Summary
Organoids mimic the physiologic and pathologic events of organs. However, no consensus on esophageal organoid (EO) culture methods has been reached. Moreover, organoid models reproducing esophageal squamous cell carcinoma (ESCC) initiation have been unavailable. Herein, we sought to develop an esophageal minimum essential organoid culture medium (E-MEOM) for culturing murine EOs and establishing an early ESCC model. We formulated E-MEOM to grow EOs from a single cell with clonal expansion, maintenance, and passage. We found that EOs cultured in E-MEOM were equivalent to the esophageal epithelium by histological analysis and transcriptomic study. Trp53 knockout and KrasG12D expression in EOs induced the development of esophageal squamous neoplasia, an early lesion of ESCC. Here we propose the new formula for EO culture with minimum components and the organoid model recapitulating ESCC initiation, laying the foundation for ESCC research and drug discovery.
Subject areas: Cancer, Biological sciences research methodologies, Methodology in biological science
Graphical abstract
Highlights
-
•
Identification of minimal components for murine EO growth and maintenance
-
•
Mouse EOs morphologically and transcriptionally recapitulate the human esophagus
-
•
Trp53 KO and KrasG12D induced esophageal neoplasia mimicking early ESCC
Cancer; Biological sciences research methodologies; Methodology in biological science
Introduction
The esophageal epithelium comprises a proliferative basal layer and differentiated suprabasal layers of epithelial cells (Whelan et al., 2018). The basal epithelium of the murine esophagus contains both proliferating stem and transit-amplifying cells that self-renew and differentiate over the lifespan of the tissue (DeWard et al., 2014). A proliferation-differentiation gradient is generated during this dynamic process over 2 weeks (Doupe et al., 2012). Recently, a three-dimensional (3D) organoid system featuring the physiologic and pathologic processes of organs has drawn tremendous attention in studying stem cells and diseases (Tuveson and Clevers, 2019). Initially, Hans Clevers' and Calvin Kuo's groups succeeded in culturing intestinal organoids (Ootani et al., 2009; Sato et al., 2009). Then, numerous laboratories adopted similar methods to grow organoids from the colon, stomach, and esophagus of both human and murine tissues (DeWard et al., 2014; Sato et al., 2011; Schumacher et al., 2015; Zhang et al., 2018b). Induced pluripotent stem cells were also shown to differentiate into specific tissue organoids (Zhang et al., 2018a). As organoids can be generated from fresh tissues from surgery, biopsy specimens, or frozen tissues, they have been proposed as a promising tool for translation into personalized medicine for cancer treatment (Li et al., 2018).
3D culture of esophageal cells was initially performed by Eric Lagasse and colleagues (DeWard et al., 2014). They established murine esophageal spheroid (henceforth termed “esophageal organoid” [EO]) from the mucosa, mimicking the stratified squamous epithelium of the esophagus by using culture medium containing advanced Dulbecco's modified Eagle's medium (DMEM)/F-12, N2 supplement, B27 supplement, GlutaMAX supplement, HEPES buffer, penicillin/streptomycin, N-acetyl-L-cysteine, gastrin, nicotinamide, SB202190 (p38 mitogen-activated protein kinase inhibitor), epidermal growth factor (EGF), Wnt3A, R-Spondin1, Noggin, and A83-01 (transforming growth factor β kinase/activin receptor-like kinase [TGF-β/ARLK] inhibitor), which was similar to that used for human and murine intestinal/colonic crypts (Sato et al., 2011) and gastric organoids (Schumacher et al., 2015). Similarly, other laboratories have used a modified culture medium to grow EOs with Rho-associated coiled-coil-containing protein kinase (ROCK) inhibitor Y-27632 (Kasagi et al., 2018; Natsuizaka et al., 2017) and FGF10 (Li et al., 2018; Sato et al., 2011), in the offsetting of depleted gastrin, nicotinamide, SB202190, A83-01, or Wnt3A (Chretien et al., 2017; Natsuizaka et al., 2017). However, there has been no comparative analysis of each component's role in EOs' initiation, growth, maintenance, and morphology.
Esophageal cancer is the eighth most common cancer and the sixth highest cause of cancer deaths globally (Torre et al., 2015). Esophageal squamous cell carcinoma (ESCC) accounts for over 80% of all esophageal cancer cases and has a poor prognosis because of a lack of symptoms at early stages (Glenn, 2001). ESCC develops from squamous dysplasia as a typical histologic precursor lesion (Wang et al., 2005). Given that early diagnosis of ESCC may lead to better outcomes (Pennathur et al., 2013), understanding the mechanisms of esophageal neoplasia is imperative. Mice treated with 4-nitroquinoline 1-oxide, a potent oral-esophageal carcinogen, for 16 weeks generate dysplastic mucosa and primary tumors (Natsuizaka et al., 2017). Ccnd1 transgenic mice combined with Trp53 KO or chemically induced carcinogenesis develop dysplasia in 3–16 months (Mueller et al., 1997; Opitz et al., 2002). However, as these models develop ESCC lesions in 4–16 months, current ESCC models are not practical for studying ESCC initiation and translation into personalized medicine.
Despite the recent advances in utilizing organoids in the context of tissue regeneration and cancer (Drost et al., 2015; Katsura et al., 2019), EOs recapitulating esophageal neoplasia remain unexplored. Herein, we sought to develop an esophageal minimum essential organoid culture medium (E-MEOM) and establish an esophageal epithelial neoplasia model pathologically relevant to ESCC initiation.
Results
Comparative analysis of EO culture media
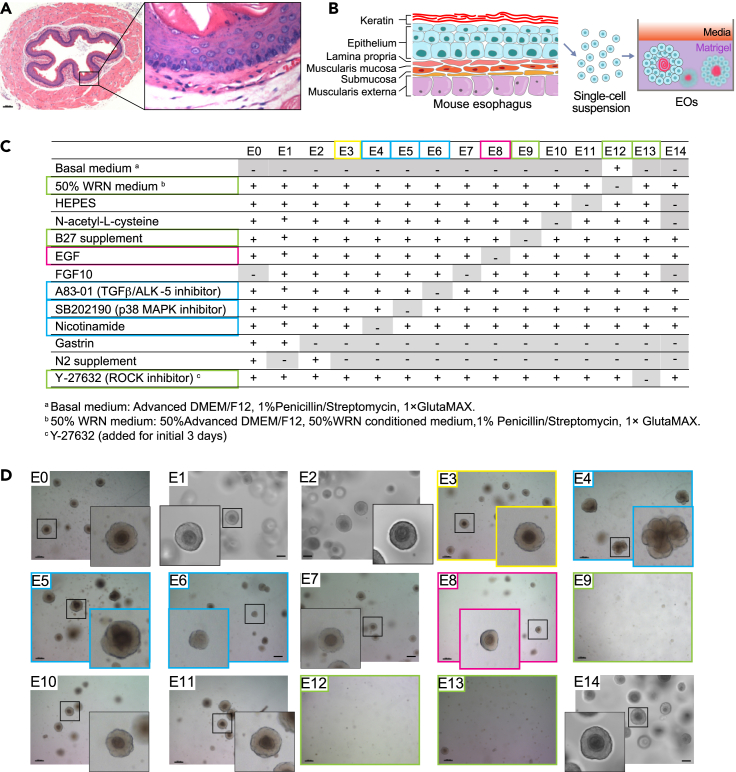
For EO culture, we dissected the esophagi from C57BL/6 mice into 0.5-cm3 pieces. The isolated esophageal epithelium was processed into a single-cell suspension with 0.05% trypsin-EDTA, seeded in Matrigel (Figures 1A and 1B). To develop E-MEOM for EO growth and maintenance, we prepared 15 different esophageal culture media (E0-E14; Figure 1C) by omitting each component of EO culture media that has previously been utilized. E0 medium, which served as a positive control, contained 50% WRN medium (50% advanced DMEM/F-12, 50% WRN [Wnt3A, R-Spondin1, and Noggin] conditioned medium, 1% penicillin/streptomycin, 1× GlutaMAX), 10 mM HEPES buffer, 1 mM N-acetyl-L-cysteine, 1 × B27 supplement, 50 ng/mL EGF, 500 nM A83-01, 10 μM SB202190, 10 mM nicotinamide, 1 × N2 supplement, 10 nM gastrin, and, for the first 3 days, 10 μM Y-27632. As N2 supplement or gastrin exclusion (E1 or E2) showed no effect on EO growth compared with E0-grown EOs, E3 medium served as the complete medium (E0 medium +200 ng/mL FGF10; without N2 and gastrin, Figures 1C and 1D; yellow box). E4 medium was complete medium without nicotinamide, E5 medium was complete medium without SB202190, E6 medium was complete medium without A83-01, E7 medium was complete medium without FGF10, E8 medium was complete medium without EGF, E9 medium was complete medium without B27, E10 medium was complete medium without N-acetyl-L-cysteine, E11 medium was complete medium without HEPES, E12 medium was complete medium without 50% WRN conditioned medium, E13 medium was complete medium without Y-27632, and E14 was complete medium without HEPES, N-acetyl-L-cysteine, and FGF10. The panel of growth conditions was evaluated by monitoring the growth of EOs cultured in each medium. We observed that the removal of B27 (E9), WRN conditioned medium (E12), or Y-27632 (E13) caused the failure of EOs to grow (Figure 1D, green boxes). Notably, the removal of either nicotinamide (E4) or A83-01 (E6) resulted in the reduction of keratinization of EOs at day 7 (d7). Conversely, SB202190 depletion (E5) led to over-keratinization and a single-cell layer of EOs at d7 (Figures 1D and S1A, blue boxes). Additionally, we found that the organoid-forming efficiencies of EOs grown in E3, E6–E8, E10, and E11 were compatible with that of EOs cultured in E0 medium. However, organoid-forming efficiencies were lower in the E4 and E5 groups compared with those in the E0 medium (Figure S1B). The size of EOs grown in E1–E7, E10, and E11 was similar to that of EOs grown in E0, whereas EOs cultured in E8 (removal of EGF) were smaller (Figure 1C, red box). These results suggest that B27, WRN conditioned medium, and Y-27632 are indispensable for the initiation of EO growth; nicotinamide, A83-01, and SB202190 likely contribute to EO differentiation such as keratinization; and EGF is essential for EO growth (Figures 1C and 1D, colored boxes). In contrast, our combinatorial approach showed that N2, gastrin, FGF10, N-acetyl-L-cysteine, and HEPES are dispensable for EO initiation, growth, and maintenance. We also tested the E14 medium, which has no N2, gastrin, FGF10, N-acetyl-L-cysteine, and HEPES from the E0 medium because they were shown to be non-essential factors for EO growth. We found that E14-grown EOs are established in similar size and efficiency compared with E0-grown EOs.
Figure 1.
Comparative analysis of esophageal culture media
(A) Photomicrographs of mouse esophagus. Transverse sectioning; scale bar, 100 μm.
(B) Illustration of mouse esophagus and experimental scheme for EO culture from primary esophageal epithelial cells.
(C) Different esophageal culture media established and tested.
(D) Morphology of EOs in different esophageal culture media. Bright-field images of EOs grown in the different culture media at 7 days (d7). Scale bars, 200 μm. The lower panels display magnified images.
Images are representative of three experiments with similar results.
Additionally, to exclude the possible effects of FBS of WRN conditioned-medium, we prepared a WRN medium containing purified Wnt3A, R-Spondin1, and Noggin rather than using WRN-conditioned medium (Figure S1D). The organoid formation of E0, E14, and WRN media was shown to be comparable. However, deficiency of any of the three factors (Wnt3A, R-Spondin1, and Noggin) resulted in slowed growth of EOs (Figures S1E and S1F). The EOs grown without Noggin showed increased keratinization, whereas EOs cultured without Wnt3A or R-Spondin1 showed decreased expansion. Thus, we identified B27, Wnt3A. R-Spondin1, Noggin, Y-27632, nicotinamide, A83-01, and SB202190 as essential factors for EO culture.
Analysis of EOs cultured in E-MEOM
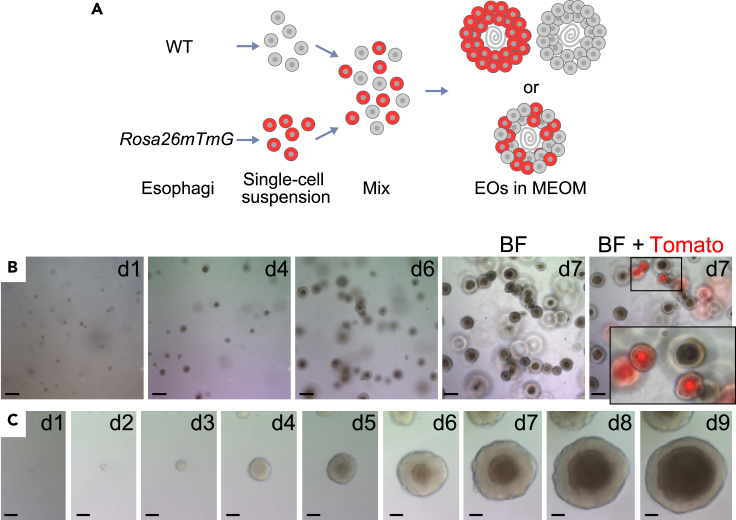
Having determined the essential components of EO culture media (Figure 1), we named the E14 as E-MEOM with the following reagents: 50% WRN medium, B27, Y-27632 (first 3 days after seeding), nicotinamide, A83-01, SB202190, and EGF. Then, we tested whether E-MEOM is as effective and compatible as the positive control medium (E0) for EO culture. First, we tested whether EOs in E-MEOM can be generated from a single esophageal epithelial cell. To this end, we utilized Rosa26mTmG mice ubiquitously expressing Tomato, a red fluorescent protein, in cells (Muzumdar et al., 2007). Two groups of single-cell suspensions of the esophageal epithelial cells isolated from wild-type (non-red; C57BL/6) and Rosa26mTmG (red; Tomato positive) mice were mixed and cultured in Matrigel with E-MEOM (Figure 2A). After the initial formation of EOs at d7, EOs were monitored for Tomato expression (Tomato+). Fluorescence microscopy showed that EOs were completely either Tomato+ or Tomato− (Figure 2B). Such mutual exclusiveness in Tomato expression suggests that EOs cultured in E-MEOM were derived from a single cell rather than the cell aggregates. Additionally, we observed the development of EOs from a single cell in E-MEOM (Figure 2C), further confirming the clonal expansion of the esophageal epithelial cells into EOs in E-MEOM. Next, we compared the initiation and growing efficiency of EOs cultured in E0 or E-MEOM medium. The forming efficiency and size of EOs grown in both E0 and E-MEOM were similar up to three passages (Figures S2A and S2B). Of note is that we were able to continuously maintain and passage EOs in E-MEOM (tested for six weeks). Next, to address whether E-MEOM is as effective as E0 medium for maintaining EOs, we performed a freezing-thawing experiment. After single-cell suspensions from EOs were frozen and thawed, EO-forming efficiency and size were equivalent in EOs cultured in either E-MEOM or E0 medium (Figures S2C–S2F). These results suggest that E-MEOM is compatible with the E0 medium with regard to clonal expansion, initiation, growth, forming efficiency, maintenance, and stocking.
Figure 2.
Analysis of EOs cultured in E-MEOM (esophageal minimum essential organoid culture medium)
(A and B) Clonal expansion and development of EOs in E-MEOM.
(A) Illustration of the experimental scheme. C57BL/6 (wild-type [WT] and Rosa26mTmG) mice were euthanized to collect the esophagi. The esophageal epithelial cells isolated from WT (no fluorescence) and Rosa26mTmG (red fluorescence; Tomato positive) mice were mixed and cultured in E-MEOM.
(B) Exclusive Tomato expression between WT (no fluorescence) and Rosa26mTmG EOs developed from single cells. Bright-field images (d1-d7) of clonal expansion of EOs in E-MEOM from the mixed single-cell suspension. Scale bars, 200 μm.
(C) E-MEOM sufficiently supports the single esophagus cell growth. Bright-field images (d1-d9) of EOs cultured in E-MEOM from a single cell. Scale bars, 50 μm.
Images are representative of three experiments with similar results.
EOs cultured in E-MEOM recapitulate the esophageal epithelium
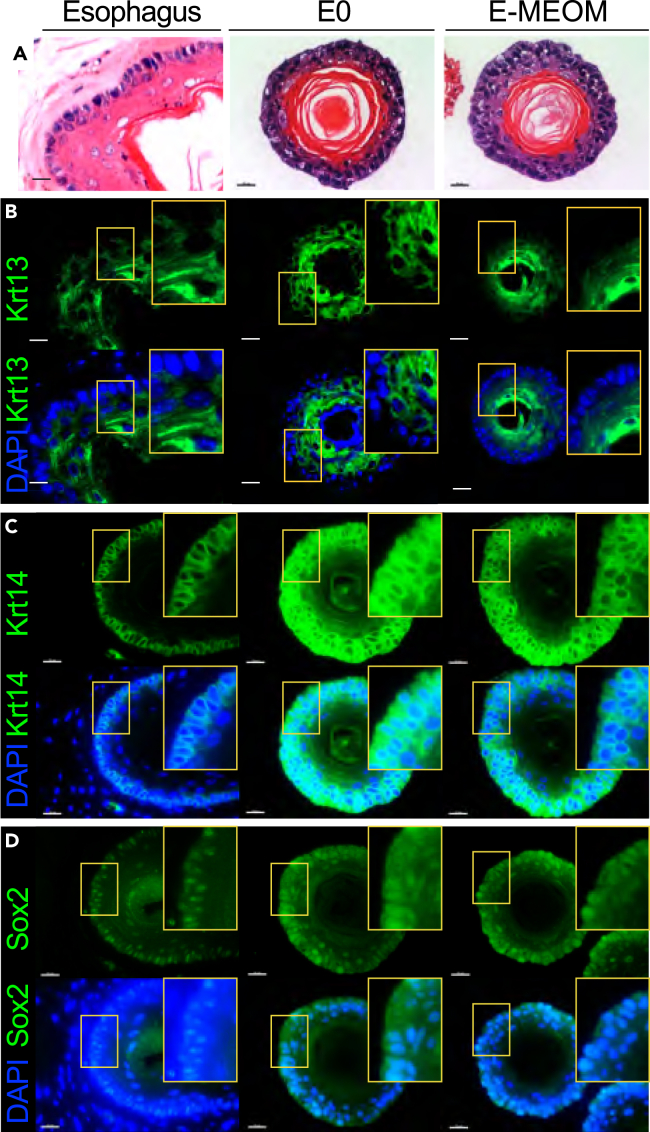
As mini-organs, EOs are expected to mimic physiologic events in the esophagus. We thus examined whether EOs in E-MEOM were morphologically similar to the normal esophageal epithelium. Hematoxylin and eosin (H&E) staining showed that the EOs cultured in E0 and E-MEOM exhibited morphological similarity to the normal esophageal epithelium, with a small basal-like cell layer outward, 2–3 large flat suprabasal-like cell layers inward, and keratinization in the lumen (Figure 3A). Next, we analyzed the expression of markers for the esophageal epithelium. Like the normal esophageal epithelial cell layer, the organoid cells showed higher expression of cytokeratin 13 (Krt13) in the differentiating cells, whereas the outer cell layer expressed cytokeratin 14 (Krt14) and Sox2 with the cell proliferation marker (Ki67) (Figures 3B–3D and S3A). Like the esophageal epithelium, EOs in E0 or E-MEOM also showed a similar β-catenin expression pattern with a rare population of cleaved caspase-3-positive cells (Figures S3B and S3C). To complement the immunohistochemical analyses, we also conducted a quantitative reverse transcriptase-PCR (qRT-PCR) analysis. EOs grown in E0 or E-MEOM expressed similar mRNA levels of Krt13, Krt14, Krt4, Ki67, Tert, and Sox2 (Figure S3D). These results suggest that EOs cultured in E-MEOM are histologically similar to the normal esophageal epithelium.
Figure 3.
Immunohistochemical analysis of EOs cultured in E-MEOM
(A) EOs cultured in E-MEOM medium recapitulate the esophageal epithelium morphology; H&E staining.
(B–D) Expression of esophageal epithelium markers in EOs cultured in E0 and E-MEOM medium; immunofluorescent staining of (B) Krt13, (C) Krt14, and (D) Sox2.
Scale bars, 20 μm. All images are representative of three experiments.
Single-cell transcriptome-based comparison of mouse EOs and human esophagus
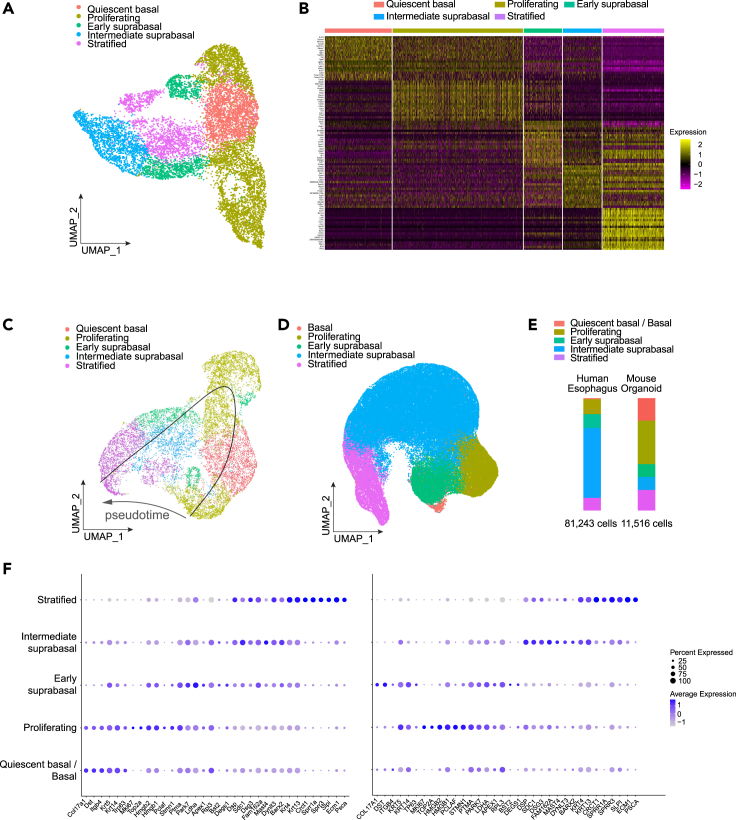
To comparatively analyze mouse EOs and human esophageal tissue, we employed single-cell RNA sequencing (scRNA-seq). We constructed 11,516 single-cell transcriptome libraries from day 8 mouse EOs and visualized the clusters using Uniform Manifold Approximation and Projection (UMAP) dimensional reduction method (Figure 4A). Cell clustering with the specific markers of unique gene expression profiles showed that our mouse EOs consist of quiescent basal, proliferating, early suprabasal, intermediate suprabasal, and stratified cells, similar to the composition of human esophageal epithelial cells (Figure 4B). Further marker gene analyses exhibited that each cell cluster was enriched with the expression of relevant marker genes (see Table S3 for marker genes of clusters information). For instance, the quiescent basal cell population showed increased expression of its marker genes, Col17a1 and Dst (Busslinger et al., 2021). Also, proliferating cells and early suprabasal cells showed high expression of Mki67, Pclaf, and Stmn1 concomitant with the previously known basal cell markers including Tp63, cytokeratin 14 (Krt14), and cytokeratin 5 (Krt5) (DeWard et al., 2014; Trisno et al., 2018). The differentiating cell markers (Sprr3, Ecm1, cytokeratin 4 [Krt4], and cytokeratin 13 [Krt13]) were enriched in the intermediate suprabasal cells and stratified cells. Next, we performed trajectory analysis to identify the direction of differentiation with two different algorithms (Slingshot and Monocle 3). Two different trajectory inferences showed the corresponding results that the quiescent basal cells or proliferating cells could differentiate to stratified cells through early suprabasal cells and intermediate suprabasal cells (Figures 4C, S4A, and S4B). Furthermore, to address whether mouse EOs recapitulate the cellular landscape of the human esophagus, we compared the single-cell transcriptional profile of EOs with that of human esophageal tissue. Given that mouse EOs do not contain mucinous and immune cells, we excluded those cells from the scRNA-seq dataset of the human esophagus and visualized the epithelial cell composition with UMAP projection (Figure 4D). Mouse EO cells and human esophageal cells are highly similar in cellular heterogeneity and marker gene expression of each cell cluster (Figures 4E and 4F). Notably, the proportion of proliferating cells was higher in mouse EO cells than that in human esophagus cells, whereas the intermediate suprabasal and stratified cells were the major populations in the human esophagus cells (Figure 4E), which might be due to the overall increased cell stemness and constitutively proliferative nature of EOs compared with human esophagus. These single-cell transcriptomic results suggest that our EO model recapitulates human esophageal epithelium with similar cell composition and transcriptional signature.
Figure 4.
Comparative analysis of murine EOs and human esophagus tissue using scRNA-seq
(A) UMAP displaying the scRNA-seq data of murine EOs. Cell clusters are indicated in different colors, and cluster names were assigned based on the marker genes’ expression.
(B) Heatmap showing the marker genes of each cell cluster. Each column represents a single cell grouped with clusters. Each row shows highly expressed genes in selected clusters.
(C) UMAP visualization of trajectory inference calculated by Slingshot algorithm. The arrows show the direction of cell differentiation. Cell clusters were distinguished by different colors.
(D) UMAP projection of human esophageal cells. The dataset was normalized with epithelial cells. Each cell cluster was named based on the marker gene expression.
(E) The proportions of each cluster of EOs and human esophagus cells are displayed in the histogram. The composition of 5 cell types of two datasets is colored for comparison.
(F) Dotplots of marker gene expression across the murine EOs and human esophageal cells in each cell cluster.
Neoplastic growth of EOs induced by KrasG12D and Trp53 KO in E-MEOM
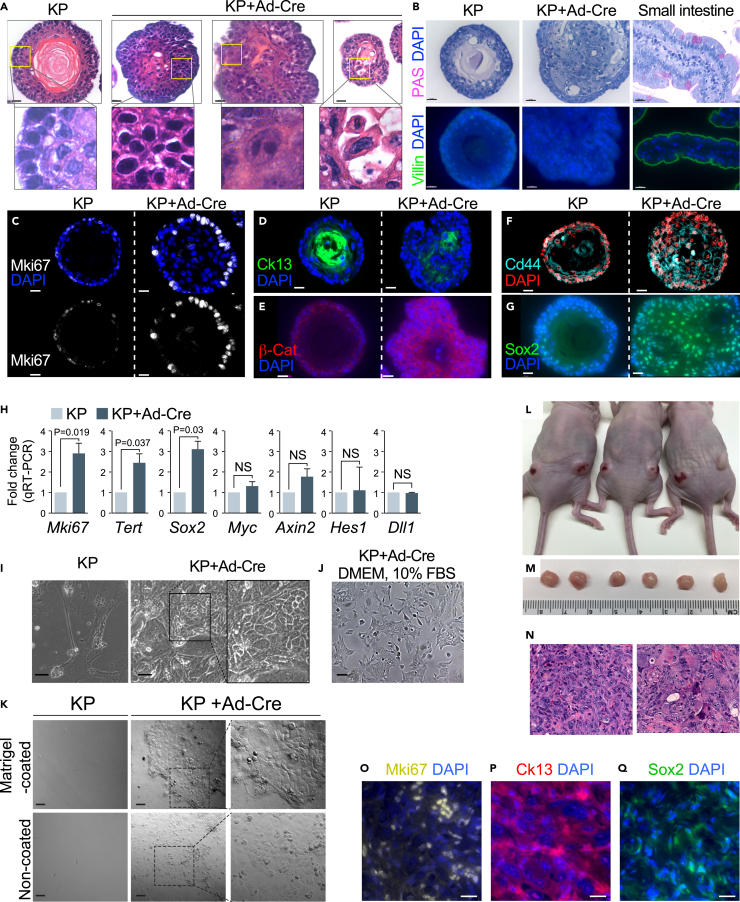
As our E-MEOM was sufficient to culture EOs from the normal primary esophageal epithelium, we asked whether EOs cultured in E-MEOM can be employed to study ESCC. Given the frequent genetic mutations of TP53 and KRAS in ESCC (Bandla et al., 2012; Smyth et al., 2017) and TP53 inactivation in early precancerous neoplasia lesions (Yamamoto et al., 2016), genetic manipulation of Trp53 and Kras likely leads to the development of neoplastic organoids. To test this hypothesis, we utilized KrasLSLG12D:Trp53floxed/floxed (KP) mice for conditional expression of oncogenic KrasG12D and KO of Trp53 alleles by Cre-LoxP recombination. Intriguingly, the EOs from KrasLSLG12D:Trp53floxed/floxed mice with Ad-Cre infection (KrasG12D:Trp53 knockout [KO]; hereafter referred to as KP+Ad-Cre) exhibited aberrant morphology, with loss of luminal keratinization and non-spherical shape, and increased growth, compared with the control groups (WT, WT + Ad-Cre, KP without Ad-Cre infection) (Figure S5). Moreover, KP+Ad-Cre organoids showed tumor-like growth in 2D and 3D culture with neoplastic characteristics of ESCC (Figures 5A–5Q). These results suggest that genetic manipulation of EOs with KrasG12D and Trp53 KO recapitulates early ESCC ex vivo (in both 3D and 2D cultures) and in vivo (mouse transplants).
Figure 5.
Recapitulation of early ESCCs by KrasG12D:Trp53 KO model
(A–I) Expression of esophageal epithelium neoplasia markers in EOs in E-MEOM.
(A) H&E staining of KP+Ad-Cre EOs displayed loss of cell polarity, increased mitotic nuclei, nuclear polymorphism, and multiple nuclei.
(B) ESCC-related neoplastic transformation. Possibility of EAC development was excluded by EAC marker immunostaining. PAS and Villin are not expressed in the KP+Ad-Cre organoids.
(C–G) Increased expression of β-catenin, Cd44, Ki67, and Sox2 in KP+Ad-Cre EOs. Immunofluorescence staining of Mki67 (C), Krt13 (D), β-catenin (E), Cd44 (F), and Sox2 (G) for KP and KP+Ad-Cre EOs cultured in E-MEOM.
(H) qRT-PCR for mRNA analysis of Ki67, Tert, Sox2, Myc, and Axin2. Experiments were performed three times.
(I–K) Neoplastic growth evaluation of KrasG12D:Trp53KO cell line in 2D culture model. (I) Bright-field images of KP+Ad-Cre versus KP cells cultured in a 24-well plate without Matrigel in EMEOM + Y-27632 (first 2 days). (J) Bright-field images of KP+Ad-Cre cells cultured in a 24-well plate in DMEM +10% FBS after three passages. (K) KP organoid cells and KP+Cre organoid cells were seeded on the Matrigel-coated or non-coated 2D culture plate. Cells were incubated for 2 weeks with medium change every 3 days, and differential interference contrast (DIC) images were taken by LSM-800.
(L–Q) In vivo tumorigenicity of KrasG12D:Trp53KO EO cells. KrasG12D:Trp53KO EO cells (5 × 106) were subcutaneously injected into nude mice and collected 4 weeks after transplantation. (L and M) Tumor images. (N) H&E staining. (N–P) Immunofluorescent staining of cell proliferation and ESCC marker. Ki67 (O), Krt13 (P), and Sox2 (Q).
Scale bars, 20 μm (A–G); 50 μm (I–K, and N–Q). All images are representative of three experiments. Error bars indicate mean ± SD. Groups were compared via two-sided unpaired t test.
Discussion
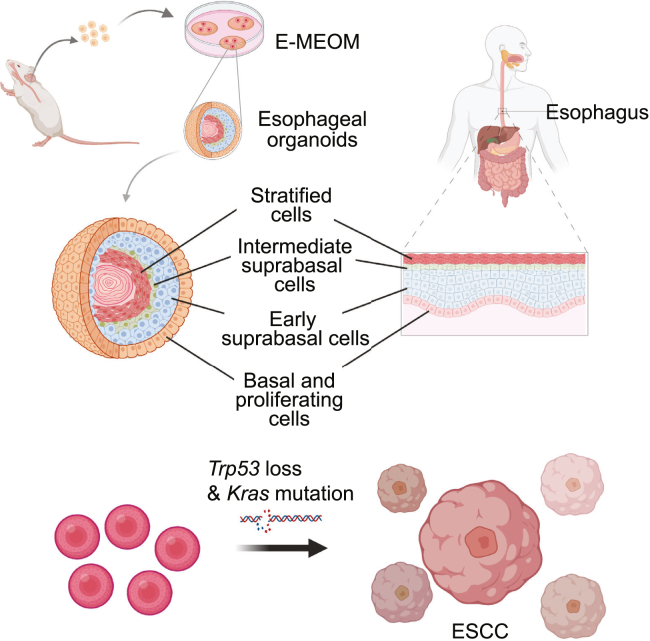
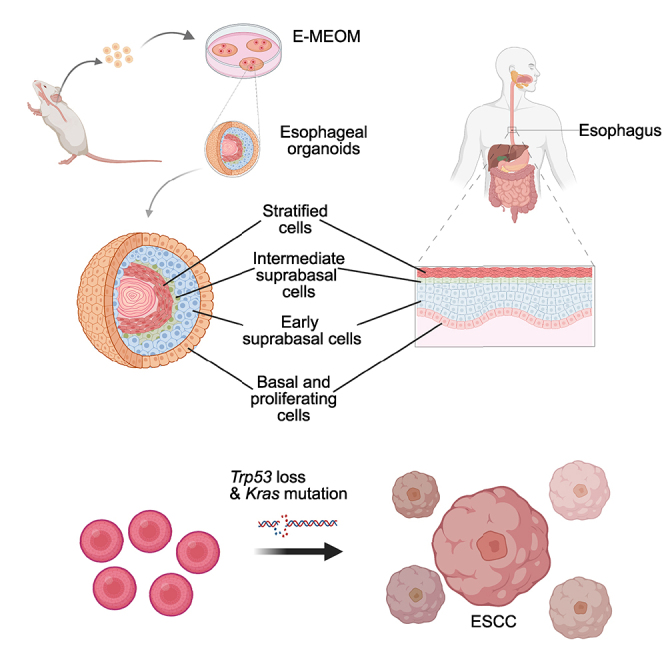
In this study, we developed E-MEOM for an efficient and economical method of EO culture and its application to study human ESCC (Figure 6). E-MEOM was sufficient for clonal expansion, initiation, growth, formation, maintenance, passaging, and stocking of EOs. EOs in E-MEOM were immunohistologically similar to the normal esophageal epithelium. Also, EOs in E-MEOM exhibited a similar cellular divergence and gene expression profile as human esophageal epithelial cells. Moreover, we established an early ESCC model by genetic manipulation of EOs in E-MEOM.
Figure 6.
Schematic concepts of EO culture system and ESCC study model
Mouse esophageal organoids are developed from single cells of esophageal tissue in E-MEOM. EOs in E-MEOM recapitulate cellular heterogeneity, differentiation, and stemness of human esophageal epithelial cells. EO culture in E-MEOM potentiates an applicable model for ESCC pathology with oncogenic Kras mutation and Trp53 deletion.
Organoids have been cultured in Matrigel with medium supplemented with growth factors (Wnt3A, R-Spondin1, Noggin, EGF, FGF10, B27, and N2), hormones (gastrin and nicotinamide), and chemical inhibitors (TGF-β/ARLK inhibitor A83-01, p38 mitogen-activated protein kinase inhibitor SB202190, and ROCK inhibitor Y-27632) (Sato et al., 2011; Schumacher et al., 2015; Whelan et al., 2018). Several groups used Noggin/R-Spondin-conditioned medium or medium with recombinant Noggin and R-Spondin protein without Wnt3A, A83-01, and SB202190 for EO culture (Giroux et al., 2017; Hisha et al., 2013; Karakasheva et al., 2020; Natsuizaka et al., 2017). However, the culture methods for EOs vary from study to study (Kasagi et al., 2018; Kijima et al., 2019; Sachdeva et al., 2021).
Given no consensus on EO culture methods, we hypothesized that there might be both essential and non-essential components for EO clonal expansion, initiation, growth, and maintenance. Having filled this knowledge gap will provide us with the opportunity to improve EO-related experiments with consistent results, recapitulating various biological and pathologic events in the esophagus. We here found that only WRN, EGF, B27, and ROCK inhibitor Y-27632 are essential for EO initiation, growth, and maintenance. Even though we did not directly compare every medium, it is likely that unlike EO cultured without Wnt3A (Giroux et al., 2017; Hisha et al., 2013; Karakasheva et al., 2020; Natsuizaka et al., 2017), EOs cultured with Wnt3A may grow relatively fast and efficiently compared with those grown without Wnt3A (Figures S1D–S1F). Also, it should be noted that R-Spondin is a Wnt agonist.
In addition, we found that nicotinamide, A83-01, and SB202190 are essential for EO differentiation. Although these components were disregarded in several studies (Karakasheva et al., 2020; Natsuizaka et al., 2017), it is clear that A83-01 and SB202190 greatly increase organoid forming efficiency and reduce incomplete organoid formation represented by abnormal keratinization (Figures 1C, S1A, and S1B). Nicotinamide is a form of vitamin B3 that inhibits sirtuins, which are involved in transcriptional regulation, metabolism, apoptosis, differentiation, and aging (Denu, 2005). EOs showed reduced keratinization without nicotinamide or A83-01, whereas the absence of SB202190 induced excessive keratinization. Additionally, nicotinamide and SB202190 are essential for EO-forming efficiency. Similar to the long-term culture of intestinal crypt organoids (Sato et al., 2011), TGF-β signaling likely drives esophageal epithelial cell differentiation. Additionally, whereas nicotinamide inhibits intestinal organoid differentiation, it promotes EO keratinization.
Our scRNA-seq data of mouse EOs showed the separated cell clusters with the same names of Proliferating, Early suprabasal, and Stratified cells. These cell clusters in the mouse esophagus also have been observed in other studies, whereas it was not observed in the human esophageal tissue scRNA dataset (Busslinger et al., 2021; Madissoon et al., 2019; Vercauteren Drubbel et al., 2021). These results indicate that there can be different lineages for cell differentiation or suggest the existence of sub-clusters. To test this, we used two different trajectory inferences to define the cell differentiation lineage. The possibility of separate cell lineage was shown in the result from Monocle 3 package (Figures S4A and S4B). With the pseudotime analysis, Quiescent basal and Proliferating cells were differentiated into Early suprabasal or Intermediate suprabasal cells in lineage 1, and Proliferating cells were differentiated into Stratified cells in the lineage 2. These different cell lineages can be originated from the distinctive structure of mouse esophageal tissue such as keratin layers compared with human esophageal tissue (Long and Orlando, 1999; Whelan et al., 2018). Also, these different cell lineage paths might indicate sub-clusters of cells that have not been identified yet. Although we have named each cluster based on the representative marker genes’ expression, the identification of new clusters or subtypes need to be further analyzed.
Although a few studies have used organoids to study EAC (Li et al., 2018), ESCC (Kijima et al., 2019), eosinophilic esophagitis (Kasagi et al., 2018), and Barrett esophagus neoplastic transformation (Liu et al., 2018), no reports have described the use of EOs in the study of ESCC precursor lesions. TP53 mutations are most frequent in ESCC (91%) (Smyth et al., 2017), which are detectable in early precancerous neoplasia lesions (Yamamoto et al., 2016). KRAS mutations are also observed in 17% of patients with ESCC (Bandla et al., 2012). Moreover, the downstream molecules of MAPK and PI3K signaling pathways are frequently dysregulated in ESCC (Lin et al., 2014). EGFR overexpression and TP53 mutations are often present in precancerous lesions, sufficient to transform esophageal epithelial cells (Okawa et al., 2007). In this context, in which ESCC lesions develop in 16 weeks to 16 months, we herein developed a new EO model pathologically similar to precancerous neoplasia of ESCC induced by KrasG12D:Trp53 KO. We cultured KP mice EOs in our E-MEOM with added Ad-Cre to induce KrasG12D:Trp53 KO mutations. Importantly, we developed high-grade neoplasia EOs, morphologically and pathologically different from WT and KP without Ad-Cre EOs. Moreover, these organoids could be transferred to 2D culture in DMEM +10% FBS and maintained well. Thus, we conclude that our KrasG12D:Trp53 KO EO model successfully mimics the pathological pattern of high-grade neoplasia of early ESCC, and we successfully developed a high-grade neoplasia cell line. Our KrasG12D:Trp53 KO EO model mimics the esophageal epithelium, whereas EOs co-cultured with fibroblasts, nerve cells, and immune cells that present in the esophagus and niche may be beneficial for studying metastasis in future studies. For example, “assembloids” composed of epithelial, stromal, and muscle cells show similar pathophysiological features of mature organs or cancer architecture (Kim et al., 2020; Marton and Pasca, 2020). Furthermore, it is necessary to apply our murine EO culture method to human EOs for diagnosis, prognosis, and drug discovery of ESCC.
Together, the E-MEOM for EO culture and a new model for murine esophageal neoplasia lay the foundation for the study of pathophysiologic mechanisms of ESCC and improvement of the prognosis of early ESCC and ESCC personalized medicine.
Limitations of the study
Although this study compared the growth media conditions and identified essential components for the murine EO culture, the finding should be translated into human esophageal organoids and ESCC initiation in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Krt13 | Abcam | Cat# ab92551; RRID:AB_2134681 |
| Rabbit anti-Krt14 | Abcam | Cat# ab7800; RRID:AB_306091 |
| Rabbit anti-Ki67 (Mki67) | Abcam | Cat# ab16667; RRID:AB_302459 |
| Rabbit anti-β-catenin | Cell Signaling Technology | Cat# 8480S; RRID:AB_11127855 |
| Rabbit anti-Sox2 | Cell Signaling Technology | Cat# 14962S; RRID:AB_2798664 |
| Rat anti-CD44 | BD Pharmingen | Cat# 550538; RRID:AB_393732 |
| Rabbit anti-Villin | Thermo Fisher Scientific. | Cat# PA5-22072; RRID:AB_11155190 |
| Biological samples | ||
| Human esophageal tissue | This paper | Fujian Medical University Union Hospital |
| Chemicals, peptides, and recombinant proteins | ||
| G-418 | Thermo Fisher Scientific. | Cat# MT30234CR |
| Hygromycin B | Thermo Fisher Scientific. | Cat# MT30240CR |
| 0.05% trypsin-EDTA | Thermo Fisher Scientific. | Cat# 25-052-CI |
| 50× B27 supplement | Thermo Fisher Scientific. | Cat# 12587010 |
| Y-27632 | Thermo Fisher Scientific. | Cat# 12541 |
| Penicillin/streptomycin | Life Technologies | Cat# 15140122 |
| 100× GlutaMAX™ | Life Technologies | Cat# 35050061 |
| 100× N2 supplement | Life Technologies | Cat# 17502-048 |
| HEPES | Sigma | Cat# H3375 |
| N-acetyl-L-cysteine | Sigma | Cat# A9165 |
| A83-01 | Sigma | Cat# SML0788 |
| SB202190 | Sigma | Cat# S7067 |
| Nicotinamide | Sigma | Cat# N0636 |
| Gastrin | Sigma | Cat# G9145 |
| Nutlin-3 | Sigma | Cat# N6287 |
| Murine recombinant EGF | PeproTech | Cat# 315-09 |
| Human recombinant FGF | PeproTech | Cat# 100-26 |
| Collagenase I | Thermo Fisher Scientific. | Cat# 17018029 |
| Critical commercial assays | ||
| Periodic Acid-Schiff (PAS) kit | Sigma | Cat# 395B-1KT |
| Deposited data | ||
| raw scRNA-seq data | This paper | GSE174577 |
| Experimental models: Cell lines | ||
| L-WRN | ATCC | CRL-3276 |
| Experimental models: Organisms/strains | ||
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | JAX:007676; RRID:IMSR_JAX:007676 |
| Mouse: B6.129P2-Trp53tm1Brn/J | The Jackson Laboratory | JAX:008462; RRID:IMSR_JAX:008462 |
| Mouse: B6.129S4-Krastm4Tyj/J | The Jackson Laboratory | JAX:008179; RRID:IMSR_JAX:008179 |
| Oligonucleotides | ||
| Primers for genotyping, qPCR, see Table S1 | This paper | NA |
| Software and algorithms | ||
| CellRanger | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| R (v4.0.3) | NA | http://www.r-project.org/ |
| R Studio (v1.4.1106) | NA | https://www.rstudio.com/ |
| Seurat (v4.0.3) | NA | https://satijalab.org/seurat/ |
| BiocManager (v1.30.16) | NA | https://www.bioconductor.org/ |
| Slingshot (v1.8.0) | NA | https://github.com/kstreet13/slingshot |
| Monocle3 | NA | http://cole-trapnell-lab.github.io/monocle-release/monocle3/ |
| ImageJ | ImageJ | https://ImageJ.nih.gov/ij/ |
| GraphPad Prism 8 | Graphpad | https://www.graphpad.com/scientificsoftware/prism/ |
Resource availability
Lead contact
Additional information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jae-Il Park (jaeil@mdanderson.org).
Materials availability
The materials will be available upon request.
Experimental model and subject details
Mice
C57BL/6, Rosa26mTmG (Jax no. 007676), Trp53floxed/floxed (Jax no. 008462), and KrasLSL-G12D/+ (008179) mice were purchased from the Jackson Laboratory. The KrasLSL-G12D/+;Trp53floxed/floxed (KP) compound strain was generated by genetic breeding. Mice were bred and housed in the Division of Laboratory Animal Resources facility at The University of Texas MD Anderson Cancer Center. All animal procedures were performed based on the guidelines of the Association for the Assessment and Accreditation of Laboratory Animal Care and institutionally approved protocols (IACUC00001141; The University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee). The study was compliant with all relevant ethical regulations regarding animal research.
Esophageal organoid culture model
The E-MEOM contained 50% WRN medium (50% Advanced DMEM/F12, 50% WRN conditioned medium, 1% penicillin/streptomycin, 1× GlutaMAX™), 1× B27, 50 ng/mL, 10 mM nicotinamide, 500 nM A83-01, 10 μM SB202190, 50 ng/mL EGF, and 10 μM Y-27632 ROCK inhibitor (first 3 days). Single esophageal epithelial cells (1000 cells) were suspended in 20 μL pre-thawed Matrigel (Corning) on ice and seeded as a droplet in the well centers of a 48-well plate. Then the plate was incubated at 37°C for 10 min to allow the Matrigel to solidify. Finally, 500 μL of different culture media (Figure 1C) were added to cover the Matrigel and incubated at 37°C with 5% CO2 to allow organoid formation. Media were changed every 1-3 days.
KrasG12D:Trp53KO EO model
Trp53floxed/floxed and KrasLSLG12D mice and WT mice were euthanized to collect the esophagi, which were digested into a single-cell suspension and seeded in Matrigel to form EOs as described above. After 7 days in culture, EOs from KP and WT mice were digested with 0.05% trypsin-EDTA at 37°C for 45 min to make a single-cell suspension. Ad-CMV-EGFP or Ad-Cre-EGFP (University of IOWA) was added to the cell suspension at 1 × 103 pfu/cell. Cells were then suspended in Matrigel to generate new organoids. After 2 days in culture, 10 μm nutlin3 was added for the selection. After 7 days in culture, EOs were collected for genotyping. Wild-type, Trp53floxed/floxed, and KO Trp53 alleles were amplified as 288 bp, 370 bp, and 612 bp, respectively. Wild-type, KrasG12D, or LSL- KrasG12D allele was amplified as 622 bp, 650 bp, and 500 bp, respectively (see Table S1 for primer information).
WRN (Wnt3A, R-spondin1, and noggin) conditioned medium
L-WRN (ATCC® CRL-3276™) cells were cultured in a 10-cm plate with culture medium (Dulbecco's modified Eagle's medium [DMEM], 0.5 mg/mL G418, 0.5 mg/mL hygromycin B, 1% penicillin/streptomycin, and 10% fetal bovine serum [FBS]). The WRN conditioned medium was made as previously described (Miyoshi and Stappenbeck, 2013). Briefly, the L-WRN cells were split 1:10 in culture medium (without G418 and hygromycin B), seeded in 10-cm plates, and incubated 3-4 days. The medium was removed, fresh medium was used to rinse the plates and discarded, and then 10 mL fresh medium was added, and the plates were incubated for 24 h. The medium was collected, centrifuged at 1000 × g for 4 min, passed through a 0.22-μm sterile filter, and stored at 4°C. This was the first batch of medium. Another 10 mL of fresh medium was added to the plates. Every 24 h, we collected second and third batches of conditioned medium, which were centrifuged, filtered, and added to the same bottle as the first batch of medium. Then, the cumulative WRN conditioned medium was aliquoted and stored at −20°C. This was a 100% WRN conditioned medium.
Method details
Esophageal tissue isolation
Mice were euthanized via CO2 inhalation, followed by cervical dislocation. The esophagi were collected in 10-cm Petri dishes with cold phosphate-buffered saline (PBS) with 1% penicillin/streptomycin and washed by gently moving the dishes back and forth until the supernatant was clear. The esophagi were opened longitudinally and washed with cold PBS with 1% penicillin/streptomycin and then dissected into 0.5-cm3 pieces with scissors. Then, the esophagi were collected in a 15-mL conical tube and digested by 0.05% trypsin-EDTA at 37°C for 60 min with vortexing every 15 min. Next, 10% FBS + DMEM was added to inactivate the trypsin, followed by vigorous pipetting ten times. Next, the cell suspension was passed through a 35-μm sterile strainer to get a single-cell suspension. Finally, the cell suspension was centrifuged down at 1000 rpm for 4 min at room temperature (RT) and re-suspended in a 50% WRN conditioned medium.
Esophageal organoids passage, freezing, and thawing
EOs were passaged every 7 days. We discarded the culture medium, removed the organoids by scraping the Matrigel with a pipette tip, digested them with 0.05% trypsin-EDTA at 37°C for 45 min, added 50% WRN conditioned medium to inactivate the trypsin, and vigorously pipetted up and down ten times. Next, we passed the cells through a 35-μm sterile filter to make a single-cell suspension and centrifuged down the cells at 1000 rpm for 4 min at RT, and they were either suspended in Matrigel to generate new organoids or suspended in Recovery Cell Culture Freezing Medium (12648010, Life Technologies) and stored in liquid nitrogen. To thaw organoids: frozen cryotubes were incubated in a 37°C water bath, and the single-cell suspensions were transferred to pre-warmed 5 mL 50% WRN conditioned medium in 15-mL centrifuge tubes, which were centrifuged down at 1000 rpm for 4 min at RT and suspended in Matrigel to generate new organoids. For live-organoid imaging and analysis, the IncuCyte® system (Sartorius) was used per the manufacturer's instructions.
Organoid-forming efficiency and size analysis
After 7 days of organoid formation by primary esophageal cells in Matrigel, the number and size of organoids (spheres larger than 50 μm in diameter) were quantified using AxioVision software (Zeiss). Organoid-forming efficiency was defined as: the number of organoids/number of cells seeded in Matrigel × 100%. The size of each organoid was analyzed by measuring the diameter under the microscope. All experiments calculated more than 50 organoids per group.
H&E, PAS, and immunofluorescence staining
All staining was performed as previously described (Jung et al., 2018). Esophagus tissue was collected and fixed in 10% formalin overnight at 4°C. EOs were removed from Matrigel using ice-cold PBS and fixed in 4% paraformaldehyde for 1 h at RT. After processing for paraffin embedding, tissue and organoid sections were mounted on glass slides. For H&E staining, sections were incubated in hematoxylin for 3–5 min, clarified for 30 s to 1 min, followed by incubating in Bluing Reagent for 1 min, and then Eosin Y for 20–40 s. For PAS staining, slides were immersed in Periodic Acid Solution for 5 min at RT, then in Schiff's Reagent for 15 min at RT, followed by Hematoxylin Solution, Gill No. 3, for 60–90 s, and finally cleared and mounted sections in xylene-based mounting media. For immunofluorescence, after blocking with 3% horse serum in PBS for 30 min at RT, sections were incubated in primary antibodies (Krt13 [1:250], Krt14 [1:250], Ki67 [1:250], β-catenin [1:500], Sox2 [1:250], cleaved caspase-3 [1:250], Villin [1:500], and CD44 [1:100]) overnight at 4°C and secondary antibody (1:250) for 1 h at RT. Sections were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen). Images were captured with the fluorescence microscope (Zeiss; AxioVision).
Gene expression analysis
Organoids grown in Matrigel were lysed, and RNAs were extracted by TRIzol (Invitrogen). RNA was quantified with the Nanodrop 2000c Spectrophotometer (Thermo Scientific), then converted to cDNAs using SuperScript II (Invitrogen) with random hexamers. PCR amplification (StepOne Real-Time PCR System, Applied Biosystems) used the following conditions: 95°C for 10 min, 95°C for 15 s (denature) and 60°C for 1 min (anneal/extend) for 40 cycles, 95°C for 15 s and 60°C for 1 min and then 95°C for 15 s (melting curve). qRT–PCR results were quantified by comparative 2−ΔΔCt methods (see Table S2 for primer information). The results were expressed as average fold change in gene expression and were normalized to the expression of Hprt.
Human esophageal organoid (hEO) culture
2 pieces of 0.5 cm3 human tissue from an endoscopic biopsy or esophageal surgery were immediately collected and washed with cold phosphate-buffered saline (PBS) with 1% penicillin/streptomycin and 1 × Fungizone (C125CB, New Cell & Molecular Biotech Co.,Ltd,China). Then, the tissue was collected in a 15-mL conical tube and digested by 3 mL HEBSS cocktail containing 100 IU/mL collagenase I (Gibco:17018029, Thermo Fisher Scientific), 10 μM Y-27632 and 1.258M CaCl2 at 37°C for 45 min and then 0.05% trypsin-EDTA at 37°C for 1 h with vortexing every 15 min. The followed process was performed with the same protocol as used for murine EOs culture. This work was approved by the Fujian Medical University Union Hospital ethics committee (2020KJT071).
KrasG12D:Trp53KO cell line 2D culture model
KP + Ad-Cre EOs were digested with 0.05% trypsin-EDTA at 37°C for 45 min to make a single-cell suspension in a 15-mL centrifuge tube. Then, the cells were centrifuged down at 1000 rpm for 4 min at RT, suspended in DMEM +10% FBS (with Y-27632 added the first 2-3 days), and seeded in a 24-well plate. Cells were passaged every 3-5 days. After the third passage, Y-27632 was removed from the culture medium. DMEM +10% FBS +10% DMSO was used for freezing cells and storing them in liquid nitrogen. For Matrigel-coated plate culture, we prepared diluted Matrigel with DMEM-F12 medium (1:30) and poured 500 μL on each well of the 24-well plate. After 1 h of incubation at 37°C, excessive Matrigel was aspirated and washed with PBS, and single-cells from organoids were seeded on the well.
Xenograft transplantation
5-week-old-immunodeficient male nude mice were maintained in the Division of Laboratory Animal Resources facility at The University of Texas MD Anderson Cancer Center. 5x106 2D-cultured KrasG12D:Trp53KO cells were injected subcutaneously into left and right dorsal flanks of 3 mice, respectively. Mice were euthanized and tumors were collected 4 weeks after injection. Excised tumors were photographed and paraffin embedded for immunostaining.
Library preparation and scRNA-seq
Organoids were collected at day 8 of the 3rd passage and digested with 0.05% trypsin-EDTA at 37°C for 30 min. After inactivating trypsin with 10% FBS DMEM, single-cell suspension was collected by passing through the 35 μm cell strainer. cDNA library was prepared with the 10x Genomics 3′ v2 kit, cDNA library was sequenced on an Illumina NovaSeq and mapped to GRCm38/mm10 genome using Cellranger v5.0.1 (Novogene). The result count matrices files were loaded into R for analysis. scRNA-seq data analysis: To analyze the scRNA-seq data, we used the Seurat R package. UMAP was used for dimensional reduction and cells were clustered into 10 groups. Each cluster was annotated based on Marker genes information and categorized into 6 groups; Quiescent basal, Proliferating, Early suprabasal, Immediate suprabasal, Stratified. Umap, heatmap, bubble plot, and violin plot were generated by Seurat toolkit DimPlot, Doheatmap, Dotplot, and VlnPlot, respectively. Trajectory inference and pseudotime analysis: We used Slingshot and Monocle3 R packages for cell lineage tracing and pseudotime inference based on the previous reports (Cao et al., 2019; Street et al., 2018). Cells were filtered, and dimensional reduction was performed following the default parameters. We generated the SingleCellExperiment dataset for Slingshot and CellDataSet object for Monocle 3 from our Seurat dataset. Both algorithms constructed a minimum spanning tree (MST) on clusters and showed trajectory inferences on UMAP projection. Public dataset: We used the public reference dataset of scRNA-seq for human esophageal cells analysis (Madissoon et al., 2019). To generate a new dataset of epithelial cells, we made the subset from the “oesophagus” dataset. Using the Seurat R package, a new dataset including only Epi_basal, Epi_dividing, Epi_stratified, Epi_suprabasal, and Epi_upper clusters was generated and renamed based on marker genes expression as Basal, Proliferating, Stratified, Early suprabasal, and Intermediate suprabasal, respectively.
Quantification and statistical analysis
The Student's t-test was used for comparisons of two groups (n ≥ 3), and one-way analysis of variance (ANOVA) statistical evaluation was used for comparisons of at least three groups (n ≥ 3). P values less than 0.05 were considered significant. Error bars indicate the standard deviation (s.d.). All experiments were performed three or more times independently under identical or similar conditions.
Acknowledgments
This work was supported by grants to the Cancer Prevention and Research Institute of Texas (RP140563 and RP200315 to J.-I.P.); Young and Middle-aged Talents Training Project of Fujian Provincial Health Technology Project (2020GGA031 to B.Z.); General Project of Fujian Natural Science Foundation (2020J011005 to B.Z.); Joint Funds for the Innovation of Science and Technology, Fujian province, China (2017Y9032 to B.Z.); and the Key Clinical Specialty Discipline Construction Program of Fujian, P.R. China (Min Wei Ke Jiao 2012 No.149 to X.W.). This work was approved by the Fujian Medical University Union Hospital ethics committee (2020KJT071). Figures were created with Biorender.com.
Author contributions
B.Z., K.P.K., and J.-I.P. conceived the work. B.Z., K.P.K., J.Z., S.J., and C.L.C. performed the experiments. B.Z. and W.L. performed the pathologic analysis. B.Z., K.P.K., M.J. K., Y.-S.J., and J.-I.P. analyzed the experimental results. K.P.K. and B.-J.K. performed scRNA-seq analysis. B.Z., X.F., and X.W. performed human EO experiments. B.Z., K.P.K., and J.-I.P. wrote the manuscript.
Declaration of interests
There are no competing financial or non-financial interests in relation to the work described.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103440.
Supplemental information
Data and code availability
Single-cell RNA-seq data have been deposited at Gene Expression Omnibus (GEO: GSE174577) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code. R packages and their algorithms were used to analyze scRNA-seq data which are listed in the key resources table. The code will be available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bandla S., Pennathur A., Luketich J.D., Beer D.G., Lin L., Bass A.J., Godfrey T.E., Litle V.R. Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann. Thorac. Surg. 2012;93:1101–1106. doi: 10.1016/j.athoracsur.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger G.A., Weusten B.L.A., Bogte A., Begthel H., Brosens L.A.A., Clevers H. Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Rep. 2021;34:108819. doi: 10.1016/j.celrep.2021.108819. [DOI] [PubMed] [Google Scholar]
- Cao J.Y., Spielmann M., Qiu X.J., Huang X.F., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien M., Giroux I., Goulet A., Jacques C., Bouchard S. Cognitive restructuring of gambling-related thoughts: a systematic review. Addict. Behav. 2017;75:108–121. doi: 10.1016/j.addbeh.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Denu J.M. Vitamin B3 and sirtuin function. Trends Biochem. Sci. 2005;30:479–483. doi: 10.1016/j.tibs.2005.07.004. [DOI] [PubMed] [Google Scholar]
- DeWard A.D., Cramer J., Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe D.P., Alcolea M.P., Roshan A., Zhang G., Klein A.M., Simons B.D., Jones P.H. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., van Jaarsveld R.H., Ponsioen B., Zimberlin C., van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H., et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- Giroux V., Lento A.A., Islam M., Pitarresi J.R., Kharbanda A., Hamilton K.E., Whelan K.A., Long A., Rhoades B., Tang Q., et al. Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J. Clin. Invest. 2017;127:2378–2391. doi: 10.1172/JCI88941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T.F. Esophageal cancer. Facts, figures, and screening. Gastroenterol. Nurs. 2001;24:271–273. [PubMed] [Google Scholar]
- Hisha H., Tanaka T., Kanno S., Tokuyama Y., Komai Y., Ohe S., Yanai H., Omachi T., Ueno H. Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci. Rep. 2013;3:3224. doi: 10.1038/srep03224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.S., Wang W., Jun S., Zhang J., Srivastava M., Kim M.J., Lien E.M., Shang J., Chen J., McCrea P.D., et al. Deregulation of CRAD-controlled cytoskeleton initiates mucinous colorectal cancer via beta-catenin. Nat. Cell Biol. 2018;20:1303–1314. doi: 10.1038/s41556-018-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakasheva T.A., Kijima T., Shimonosono M., Maekawa H., Sahu V., Gabre J.T., Cruz-Acuna R., Giroux V., Sangwan V., Whelan K.A., et al. Generation and characterization of patient-derived head and neck, oral, and esophageal cancer organoids. Curr. Protoc. Stem Cell Biol. 2020;53:e109. doi: 10.1002/cpsc.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasagi Y., Chandramouleeswaran P.M., Whelan K.A., Tanaka K., Giroux V., Sharma M., Wang J., Benitez A.J., DeMarshall M., Tobias J.W., et al. The esophageal organoid system reveals functional interplay between notch and cytokines in reactive epithelial changes. Cell Mol. Gastroenterol. Hepatol. 2018;5:333–352. doi: 10.1016/j.jcmgh.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Kobayashi Y., Tata P.R., Hogan B.L.M. IL-1 and TNFalpha contribute to the inflammatory niche to enhance alveolar regeneration. Stem Cell Rep. 2019;12:657–666. doi: 10.1016/j.stemcr.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima T., Nakagawa H., Shimonosono M., Chandramouleeswaran P.M., Hara T., Sahu V., Kasagi Y., Kikuchi O., Tanaka K., Giroux V., et al. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol. Gastroenterol. Hepatol. 2019;7:73–91. doi: 10.1016/j.jcmgh.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Choi S., Kang B., Kong J., Kim Y., Yoon W.H., Lee H.R., Kim S., Kim H.M., Lee H., et al. Creation of bladder assembloids mimicking tissue regeneration and cancer. Nature. 2020;588:664–669. doi: 10.1038/s41586-020-3034-x. [DOI] [PubMed] [Google Scholar]
- Li X., Francies H.E., Secrier M., Perner J., Miremadi A., Galeano-Dalmau N., Barendt W.J., Letchford L., Leyden G.M., Goffin E.K., et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018;9:2983. doi: 10.1038/s41467-018-05190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.C., Hao J.J., Nagata Y., Xu L., Shang L., Meng X., Sato Y., Okuno Y., Varela A.M., Ding L.W., et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cheng Y., Abraham J.M., Wang Z., Wang Z., Ke X., Yan R., Shin E.J., Ngamruengphong S., Khashab M.A., et al. Modeling Wnt signaling by CRISPR-Cas9 genome editing recapitulates neoplasia in human Barrett epithelial organoids. Cancer Lett. 2018;436:109–118. doi: 10.1016/j.canlet.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.D., Orlando R.C. Esophageal submucosal glands: structure and function. Am. J. Gastroenterol. 1999;94:2818–2824. doi: 10.1111/j.1572-0241.1999.1422_b.x. [DOI] [PubMed] [Google Scholar]
- Madissoon E., Wilbrey-Clark A., Miragaia R.J., Saeb-Parsy K., Mahbubani K.T., Georgakopoulos N., Harding P., Polanski K., Huang N., Nowicki-Osuch K., et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21:1. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton R.M., Pasca S.P. Organoid and assembloid technologies for investigating cellular crosstalk in human brain development and disease. Trends Cell Biol. 2020;30:133–143. doi: 10.1016/j.tcb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A., Odze R., Jenkins T.D., Shahsesfaei A., Nakagawa H., Inomoto T., Rustgi A.K. A transgenic mouse model with cyclin D1 overexpression results in cell cycle, epidermal growth factor receptor, and p53 abnormalities. Cancer Res. 1997;57:5542–5549. [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Natsuizaka M., Whelan K.A., Kagawa S., Tanaka K., Giroux V., Chandramouleeswaran P.M., Long A., Sahu V., Darling D.S., Que J., et al. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat. Commun. 2017;8:1758. doi: 10.1038/s41467-017-01500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa T., Michaylira C.Z., Kalabis J., Stairs D.B., Nakagawa H., Andl C.D., Johnstone C.N., Klein-Szanto A.J., El-Deiry W.S., Cukierman E., et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz O.G., Harada H., Suliman Y., Rhoades B., Sharpless N.E., Kent R., Kopelovich L., Nakagawa H., Rustgi A.K. A mouse model of human oral-esophageal cancer. J. Clin. Invest. 2002;110:761–769. doi: 10.1172/JCI15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennathur A., Gibson M.K., Jobe B.A., Luketich J.D. Oesophageal carcinoma. The Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- Sachdeva U.M., Shimonosono M., Flashner S., Cruz-Acuna R., Gabre J.T., Nakagawa H. Understanding the cellular origin and progression of esophageal cancer using esophageal organoids. Cancer Lett. 2021;509:39–52. doi: 10.1016/j.canlet.2021.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Aihara E., Feng R., Engevik A., Shroyer N.F., Ottemann K.M., Worrell R.T., Montrose M.H., Shivdasani R.A., Zavros Y. The use of murine-derived fundic organoids in studies of gastric physiology. J. Physiol. 2015;593:1809–1827. doi: 10.1113/jphysiol.2014.283028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth E.C., Lagergren J., Fitzgerald R.C., Lordick F., Shah M.A., Lagergren P., Cunningham D. Oesophageal cancer. Nat. Rev. Dis. primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Trisno S.L., Philo K.E.D., McCracken K.W., Cata E.M., Ruiz-Torres S., Rankin S.A., Han L., Nasr T., Chaturvedi P., Rothenberg M.E., et al. Esophageal organoids from human pluripotent stem cells delineate Sox2 functions during esophageal Specification. Cell Stem Cell. 2018;23:501–515 e507. doi: 10.1016/j.stem.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson D., Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- Vercauteren Drubbel A., Pirard S., Kin S., Dassy B., Lefort A., Libert F., Nomura S., Beck B. Reactivation of the Hedgehog pathway in esophageal progenitors turns on an embryonic-like program to initiate columnar metaplasia. Cell Stem Cell. 2021;28:1411–1427 e1417. doi: 10.1016/j.stem.2021.03.019. [DOI] [PubMed] [Google Scholar]
- Wang G.Q., Abnet C.C., Shen Q., Lewin K.J., Sun X.D., Roth M.J., Qiao Y.L., Mark S.D., Dong Z.W., Taylor P.R., et al. Histological precursors of oesophageal squamous cell carcinoma: results from a 13 year prospective follow up study in a high risk population. Gut. 2005;54:187–192. doi: 10.1136/gut.2004.046631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan K.A., Muir A.B., Nakagawa H. Esophageal 3D culture systems as modeling tools in esophageal epithelial pathobiology and personalized medicine. Cell Mol. Gastroenterol. Hepatol. 2018;5:461–478. doi: 10.1016/j.jcmgh.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Wang X., Bertrand D., Kern F., Zhang T., Duleba M., Srivastava S., Khor C.C., Hu Y., Wilson L.H., et al. Mutational spectrum of Barrett's stem cells suggests paths to initiation of a precancerous lesion. Nat. Commun. 2016;7:10380. doi: 10.1038/ncomms10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.R., Koido M., Tadokoro T., Ouchi R., Matsuno T., Ueno Y., Sekine K., Takebe T., Taniguchi H. Human iPSC-derived posterior gut progenitors are expandable and capable of forming gut and liver organoids. Stem Cel. Rep. 2018;10:780–793. doi: 10.1016/j.stemcr.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.C., Yang Y., Jiang M., Huang S.X., Zhang W.W., Al Alam D., Danopoulos S., Mori M., Chen Y.W., Balasubramanian R., et al. 3D modeling of esophageal development using human PSC-derived basal progenitors reveals a critical role for notch signaling. Cell Stem Cell. 2018;23:516. doi: 10.1016/j.stem.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at Gene Expression Omnibus (GEO: GSE174577) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code. R packages and their algorithms were used to analyze scRNA-seq data which are listed in the key resources table. The code will be available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.