletal muscle has remarkable regenerative ability after injury. Mesenchymal fibro-adipogenic progenitors (FAPs) are necessary, active participants during this repair process, but the molecular signatures of these cells and their functional relevance remain largely unexplored. Here, using a lineage tracing mouse model (Gli1-CreER Tomato), we demonstrate that Gli1 marks a small subset of muscle-resident FAPs with elevated Hedgehog (Hh) signaling. Upon notexin muscle injury, these cells preferentially and rapidly expanded within FAPs. Ablation of Gli1+ cells using a DTA mouse model drastically reduced fibroblastic colony-forming unit (CFU-F) colonies generated by muscle cells and impaired muscle repair at 28 days. Pharmacologic manipulation revealed that Gli1+ FAPs rely on Hh signaling to increase the size of regenerating myofiber. Sorted Gli1+ FAPs displayed superior clonogenicity and reduced adipogenic differentiation ability in culture compared to sorted Gli1− FAPs. In a glycerol injury model, Gli1+ FAPs were less likely to give rise to muscle adipocytes compared to other FAPs. Further cell ablation and Hh activator/inhibitor treatments demonstrated their dual actions in enhancing myogenesis and reducing adipogenesis after injury. Examining single-cell RNA-sequencing dataset of FAPs from normal mice indicated that Gli1+ FAPs with increased Hh signaling provide trophic signals to myogenic cells while restrict their own adipogenic differentiation. Collectively, our findings identified a subpopulation of FAPs that play an essential role in skeletal muscle repair.
Keywords: FIBRO-ADIPOGENIC PROGENITORS (FAPs), MUSCLE REGENERATION, HEDGEHOG SIGNALING, MUSCLE INJURY, INTRAMUSCULAR ADIPOGENESIS, MYOGENIC REGULATORS
Introduction
Skeletal muscle is the most abundant tissue and displays remarkable regenerative capacity after injury. Its architecture is characterized by a very particular and well-described arrangement of myofibers, muscle stem cells (MuSCs, also called satellite cells), interstitial cells, nerves, blood vessels, and associated connective tissue.(1) Giving rise to myofibers, MuSCs are indispensable for muscle growth, repair, and regeneration.(2) Recently, a new type of muscle-resident mesenchymal progenitors, referred to as fibro-adipogenic progenitors (FAPs), was identified and found to be crucial in supporting the maintenance and regeneration of skeletal muscles following injury.(3,4)
In healthy muscle, FAPs reside within the muscle interstitium, between muscle fibers, often close to blood vessels but outside the basal lamina.(3,4) Different from the myogenic origin of MuSCs, FAPs are capable of differentiation into fibroblasts, adipocytes, and possibly osteoblasts, but not myoblasts.(3,5,6) They highly express several mesenchymal stem cell markers, such as Sca1, CD34, and PDGFRα, and thus, in a broad sense, are considered mesenchymal progenitors.(4,7) Shortly after muscle injury, FAPs quickly proliferate to provide a favorable trophic support for skeletal muscles.(3,4,8) Improper FAP decisions during regeneration impair myogenesis.(9,10) For example, an abnormal existence of FAPs in chronic muscle injuries is associated with intramuscular adipocytic tissue accumulation, inflammation and fibrosis formation.(3,8)
Despite the clear evidence of the importance of FAPs in performing different activities in muscle diseases, our understanding of their exact contribution to regenerating muscles is limited by the fact that they are functionally heterogeneous. Here, we identified Gli1 as a marker for a subset of FAPs that is preferentially expanded after injury to support muscle repair. Further characterization revealed that compared to Gli1− (negative) FAPs, Gli1+ (positive) FAPs are more clonogenic but possess reduced adipogenic differentiation potential. Genetic cell ablation and pharmacological manipulation suggested an important role of these cells in regulating muscle regeneration and adiposity. Computational analysis further revealed their orchestral actions in promoting muscle injury repair. Thus, our results unveil a vital subpopulation of FAPs that utilize Hedgehog (Hh)/Gli1 signaling to coordinate muscle healing and prevent intramuscular fat deposition.
Materials and Methods
Mice
Gli1-CreER Rosa-tdTomato (Gli1ER/Td) mice were generated by breeding Gli1-CreER mice (Jackson Laboratory, Bar Harbor, ME, USA; #007913; 129S6/SvEvTac)(11) with Rosa-tdTomato mice (Jackson Laboratory; #007909; 129S6/SvEvTac x C57BL/6NCrl). Only male mice were used in this project. For cell ablation experiments, we bred Gli1ER/Td mice with Rosa-DTA mice (Jackson Laboratory, #009669; B6.129P2) to generate Gli1ER/Td/DTA mice. To induce Td expression or ablate Td+ cells, Gli1ER/Td or Gli1ER/ Td/DTA mice received Tamoxifen (Tam; Sigma, St. Louis, MO, USA; #T5648) injections at 75 mg/kg for 5 days at indicated ages. In accordance with the standards for animal housing, mice were group housed at 23°C to 25°C with a 12-hours light/dark cycle and allowed free access to water and standard laboratory chow. All animal work performed in this report was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania.
Skeletal muscle injuries
To induce muscle injury, 10 μL Notexin (10 μg/mL; Accurate Chemical, Westbury, NY, USA; #TXL8104–100) was injected into male mouse tibialis anterior (TA) muscle with an insulin syringe to induce acute muscle injury, as we described.(12,13) To induce fatty degeneration in skeletal muscle, 50 μL 50% vol/vol glycerol (Sigma; #G5516) in water was injected into mouse TA muscle, as described.(14) For Hh activator or inhibitor treatment, 10 μL 1mM purmorphamine (PUR; Sigma; #SML0868) or 10mM GANT61 (Selleckchem, Houston, TX, USA; #S8075) were injected into the TA muscle 2 days after injury.
Immunohistochemistry
TA muscle samples collected at indicated time points were fixed in 4% paraformaldehyde (PFA), immersed into 30% sucrose at 4°C overnight, and embedded with optimal cutting temperature compound (OCT compound). Tissue was sliced to generate 10-μm-thick cryosections for hematoxylin and eosin (H&E) staining or immunofluorescence staining. For immunofluorescence staining, sections were blocked by 3% bovine serum albumin (BSA) for 30 min and incubated with rat anti-Sca1 (BioLegend, San Diego, CA, USA; #122501), mouse anti-PDGFRα (Santa Cruz, Dallas, TX, USA; #SC-398206), or rabbit anti-Perilipin (Cell Signaling, Danvers, MA, USA; #9349) antibodies at 4°C overnight.
Secondary antibodies, such as Alexa Fluor 488 anti-rat (Abcam, Cambridge, UK; #ab150157), and Alexa Fluor 647 anti-mouse (Abcam; #ab150115), or anti-rabbit (Abcam; #ab150079) were then added to sections for 1 h. To label the extracellular matrix, sections were stained with CF488 Wheat Germ Agglutinin (WGA; Biotium, Inc. Hayward, CA, USA; #29022) for 1 h.
Proliferation assay
For the 5-ethynyl-2’-deoxyuridine (EdU) labeling experiment, mice were injected with 1.6 mg/kg EdU (Invitrogen, Carlsbad, CA, USA; A10044) 1 day before harvesting. Isolated muscles were cryopreserved and sectioned at 10 μm thickness. EdU detection was carried out according to the manufacturer’s instructions (ThermoFisher Scientific, Waltham, MA, USA; Click-iT EdU Alexa Fluor 647 Imaging Kit; #D3822).
Fiber analysis and quantification
For the fiber analyses, images were enumerated and quantitated using ImageJ software (NIH, Bethesda, MD, USA; https://imagej.nih.gov/ij/).The Trainable Weka Segmentation (TWS) function was used to measure individual WGA-stained myofiber cross-sectional areas. First, a TWS classifier was trained to recognize and distinguish between the muscle fiber boundaries (WGA staining) and intrafiber space after a few simple manual annotations that instantiate these two features. Then, it was used to predict the muscle fiber boundary and intrafiber space in the rest area. The results of this prediction were represented as a probability rendering of the original image, where darker pixels represent higher probabilities for the boundary, and lighter pixels represent lower probabilities. The intrafiber space was quantified using Analyze Particles function. For each muscle sample, three sections, spaced 500 μm apart with the central one at the mid-belly of the TA, were selected for quantification of myofibers in a central muscle region of 0.64 mm2.
Fluorescence-activated cell sorting analysis of muscle cells
Hindlimb muscles (quadriceps, gastrocnemius, and tibialis anterior) were dissected and minced with scissors and then enzymatically dissociated with 0.1% collagenase (Sigma; #C9891) and 4.8 IU/mL dispase (Roche, Basel, Switzerland; #4942078001) in Dulbecco’s modified Eagle medium (DMEM) using the gentle-MACs system (Miltenyi Biotech, Bergisch Gladbach, Germany; #130093235). The cell slurry was pulled through a 21G needle until all remaining muscle tissue was broken apart, after which the cell solution was filtered through a 40-μm cell strainer. Red blood cells were eliminated with ACK Lysing buffer (Thermo-Fisher Scientific; #A1049201). Cells were stained with fluorescent labeled primary antibodies, including fluorescein isothiocyanate (FITC) anti-CD45 (BioLegend; #103107), FITC anti-CD31 (BD, Franklin Lakes, NJ, USA; #561813), FITC anti-CD11b (BioLegend; #101205), Brilliant Violet 711 anti-Sca1 (BioLegend; #108131), allophycocyanin (APC) anti-Itga7 (AbLab, Vancouver, BC, Canada; #67–0010-10), and Brilliant Violet 421 anti-CD34 antibodies (BD; #562608) for 60 min at 4°C, washed in flow buffer (2% fetal bovine serum [FBS] in phosphate-buffered saline [PBS]). Cells were then analyzed using a BD Biosciences LSR II flow cytometer or sorted using a BD Biosciences FACSAria II with a 100-μm nozzle. Strict gating schemes were determined and applied based on no antibody or fluorescence minus one control.
Cell culture assays
For the CFU-F assay, unsorted and sorted hindlimb muscle cells were plated at 3 × 104 cells/T25 and 3 × 103 cells/T25 flask, respectively. Cells were cultured in growth medium (α Eagle minimal essential medium [α-MEM] supplemented with 15% FBS, 0.1% β-mercaptoethanol, 20mM glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin) for 7 days before counting CFU-F number. Brightfield and fluorescent images of CFU colonies were taken at day 7 by fluorescence inverted widefield microscopy (Nikon Eclipse, Melville, NY, USA; #TE2000-U). Bone marrow mesenchymal progenitors (BM MPs) were obtained by culturing BM cells flushing out from long bones in the growth medium. Once confluent, BM MPs or muscle FAP cells were switched to adipogenic medium (DMEM with 10% FBS, 10 ng/mL triiodothyronine, 1μM rosiglitazone, 1μM dexamethasone, 10 μg/mL insulin, 100 IU/mL penicillin, and 100 μg/mL streptomycin) for 7 days or osteogenic medium (α-MEM supplemented with 10% FBS, 0.1% β-mercaptoethanol, 0.05mM ascorbic acid-2-phosphate, 10mM β-glycerophosphate, 0.1nM dexamethasone, 20mM glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin) for 14 days. For Bodipy staining, cells were first fixed by 4% PFA for 10 min and then incubated with Bodipy dye (ThermoFisher Scientific; #D3922) for 1 h at room temperature (RT).
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA from freshly sorted cells or cells cultured in differentiation medium was collected in Tri Reagent (Sigma; #T9424) for RNA purification. A High-capacity Reverse Transcription Kit (Applied BioSystems, Inc., Foster City, CA, USA; #4368814) was used to reverse transcribe messenger RNA (mRNA) into complementary DNA (cDNA). The power SYBR Green PCR Master Mix Kit (Applied BioSystems, Inc.; #4367659) was used for quantitative real-time polymerase chain reaction (qRT-PCR). The primer sequences for the genes used in this study are listed in Table S1.
Single-cell RNA-sequencing analysis of muscle cells
Prealigned and filtered single-cell RNA sequencing (RNA-seq) matrix files were acquired from GEO: GSE110878 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110878). Seurat package V3(15) for filtering, variable gene selection, dimensionality reduction analysis, and clustering. Briefly, doublets or cells with poor quality (genes >4000, genes <500, or >5% genes mapping to mitochondrial genome) were excluded. Expression was natural log transformed and normalized for scaling the sequencing depth to a total of 1 × 104 molecules per cell, followed by regressing out the number of Unique Molecular Identifiers (UMIs) and percent of mitochondrial genes, and identifying the variable genes by using the FindVariableFeatures function. Selection of principal component analysis (PCA) was performed using the JackStraw function. Statistically significant PCs were selected as input for t-Distributed Stochastic Neighbor Embedding (tSNE). For subclustering, we repeated the same procedure of finding variable genes, dimensionality reduction, and clustering. Different resolutions for clustering have been tested to demonstrate the robustness of clusters. MAGIC(16) was used to impute the dropout values. To computationally delineate the trajectory of FAP cells and order them in pseudotime, we used the algorithms implemented in the Monocle 2 package.(17) We ordered cells by selecting genes with high dispersion across cells and using a parameter of “mean_expression ≥ 0.05 & dispersion_empirical ≥ 1 * dispersion_fit”. Lists of genes were selected for dimensional reduction to generate the trajectory reconstruction using the nonlinear reconstruction algorithm DDRTree. Differential expressed genes (DEGs) were identified using the FindMarkers of the “RNA” assay. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using the clusterProfiler package.(18)
Statistical analysis
All data except myofiber size analysis are presented as box plots, either median with interquartile range where whiskers indicate minimum to maximum or median with a box indicating minimum to maximum. Data of myofiber size are presented as bar graphs with means ± standard error of the mean (SEM). For the comparison of two groups, statistical analysis was performed using unpaired, two-tailed Student’s t test. For the comparison of multiple groups, we used ANOVA with Turkey’s post-test (Graph Pad Prism; GraphPad Software, Inc., La Jolla, CA, USA). For assays using primary cells, experiments were repeated independently at least three times and representative data were shown here. Values of p < .05 were considered statistically significant.
Data and code availability
A published sequence data has been used in this work (GEO: GSE110878). Software used to analyze the data is either freely or commercially available.
Results
Gli1 marks a small subset of FAPs within skeletal muscles
To comprehensively assess the Gli1+ cells in skeletal muscles, we crossed Gli1-CreER(11) mice to a tdTomato reporter(19) for lineage tracing. To induce gene recombination, 2-month-old Gli1ER/Td mice were pulsed with five daily Tam injections. We observed similar levels of recombination using either three or five Tam injections, based on the number of Td+ cells in muscle, suggesting that this protocol maximally labels Gli1+ cells. We then analyzed the location of Td+ cells in TA in combination with fluorescently conjugated WGA staining to visualize muscle fiber outlines.(20) Interestingly, Td+ cells were exclusively located in the interstitial area of myofibers in skeletal muscles (Fig. 1A), where FAPs are known to reside.(7) To investigate whether Td+cells co-localize with FAPs, we performed staining with the FAP markers Sca1 and PDGFRα and found that the majority of Td+ cells (157/209; 75%) are FAPs (Sca1+PDGFRα+, designated herein as Td+FAPs) (Fig. 1B,C). Meanwhile, only ∼11.5% of FAPs (157/1366) were Td+, suggesting that Gli1+ resident cells are a small subset of the FAP population. To validate and further quantify the Gli1+ population in the FAP fraction, we next performed flow cytometry on mononucleated muscle cells and focused on the analysis of the lineage negative (CD31−CD45−CD11b−) population, which was previously shown to be enriched for FAPs.(4) In line with our histology data, 79% of Td+ cells were Lin−Sca1+CD34+Itga7− FAPs (Fig. 1D,E) and 17% of Lin−Sca1+CD34+Itga7− FAPs were Td+ (Fig. 1F,G).
Fig 1.

Gli1 marks a small subset of resident FAPs within skeletal muscles. (A) Representative fluorescent images of TA muscles from Gli1ER/Td mice with WGA staining show Td+ cells (arrows) are located in the interstitial area of myofibers. Mice at 2 months of age received Tam injections for 5 days and muscle was collected 7 days later. Panel d is a magnified image from the outlined area in panel c. Red: Td; blue: DAPI; green: WGA. Scale bar: 50 μm. (B) Td+ cells co-express FAP markers Sca1 (green) and PDGFRα (white). Panel d is a magnified image from the outlined area in panel c. Arrows point to PDGFRα+Sca1+Td+ cells (Td+ FAPs) and arrow heads point to PDGFRα+Sca1+Td− cells (Td− FAPs). Scale bar: 50 μm. (C) Venn diagram of Sca1+, PDGFRα+, and Td+ cells in TA muscles. Quantification reveals that Td+ cells constitute a small portion of FAP cells (Td+ FAPs highlighted with a red grid). n = 5 mice. (D) Gating strategy of flow analysis studying the percentage of FAPs in Td+ cells. (E) Quantification reveals that the majority of Td+ cells are FAPs (Lin−Sca1+CD34+Itga7−). n = 4 mice/group. (F) Gating strategy of flow analysis studying the percentage of Td+ cells in FAPs. (G) Quantification reveals that a small subset of FAPs (Lin−Sca1+CD34+Itga7−) are Td+. n = 4 mice/group. (H) qRT-PCR analysis showed increased expression levels of Hh-regulated genes in FACS-sorted Td+ FAPs compared to Td- FAPs. n = 3 mice/group.
Based on the location of the Td+ FAPs in skeletal muscles, it is highly unlikely that they are MuSCs. To definitively exclude the possibility that Gli1+ cells comprise a majority of the MuSC population, we sorted the Td+ cells based on standard MuSC markers(13,21) and confirmed that the Td+ cells lack MuSC markers (Lin−Sca1−CD34+Itga7+) (Fig. S1). To verify that the Hh pathway is active in Td+ FAPs cells, we investigated the expression of known Hh genes by qRT-PCR and demonstrated significant increase of Gli1, Gli2, Hhip, and Smo expression levels in Td+ FAPs compared to Td− FAPs (Fig. 1H). Altogether, these results suggest that Gli1+ cells are mesenchymal progenitors residing within skeletal muscle and comprise a subset of the FAP population with elevated Hh signaling.
Gli1+ FAPs are dynamically expanded after acute injury
To investigate the inherent in vivo behavior of skeletal muscle-resident Gli1+ cells during muscle injury, Gli1ER/Td mice received an intramuscular injection of notexin (NTX), a myotoxin that is widely used to study muscle regeneration after acute injury.(22) Following injury, muscles were collected at 3, 6, and 9 days. Previous studies have indicated that FAP expansion after this type of injury peaks around day 3,(23) and we utilized this time point to compare Gli1-labeled (Td+) cells with the typical behavior of FAPs. To this end, histological analysis showed that severe muscle degeneration occurs at this time point (Fig. 2A). Although enumeration of Td+ cells in uninjured muscle yielded 167 ± 21 cells/muscle area, their density in the necrotic muscle tissues at day 3 postinjury drastically increased to 731 ± 67 cells/muscle area (Fig. 2A,B). Additionally, flow cytometry analysis of injured muscles demonstrated that the number of total FAPs, as well as the Td+ subpopulation greatly increased 3 days postinjury (Fig. S2). Assessment of the Td+ FAP population revealed a change in cell percentage from 17% before injury to 40% after injury (Fig. 2C,D), indicating that within FAPs, Gli1+ cells rapidly expanded. By day 6 postinjury, small regenerating myofibers with centralized nuclei appeared, with enlarged interstitial spaces and the presence of inflammatory cellular infiltration. By day 9 postinjury, muscle exhibited a greater proportion of regenerated myotubes of varying diameters with a better organization. During this time course of repair, Td+ cells gradually decreased and reached the baseline level around day 9 (Fig. 2A,B,D). To validate the preferential expansion of Td+ cells in FAPs, we assessed their in vivo proliferation capacity following injury by administration of EdU 1 day before tissue collection (Fig. 2E). Muscle sections were then co-stained with PDGFRα to mark FAPs. Strikingly, at day 3 postinjury, Td+PDGFRα+ (Td+ FAPs) cells displayed significantly higher EdU incorporation compared to Td−PDGFRα+ (Td− FAPs) cells (Fig. 2F,G), confirming that FAP expansion after injury is primarily contributed by Gli1+ FAPs. In summary, our results show that compared with Gli1− FAPs, the Gli1+ cells are a more distinct dynamic subpopulation of FAPs that expands rapidly in skeletal muscles in response to injury.
Fig 2.

The Gli1+ FAPs subpopulation preferentially expand in response to acute muscle injury. (A) Representative images of H&E staining (top panel) and fluorescent Td+ cells (red, bottom panel) in TA muscles at day 0, 3, 6 and 9 post NTX-induced muscle injury. Mice at 2 months of age were injected with Tam for 5 days followed by an NTX injury 7 days later. Scale bar: 50 μm. (B) Td+ cells in muscle at different time points post injury were counted. MA: muscle area. n = 4 mice/group. (C) Flow analysis of Td+ cells in total FAPs at day 0 and day 3 after injury. (D) Quantification of the percentage of Td+ cells within FAPs at day 0, 3, 6, 9, and 14 after injury. n = 3 mice/group. (E) Schematic plot of in vivo proliferation assay. Two-month old Gli1ER/Td mice received Tam injections for 5 days, an intramuscular injection of NTX 7 days later, and an EdU injection 9 days later. Muscle tissues were harvested at 3 days postinjury. (F) Representative TA muscle image at day 3 post injury show PDGFRα staining (green), Td (red), DAPI (blue), and EdU staining (white). Arrows point to EdU+PDGFRα+Td+ (proliferating Td+ FAPs) cells and an arrowhead points to EdU+PDGFRα+Td− (proliferating Td− FAPs) cells. Scale bar: 50 μm. (G) Quantification shows that Td+ FAPs have greater EdU incorporation than Td− FAPs. n = 3 mice/group.
Gli1+ cell ablation impairs skeletal muscle regeneration
To access the impact of Gli1+ cells on skeletal muscle regeneration, we depleted these cells using a mouse model overexpressing DTA (diphtheria toxin A-subunit).(24) For this purpose, we constructed Gli1ER/Td/DTA mice and subjected them to Tam induction at 2 months of age, while Gli1ER/Td mice served as controls. One week after the last injection, histology revealed a 77% decrease of Td+ cells in uninjured TA muscle of DTA mice (Fig. 3A,B), whereas flow cytometry similarly revealed an 70% decrease of Td+ cells in FAPs (Fig. S3A,B), confirming the success of cell ablation. CFU-F is an assay widely used as a functional method to quantify mesenchymal progenitors,(25) which encompass FAPs. We performed the CFU-F assay to determine the consequences of the depletion of Gli1-expressing FAPs and assess the competence of remaining cells to form colonies. Interestingly, ablation of Td+ cells drastically reduced the CFU-F colonies derived from muscle cells with a major effect on Td+ CFU-Fs (Fig. 3C,D), indicating the importance of Gli1+ cells in the expansion capacity of FAPs.
Fig 3.
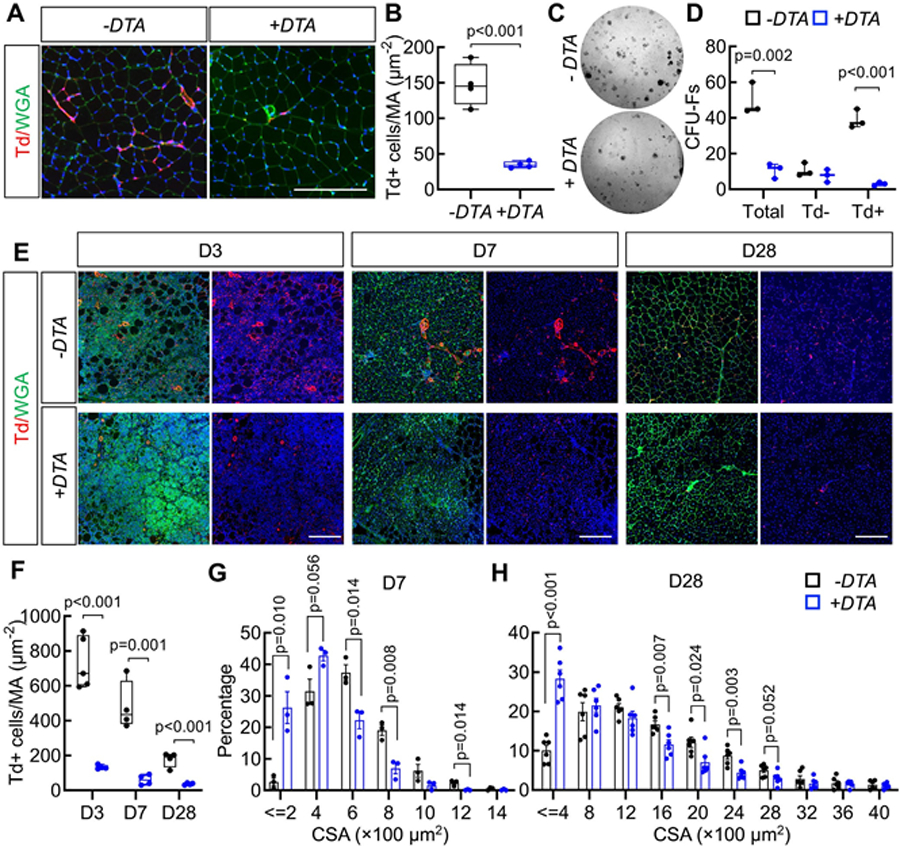
Genetic ablation of Gli1+ cells causes delayed muscle regeneration. (A) Representative fluorescent images of TA muscles from Gli1ER/Td and Gli1ER/ Td/DTA mice show Td+ cells (red), DAPI (blue), and WGA (green) staining. Mice at 2 months of age received Tam for 5 days and tissue was harvested 1 week later for analysis. Scale bar: 200 μm. (B) Quantification reveals a significant decrease of Td+ cells in Gli1ER/Td/DTA muscle compared to Gli1ER/ Td muscle. n = 4 mice/group. (C) Representative image of CFU-F colonies from Gli1ER/Td and Gli1ER/Td/DTA muscle cells. (D) The number of CFU-Fs per 3 × 104 digested muscle cells was quantified. n = 3 mice/group. (E) Representative fluorescent images of Gli1ER/Td and Gli1ER/Td/DTA muscle at day 3, 7, and 28 days post-NTX injury. Red: Td+ cells; blue: DAPI; green: WGA. At each time point, the left panel contain all three colors and the right panel contain red and blue signals only. Scale bar: 200 μm. (F) Quantification reveals significant reduction in Td+ muscle cells after Tam injections over the repair process. Note that the remaining Td+ muscle cells (escapers, blue bars) in Gli1ER/Td/DTA mice still expanded at day 3 after injury but were drastically less than those in Gli1ER/Td muscle. (G) Myofiber CSA of Gli1ER/Td and Gli1ER/Td/DTA mice is plotted as the percentage of fibers with discrete sizes at day 7 post-NTX injury. n = 3 mice/group. (H) Myofiber CSA of Gli1ER/Td and Gli1ER/Td/DTA mice is plotted as the percentage of fibers with discrete sizes at day 28 post-NTX injury. n = 5 mice/group. CSA, cross-sectional area.
We next investigated the effects of Gli1 cell ablation in an acute injury model. Following Tam induction, mice were subjected to NTX-induced injury and the regenerating muscles were analyzed at 3, 7, and 28 days after injury. Muscle Td+ cells were drastically reduced after Tam injections at all time points in Gli1ER/Td/DTA mice (Fig. 3E,F). The remaining non-recombined (escapers) Td+ FAPs cells still expanded at day 3 after injury but their density was 82% less than those in Gli1ER/Td muscle, and those cells were almost completely disappeared at day 7 and day 28 (Fig. 3E,F). Flow analysis also revealed that ablation of Gli1+ cells reduce the percentage and number of total FAPs by 42.3% and 48.4%, respectively, at day 0 before injury, and by 35.1% and 39.2%, respectively, at day 28 after injury (Fig. S3C). Because FAPs are important for proper muscle regeneration,(4,6,26) we quantified the cross-sectional area (CSA) of myofibers at day 7 and day 28 postinjury. The size distribution graph clearly shows that Gli1ER/Td/DTA mice display much smaller myofibers, compared to Gli1ER/Td mice at those time points (Fig. 3G,H, Fig. S3D–F). As a negative control, vehicle (corn oil) injections did not induce Td expression (Fig. S3D), nor did it affect muscle regeneration (Fig. S3F). Overall, these data clearly establish the requirement of Gli1+ FAPs in the regeneration of skeletal muscle.
Pharmacological activation of Hh signaling after acute injury increases muscle fiber size
Given that Gli1 is a transcription factor in the Hh signaling pathway,(27) and Td+ FAPs are indeed associated with elevated Hh signaling (Fig. 1H), we next studied whether activated Hh signaling could accelerate muscle repair. To this end, we administered PUR, a known agonist of Hh signaling that targets Smoothened,(28) into Gli1ER/Td mice once weekly, following Tam induction and NTX injury (Fig. 4A). Both H&E staining and WGA staining showed improved myofiber regeneration at 30 days after injury (Fig. 4B). This finding was accompanied by remarkable increases of Td+ FAPs cells at day 4 and day 30 after injury (Fig. 4C). Quantification of the cross-sectional areas of regenerating fibers at day 30 showed a shift toward larger myofiber size in the PUR-treated cohort, compared to dimethylsulfoxide (DMSO)-treated groups (Fig. 4D), as well as an increase of total average sizes of myofibers (Fig. S4A). Interestingly, PUR did not improve muscle regeneration in Gli1ER/Td/DTA mice with Tam induction and NTX injury, suggesting that it acts through Gli1+ cells only (Fig. S4B). Taken together, these data demonstrate that the function of Gli1+ FAPs in muscle repair relies on Hh/Gli1 signaling to increase the size of regenerating myofibers.
Fig 4.
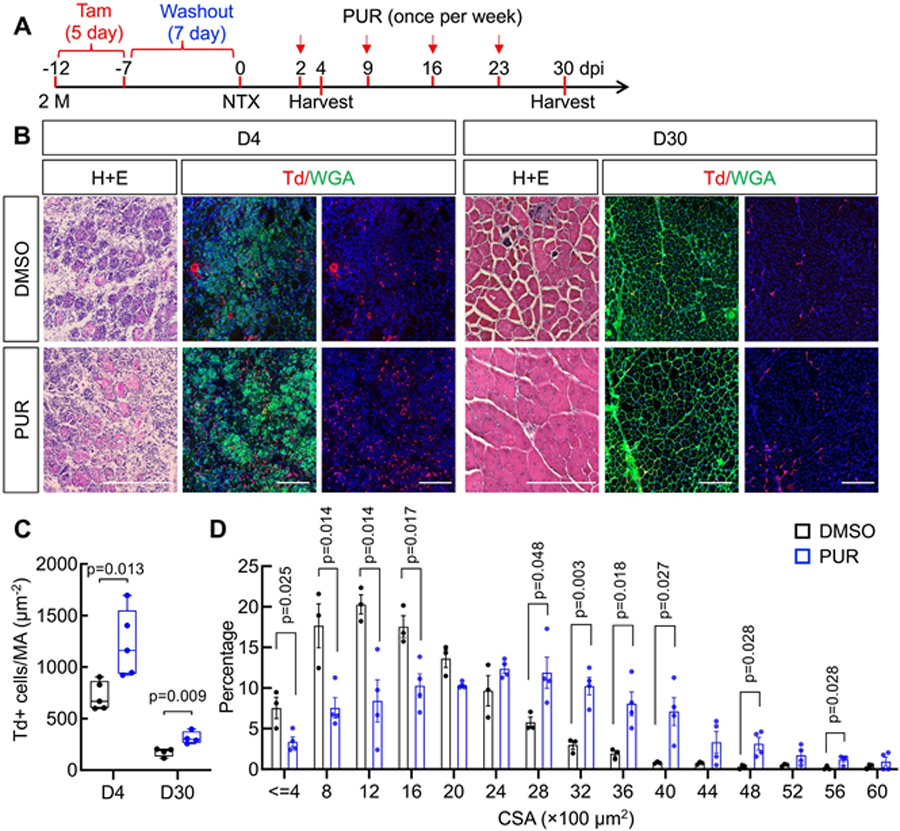
Activation of Hh signaling increases fiber size after acute muscle injury. (A) Schematic plot of treating muscle NTX injury with a Hh activator. Two-month-old Gli1ER/Td mice received Tam injections for 5 days and an intramuscular injection of NTX 7 days later. Two days after injury, mice were weekly injected with vehicle (DMSO) or PUR. Tissues were harvested at day 4 and day 30 postinjury. (B) Representative H&E staining and luorescent images of TA muscles from DMSO-treated or PUR-treated mice at day 4 and day 30 post-NTX injury. Red: Td+ (red); green: WGA. At each time point, the middle panel contain both color and the right panel contain red color only. Scale bar: 200 μm. (C) Td+ muscle cells were counted at day 4 and day 30 postinjury. n =5 mice/group. (D) Myofiber CSA of DMSO-treated or PUR-treated mice is plotted as percentage of discrete fiber sizes. n = 4 mice/group. CSA, cross-sectional area.
Gli1+ muscle resident cells have superior clonogenicity in vitro and reduced adipogenic capability
To confirm that Gli1-expressing cells exhibit properties associated with FAPs, we next queried their growth and differentiation potential in vitro. Upon culturing the hindlimb muscle cells, Td+cells increased from 7% in freshly isolated muscle cells to 40% when cells became confluent (Fig. S5A,B). Upon seeding, enzymatically released fractionated muscle cells were capable of generating CFU-F colonies in culture (Fig. S5C). Interestingly, although Td− and Td+ colonies were both observed, the number of Td+ colonies was four times more than the number of Td− colonies (Fig. S5C,D). To further investigate the in vitro properties of these cells, we used fluorescence-activated cell sorting (FACS) to isolate Td+ FAPs and Td− FAPs and seeded them separately for CFU-F assays. Similar to the results from the unfractionated experiments, we found that Td+ FAPs have a much higher CFU-F frequency than Td− FAPs (Fig. 5A,B). Taken together, these findings indicate that while Td− FAPs far exceed Td+ FAPs in the number, the population of Td+ FAPs exhibits higher clonogenicity potential in vitro.
Fig 5.

Gli1+ muscle resident cells have high clonogenicity and reduced adipogenesis in vitro. (A) CFU-F assay of Td− FAPs and Td+ FAPs sorted from Gli1ER/Td hindlimb muscle. (B) The number of CFU-Fs from 3 × 103 sorted Td− FAPs and Td+ FAPs was counted. n = 3 mice. (C) Representative images of osteogenic differentiation (top panel, Alizarin red staining) and adipogenic differentiation (bottom panel, Oil red staining) of BM MPs, Td+ FAPs, and Td− FAPs. Scale bar (top): 200 mm. Scale bar (bottom): 200 μm. (D) qRT-PCR analysis of osteogenic markers in cells after 2 weeks of culturing in osteogenic medium. (E) qPCR analysis of adipogenic markers in cells after 1 week of culturing in adipogenic medium.
Muscle FAPs are shown to be capable of multilineage differentiation upon induction in culture.(26) To further test the potency of Td+ FAPs, we queried their lineage differentiation upon exposure to conditions promoting either adipogenesis or osteogenesis. BM MPs, whose differentiation abilities are well documented,(29) served as a positive control. Both sorted Td+ FAPs and Td− FAPs had less potential to become osteogenic cells compared to BM MPs as shown by Alizarin Red staining and osteogenic marker gene expression after 2 weeks of culture in osteogenic medium (Fig. 5C,D). Interestingly, as judged by Oil red O staining and adipogenic marker expression after 1 week of culture in adipogenic medium, Td+ FAP cells exhibited decreased adipogenic differentiation potential than Td− FAPs (Fig. 5C,E). Similarly, Bodipy staining confirmed the reduced potential of Td+ FAPs cells to become mature adipocytes (Fig. S5E,F). Together, these results suggest that the Gli1+ FAPs have reduced adipogenic differentiation capacity in vitro.
Gli1+ FAPs regulate intramuscular adipogenesis and myofiber formation in response to injury
To further investigate our in vitro findings, we next adopted an in vivo glycerol injury model. This system exhibits significant fat infiltration(14) and is useful in determining the in vivo adipogenic properties of Td+ FAPs and to study the role of Gli1+ cells in muscle adiposity (Fig. 6A). In young uninjured mice, very few muscle Perilipin+ adipocytes were detected, and staining was confined to Td− cells in Gli1ER/Td mice after Tam induction (Fig. 6B). After glycerol injection, Perilipin+ cells gradually increased with an interstitial location between myofibers (Fig. 6B). Muscle adipocytes are derived from FAPs.(3) Interestingly, Td+ cells constituted 7.4% of Perilipin+ adipocytes in muscle sections, much lower than 17.3% of Td+ cells in FAPs as analyzed by FACS. These data, in accordance with our in vitro data (Fig. 5), further showed that Td+ FAPs have reduced capacity in vivo to evolve into adipocytes after injury.
Fig 6.
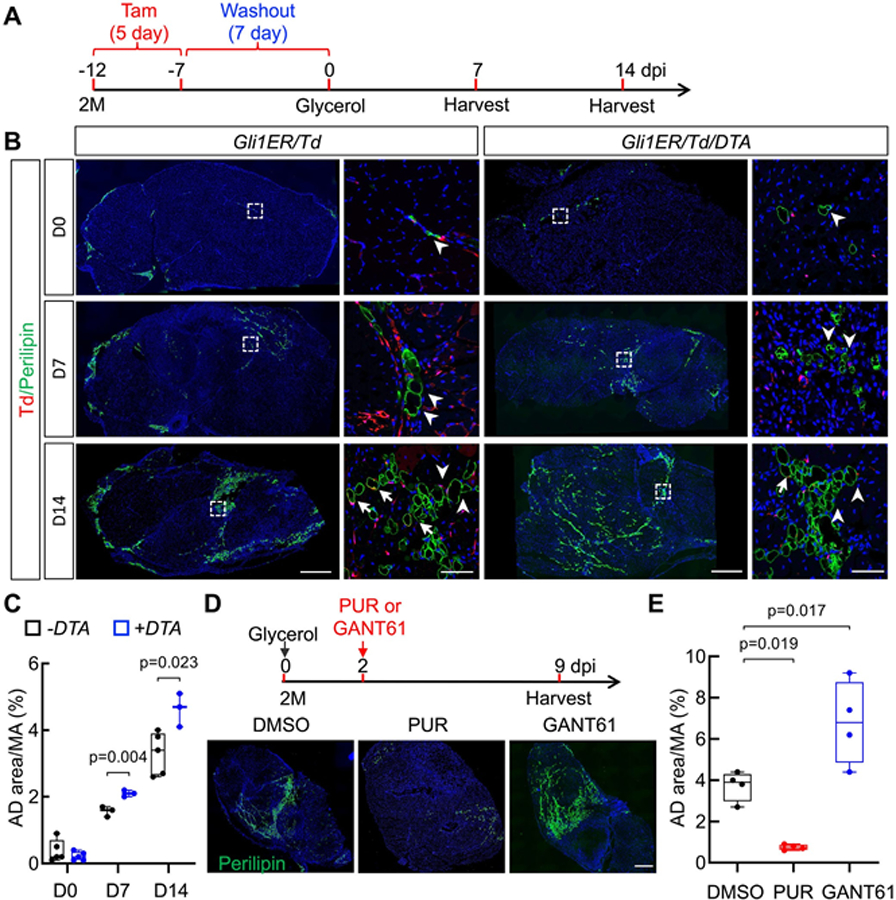
Gli1+ FAPs suppress intramuscular adipogenesis and improve myofiber size in response to glycerol injury. (A) Schematic plot of a mouse glycerol injury model. Two-month-old Gli1ER/Td and Gli1ER/Td/DTA mice received Tam injections for 5 days and an intramuscular injection of glycerol 7 days later. Tissues were harvested at day 7 and day 14 postinjury. (B) Representative immunofluorescence images of TA muscle from Gli1ER/Td and Gli1ER/Td/DTA mice at day 0, 7, and 14 post-glycerol injury with Perilipin staining. Under each mouse genotype, the right panel (scale bar: 50 μm) is the magnified area in the left panel (scale bar: 500 μm). Arrows point to Perilipin+Td+ cells and arrowheads point to Perilipin+Td− cells. (C) Quantification of adipocyte area reveals that muscle adiposity is increased in Gli1ER/Td/DTA mice. (D) Upper panel: Schematic plot of treating glycerol-injured mice with Hh regulators. Two-month-old mice received Tam injections for 5 days and an intramuscular injection of glycerol 7 days later. Two days later, mice received a DMSO, PUR, or GANT61 injection. Tissues were harvested at day 9 post-glycerol injection. Lower panel: Representative fluorescent images of TA muscle after var- ious treatments. Green: Perilipin; Blue: DAPI. Scale bar: 500 μm. (E). Quantification of muscle AD area after treatments. n = 4 mice/group. AD, adipocyte; MA, muscle area.
This remarkable result prompted us to combine fate tracing and ablation or Hh signaling manipulation studies in the glycerol injury model. Depletion of Gli1+ FAPs in Gli1ER/Td/DTA mice increases Perilipin+ area within skeletal muscles at day 7 and day 14 after injury (Fig. 6B,C). We then asked whether pharmacological alteration of Hh signaling affects muscle adiposity after glycerol injury. In line with our genetic depletion experiment, treating mice with a Hh inhibitor, GANT61,(30) caused 78% more adipocytes in muscle at day 9 after glycerol injection, whereas treating mice with a Hh activator, PUR, almost abolished adipocyte infiltration (Fig. 6D,E and Fig. S6A).
These data (Fig. 6) demonstrate that activation of Hh signaling diminishes intramuscular adipogenesis, while Gli1+ genetic cell ablation or Hh inhibition induces extensive adipogenesis. In corroboration, qRT-RCR analysis of muscle tissue exhibited increased expression of the mature adipocyte markers Adipoq, Lpl, Pparg, and Cebpa, when mice were treated with Hh inhibitor (Fig. S6B). In contrast, the Hh activator was able to attenuate the increase of mature adipocyte marker expression (Fig. S6B), suggesting that Gli1 signaling negatively affects adipogenesis after glycerol muscle injury. qRT-PCR analysis further confirmed that PUR indeed upregulates the expression of Hh signaling components in muscle cells and GANT61 suppresses their expression (Fig. S6C). Interestingly, quantification of cross-sectional area at day 9 after glycerol injury demonstrated that inhibition of Hh signaling (GANT61) reduced average myofiber area (Fig. S6D) and shifted toward smaller fibers (Fig. S6E), whereas activation (PUR) increases myofiber area (Fig. S6D) and shifted fiber distribution toward larger myofibers (Fig. S6E). Altogether, these data demonstrate that Gli1 signaling has a dual action by enhancing myogenesis and reducing adipogenesis in response to muscle injury.
The subcluster of FAPs with high Hh signaling express high myogenic regulators and low adipogenic regulators
To fully understand the Gli1+ FAP-dependent regulation of adipogenesis and myogenesis, we analyzed a single cell RNA-sequencing (scRNA-seq) dataset previously performed on mononucleated mouse skeletal muscle cells.(31) Among 10 identified cell clusters, FAP is a distinct one that was clearly separated from hematopoietic cells, endothelial cells, mural cells, and MuSCs (Fig. 7A), in agreement with previous assessments.(31) Interestingly, the expression of Gli1 was positively correlated with the expression of FAPs markers, Pdgfra, Cd34, and Ly6a (Fig. S7A) but negatively correlated MuSC markers, Pax7, Pax3, and Myod1 (Fig. S7B). Further analysis of FAPs divided them into five clusters (Fig. 7B), among which clusters 4 and 5 express Gli1 at a much higher level than clusters 1 to 3 (Fig. 7C). Pseudotime trajectory analysis placed cluster 1 and 2 at one end, cluster 3 at the branch point, and clusters 4 and 5 at opposite two ends (Fig. 7D), distinguishing clear separation between Gli1− (1–3) and Gli1+ (4–5) clusters. An examination of Hh pathway components showed that Hh signaling is more activated in Gli1+ cells than Gli1− cells (Fig. S7C), indicating that the Hh pathway is active in FAPs expressing high levels of Gli1.
Fig 7.
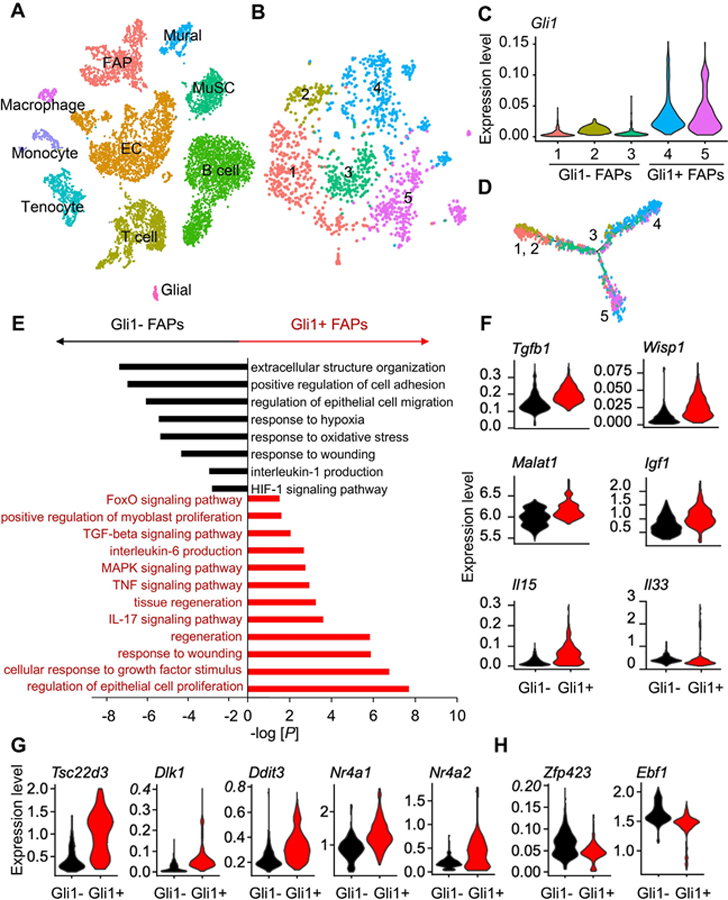
Single cell RNA-seq analysis indicates that Gli1+ cells express myogenic and anti-adipogenic regulators. (A) tSNE plot of mouse mononucleated muscle cells reveals a distinct FAP cluster. (B) Subclustering of FAPs generates 5 subclusters. (C) Subclusters 4 and 5 express Gli1 at a higher level compared to subclusters 1, 2, and 3. (D) Pseudotime trajectory plot of FAP subclusters by Monocle V2. (E) GO term and KEGG pathway analyses of DEGs upregulated in Gli1− FAPs and Gli1+ FAPs. (F) Violin plots of myogenic factor genes in Gli1− FAPs and Gli1+ FAPs. (G) Violin plots of anti-adipogenic genes in Gli1− FAPs and Gli1+ FAPs. (H) Violin plots of pro-adipogenic genes in Gli1− FAPs and Gli1+ FAPs.
GO term and KEGG pathway analyses of differentially expressed genes (DEGs) between Gli1+ and Gli1− FAP clusters revealed that Gli1+ FAPs are more metabolically active and more related to tissue regeneration (Fig. 7E). In particular, interleukin-6 (IL6) production and transforming growth factor beta (TGF-β) signaling pathway, both known positive regulators of myoblast proliferation and tissue regeneration,(8,32) are enriched in the Gli1+ FAP population, suggesting that Gli1+ FAPs are likely to secrete factors that influence myogenesis. It is well known that FAPs secrete factors are required for MuSC expansion and commitment.(6,33) To complement this analysis and further characterize the nature of the specific factors produced in Gli1+ FAPs, we investigated known molecules that are important in the crosstalk between muscle cells and FAPs. Indeed, Gli1+ FAPs express higher levels of Tgfb1, Wisp1, Malat1, Igf1, Il15, and Il33, compared with Gli1− FAPs (Fig. 7F). qPCR analysis of sorted Td+ FAPs and Td− FAPs validated the significant increase of Il6, Igf1, and Wisp1 expression in noninjured or day 3 post-NTX injured Td+ FAPs (Fig. S7D), suggesting a role of Gli1+ FAPs in supporting myogenesis by differentially expressing prominent myogenic factors.
Reflecting their adipogenic potential, FAPs can also give rise to ectopic adipocytes.(3) Interestingly, we noticed that some of the regulators we identified to be highly regulated in Gli1+ FAPs, such as Il15 and Wisp1, are also involved in inhibiting FAPs differentiation into adipocytes.(34,35) Further analysis revealed increased expression of anti-adipogenic regulators, such as Tsc22d3, Dlk1, Ddit3, Nr4a1, and Nr4a2 (Fig. 7G), and reduced expression of pro-adipogenic regulators, such as Zfp423 and Ebf1 (Fig. 7H). Taken together, our data suggest that Gli1+ FAPs subpopulation releases myogenic signals and shifts toward anti-adipogenic regulation that together influences both fiber size regulation and intermuscular fat determination.
Discussion
FAPs are a multipotent muscle resident population that are able to regulate skeletal muscle regeneration in normal and diseased conditions.(4,8) Since their initial discovery(3,4,9) as a supportive cell type in muscle regeneration, FAPs have been shown to have a remarkable phenotypic diversity. In fact, they orchestrate a plethora of processes and are a major source of fibroblasts and adipocytes in diseased muscles(7,36) and are capable of undergoing multilineage differentiation.(5) This incredible plasticity makes it difficult to understand their exact function and distinguish the origin of their multipotency. Accordingly, a previous study reported that Tie2 and Vcam1 are differentially expressed in FAPs in response to acute injury or diseased environment.(23) Another study identified Hic1 expression in skeletal muscle-resident mesenchymal progenitor cells that acquire a myofibroblast-like phenotype to repair myotendinous junction upon injury.(37) In other cases, although FAPs and similar mesenchymal-like cells in the interstitial space of the muscle share the same cell surface markers, they have different functional potentials,(38,39) suggesting that there is a necessity to further identify additional markers in order to better define and determine the biological role of FAPs subpopulations.
In this study, we identified for the first time that Gli1, a crucial mediator of Hh signaling, labels a subset of FAPs that influence myogenesis and adipogenesis in response to muscle injury (Fig. 8). Gli1 is an integral effector protein of the Hh pathway,(27) a fundamental pathway that maintains adult stem and progenitor cells in various organs, such as brain, skin, bladder, teeth, and others.(40) Following injury, Hh signaling can trigger stem cells and other resident cells to participate in repair, and therefore, Hh upregulation is viewed not only as a natural response to injury but also as a way to stimulate tissue repair by activating stem cells.(40) Recently, our work and others on Gli1ER/Td mice revealed Gli1+ cells as mesenchymal progenitors in bone periosteum that contribute to fracture healing,(41,42) indicating an important role of Hh/Gli1 signaling in musculoskeletal injury repair. Although the Hh pathway was reported to initiate early activation of the myogenic program during skeletal muscle development,(43) it is generally considered silent in postnatal life. However, our data demonstrate that Gli1 is an in vivo marker for a discrete population of resident FAPs with a high Hh signaling during muscle homeostasis. Our genetic fate tracing indicates that muscle resident Gli1+ FAPs undergo more dynamic proliferation after injury than Gli1− FAPs. Around 3 days postinjury, Gli1+ FAPs reach a maximum in number and then quickly return to baseline levels by day 9, mirroring the reported proliferative dynamics of FAPs(8) and suggesting that this precise temporal increase is critical for proper muscle regeneration. Flow cytometry and EdU proliferation assays also indicated that Gli1+ FAPs are the major population being activated after muscle damage. Furthermore, we showed that genetic ablation of Gli1+ cells drastically reduced total FAP numbers, and is sufficient to impair muscle regeneration and reduces the size of newly forming myofibers after injury. Therefore, although they are only a small subset of FAPs, Gli1+ FAPs are the major component of FAPs that are regulated by Hh signaling for supporting myogenesis of skeletal muscles after injury.
Fig 8.
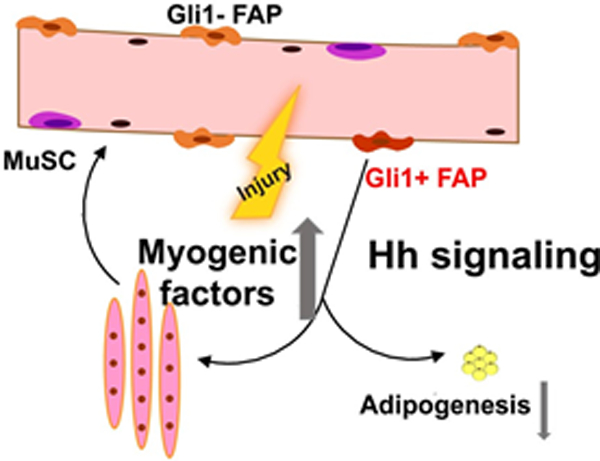
A schematic diagram depicts the role of Gli1+ FAPs in myogenesis and their adipogenic fate in response to muscle injury.
During muscle regeneration, FAP expansion is critical to sustain MuSC differentiation and maintenance of the stem cell pool in a paracrine manner.(6) In physiological conditions, a number of studies have combined in vivo analysis with ex vivo assays to suggest that FAPs support MuSC differentiation by secretion of myogenic factors.(4,35) Interestingly, we found that Gli1+ FAPs increase production of Igf1 and Il6, which have been shown to promote MuSC function and muscle regeneration.(4,32,44) Our computational analysis also detected upregulation of Tgfb1 and Il33, which block tumor necrosis factor (TNF)-induced FAPs apoptosis(8) and promote muscle repair,(45) respectively. Interestingly, TGF-β has been also suggested to regulate fatty infiltration by promoting FAP accumulation,(46) implying that there is often an association between muscle repair and adipocyte regulation by FAPs. Our findings on the differential expression of several other genes, such as Il-15 and Wisp1 in Gli1+ FAPs are important to consider in light of recent studies that implicate a role of these molecules in stimulating muscle cell expansion and inhibiting intermuscular adipogenesis.(34,35,47) An important question arising from these observations is whether Gli1+ FAPs also contribute to the adipocyte differentiation. Our single-cell analysis revealed that these cells upregulate several genes associated with an anti-adipogenic fate and downregulate genes associated with an adipogenic fate, suggesting that the expression of Gli1 in FAPs shifts cell fate toward a reduced capacity of adipocyte differentiation.
Muscle regeneration associates with a transient ectopic adipocyte accumulation(3,4,35) as well as fibrosis deposition, especially under disease conditions.(48) Although the fibrogenic effect of FAPs has been studied extensively both in vitro and in vivo,(1,8,10,37,47,49,50) their role in adipogenic regulation in vivo is less understood.(3,8,36,51) In bone marrow and heart, Gli1+ mesenchymal progenitors become myofibroblasts and contribute to fibrosis and dysfunction.(52,53) Our data demonstrate a different, previously unrecognized role of Gli1 for skeletal muscle, in regulating regeneration by repressing adipogenic differentiation. In addition to their transcriptional regulation in our single-cell analysis, we further demonstrate that Gli1+ FAPs have an increased capacity for clonal expansion and a concomitant reduction in adipogenic fate decisions in vitro. These results are supported by our in vivo experiments using a muscle glycerol injury model with significant fat infiltration. By combining fate tracing with cell ablation studies, we found that elimination of Gli1+ FAPs increases the number of adipocytes within muscles. We also observed that treating mice with a Hh activator reduced intramuscular adipocytes and improved muscle healing after injury, whereas treatment with a Hh inhibitor had opposite effects. One limitation of these agents is that they can work both on Gli1+ and Gli1− FAPs, as well as on other cell types, such as endothelial cells in the interstitium and myoblasts. Thus, we cannot rule out the effects of Hh signaling on other cell types or mature myofibers, since pro-myogenic and anti-adipogenic effects of Hh activation might involve both cell-autonomous and cell-nonautonomous mechanisms. Although Gli1+ FAPs exhibit an intrinsically weak adipogenic differentiation ability, these cells could affect the fat infiltration through additional mechanisms, such as suppressing adipogenesis in the rest of FAPs via secreting unknown factors. Moreover, our in vitro differentiation assay and the in vivo cell ablation experiment clearly indicate that direct action of Hh signaling on Gli1+ FAPs plays an important role in muscle repair.
In summary, we discovered a subpopulation of FAPs with a high level of Hh signaling and provided a model in which the Gli1+ FAPs cells respond and participate in muscle regeneration. Our work strongly supports a model in which a distinct subpopulation of FAPs with a high expression level of Gli1 ensures efficient muscle regeneration by regulating myofiber size and preventing adipogenic fate (Fig. 8) that together promote the proper regenerative function of skeletal muscles. Failure of this mechanism leads to defective tissue repair, including decreased myofiber size and intramuscular accumulation of fat. Consistent with a previous study showing that Hh signaling regulates adipogenesis of FAPs and muscle regeneration,(36) our findings discover the underlying mechanism and support the notion that pharmacological perturbation of Hh signaling in FAPs can be therapeutically applied to boost muscle regeneration.
Supplementary Material
Acknowledgments
This work was supported by NIH grants NIH/NIAMS R01AR066098, R21AR074570 to LQ, startup funds from the Perelman School of Medicine, the McCabe Award, the Muscular Dystrophy Association (MDA-603883), NASA (18-FG_ind_2–0022), the NIH (R01HL146662–01A1) and the AHA (17GRNT33671042) to FM, and NIH P30AR069619 to PCMD. We thank the Stemmler Animal Facility staff, the Penn Flow Cytometry Core, and the Penn Center for Musculoskeletal Disorders (PCMD) Histology Core at the McKay Laboratory for their assistance.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
The authors declare no competing financial interests or potential conflicts of interest.
Authors’ roles: LQ, FM, LY, and EDT designed the study and interpreted data. LY performed animal experiments. LY, EDT, and LZ performed FACS sorting and flow cytometry analysis. LY, SM, EA, and LW performed histology and imaging analysis. LY and SM performed cell culture and qRT-PCR experiments. LY analyzed scRNA-seq data. EDT and SY provided technical support and consultation. LQ and FM wrote the manuscript with input from LY and EDT.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1002/jbmr.4265.
References
- 1.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 2011;138(17):3625–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev 2013;93(1):23–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol 2010;12(2):143–152. [DOI] [PubMed] [Google Scholar]
- 4.Joe AW, Yi L, Natarajan A, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol 2010;12(2):153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wosczyna MN, Biswas AA, Cogswell CA, Goldhamer DJ. Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. J Bone Miner Res 2012;27(5):1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wosczyna MN, Rando TA. A muscle stem cell support group: coordinated cellular responses in muscle regeneration. Dev Cell 2018;46(2): 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uezumi A, Fukada S, Yamamoto N, et al. Identification and characterization of PDGFRalpha+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis 2014;5:e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemos DR, Babaeijandaghi F, Low M, et al. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat Med 2015;21(7):786–794. [DOI] [PubMed] [Google Scholar]
- 9.Mozzetta C, Consalvi S, Saccone V, et al. Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol Med 2013;5(4):626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiore D, Judson RN, Low M, et al. Pharmacological blockage of fibro/adipogenic progenitor expansion and suppression of regenerative fibrogenesis is associated with impaired skeletal muscle regeneration. Stem Cell Res 2016;17(1):161–169. [DOI] [PubMed] [Google Scholar]
- 11.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 2004; 118(4):505–516. [DOI] [PubMed] [Google Scholar]
- 12.Tichy ED, Sidibe DK, Greer CD, et al. A robust Pax7EGFP mouse that enables the visualization of dynamic behaviors of muscle stem cells. Skelet Muscle 2018;8(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tichy ED, Sidibe DK, Tierney MT, et al. Single stem cell imaging and analysis reveals telomere length differences in diseased human and mouse skeletal muscles. Stem Cell Rep 2017;9(4):1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisani DF, Bottema CD, Butori C, Dani C, Dechesne CA. Mouse model of skeletal muscle adiposity: a glycerol treatment approach. Biochem Biophys Res Commun 2010;396(3):767–773. [DOI] [PubMed] [Google Scholar]
- 15.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177(7):1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijk D, Sharma R, Nainys J, et al. Recovering gene interactions from single-cell data using data diffusion. Cell 2018;174(3):716–29.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16(5): 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13(1):133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostrominova TY. Application of WGA lectin staining for visualization of the connective tissue in skeletal muscle, bone, and ligament/tendon studies. Microsc Res Tech 2011;74(1):18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature 2008; 456(7221):502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris JB, MacDonell CA. Phospholipase A2 activity of notexin and its role in muscle damage. Toxicon 1981;19(3):419–430. [DOI] [PubMed] [Google Scholar]
- 23.Malecova B, Gatto S, Etxaniz U, et al. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat Commun 2018;9(1):3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Wei H, Guo X, et al. Functional verification of the diphtheria toxin A gene in a recombinant system. J Anim Sci Biotechnol 2012;3 (1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penfornis P, Pochampally R. Colony forming unit assays. Methods Mol Biol 2016;1416:159–169. [DOI] [PubMed] [Google Scholar]
- 26.Uezumi A, Ikemoto-Uezumi M, Tsuchida K. Roles of nonmyogenic mesenchymal progenitors in pathogenesis and regeneration of skeletal muscle. Front Physiol 2014;5:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 2006;133(1):3–14. [DOI] [PubMed] [Google Scholar]
- 28.Sinha S, Chen JK. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol 2006;2(1):29–30. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J, Siclari VA, Qin L. Isolating endosteal mesenchymal progenitors from rodent long bones. Methods Mol Biol 2015;1226:19–29. [DOI] [PubMed] [Google Scholar]
- 30.Geng L, Lu K, Li P, et al. GLI1 inhibitor GANT61 exhibits antitumor efficacy in T-cell lymphoma cells through down-regulation of p-STAT3 and SOCS3. Oncotarget 2017;8(30):48701–48710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giordani L, He GJ, Negroni E, et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol Cell 2019;74(3):609–21.e6. [DOI] [PubMed] [Google Scholar]
- 32.Madaro L, Passafaro M, Sala D, et al. Denervation-activated STAT3-IL-6 signalling in fibro-adipogenic progenitors promotes myofibres atrophy and fibrosis. Nat Cell Biol 2018;20(8):917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biferali B, Proietti D, Mozzetta C, Madaro L. Fibro-adipogenic progenitors cross-talk in skeletal muscle: the social network. Front Physiol 2019;10:1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang X, Yang MY, Shi YX, et al. Interleukin-15 facilitates muscle regeneration through modulation of fibro/adipogenic progenitors. Cell Commun Signal 2018;16(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukjanenko L, Karaz S, Stuelsatz P, et al. Aging disrupts muscle stem cell function by impairing matricellular WISP1 secretion from fibroadipogenic progenitors. Cell Stem Cell 2019;24(3):433–46.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopinke D, Roberson EC, Reiter JF. Ciliary hedgehog signaling restricts injury-induced adipogenesis. Cell 2017;170(2):340–51.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott RW, Arostegui M, Schweitzer R, Rossi FMV, Underhill TM. Hic1 defines quiescent mesenchymal progenitor subpopulations with distinct functions and fates in skeletal muscle regeneration. Cell Stem Cell 2019;25(6):797–813.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu N, Garry GA, Li S, et al. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol 2017;19(3):202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pannerec A, Formicola L, Besson V, Marazzi G, Sassoon DA. Defining skeletal muscle resident progenitors and their cell fate potentials. Development 2013;140(14):2879–2891. [DOI] [PubMed] [Google Scholar]
- 40.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 2014;141(18):3445–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, He G, Lee W-C, McKenzie JA, Silva MJ, Long F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat Commun 2017;8(1):2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Tower RJ, Chandra A, et al. Periosteal mesenchymal progenitor dysfunction and extraskeletally-derived fibrosis contribute to atrophic fracture nonunion. J Bone Miner Res 2019;34(3):520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu JK, McGlinn E, Harfe BD, Kardon G, Tabin CJ. Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev 2012;26(18):2088–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forcina L, Miano C, Scicchitano BM, Musaro A. Signals from the niche: insights into the role of IGF-1 and IL-6 in modulating skeletal muscle fibrosis. Cells 2019;8(3):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuswanto W, Burzyn D, Panduro M, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin 33-dependent accumulation of regulatory T cells. Immunity 2016; 44(2):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uezumi A, Ito T, Morikawa D, et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 2011;124(Pt 21):3654–3664. [DOI] [PubMed] [Google Scholar]
- 47.Heredia JE, Mukundan L, Chen FM, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell 2013;153(2):376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mann CJ, Perdiguero E, Kharraz Y, et al. Aberrant repair and fibrosis development in skeletal muscle. Skelet Muscle 2011;1(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soliman H, Paylor B, Scott RW, et al. Pathogenic potential of Hic1-expressing cardiac stromal progenitors. Cell Stem Cell 2020;26 (2):205–20.e8. [DOI] [PubMed] [Google Scholar]
- 50.Soliman H, Rossi FMV. Cardiac fibroblast diversity in health and disease. Matrix Biol 2020;91–92:75–91. [DOI] [PubMed] [Google Scholar]
- 51.Arrighi N, Moratal C, Clement N, et al. Characterization of adipocytes derived from fibro/adipogenic progenitors resident in human skeletal muscle. Cell Death Dis 2015;6:e1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kramann R, Schneider RK, DiRocco DP, et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 2015;16(1):51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider RK, Mullally A, Dugourd A, et al. Gli1(+) mesenchymal stromal cells are a key driver of bone marrow fibrosis and an important cellular therapeutic target. Cell Stem Cell 2017;20(6): 785–800.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A published sequence data has been used in this work (GEO: GSE110878). Software used to analyze the data is either freely or commercially available.


