Summary
In cells, segregation allows for diverse biochemical reactions to take place simultaneously. Such intricate regulation of cellular processes is achieved through the dynamic formation and disassembly of membraneless organelles via liquid-liquid phase separation (LLPS). Herein, we demonstrate the light-controlled formation and disassembly of liquid droplets formed from a complex of polylysine (pLys) and arylazopyrazole (AAP)-conjugated single-stranded DNA. Photoswitchablility of droplet formation was also shown to be applicable to the control of chemical reactions; imine formation and a DNAzyme-catalyzed oxidation reaction were accelerated in the presence of droplets. These outcomes were reversed upon droplet disassembly. Our results demonstrate that the photoswitchable droplet formation system is a versatile model for the regulation of reactions through dynamic LLPS.
Subject areas: Imaging methods in chemistry, Biochemistry, Cell biology
Graphical abstract
Highlights
-
•
Incorporating AAP enabled light-controlled droplet formation with ssDNA and pLys
-
•
Droplets were reversibly formed or disassembled without altering sample composition
-
•
Photoswitchability depended on sequence and ionic interactions but not flexibility
-
•
Photoswitchable droplet formation accelerated uncatalyzed and catalyzed reactions
Imaging methods in chemistry; Biochemistry; Cell biology
Introduction
Cells are often described as mini-factories, with a multitude of product-generating processes occurring within the confines of their plasma membranes. Akin to factories which assign different areas of their floor space to different lines so as to avoid confusion in production, cells too are able to segregate their cellular volume into compartmentalized membraneless organelles through liquid-liquid phase separation (LLPS) (Banani et al., 2017; Mitrea and Kriwacki, 2016).
LLPS results in the formation of dense phases enriched in specific biomolecules, typically proteins and nucleic acids (Kamimura and Kanai, 2021). By concentrating specific collections of proteins and nucleic acids, LLPS enables cells to regulate biochemical reactions within the droplets or coacervates formed (Kohata and Miyoshi, 2020; Lyon et al., 2021; Stroberg and Schnell, 2018). As the mechanisms involved in droplet formation and LLPS-controlled reactions remain unclear, researchers have employed numerous model systems to recreate and study LLPS in vitro. One such example is the ATP-polylysine (pLys) droplets developed by Koga and co-workers (Koga et al., 2011). When ABTS oxidation and glucose phosphorylation were carried out in the presence of the coacervates, accelerated kinetics were observed. More complex cascade reactions have been carried out in PEG-dextran droplets reported by Kojima and colleagues (Kojima and Takayama, 2018). In addition, gene expression, i.e., transcription and translation, has been shown to occur more rapidly in (carboxymethyl-dextran)-pLys droplets (Tang et al., 2015). Despite the advances made in these endeavors, systematic programming of droplet properties through methodical modification of their components remains cumbersome.
Programmable model systems leveraging electrostatic interactions between oppositely-charged polyelectrolytes have also been developed (Alshareedah et al., 2019). Of these, systems involving DNA and pLys have been reported on extensively (Lu and Spruijt, 2020; Shakya and King, 2018; Vieregg et al., 2018). Systematic changes in DNA sequence have shown that droplet formation is highly dependent on DNA local flexibility among other factors (André and Spruijt, 2018; Sato et al., 2020; Shakya and King, 2018), thus enabling the programming of droplet properties and architectures through DNA sequence modification.
Although the models described above have been designed to be stable, phase-separated compartments in cells are dynamic, assembling and disassembling based on the cellular environment (Alberti, 2017; Anderson and Kedersha, 2006; Smith et al., 2016). To simulate the dynamic behavior of cellular coacervates, photoswitches have been utilized. A variety of photoswitches which have altered affinity to double-stranded DNA (dsDNA) upon light irradiation and subsequent cis-trans isomerization are available (Adam et al., 2018; Asanuma et al., 2007; Deiana et al., 2016; Lubbe et al., 2017, 2018). By controlling the affinity of a cationic azobenzene photoswitch to dsDNA, Martin and co-workers have demonstrated light-modulated droplet formation (Martin et al., 2019). However, droplet assembly and disassembly based on the affinity between dsDNA and a photoswitch limits its degree of programmability.
Photoswitches directly conjugated to DNA have also been reported. Developed by Adam et al., the arylazopyrazole (AAP) photoswitch exhibits longer isomerization wavelength and improved switching efficiency as compared to azobenzene (Adam et al., 2018). By directly conjugating AAP to single-stranded DNA (ssDNA), a wider range of droplet properties, and by extension architectures, could be made accessible. This could be done through sequence modifications, analogous to the sequence effects exhibited by membraneless organelles consisting of intrinsically disordered proteins and RNAs (Martin and Mittag, 2018; Saar et al., 2021; Schuster et al., 2020).
By exploiting AAP-conjugated ssDNA, herein, we demonstrate the light-controlled formation and disassembly of DNA-pLys droplets. Systematic investigation of DNA sequence revealed that the photoswitchable droplet formation is influenced by sequence effects. Through real-time fluorescence measurements, we show that photoswitchable droplet formation allows for light-controlled improvements in condensation reaction rates and increase in kinetics of a DNAzyme-catalyzed oxidation reaction. Together, our results illustrate a dynamic phase separation model system, which if further refined, may shed light on the control of reactions through droplet formation in protocells.
Results
Incorporation of a photoswitch into ssDNA allows for light-controlled droplet formation
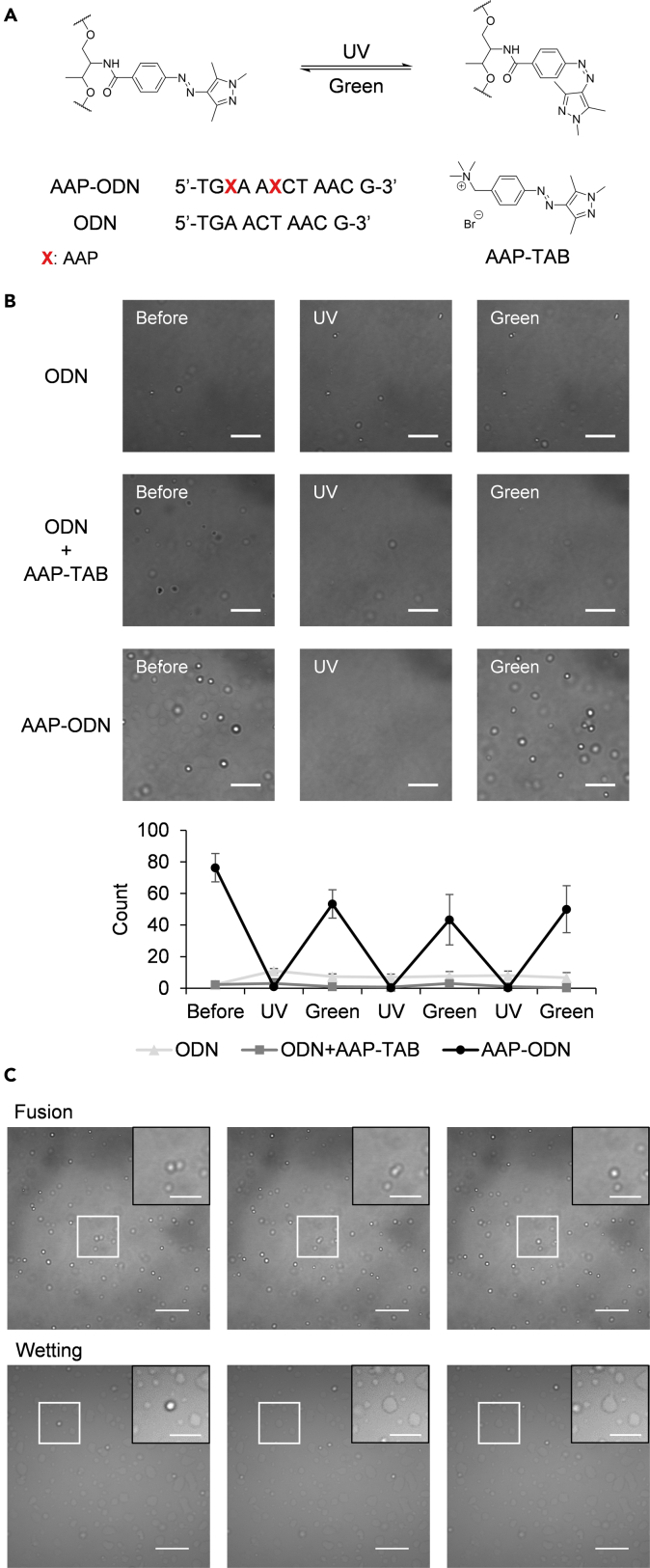
We first asked whether ssDNA-pLys complexes demonstrate light-controllable phase separation in the presence of a photoswitch. To this end, we designed the following sequences shown in Figure 1A. Microscopy images of solutions containing the DNA oligonucleotide, ODN, revealed that it was able to form micrometre-sized droplets upon complexation with pLys (Figure 1B; Table S1). The samples were then irradiated alternately with UV and green light. To quantify the extent of coacervation after each bout of irradiation, droplet counts were obtained for each image using ImageJ (supplemental information and Figure S1D) (Grishagin, 2015). As expected for ODN, UV and subsequent green light irradiation did not elicit an observable change in droplet count in response to the different wavelengths of light.
Figure 1.
Incorporation of arylazopyrazole (AAP) moieties into ssDNA allows for photoswitchable droplet formation
(A) AAP is able to undergo trans-to-cis isomerization under UV irradiation and reverts to its trans isomer upon irradiation of green light. AAP was incorporated into two positions in the native ODN sequence to give the modified AAP-ODN sequence. The structure of the positively charged AAP monomer, AAP-TAB, is as given.
(B) Microscopy images and droplet counts of mixtures of the various DNA sequences (1.85 mM phosphate) with pLys (2.5 mM amine) (scale bar: 10 μm). Although ODN and ODN + AAP-TAB exhibited limited droplet formation, only AAP-ODN showed significant photoswitching ability. Error bars represent standard deviation for three independent experiments. Irradiation was conducted in situ and the same field of view was maintained within each experiment.
(C) Droplets formed from AAP-ODN and pLys mixtures exhibited fusion and wetting behaviors which are typical of liquid droplets (scale bar: 20 μm; scale bar for zoomed inset: 10 μm).
See also Figure S1.
We next examined the effect of the addition of AAP-TAB (Scheme S1), a cationic AAP monomer, on droplet formation. Martin et al. have previously reported that complexes comprising dsDNA and the tetraamine salt of azobenzene (Azo-TAB) can result in the formation of photoswitchable coacervates (Martin et al., 2019). However, unlike the case for dsDNA, the ssDNA-pLys droplets did not exhibit light-modulated phase separation upon addition of AAP-TAB (Figure 1B). Whereas changes in affinity of Azo-TAB to dsDNA provides an impetus for the photoswitchability of dsDNA droplets, ssDNA lacks the differences in affinity toward the cis and trans isomers of AAP-TAB (Diguet et al., 2010; Le Ny and Lee, 2006; Venancio-Marques et al., 2014). In addition, although the positively-charged Azo-TAB allows for complexation of dsDNA, the same function is achieved with pLys in ssDNA-pLys droplets, which further reduces the influence of AAP-TAB on coacervation.
To overcome these challenges, we generated an AAP-containing DNA sequence, AAP-ODN, by incorporating AAP via its phosphoramidite directly into DNA through solid-phase oligonucleotide synthesis (Table S1). Remarkably, the molecular-scale changes in the structure of AAP in response to light irradiation were sufficient to induce micro-scale level assembly and disassembly of the droplets (Figures 1B, S1A, and S1B; Videos S1 and S2). By covalently tethering the AAP moiety to DNA, the effects of cis-trans isomerization of AAP may have been more directly translated into changes in DNA properties. The photoswitchability of droplet formation was highly repeatable with photoswitching observed after at least three cycles of UV and green light irradiation. The incorporation of AAP also resulted in better droplet formation ability although the reason for the improvement remains unclear.
Closer examination of the droplets formed from AAP-ODN and pLys confirmed that the droplets exhibited liquid-like behavior. Droplets were observed to coalesce on contact and over time were seen to wet the surface of a glass slide (Figure 1C).
Photoswitchability of ssDNA-pLys is independent of local flexibility
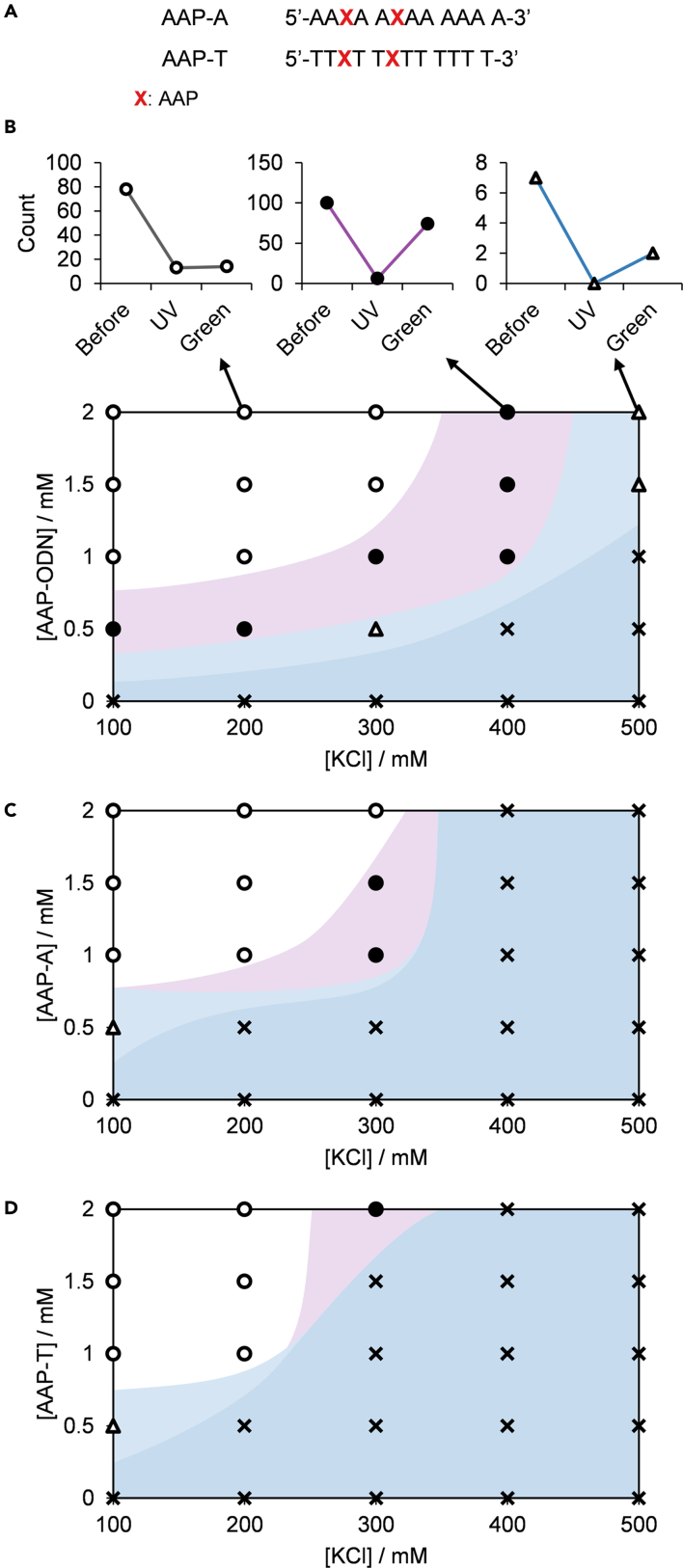
As this is the first report demonstrating light-modulated phase separation with ssDNA, we were determined to elucidate the potential mechanisms for the visible micro-scale changes induced by molecular-scale cis-trans isomerization and structure perturbation. One possibility is that cis-trans isomerization of AAP results in changes in local flexibility of AAP-ODN. Differences in local flexibility have been linked to changes in phase separation ability. The more flexible poly(thymine) sequence, for instance, has been shown to induce phase separation more readily than poly(adenine) (Mills et al., 1999; Shakya and King, 2018; Sim et al., 2012). To determine whether local flexibility may have played a role in the photoswitchability of (AAP-ODN)-pLys droplets, AAP-containing poly(adenine) (AAP-A) and poly(thymine) (AAP-T) DNA sequences were constructed (Figure 2A; Table S1). We hypothesized that if changes in local flexibility were crucial to photoswitchability, the phase diagram of AAP-T should contain more phase-separated states, i.e. conditions at which droplets are observed.
Figure 2.
Sequence effects on light-modulated droplet formation—photoswitchability of droplets is independent of local flexibility
(A) Sequences of AAP-A and AAP-T. Phase diagrams of (B) AAP-ODN, (C) AAP-A and (D) AAP-T depicting range of photoswitchable droplet behavior in mixtures with pLys (2.5 mM amine). Empty circles represent non-photoswitchable phase-separated states; filled circles represent photoswitchable phase-separated states; empty triangles represent borderline states; crosses represent non-phase-separated states (dispersed states). Plot areas are shaded only as a visual guide.
See also Figures S2 and S3.
The phase diagrams of AAP-ODN, AAP-A, and AAP-T were thus determined (Figures 2B–2D, S2, and S3). Non-photoswitchable phase-separated states were defined as conditions at which droplets did not reassemble upon green photoirradiation; photoswitchable phase-separated states were defined as conditions at which droplet counts recovered by at least 50% after green photoirradiation; borderline states were defined as conditions at which fewer than 10 droplets were observed, and non-phase-separated states were defined as conditions at which no droplets were observed. Phase-separated states thus refer to non-photoswitchable and photoswitchable phase-separated states as well as borderline states. Surprisingly, AAP-A and AAP-T were observed to have similar numbers of phase-separated states in contrast to AAP-ODN, which had the highest number of phase-separated states. The high number of phase-separated states for AAP-ODN was also observed when cross-referenced with the phase diagram of AAP-ODN constructed from turbidity measurements (Figures S2B and S2C).
Photoswitchability of droplet formation occurs when there is a disparity between the droplet formation abilities of the cis and trans forms of a DNA sequence at a given condition. The similarity of droplet formation ability between AAP-A and AAP-T therefore seemed to suggest that changes in local flexibility were unlikely to play a significant role in the photoswitchability of droplet formation. This contradicts a previous report on the importance of low persistence lengths on phase separation (Shakya and King, 2018). It should, however, be noted that sequences used in this study are significantly shorter (10-mer) compared to the 20-mer sequences employed in the previous report, which may account for the apparent independence of phase separation ability on local flexibility. Furthermore, the high droplet formation ability exhibited by AAP-ODN suggests that there are other more pertinent factors at play and that elucidating those factors is key to developing droplet programmability.
Ionic interactions play an important role for phase separation and photoswitchability
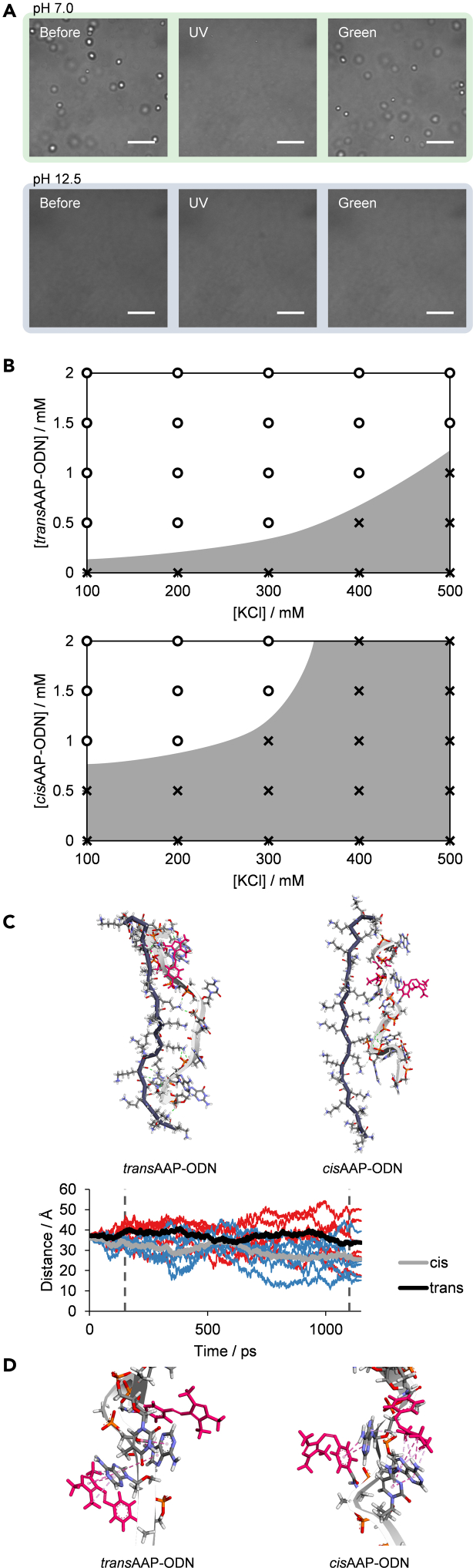
From the phase diagrams, an increase in KCl concentration decreases the propensity for droplet formation (Figures 2B–2D). As an increase in salt concentration decreases the strength of electrostatic interactions and hydrogen bonds, this suggests that these two forces may be important for photoswitchability (Kříž et al., 2013; Vieregg et al., 2018; Wang and Schlenoff, 2014; Zhang et al., 2015). To further substantiate this observation, buffer pH was increased from pH 7.0 to pH 12.5. As expected, although droplets were present at pH 7.0, droplet formation was not observed at pH 12.5 (Figure 3A). As pLys has a pKa of around 10 (Dos et al., 2008), the increase in pH significantly reduced the positive charges on pLys and thus its interaction with AAP-ODN. This further corroborates the importance of electrostatic interactions between pLys and ssDNA for phase separation.
Figure 3.
Electrostatic interactions account for phase separation and photoswitching abilities
(A) Photoswitchable droplet formation at pH 7.0 in contrast to dispersed states at pH 12.5 (20 mM buffer concentration) (scale bar: 10 μm).
(B) Phase diagrams illustrating phase-separated states of trans and cisAAP-ODN. Empty circles represent phase-separated states; crosses represent non-phase-separated states (dispersed states). Plot areas are shaded only as a visual guide.
(C) Images of the energy minimum structures of trans and cisAAP-ODN with 20-mer pLys obtained through molecular dynamics simulation. End-to-end distance with simulation time is plotted; red and blue lines are distances from individual runs of trans and cisAAP-ODN respectively, with a total of six runs each; black and gray lines represent averages of the respective runs; dashed lines bound production runs.
(D) Interactions of AAP with neighboring bases in AAP-ODN. AAP is indicated in pink and hydrophobic interactions are shown as dashed lines.
See also Figure S2D.
Further inspection of the phase diagrams showed that transAAP-ODN and cisAAP-ODN displayed a large difference in phase separation ability, accounting for the large number of photoswitchable states of AAP-ODN (Figure 3B). However, cis-trans isomerization of AAP does not affect the net charge of AAP-ODN. To examine whether AAP-induced structural changes in DNA could have been translated into local changes in the extent of electrostatic interactions, molecular dynamic simulations of trans and cisAAP-ODN with 20-mer pLys were conducted (Table S2). The energy minimum structures revealed that transAAP-ODN was able to interact more extensively with pLys as compared to cisAAP-ODN (Figure 3C). Unlike cisAAP-ODN, transAAP-ODN was able to form amine-phosphate interactions with pLys along its length, whereas cisAAP-ODN was more compact. This was also reflected in the end-to-end distances of trans and cisAAP-ODN, with cisAAP-ODN generally exhibiting shorter distances during the simulation (Figure 3C). The compaction of cisAAP-ODN appeared to be the result of hydrophobic interactions between the pyrazole ring of AAP with distal bases (Figures 3D and S2D). This compaction of DNA as a result of trans-to-cis isomerization thus leads to fewer electrostatic interactions with pLys, and the lower phase separation ability observed in cisAAP-ODN.
Taken together, the results suggest that AAP-ODN was able to form coacervate droplets with pLys mainly because of electrostatic interactions between the two species. Irradiation of the droplets with UV light resulted in trans-to-cis isomerization of AAP. Unlike transAAP-ODN, cisAAP-ODN was compacted and thus had less extensive electrostatic interactions with pLys. At certain conditions, this difference in phase separation ability meant that transAAP-ODN was able to form droplets while cisAAP-ODN remained in a dispersed state. This disparity was reflected as the photoswitchability of droplet formation.
Photoswitchable droplet formation allows for light-modulation of imine formation
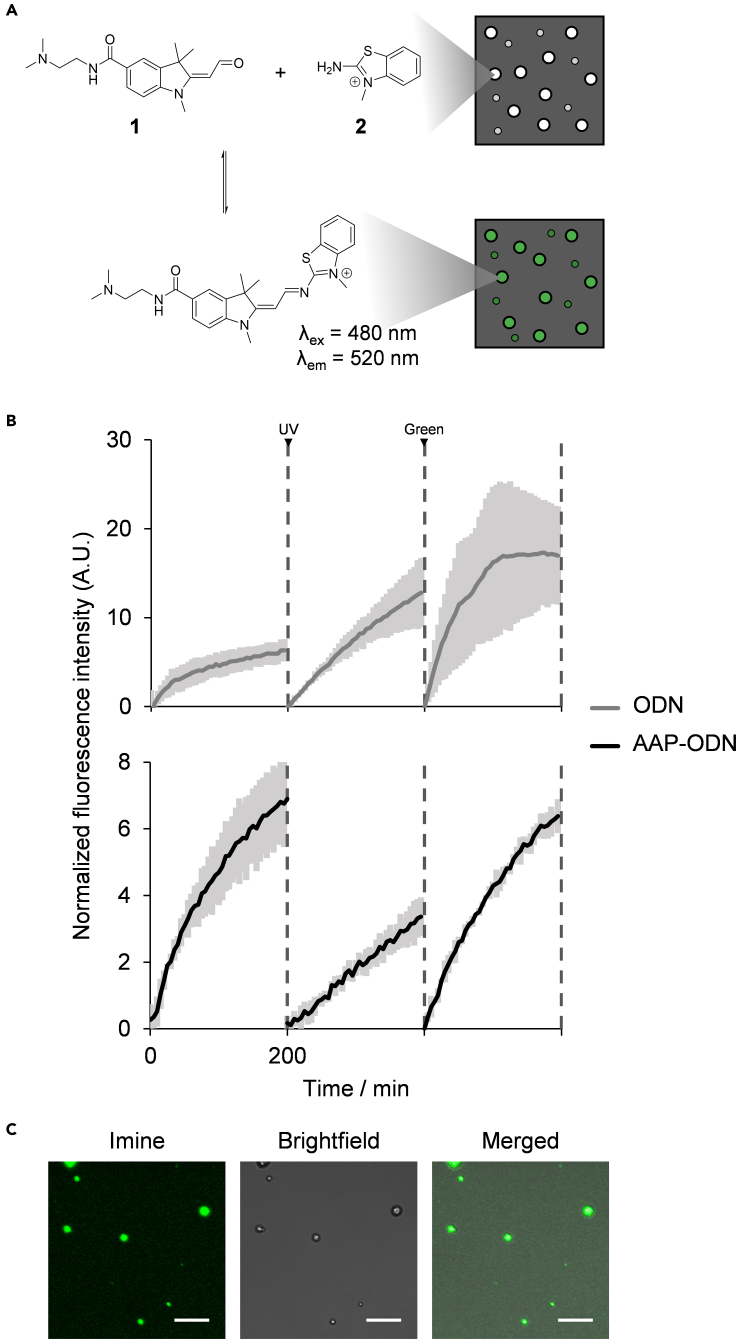
Having established a system allowing for light-controllable phase separation, we hoped to mimic rudimentary reaction control to demonstrate the applicability of our system. Condensation reactions, which result in the covalent linkage of molecules with the release of water, feature heavily in biochemical processes; however, they proceed unfavorably in aqueous environments (Kaufmann, 2009; do Nascimento Vieira et al., 2020). In a prebiotic world, phase separation has been proposed to act as a bioreactor, driving condensation and allowing for the non-enzymatic RNA polymerization (Guo et al., 2021; Poudyal et al., 2018). Here, we demonstrate that the condensation of an amine and aldehyde to form an imine can be controlled through photoswitchable droplet formation.
Although imine formation from aldehyde 1 and amine 2 is highly disfavored in dispersed state, upon micelle formation, the equilibrium of the reaction shifts toward the fluorescent imine product (λex = 480 nm; λex = 520 nm) (Figure 4A) (Meguellati et al., 2013). In contrast, the rate of the reaction barely increases, if at all, after micelle formation. To study the change in kinetics after micelle formation, prior studies have relied on increasing the surfactant concentration above a critical value in order to induce the formation of micelles. However, by doing so, it is difficult to separate the effects of an increase in surfactant concentration, and micelle formation on reaction kinetics. By employing our photoswitchable droplet formation system, the change in reaction kinetics upon droplet formation can be studied independently of the concentration of the components inducing droplet formation, i.e., AAP-ODN and pLys.
Figure 4.
Control of a condensation reaction resulting in imine formation through photoswitchable droplet formation
(A) Condensation of aldehyde 1 and amine 2 to form a fluorescent imine is expected to be favored within droplets; photocontrol of imine formation could be achieved through UV and green light irradiation.
(B) Emission intensity at 520 nm before and after UV (365 nm) and green light (520 nm) irradiation with each measurement taken for 200 min. Minimum intensities for each iteration were adjusted to zero. Dark lines represent average values and shaded areas represent standard deviation for three independent experiments.
(C) Fluorescence image of AAP-ODN-pLys droplets after imine formation (scale bar: 10 μm).
See also Figure S4.
We thus sought to observe how the presence or absence of droplets affects the rate of imine formation by tracking the emission intensity of the fluorescent product over time. In the presence of AAP-ODN and pLys, a visible difference in reaction rate was observed before (kobs = 0.060 ± 0.006 min−1) and after UV irradiation (kobs = 0.016 ± 0.001 min−1) (Figure 4B). The rate of reaction was recovered after green light irradiation (kobs = 0.050 ± 0.003 min−1), suggesting that the presence of droplets was able to accelerate imine formation. In contrast, the reaction rate remains high even after UV irradiation when non-photoswitchable droplets formed from ODN and pLys were present.
Microscopy images of (AAP-ODN)-pLys droplets after imine formation revealed that the fluorescent product was located within the droplets (Figure 4C). This was corroborated by HPLC absorbance data of the bulk mixture and of the dilute phase, which indicated that reactants were also concentrated within droplets; only 84 ± 4% of aldehyde 1 and 96 ± 4% of amine 2 were present in the dilute phase with respect to the bulk mixture (Figure S4A). In addition, given previous reports showing that aldehyde 1 and amine 2 are depleted from the dilute phase and concentrated within separated compartments in a two-phase system (Meguellati et al., 2013; Wilson et al., 2020), these suggest that imine formation was favored inside droplets as compared to the dilute phase. The increase in local concentration of reactants within the droplets could thus account for the enhanced reaction rates observed when droplets were present.
Interestingly, the observable difference in reaction rates upon droplet formation is in contrast to previous data showing that reaction rate hardly changes upon micelle formation (Meguellati et al., 2013). To ensure that the difference observed was not because of the dissimilar absorbance profiles of cis and transAAP, the reaction was conducted in the presence of AAP-ODN without pLys (Figure S4B). Unlike when pLys was present, the rate of reaction could not be controlled through light irradiation, implying that the presence of droplets was indeed necessary for the photocontrol of reaction kinetics. This also suggested that micelles and droplets may facilitate the imine formation reaction in distinct ways, although more work has to be done to confirm this.
On the whole, by controlling droplet formation through light irradiation, we were able to illustrate how phase separation may favor the condensation of molecules.
Catalyzed oxidation can be controlled through photoswitchable droplet formation
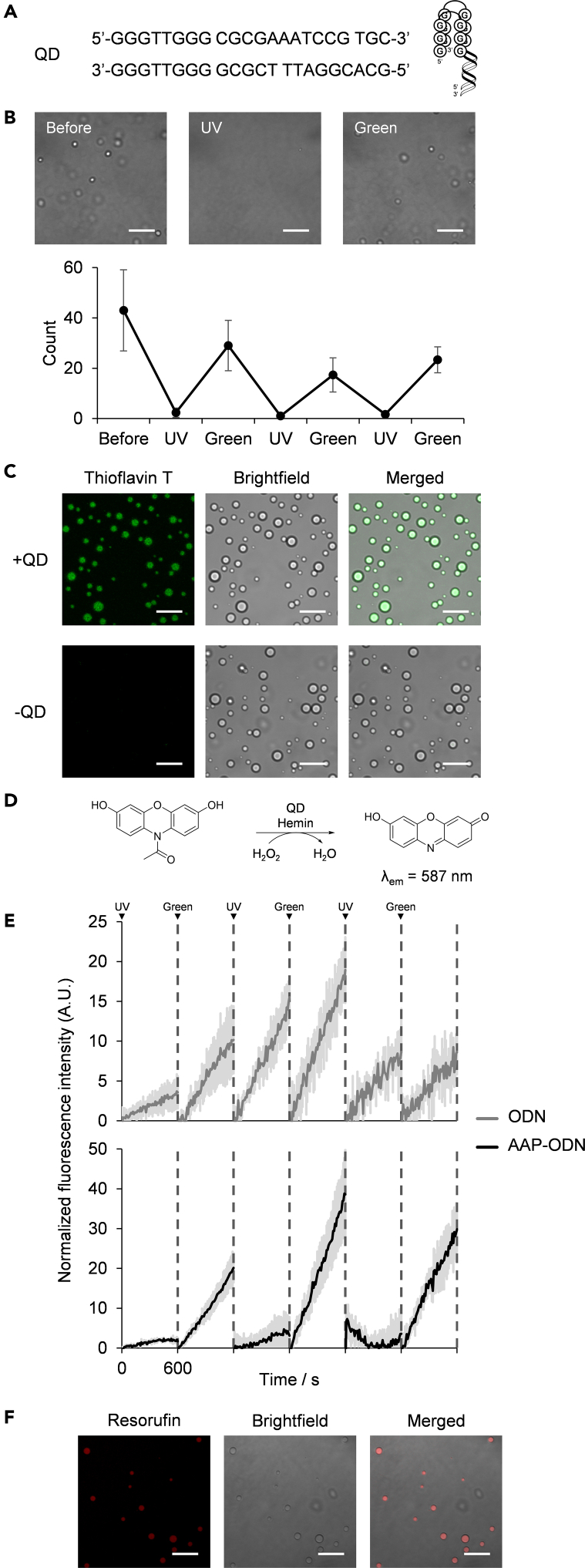
In extent cells, however, biomolecular reactions are typically catalyzed by enzymes. Transiently phase separated cellular compartments concentrate enzymes and auxiliary factors within them, enabling the regulation of genome and other cellular activities (Case et al., 2019; Razin and Gavrilov, 2020). To recreate an elementary reaction control system through the sequestration of a catalyst and its substrate within a phase separated compartment, we introduced a quadruplex-duplex (QD)-hemin complex as a DNA catalyst to our photoswitchable droplets (Figure 5A). Notably, addition of the DNAzyme did not adversely affect photoswitchable droplet formation. As with droplets containing only AAP-ODN and pLys, the QD-hemin-containing droplets exhibited good photoswitching ability (Figure 5B). In addition, when the samples were doped with thioflavin T (ThT)—an established fluorescence light-up probe for quadruplex structures (De La Faverie et al., 2014)—fluorescence microscopy images revealed that the QD-hemin complexes were sequestered within the droplets (Figure 5C).
Figure 5.
Control of an oxidation reaction catalyzed by a DNA quadruplex-duplex-hemin complex through photoswitchable droplet formation
(A) Sequences of quadruplex-duplex (QD)-forming oligonucleotide pairs and an illustration of the QD structure.
(B) Images and droplet counts of photoswitchable droplet formation of AAP-ODN and pLys in the presence of QD (1 μM) (scale bar: 10 μm).
(C) Fluorescence images of ThT (10 μM)-stained QD (1 μM) in droplets formed from AAP-ODN and pLys (scale bar: 10 μm).
(D) Reaction scheme of the oxidation of Amplex Red to resorufin catalyzed by QD complexed with hemin.
(E) Fluorescence intensity at 587 nm after alternate UV (365 nm) and green light (520 nm) irradiation of (AAP-ODN)-pLys droplets containing the QD-hemin complex (0.1 μM). Each measurement was taken for 600 s and minimum intensities were set to zero. Dark lines represent average values and shaded areas represent standard deviation for three independent experiments.
(F) Fluorescence images of the Amplex Red oxidation product, resorufin, in the presence of (AAP-ODN)-pLys droplets after three irradiation cycles (scale bar: 10 μm).
See also Figure S5.
Encouraged by the results, we proceeded to investigate whether reaction kinetics can be controlled by photoswitchable droplet formation. The introduced QD-hemin complex contains a G-quadruplex (G4) domain and G4-hemin complexes have been reported as horseradish peroxidase mimics (Golub et al., 2015; Travascio et al., 1998; Yang et al., 2011). We thus anticipated that QD-hemin complexes would also be able to act as horseradish peroxidase mimics. To track the activity of the QD-hemin complex, Amplex Red was used as a substrate. Upon oxidation by the QD-hemin complex in the presence of H2O2, a highly fluorescent product, resorufin, is formed (Figure 5D). Monitoring the emission intensity of resorufin with respect to time would thus enable us to correlate oxidation rate with droplet formation. After UV (365 nm) irradiation and droplet disassembly, oxidation proceeds slowly (kobs = 0.003 ± 0.007 s−1) in the (AAP-ODN)-pLys droplets (Figure 5E). However, upon green light (520 nm) irradiation and reassembly of the droplets, the rate of oxidation rose dramatically (kobs = 0.05 ± 0.02 s−1). This change in oxidation rate was repeatable for at least three cycles. In contrast, such switches in oxidation rate were not observed for droplets comprising ODN and pLys, as well as solutions containing AAP-TAB (Figures 5E and S5).
Fluorescence microscopy images corroborated the importance of droplet formation in accelerating the DNAzyme-catalyzed oxidation of Amplex Red (Figure 5F). Images of the droplets revealed that the oxidation product, resorufin, was present within the droplets, suggesting that photoswitchable droplet formation could control the kinetics of a catalyzed reaction. Real-time fluorescence analysis of a condensation and oxidation reaction thus illustrated how photoswitchable droplet formation allows for light-controlled shifts in reaction kinetics.
Discussion
This work presents the first report of droplet formation control using ssDNA conjugated to a photoswitch. Photoswitches are typically employed with dsDNA as dsDNA is more rigid and has a more defined conformation as compared to ssDNA (Lubbe et al., 2017; Szymański et al., 2013). However, we have shown that photoswitches can also be effectively used to control the range of conformations available to ssDNA. This allowed us to control the formation of droplets with irradiations of different wavelengths. The use of ssDNA instead of dsDNA also permits higher programmability of droplet properties through modifications of the ssDNA sequence. Furthermore, as complex layered droplet structures with distinct microcompartments have been reported to form when differences in surface tensions are present (Feric et al., 2016; Jo and Jung, 2020; Lu and Spruijt, 2020), the use of ssDNA may allow for tuning of such properties through sequence engineering and the incorporation of artificial nucleosides.
Even more remarkable is the high degree of dependence of droplet formation on the geometric orientations of monomeric units in DNA; the same AAP moiety presented in its cis or trans isomer can result in vastly different outcomes in terms of phase separation. As the interaction between pLys and ssDNA has mainly been attributed to electrostatic interactions between the charged amine groups of pLys and the phosphate backbone of DNA (Elder et al., 2011; Vieregg et al., 2018), it is noteworthy that a simple cis-trans isomerization of units not involved in the binding can cascade down to influence the interactions between pLys and ssDNA. Although Perry and co-workers have reported precipitate or droplet formation depending on the chirality (racemic, L or D) of the pair of polyelectrolytes involved (Perry et al., 2015), the differences in chirality are exhibited throughout the polyelectrolyte structures unlike the localized structural changes observed in AAP-ODN. In a broader context, this suggests that perhaps post-transcriptional modifications of RNA may have a more far-reaching effect on phase separation in cells than previously thought (Drino and Schaefer, 2018; Peng et al., 2020).
The development of the photoswitchable droplet system presented in this work also enabled us to investigate the effects of droplet formation on reactions while keeping all conditions unchanged. By using a photoswitch, droplet formation is decoupled from other confounding factors including bulk salt and polyelectrolyte concentrations, as well as temperature. Through photoswitchable droplet formation, we demonstrated that droplet formation alone can induce an acceleration in kinetics in uncatalyzed as well as catalyzed reactions. Moreover, as the irradiation of light alone controls the formation and disassembly of the droplets, photoswitchable droplet formation also allows for a higher degree of control over droplet formation.
In summary, we successfully designed and implemented a photoswitchable droplet formation system for the control of reactions. Mixtures of pLys and AAP-conjugated ssDNA, AAP-ODN, were shown to form droplets. These droplets were disassembled upon UV irradiation and reassembled upon green light irradiation. Through molecular dynamics simulations, we proposed that cis-trans isomerization of AAP can alter the extent of electrostatic interactions between AAP-ODN and pLys, thus allowing for photoswitchable droplet formation. This dynamic control of droplet formation through conjugation of a photoswitch serves as an improvement over static systems in reflecting the transient and dynamic LLPS observed in cells. Photoswitchable droplet formation was also demonstrated to accelerate the formation of an imine product in an uncatalyzed condensation reaction. Furthermore, reaction kinetics were also improved upon droplet formation for a QD-hemin catalyzed oxidation reaction. Taken together, photoswitchable droplet formation allowed us to examine the effect of droplet formation on reactions without a change in sample composition. Now, we turn to expansion of the scope of the photoswitchable droplet formation system. Further work is underway to investigate more complex reactions, such as cascade reactions, and more biologically relevant processes, for example gene expression.
Limitations of the study
Notably, for photoswitching to occur, a degree of droplet instability is necessary. For this system to be applied in a general manner, a balance has to be achieved to allow for both photoswitchability as well as a certain measure of stability once droplets have been formed. Preliminary investigations suggest that under the current conditions, protein concentrations around 1 μM and nucleotide triphosphate concentrations around 160 μM are compatible with photoswitchable droplet formation (Figure S1C). Although the system remains crude at the moment, further refinement would allow for a wide-scale investigation of the effect of droplet formation on individual reactions without a need to rely on other factors, such as enzyme activity (Aumiller and Keating, 2016), for the assembly and disassembly of droplets.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 2-Cyanoethyl diisopropylchlorophosphoramidite | Wako Chemicals | Cat#327-75411 |
| Poly-L-lysine hydrobromide (MW 30,000-70,000) | Sigma-Aldrich | Cat#P9155 |
| Poly-L-lysine-FITC label (MW 30,000-70,000) | Sigma-Aldrich | Cat#P3069 |
| DMT-dC(bz) phosphoramidite | Sigma-Aldrich | Cat#C111030 |
| DMT-dT phosphoramidite | Sigma-Aldrich | Cat#T111030 |
| DMT-dA(bz) phosphoramidite | Sigma-Aldrich | Cat#A111030 |
| DMT-dG(ib) phosphoramidite | Sigma-Aldrich | Cat#G111030 |
| OxiRedTM | BioVision | Cat#1572 |
| Caffeine anhydrous | Nacalai Tesque | Cat#06712-42 |
| Riboflavin 5’-monophosphate sodium salt | TCI | Cat#R0249 |
| Oligonucleotides | ||
| AAP-ODN: TGXAAXCTAACG (X = AAP) | This paper | NA |
| ODN: TGAACTAACG | Sigma Genosys | NA |
| AAP-A: AAXAAXAAAAAA | This paper | NA |
| AAP-T: TTXTTXTTTTTT | This paper | NA |
| QD: (Forward) GGGTTGGGCGCGAAATCCGTGC (Reverse) GGGTTGGGGCGCTTTAGGCACG |
This paper | NA |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| ImageJ droplet counting macro | This paper | https://doi.org/10.7910/DVN/NW2RU1 |
| Molecular operating environment | Chemical Computing Group | https://www.chemcomp.com/Products.htm |
| ChemDraw 19.1 | PerkinElmer | https://www.perkinelmer.com/category/chemdraw |
| BIOVIA discovery studio visualizer | Dassault Systemes | https://discover.3ds.com/discovery-studio-visualizer-download |
| FV10-ASW 4.2 Viewer | Olympus | https://www.olympus-lifescience.com/es/support/downloads/#!dlOpen=%23detail847249651 |
| Other | ||
| Glass slide (24×60 mm No.1, thickness 0.13~0.17 mm) | Matsunami Glass | Cat#C024601 |
| Glass cover slip (18×18 mm No.1, thickness 0.13~0.17 mm) | Matsunami Glass | Cat#C218181 |
| Glen-Pak DNA Purification Cartridge | Glen Research | Cat#60-5200-xx |
| Olympus Laser Scanning Biological Microscope FV1200 IX83 | Olympus | https://www.olympus-lifescience.com/en/laser-scanning/fv1200/ |
| ZUV-C30H controller | Omron | https://www.fa.omron.co.jp/products/family/1876/download/catalog.html |
| ZUV-H30M C head unit | Omron | https://www.fa.omron.co.jp/products/family/1876/download/catalog.html |
| HM-3 monochromatic light source | JASCO | NA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Soyoung Park (spark@ifrec.osaka-u.ac.jp).
Materials availability
All other data supporting findings of this study are available within the article and the supplemental information or from the lead contact upon reasonable request.
Method details
Materials
2-Cyanoethyl diisopropylchlorophosphoramidite was received from Wako Chemicals and used without further purification. Poly-L-lysine hydrobromide (MW 30,000-70,000) and poly-L-lysine-FITC label (MW 30,000-70,000) were purchased from Sigma-Aldrich Chemicals Co. and used as received. OxiRedTM probe (also marketed and more commonly known as Amplex RedTM) was purchased from BioVision and used as received. N,N-diisopropylethylamine (DIPEA) was purchased from Nacalai and used as received. Nuclease-free water was purchased from Life Technologies Corporation and used as received. All other chemicals and solvents were purchased from Sigma-Aldrich Chemicals Co., Wako Pure Chemical Ind. Ltd., TCI, or Kanto Chemical Co. Inc. and used without further purification. Glass slides (24 × 60 mm, thickness 0.13-0.17 mm) and cover slips (18 × 18 mm, thickness 0.12–0.17 mm) were purchased from Matsunami Glass and used as received.
Glen-Pak™ DNA and RNA cartridges columns were purchased at Glen Research. ODN was obtained from Sigma Genosys. Water was deionized (specific resistance of ∼18.2 MW cm at 25°C) by a Milli-Q system (Millipore Corp.). All reactions were carried out under an argon atmosphere unless otherwise stated.
Synthesis and characterization of AAP-TAB
Compound 2 (Scheme S1) was synthesized according to the synthetic route reported by Adam and co-workers (Adam et al., 2018). To compound 2, NBS and AIBN were added, and the mixture was dissolved in CCl4 before refluxing for 4 h. The precipitate was filtered and washed with CH2Cl2 and H2O, and finally purified by silica gel column chromatography to obtain compound 3. Me3N in MeOH was subsequently added to intermediate 3 before the addition of EtOH. The reaction mixture was stirred at 50°C for 3 days. The resulting precipitate was filtered, washed with ether and finally dried under vacuum to afford AAP-TAB.
1H NMR (500 MHz, CDCl3): δ 7.80 (d, JHH = 6.9 Hz, 2H), 7.74 (d, JHH = 6.9 Hz, 2H), 5.11 (t, JHH = 12.6 Hz, 2H), 3.78 (s, 3H), 3.43 (s, 9H), 5.57 (s, 3H), 2.48 (s, 3H). 13C NMR (125 MHz, D2O): δ 156.7, 145.2, 143.5, 137.0, 136.3, 131.0, 124.5, 71.6, 55.0, 37.8, 15.2, 11.8. HRMS (ESI-TOF) calculated for C16H24BrN5 [M-Br]+ 286.2027, found 286.2027.
Oligonucleotide synthesis
AAP-containing oligonucleotides, AAP-ODN, AAP-A and AAP-T, were synthesized on solid support using DMTr-protected AAP phosphoramidite and commercially available O5′-dimethoxytrityl-2′-deoxyribonucleoside O3′-phosphoramidites (Sigma-Aldrich Chemicals Co.). The detailed synthetic route and characterization data for DMTr-protected AAP were reported by Adam and co-workers (Adam et al., 2018). DMTr-protected AAP phosphoramidite was prepared by dropwise addition of DIPEA at 0°C to a CH2Cl2 solution of DMTr-protected AAP. Phosphoramidochloridous acid was subsequently added at 0°C and after 10 min, the mixture was warmed to room temperature and stirred for 1 h. After removal of the solvent, the residue was dissolved in MeCN and used immediately without further purification for solid phase DNA synthesis. Solid-phase oligonucleotide synthesis was performed on an ABI DNA synthesizer (Applied Biosystem, Foster City, CA). The modified phosphoramidite (DMTr-protected AAP) was incorporated into the oligonucleotides through coupling for 10 min. The coupling yields of AAP phosphoramidite were around 10% less than those obtained with standard phosphoramidite building blocks.
Cleavage from the solid support and deprotection were accomplished with 50:50 of MeNH2 in 40 wt.% in water and NH3 in 28 wt.% in water at room temperature for 15 min and then at 65°C for 15 min. The synthesized oligonucleotides were eluted from Glen-Pak™ DNA purification cartridges and purification steps were performed as per procedure. The final elution was subjected to normal-phase HPLC purification (2%–30% ACN in 50 mM TEAA (pH 7.0) buffer, flow rate of 3.0 mL/min). After purification by HPLC, the products were confirmed by MALDI-TOF MS using a Bruker microflex-KSII (Bruker Corporation, Billerica, MA) and the purities were checked by HPLC (Table S1). DNA concentrations were determined using NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE); nearest-neighbour and individual-bases molar extinction coefficients of the nucleotides were used in the calculations. The purchased sequences given in Table S1 were obtained from Sigma Genosys.
For HPLC analysis, COSMOSIL 5C18 AR-II (Nacalai Tesque, Inc., Kyoto, 150 × 4.6 mm id), a linear gradient of 2%–30% acetonitrile (in 50 mM TEAA (pH 7.0) buffer) over 30 min at a flow rate of 1.0 mL/min and detection at 254 nm were used.
Microscope observation
Samples containing the oligonucleotides in Na3PO4 buffer (pH 7.0) containing KCl were prepared. The solutions were annealed by heating at 95°C followed by slow cooling to room temperature over 3 h before the addition of pLys in pH 7.5 TE buffer (final amine concentration of 2.5 mM; final phosphate concentration of 1.85 mM unless otherwise stated, final KCl concentration of 400 mM unless otherwise stated; final buffer concentration of 5 mM unless otherwise stated). For samples containing QD sequences, 1 μM of each strand was added prior to annealing and the sample was incubated for 1 h at room temperature after the addition of hemin. Immediately after the addition of pLys, 5 μL of the mixture was loaded onto a glass slide and covered with a greased coverslip, then incubated for 5 min before imaging. For fluorescence microscopy images, the glass slides and coverslips were passivated with 3.5% bovine serum albumin (Promega Corporation) in PBS buffer for 15 min, washed with deionized water and air-dried before use.
Samples were imaged with Olympus Laser Scanning Biological Microscope FV1200 IX83 and its associated FV10-ASW 4.2 software. For photoswitching experiments, the in-built Olympus U-HGLGPS mercury lamp and FUW or FGW fluorescent mirror unit was used for UV or green light irradiation respectively. Samples were irradiated with UV or green light for 10 min or 5 min respectively before images were taken. Images were taken from the same field of view for each sample. Droplet counts were obtained using ImageJ (the macro code used is supplied in the supplemental information) with output images visually inspected to verify the validity of droplet counts.
For time-lapse images taken during UV irradiation, the sample was irradiated from t = 9s 79ms (365 nm, ZUV-C30H system equipped with a ZUV-H30M head unit (Omron)) while scanning with a 635 nm laser at a frame rate of 2.218 s. For time-lapse images taken during green light irradiation, scanning with a 473 nm laser at a frame rate of 2.218 s was sufficient for droplet formation.
Characterization of droplets
Fluorescence microscopy images were obtained as described previously to determine the localizations of AAP-ODN and pLys in droplet samples (Figure S1B). For the sample doped with FAM-ODN, final phosphate concentrations of AAP-ODN and FAM-ODN were 1.48 mM and 0.37 mM respectively. For the sample doped with FITC-pLys, final amine concentrations of pLys and FITC-pLys were 1.5 mM and 1.0 mM respectively.
Absorption spectra of an AAP-ODN sample obtained after UV and green light irradiation showed the presence of cis and trans isomers respectively (Figure S1A). Absorption was measured using NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE). UV irradiation (365 nm) was carried out with ZUV-C30H system equipped with a ZUV-H30M head unit (Omron) for 2 min. After UV irradiation, the sample was left in the dark for 30 min at room temperature and its absorption spectrum was measured again. Green light irradiation was carried out at 520 nm with a HM-3 monochromatic light source (JASCO) for 1 min.
Owing to the low contrast nature of the wetting process, the contrast of the zoomed insets of Figure 1C bottom were enhanced. All other brightfield confocal microscope images were taken under the same settings.
To determine the concentration of aldehyde 1 in the dilute phase as compared to the bulk, samples containing (AAP-ODN)-pLys were prepared as described previously. After addition of pLys, aldehyde 1 was added to a final concentration of 15 mM and an aliquot was taken from the sample. Riboflavin 5′-monophosphate (FMN) was then added as an internal sample and the final solution was analyzed using HPLC. For HPLC analysis, COSMOSIL 5C18 AR-II (Nacalai Tesque, Inc., Kyoto, 150 × 4.6 mm id), a linear gradient of 2%–60% acetonitrile (in 50 mM TEAA (pH 7.0) buffer) over 30 min at a flow rate of 1.0 mL/min and detection at 340 nm were used. The remaining (AAP-ODN)-pLys sample was then centrifuged at 16 000 ×g for 20 min (Hitachi himac CT15RE). An aliquot was taken from the supernatant, to which FMN was added as an internal standard. The final solution was finally analysed using HPLC. The areas corresponding aldehyde 1 were normalized based on the areas of FMN and compared. The same was done for amine 2 using caffeine as an internal standard and a detection wavelength of 270 nm.
Phase diagrams
Droplet counts of samples were obtained using the droplet count procedure detailed above (Figures S2 and S3). Samples were deemed to be capable of phase separation if droplets were formed prior to irradiation and were photoswitchable if there was more than 50% recovery in droplet count after green light irradiation. Samples with droplet counts of less than or equal to 10 were considered to be borderline. All DNA concentrations given refer to phosphate concentrations.
Molecular dynamics simulation
Energy minimizations and atomistic molecular dynamics (MD) simulations of the oligonucleotide-pLys systems were performed with explicit water molecules and 0.4 M KCl using the Amber14:EHT force field implemented in Molecular Operating Environment (MOE). Solvation was conducted in a TIP3P-model periodic box with a 10 Å margin between the solutes and the cell. Oligonucleotide and 20-mer pLys structures were built with the in-built nucleic acid and protein builders. AAP was built within MOE and energy minimized in gas phase before being appended to the oligonucleotide structure. The oligonucleotide and pLys structures were energy minimized in the gas phase before being solvated and then energy minimized again. For the oligonucleotides, six unrestrained NAMD trials of 1.15 ns were performed as per the protocol given in Table S2 and the minimum energy oligonucleotide structures were obtained from the production runs. End-to-end distances were obtained for the duration of the simulations by calculating the linear distances between C5′ of the 5′ end and C3′ of the 3′ end. The energy minimized oligonucleotide and pLys structures were aligned along their coordinate axes (to reduce the size of the cell) and the system was solvated. For each system, three unrestrained NAMD simulation trials of 1.15 ns were performed with the protocol given in Table S2 (Figure S2D). The minimum energy oligonucleotide-pLys system was finally obtained from the production run.
Imine formation
Aldehyde 1 and amine 2 were synthesized following the procedure reported by Meguellati and co-workers (Meguellati et al., 2013), and stock solutions of aldehyde 1 in H2O and amine 2 in DMSO were prepared. Fluorescence measurements were obtained using 3 mm path length JASCO FMH-857 quartz microcells on a JASCO FP-8300 spectrofluorometer equipped with a JASCO PAC-743R thermocontrolled cell changer and a JASCO CTU-100. Emission at 520 nm was recorded every 5 min with an excitation wavelength of 480 nm. Temperature was kept constant at 30°C. For subsequent measurements, samples were irradiated with UV (365 nm) light (ZUV-C30H system equipped with a ZUV-H30M head unit (Omron)) for 5 min or UV light for 5 min then green (520 nm) light for 2 min. Due to its degradation at 365 nm irradiation, aldehyde 1 was refreshed after each irradiation cycle. Minimum emission intensities were set to zero before average values and SD were obtained.
QD-hemin-catalysed oxidation
Fluorescence measurements were obtained using 3 mm path length JASCO FMH-857 quartz microcells on a JASCO FP-8300 spectrofluorometer equipped with a JASCO PAC-743R thermocontrolled cell changer and a JASCO CTU-100. Emission at 587 nm was recorded every 10 s with an excitation wavelength of 480 nm. Temperature was kept constant at 30°C. Samples containing QD-hemin complex were prepared as described previously (final AAP-ODN phosphate concentration of 1.85 mM; final QD concentration of 0.1 μM; final hemin concentration of 0.4 μM; final KCl concentration of 400 mM; final buffer concentration of 5 mM). The sample was then subjected to UV (365 nm) irradiation with ZUV-C30H system equipped with a ZUV-H30M head unit (Omron) for 5 min before addition of H2O2 (25 μM) and Amplex Red (25 μM). After equilibration at 30°C for 10 min, fluorescence was measured for 600 s. After measurement, the sample was again irradiated with UV light for 5 min to completely react the unreacted Amplex Red (Zhao et al., 2012). The same process was then repeated for green light irradiation (520 nm, 1 min, HM-3 monochromatic light source (JASCO)) and subsequent cycles. Minimum intensities were set to zero before average values and SD were obtained.
Quantification and statistical analysis
All statistical details of experiments can be found in the figure legends.
Acknowledgments
We are grateful to Ji Hye Yum (Graduate School of Science, Kyoto University) for additional synthesis of DMTr-protected AAP and AAP-TAB. We would like to thank Karin Nishimura (Graduate School of Engineering, Kyoto University) for technical assistance in obtaining the mass spectra of synthesized compounds. This work was supported by JSPS KAKENHI (grant numbers 20H05936 and 21H04705) and by AMED under JP21am0101101 (Platform Project for Supporting Drug Discovery and Life Science Research (BINDS)) and NIH award RO1CA236350 to H. S. We also thank the KAKENHI program (Grant-in-Aid for scientific research C, 18K05315) for support to S. P.
Author contributions
S.P., W.A.W. and H.S. conceptualized and designed the research. S.P. and W.A.W. synthesized the compounds; W.A.W. developed the methodology, performed the research and analyzed the data; W.A.W. wrote the paper. S.P. and H.S. supervised the work.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103455.
Contributor Information
Hiroshi Sugiyama, Email: hs@kuchem.kyoto-u.ac.jp.
Soyoung Park, Email: spark@ifrec.osaka-u.ac.jp.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. All original code is available in this paper's supplemental information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adam V., Prusty D.K., Centola M., Škugor M., Hannam J.S., Valero J., Klöckner B., Famulok M. Expanding the toolbox of photoswitches for DNA nanotechnology using arylazopyrazoles. Chemistry. 2018;24:1062–1066. doi: 10.1002/chem.201705500. [DOI] [PubMed] [Google Scholar]
- Alberti S. The wisdom of crowds: Regulating cell function through condensed states of living matter. J. Cell Sci. 2017;130:2789–2796. doi: 10.1242/jcs.200295. [DOI] [PubMed] [Google Scholar]
- Alshareedah I., Kaur T., Ngo J., Seppala H., Kounatse L.A.D., Wang W., Moosa M.M., Banerjee P.R. Interplay between short-range attraction and long-range repulsion controls reentrant liquid condensation of ribonucleoprotein-RNA complexes. J. Am. Chem. Soc. 2019;141:14593–14602. doi: 10.1021/jacs.9b03689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André A.A.M., Spruijt E. Rigidity rules in DNA droplets: Nucleic acid flexibility affects model membraneless organelles. Biophys. J. 2018;115:1837–1839. doi: 10.1016/j.bpj.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma H., Liang X., Nishioka H., Matsunaga D., Liu M., Komiyama M. Synthesis of azobenzene-tethered DNA for reversible photo-regulation of DNA functions: hybridization and transcription. Nat. Protoc. 2007;2:203–212. doi: 10.1038/nprot.2006.465. [DOI] [PubMed] [Google Scholar]
- Aumiller W.M., Keating C.D. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case L.B., Zhang X., Ditlev J.A., Rosen M.K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiana M., Pokladek Z., Olesiak-Banska J., Młynarz P., Samoc M., Matczyszyn K. Photochromic switching of the DNA helicity induced by azobenzene derivatives. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep28605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diguet A., Mani N.K., Geoffroy M., Sollogoub M., Baigl D. Photosensitive surfactants with various hydrophobic tail lengths for the photocontrol of genomic DNA conformation with improved efficiency. Chemistry. 2010;16:11890–11896. doi: 10.1002/chem.201001579. [DOI] [PubMed] [Google Scholar]
- Dos A., Schimming V., Tosoni S., Limbach H.H. Acid-base interactions and secondary structures of poly-l-lysine probed by15N and13C solid state NMR and ab initio model calculations. J. Phys. Chem. B. 2008;112:15604–15615. doi: 10.1021/jp806551u. [DOI] [PubMed] [Google Scholar]
- Drino A., Schaefer M.R. RNAs, phase separation, and membrane-less organelles: Are post-transcriptional modifications modulating organelle dynamics? BioEssays. 2018;40:1800085. doi: 10.1002/bies.201800085. [DOI] [PubMed] [Google Scholar]
- Elder R.M., Emrick T., Jayaraman A. Understanding the effect of polylysine architecture on DNA binding using molecular dynamics simulations. Biomacromolecules. 2011;12:3870–3879. doi: 10.1021/bm201113y. [DOI] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/J.CELL.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E., Albada H.B., Liao W.-C., Biniuri Y., Willner I. Nucleoapzymes: Hemin/G-quadruplex DNAzyme–aptamer binding site conjugates with superior enzyme-like catalytic functions. J. Am. Chem. Soc. 2015;138:164–172. doi: 10.1021/JACS.5B09457. [DOI] [PubMed] [Google Scholar]
- Grishagin I.V. Automatic cell counting with ImageJ. Anal. Biochem. 2015;473:63–65. doi: 10.1016/j.ab.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Guo W., Kinghorn A.B., Zhang Y., Li Q., Poonam A.D., Tanner J.A., Shum H.C. Non-associative phase separation in an evaporating droplet as a model for prebiotic compartmentalization. Nat. Commun. 2021;12:3194. doi: 10.1038/s41467-021-23410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y., Jung Y. Interplay between intrinsically disordered proteins inside membraneless protein liquid droplets. Chem. Sci. 2020;11:1269–1275. doi: 10.1039/C9SC03191J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y.R., Kanai M. Chemical insights into liquid-liquid phase separation in molecular biology. Bull. Chem. Soc. Jpn. 2021;94:1045–1058. doi: 10.1246/BCSJ.20200397. [DOI] [Google Scholar]
- Kaufmann M. On the free energy that drove primordial anabolism. Int. J. Mol. Sci. 2009;10:1853–1871. doi: 10.3390/ijms10041853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga S., Williams D.S., Perriman A.W., Mann S. Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 2011;3:720–724. doi: 10.1038/nchem.1110. [DOI] [PubMed] [Google Scholar]
- Kohata K., Miyoshi D. RNA phase separation–mediated direction of molecular trafficking under conditions of molecular crowding. Biophys. Rev. 2020;123:669–676. doi: 10.1007/S12551-020-00696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T., Takayama S. Membraneless compartmentalization facilitates enzymatic cascade reactions and reduces substrate inhibition. ACS Appl. Mater. Inter. 2018;10:32782–32791. doi: 10.1021/acsami.8b07573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kříž Z., Klusák J., Krištofíková Z., Koča J. How Ionic strength affects the conformational behavior of human and rat beta amyloids - A computational study. PLoS One. 2013;8:62914. doi: 10.1371/journal.pone.0062914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Faverie A.R., Guédin A., Bedrat A., Yatsunyk L.A., Mergny J.L. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic Acids Res. 2014;42:e65. doi: 10.1093/nar/gku111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Spruijt E. Multiphase complex coacervate droplets. J. Am. Chem. Soc. 2020;142:2905–2914. doi: 10.1021/jacs.9b11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbe A.S., Szymanski W., Feringa B.L. Recent developments in reversible photoregulation of oligonucleotide structure and function. Chem. Soc. Rev. 2017;46:1052–1079. doi: 10.1039/c6cs00461j. [DOI] [PubMed] [Google Scholar]
- Lubbe A.S., Liu Q., Smith S.J., De Vries J.W., Kistemaker J.C.M., De Vries A.H., Faustino I., Meng Z., Szymanski W., Herrmann A., et al. Photoswitching of DNA hybridization using a molecular motor. J. Am. Chem. Soc. 2018;140:5069–5076. doi: 10.1021/jacs.7b09476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon A.S., Peeples W.B., Rosen M.K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 2021;22:215–235. doi: 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E.W., Mittag T. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry. 2018;57:2478–2487. doi: 10.1021/acs.biochem.8b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N., Tian L., Spencer D., Coutable-Pennarun A., Anderson J.L.R., Mann S. Photoswitchable phase separation and oligonucleotide trafficking in DNA coacervate microdroplets. Angew. Chem. Int. Ed. 2019;58:14594–14598. doi: 10.1002/anie.201909228. [DOI] [PubMed] [Google Scholar]
- Meguellati K., Fallah-Araghi A., Baret J.C., El Harrak A., Mangeat T., Marques C.M., Griffiths A.D., Ladame S. Enhanced imine synthesis in water: From surfactant-mediated catalysis to host-guest mechanisms. Chem. Commun. 2013;49:11332–11334. doi: 10.1039/c3cc46461j. [DOI] [PubMed] [Google Scholar]
- Mills J.B., Vacano E., Hagerman P.J. Flexibility of single-stranded DNA: Use of gapped duplex helices to determine the persistence lengths of Poly(dT) and Poly(dA) J. Mol. Biol. 1999;285:245–257. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- Mitrea D.M., Kriwacki R.W. Phase separation in biology; Functional organization of a higher order short linear motifs - The unexplored frontier of the eukaryotic proteome. Cell Commun. Signal. 2016;14:1–20. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Nascimento Vieira A., Kleinermanns K., Martin W.F., Preiner M. The ambivalent role of water at the origins of life. FEBS Lett. 2020;594:2717–2733. doi: 10.1002/1873-3468.13815. [DOI] [PubMed] [Google Scholar]
- Le Ny A.L.M., Lee C.T. Photoreversible DNA condensation using light-responsive surfactants. J. Am. Chem. Soc. 2006;128:6400–6408. doi: 10.1021/ja0576738. [DOI] [PubMed] [Google Scholar]
- Peng L., Li E.M., Xu L.Y. From start to end: Phase separation and transcriptional regulation. Biochim. Biophys. Acta - Gene Regul. Mech. 2020;1863:194641. doi: 10.1016/j.bbagrm.2020.194641. [DOI] [PubMed] [Google Scholar]
- Perry S.L., Leon L., Hoffmann K.Q., Kade M.J., Priftis D., Black K.A., Wong D., Klein R.A., Pierce C.F., Margossian K.O., et al. Chirality-selected phase behaviour in ionic polypeptide complexes. Nat. Commun. 2015;6:1–8. doi: 10.1038/ncomms7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudyal R.R., Pir Cakmak F., Keating C.D., Bevilacqua P.C. Physical principles and extant biology reveal roles for RNA-containing membraneless compartments in origins of life chemistry. Biochemistry. 2018;57:2509–2519. doi: 10.1021/acs.biochem.8b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S.V., Gavrilov A.A. The role of liquid–liquid phase separation in the compartmentalization of cell nucleus and spatial genome organization. Biochemistry. 2020;85:643–650. doi: 10.1134/S0006297920060012. [DOI] [PubMed] [Google Scholar]
- Saar K.L., Morgunov A.S., Qi R., Arter W.E., Krainer G., Lee A.A., Knowles T.P.J. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2019053118. e2019053118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Sakamoto T., Takinoue M. Sequence-based engineering of dynamic functions of micrometer-sized DNA droplets. Sci. Adv. 2020;6:eaba3471. doi: 10.1126/sciadv.aba3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B.S., Dignon G.L., Tang W.S., Kelley F.M., Ranganath A.K., Jahnke C.N., Simpkins A.G., Regy R.M., Hammer D.A., Good M.C., et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11421–11431. doi: 10.1073/pnas.2000223117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya A., King J.T. DNA local-flexibility-dependent assembly of phase-separated liquid droplets. Biophys. J. 2018;115:1840–1847. doi: 10.1016/j.bpj.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim A.Y.L., Lipfert J., Herschlag D., Doniach S. Salt dependence of the radius of gyration and flexibility of single-stranded DNA in solution probed by small-angle x-ray scattering. Phys. Rev. E - Stat. Nonlinear, Soft Matter Phys. 2012;86:021901. doi: 10.1103/PhysRevE.86.021901. [DOI] [PubMed] [Google Scholar]
- Smith J., Calidas D., Schmidt H., Lu T., Rasoloson D., Seydoux G. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. ELife. 2016;5:e21337. doi: 10.7554/eLife.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroberg W., Schnell S. Do cellular condensates accelerate biochemical reactions? Lessons from microdroplet chemistry. Biophys. J. 2018;115:3–8. doi: 10.1016/j.bpj.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymański W., Beierle J.M., Kistemaker H.A.V., Velema W.A., Feringa B.L. Reversible photocontrol of biological systems by the incorporation of molecular photoswitches. Chem. Rev. 2013;113:6114–6178. doi: 10.1021/cr300179f. [DOI] [PubMed] [Google Scholar]
- Tang D.T.Y., Van Swaay D., DeMello A., Ross Anderson J.L., Mann S. In vitro gene expression within membrane-free coacervate protocells. Chem. Commun. 2015;51:11429–11432. doi: 10.1039/c5cc04220h. [DOI] [PubMed] [Google Scholar]
- Travascio P., Li Y., Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998;5:505–517. doi: 10.1016/S1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- Venancio-Marques A., Bergen A., Rossi-Gendron C., Rudiuk S., Baigl D. Photosensitive polyamines for high-performance photocontrol of DNA higher-order structure. ACS Nano. 2014;8:3654–3663. doi: 10.1021/nn500266b. [DOI] [PubMed] [Google Scholar]
- Vieregg J.R., Lueckheide M., Marciel A.B., Leon L., Bologna A.J., Rivera J.R., Tirrell M.V. Oligonucleotide-peptide complexes: Phase control by hybridization. J. Am. Chem. Soc. 2018;140:1632–1638. doi: 10.1021/jacs.7b03567. [DOI] [PubMed] [Google Scholar]
- Wang Q., Schlenoff J.B. The polyelectrolyte complex/coacervate continuum. Macromolecules. 2014;47:3108–3116. doi: 10.1021/ma500500q. [DOI] [Google Scholar]
- Wilson K.R., Prophet A.M., Rovelli G., Willis M.D., Rapf R.J., Jacobs M.I. A kinetic description of how interfaces accelerate reactions in micro-compartments. Chem. Sci. 2020;11:8533–8545. doi: 10.1039/D0SC03189E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Fang C., Mei H., Chang T., Cao Z., Shangguan D. Characterization of G-quadruplex/hemin peroxidase: Substrate specificity and inactivation kinetics. Chemistry. 2011;17:14475–14484. doi: 10.1002/chem.201101941. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yang Y., Tang X., Chen Y., You Y. Effects of ionic strength on chemical forces and functional properties of heat-induced myofibrillar protein gel. Food Sci. Technol. Res. 2015;21:597–605. doi: 10.3136/fstr.21.597. [DOI] [Google Scholar]
- Zhao B., Summers F.A., Mason R.P. Photooxidation of amplex red to resorufin: Implications of exposing the amplex red assay to light. Free Radic. Biol. Med. 2012;53:1080–1087. doi: 10.1016/j.freeradbiomed.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. All original code is available in this paper's supplemental information. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.