Summary
Exosomes are important for cell–cell communication. Deficiencies in the human dihydroceramide desaturase gene, DEGS1, increase the dihydroceramide-to-ceramide ratio and cause hypomyelinating leukodystrophy. However, the disease mechanism remains unknown. Here, we developed an in vivo assay with spatially controlled expression of exosome markers in Drosophila eye imaginal discs and showed that the level and activity of the DEGS1 ortholog, Ifc, correlated with exosome production. Knocking out ifc decreased the density of the exosome precursor intraluminal vesicles (ILVs) in the multivesicular endosomes (MVEs) and reduced the number of exosomes released. While ifc overexpression and autophagy inhibition both enhanced exosome production, combining the two had no additive effect. Moreover, DEGS1 activity was sufficient to drive ILV formation in vitro. Together, DEGS1/Ifc controls the dihydroceramide-to-ceramide ratio and enhances exosome secretion by promoting ILV formation and preventing the autophagic degradation of MVEs. These findings provide a potential cause for the neuropathy associated with DEGS1-deficient mutations.
Subject areas: Cell biology, Functional aspects of cell biology
Graphical abstract

Highlights
-
•
An in vivo system was developed for observing exosome production in Drosophila
-
•
Dihydroceramide desaturase (DEGS1/Ifc) promotes exosome production at two steps
-
•
DEGS1/Ifc drives membrane invagination for the formation of intraluminal vesicles
-
•
DEGS1/Ifc inhibits autophagic degradation of MVEs and increases exosome release
Cell biology; Functional aspects of cell biology
Introduction
Exosomes are secreted vesicles of approximately 100 nm in diameter that mediate cell–cell communication via the transport of proteins, RNAs, and lipid cargos. In the nervous system, exosomes contribute to the formation and maintenance of myelin sheath (Kramer-Albers et al., 2007). Exosomes originate from the invagination of the endosomal membrane in multivesicular endosomes (MVEs) to form intraluminal vesicles (ILVs) (Raposo and Stoorvogel, 2013). ILVs can form by a mechanism involving the endosomal sorting complexes required for transport (ESCRT) complex, which consists of more than 20 proteins that recruit ubiquitinated protein and other cargos and facilitate the budding and scission of the membrane into the MVE lumen to form ILVs (Katzmann et al., 2001; Adell et al., 2014). ILVs can also form via an ESCRT-independent, ceramide (Cer)-dependent mechanism. Trajkovic et al. (2008) show that the inhibition of neutral sphingomyelinase blocks the conversion of sphingomyelin to Cer and reduces exosome release from oligodendrocytes, suggesting that Cer promotes membrane inward budding for ILV formation. When the MVE membrane fuses with the plasma membrane, ILVs are released to the extracellular space as exosomes. Alternatively, ILV-containing MVEs may be targeted for autophagic-lysosomal degradation. Indeed, inhibition of lysosome–autophagosome fusion by bafilomycin A1 has been shown to increase the secretion of α-synuclein in the exosomes (Poehler et al., 2014; Alvarez-Erviti et al., 2011), suggesting that cells may use exosomes as a protective mechanism to remove protein aggregates during lysosomal or autophagic dysfunction. Although an ubiquitin-like post-translational modification called ISGylation has been described to promote MVE degradation and thus block exosome secretion (Villarroya-Beltri et al., 2016), little is known regarding the effect of membrane lipid composition on the fate of MVEs.
The final step of Cer de novo synthesis is catalyzed by the enzyme dihydroceramide desaturase (DEGS1 in humans and Ifc in Drosophila) which inserts a 4,5-trans-double bond into the sphingolipid backbone of dihydroceramide (dhCer) to produce Cer. This structural difference has a profound effect on the biophysical properties of Cer; therefore, the relative abundance of Cer versus dhCer may affect the biological properties of cellular membranes (Li et al., 2002). Indeed, genetic or drug inhibition of DEGS1/Ifc has been shown to cause significant accumulation of dhCer which increases reactive oxygen species, induces autophagy and ER stress, causes cell-cycle arrest, and even results in cell death in experimental models (Lee et al., 2012). In humans, variants of the dhCer desaturase gene, DEGS1, have recently been linked to hypomyelinating leukodystrophy and systemic neuropathy (Dolgin et al., 2019; Karsai et al., 2019; Pant et al., 2019). These reports not only confirm an essential role for DEGS1 in the human nervous system but also show that loss-of-function mutation of DEGS1cause significant accumulation of dhCer and an overall shift of cellular sphingolipid pool toward the dihydro forms (Karsai et al., 2019). However, the underlying cause of neuropathy associated with loss of DEGS1 activity remains to be investigated.
Our previous study shows that eye-specific knockout of Drosophila ifc leads to activity-dependent neurodegeneration, supporting a conserved function in the nervous system. We also show that Ifc can function cell non-autonomously, possibly mediated by exosomes (Jung et al., 2017). Although the cellular level of dhCer is low, several studies report the presence of dhCer in the exosome membrane (Brzozowski et al., 2018; Podbielska et al., 2016; Vallabhaneni et al., 2015). Considering ∼30% of cellular Cer is estimated to be produced via the de novo synthesis pathway (Lee et al., 2012), whether DEGS1 activity and dhCer level affect exosome formation and MVE degradation to determine exosome production warrants further study.
In the present study, we developed an assay to observe exosome secretion in vivo by expressing exosome markers in the dorsal half of Drosophila eye imaginal discs. This system allowed the quantification of dorsally made exosomes in response to genetic manipulations on the ventral half of the eye imaginal discs. We also used the giant unilamellar vesicle (GUV) system to examine if human DEGS1 activity is sufficient to drive the formation of ILVs in vitro. Our findings demonstrate an important role for DEGS1/Ifc in regulating both the formation of intraluminal vesicles and the autophagic degradation of MVEs, providing a possible mechanism underlying its function in the nervous system.
Results
Knocking out ifc reduces the number of exosomes in vivo
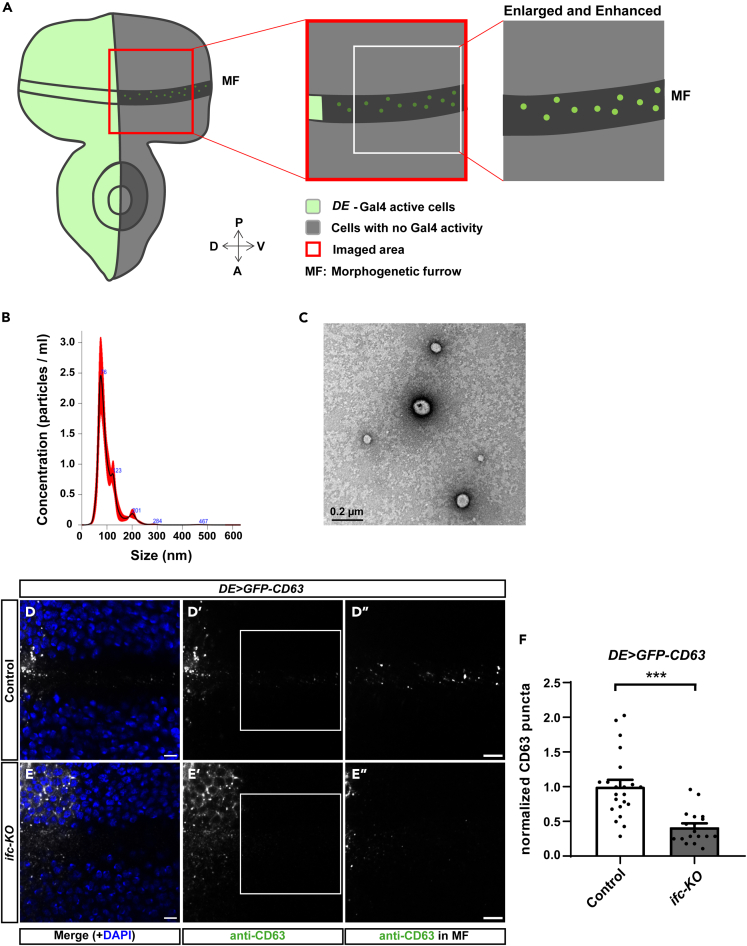
To evaluate the role of ifc in exosome biogenesis, we established an in vivo system with the Drosophila eye imaginal discs that have been shown to secrete exosomes (Gross et al., 2012). We utilized the Dorsal-Eye Gal4 (DE-GAL4) expression system (Morrison and Halder, 2010) to drive the expression of the exosome marker GFP-CD63 (Panakova et al., 2005) restricted to the dorsal half of the disc. We were able to detect the presence of GFP-CD63 puncta in the acellular space above the morphogenetic furrow marked by the lack of DAPI staining on the ventral side of the eye imaginal discs (illustrated in Figure 1A), indicating the movement of GFP-CD63+ extracellular vesicles moving along the dorsal-ventral axis. We collected particles released by the discs and determined their sizes by nanoparticle tracking analysis. The average diameter of the secreted vesicles was 113.8 ± 14.3 nm (Figure 1B), consistent with the size range of exosomes. The morphology of the collected vesicles was also consistent with that of exosomes by transmission electron microscopy (TEM) (Figure 1C). Together, these findings show that the labeled puncta exhibit main characteristics of exosomes based on marker proteins, size distribution, and dorsal-ventral movement (Thery et al., 2018), supporting the use of GFP-CD63 as a surrogate to monitor exosome production in the system of Drosophila eye imaginal discs.
Figure 1.
Knocking out ifc reduces the number of released exosomes in vivo
(A) A schematic illustration of the system for observing the release of exosomes. Dorsal-Eye Gal4 (DE-GAL4) drives the expression of UAS-GFP-CD63 in the dorsal half of the eye imaginal disc (green). The secreted exosomes that move toward the ventral side alone, the morphogenetic furrow (MF) into the ventral side where images were taken (red box). The intensity and contrast were linearly enhanced for the quantification of fluorescent puncta in the MF within the region of interest (ROI, white box).
(B) The representative size distribution of exosomes collected from approximately 50 eye imaginal discs was determined by nanoparticle tracking analysis using Nanosight NS300.
(C) Representative image showing the morphology of exosomes by transmission electron microscopy. Scale bar: 0.2 μm.
(D and E) Representative confocal images of Drosophila eye imaginal discs with dorsally expressed GFP-CD63 in FRT control (D-D″) and ifc-KO (E-E″) showing signals merged with DAPI in blue (D,E), GFP-CD63 detected by anti-CD63 staining in white (D′,E′), and enlarged images of the ROIs with linearly enhanced brightness and contrast to show the GFP-CD63 puncta in the MF (D″,E″). Scale bars: 10 μm.
(F) Quantification of GFP-CD63 puncta. The bar chart shows the mean ± SEM of at least three independent experiments. ∗∗∗p < 0.001 (Student's t-test).
See also Figures S1 andS2.
To circumvent the larval lethality of systemic ifc-KO, we used the FLP-FRT system to clonally knock out ifc in the eye imaginal discs (Jung et al., 2017). The chromosome in the opposite of ifc-KO contained the recessive cell lethal mutation cl, resulting in the elimination of cells that were not ifc-KO thus creating the homozygous ifc-KO eye imaginal discs. Knocking out ifc did not affect the expression levels of GFP-CD63 in the dorsal region where GFP-CD63 was expressed under DE-GAL4 (Figures S1A–S1C). Upon quantification, we found that the number of CD63 puncta detected away from the dorsal compartment was significantly reduced in the ifc-KO eye imaginal discs in comparison with the FRT controls (Figures 1D–1F). We further utilized this system to express the fusion protein of the flotillin protein Flo2 and RFP as a second exosome marker (Bischoff et al., 2013). Consistent with the results observed with GFP-CD63, ifc-KO reduced the number of Flo2-RFP puncta in the ventral compartment (Figures S1D–S1F). Because ifc-KO leads to a drastic accumulation of dhCer, the decrease in exosomes produced by ifc-KO clones suggests that the accumulation of dhCer suppresses exosome secretion.
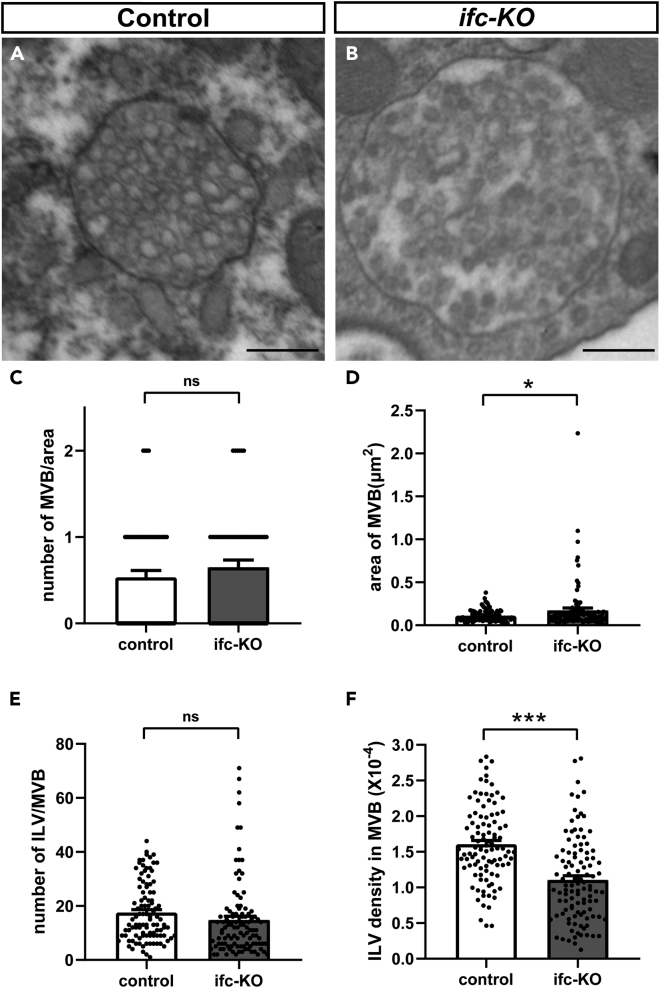
We then examined the morphology of MVE by analyzing the ultrastructural images of MVEs in the control and ifc-KO photoreceptors by TEM. The control MVEs typically contained densely packed ILVs (Figure 2A). In comparison, the MVEs in ifc-KO cells appeared dilated (Figure 2B). Indeed, while the numbers of MVE per view field were similar between the control and mutant photoreceptors (Figure 2C, p = 0.3213), the size of the MVEs was significantly larger in ifc-KO photoreceptors (Figure 2D, p = 0.0226). Moreover, the average number of ILVs in each MVE was slightly reduced in ifc-KO, although the difference from the control was not statistically significant (Figure 2E, p = 0.1153). However, when the size of MVEs was taken into consideration, we found that the density of ILVs was significantly reduced in the mutant MVEs compared with that of the control (Figure 2F, p < 0.0001). Together, these data show that ifc-KO affects the morphology of MVEs.
Figure 2.
Knocking out ifc reduces the density of intraluminal vesicles (ILVs) inside of multivesicular endosomes (MVEs)
(A and B) Representative transmission electron microscopy (TEM) images of the control (A) and ifc-KO photoreceptors (B) after 3 days of light stimulation. Scale bars: 250 nm.
(C–F) Quantification of TEM images including the number of MVEs per field (C), the area of MVEs (D), the number of ILVs per MVE (E), and the number of ILVs per unit area of MVEs (F). The bar charts show the mean ± SEM of at least three independent experiments. ∗∗∗p < 0.001; ∗p < 0.05; ns: Not significant (Student's t-test).
Enhancing Ifc activity increases the number of exosomes
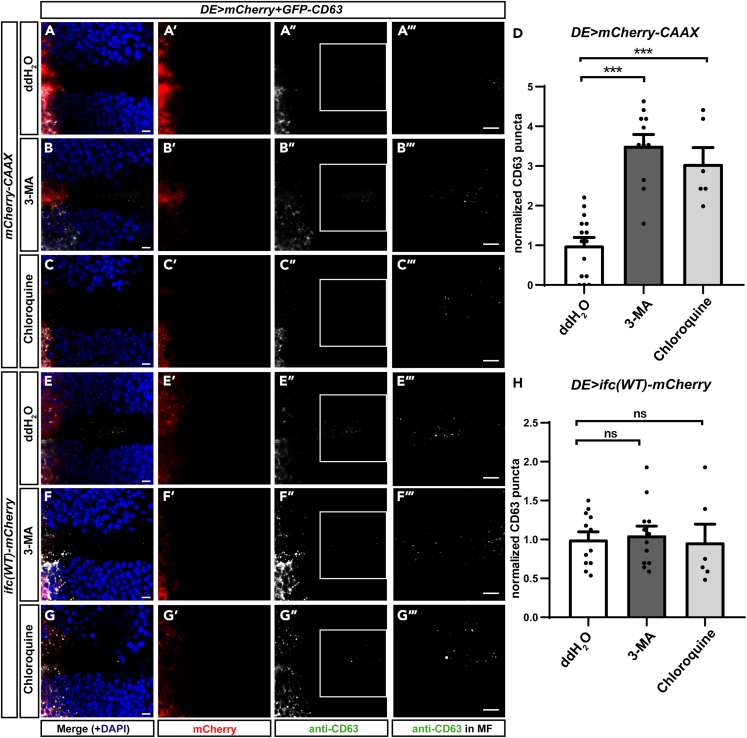
To determine whether enhanced ifc activity has the opposite effect of ifc-KO, we tested the effect of overexpressing wild-type Ifc with a C-terminal mCherry tag (ifc(WT)-mCherry) which we have shown to have the ability to rescue the ifc-KO phenotype (Jung et al., 2017). Overexpression of ifc(WT)-mCherry increased the number of GFP-CD63 puncta compared with the UAS control of mCherry-CAAX overexpression (Figures 3A, 3B, and 3D). We performed live imaging of ex vivo eye imaginal discs using spinning disc confocal microscopy and light sheet microscopy and found that, in real-time, ifc(WT)-mCherry produced more GFP-CD63 puncta than the mCherry-CAAX control (Videos S1, S2, S3, and S4). Consistently, overexpression of the ifc(WT)-mCherry also increased the number of extracellular puncta labeled with another exosome marker TSG101-HA (Thery et al., 2001) (Figure S2). These results suggest that an increased level of Ifc promotes exosome production.
Figure 3.
Enhancing Ifc activity increases the number of exosomes
(A–C) Representative confocal images of Drosophila eye imaginal discs with dorsally expressed GFP-CD63 with the UAS control mCherry-CAAX (A-A‴), ifc(WT)-mCherry (B-B‴), or ifc(C3mut)-mCherry (C-C‴) showing merged images with DAPI in blue (A,B,C), mCherry fluorescence in red (A′,B′,C′), anti-CD63 staining in white (A″,B″,C″), and enlarged images of the ROI (white box) with linearly enhanced brightness and contrast to reveal GFP-CD63 puncta in the MF (A‴,B‴,C‴). Scale bars: 10 μm.
(D) Quantification of GFP-CD63 puncta. The bar chart shows the mean ± SEM of at least three independent experiments. ∗∗∗p < 0.001; ns: Not significant (Student's t-test).
See also Figure S3.
To investigate whether the catalytic activity of Ifc was required, we mutated one of the highly conserved histidine boxes that are required for the enzyme activity of Ifc (Figure S3A) (Ternes et al., 2002). The mutation did not affect the protein level as we showed by western blot that the level of Ifc(C3mut)-mCherry was comparable with that of Ifc(WT)-mCherry (Figures S3B and S3C). Sphingolipidomics results showed that ifc(C3mut)-mCherry failed to reverse the increase of dhCer-to-Cer ratio in ifc-KO, suggesting that the C3 mutation impaired its catalytic activity (Figure S3D). Overexpression of ifc(C3mut)-mCherry in ifc-KO eye imaginal discs failed to increase the number of GFP-CD63 puncta released to the ventral side in comparison with the mCherry-CAAX control (Figures 3A, 3C, and 3D), suggesting that the desaturase activity of Ifc is required to promote the production of exosomes. These results show that the activity of DEGS1/Ifc to convert dhCer to Cer is required to drive exosome formation in vivo and demonstrate a correlation between the level and activity of Ifc with exosome production.
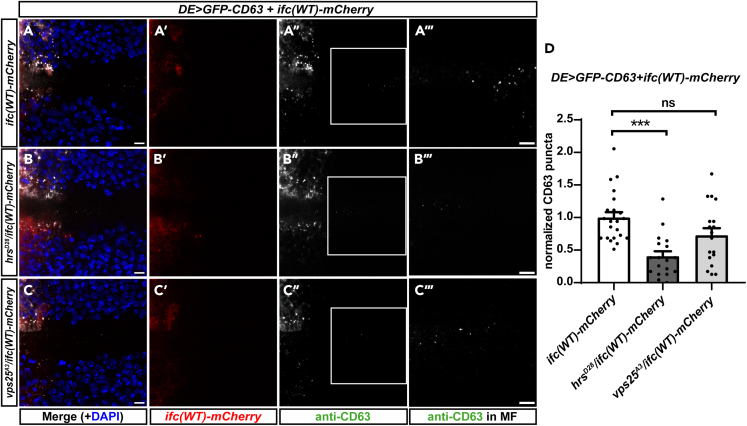
Ifc interacts with ESCRT genes to regulate exosome production
Because the role of ESCRT proteins in regulating the process of ILV formation is well studied, we investigated the genetic interactions between ifc and the ESCRT-0 gene hrs and the ESCRT-II gene vps25. Because homozygous mutations of hrsD28 and vps25A3 are both lethal (Lloyd et al., 2002; Vaccari and Bilder, 2005), we analyzed whether Ifc overexpression genetically interacts with the heterozygous mutations of these two ESCRT genes to regulate exosome production. As expected, the exosomes detected in hrsD28/+ and vps25A3/+ were significantly fewer than that in control (Figure S4). We found that overexpression of ifc(WT)-mCherry significantly rescued exosome production in vps25A3/+ but not in hrsD28/+ (Figure 4). The result indicates that, for exosome formation, overexpression of ifc can compensate for the partial loss of vps25 but not hrs. Because ESCRT-0 (Hrs) is important for the recruitment of ubiquitinated cargos while the ESCRT-II (Vps25) complex is necessary for the initiation of membrane invagination and the recruitment of the ESCRT-III complex (Wollert and Hurley, 2010), this finding supports a role for Ifc in the inward budding of MVE membrane during ILV biogenesis but possibly not in cargo recruitment.
Figure 4.
Ifc genetically interacts with ESCRT genes to regulate exosome production
(A–C) Confocal images of Drosophila eye imaginal discs with dorsally expressed GFP-CD63 and ifc(WT)-mCherry in the control (A), hrsD28/+ (B), and vps25A3/+ (C) showing merged images with DAPI in blue (A,B,C), mCherry fluorescence in red (A′,B′,C′), anti-CD63 staining in white (A″,B″,C″), and enlarged images of the ROI (white box) with linearly enhanced brightness and contrast to reveal GFP-CD63 puncta in the MF in the ROIs (A‴,B‴,C‴). Scale bars: 10 μm.
(D) Quantification of GFP-CD63 puncta. The bar chart shows the mean ± SEM of at least three independent experiments. ∗∗∗p < 0.001; ns: Not significant (one-way ANOVA).
See also Figure S4.
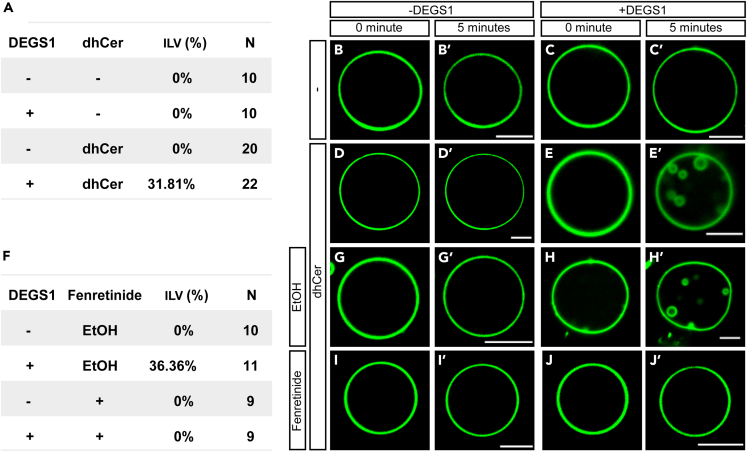
DEGS1 drives the formation of ILVs in GUVs in vitro
To evaluate if dhCer desaturase is sufficient to drive ILV formation, we resorted to the in vitro model membrane system of GUV. After the generation of GUVs, we tracked the morphology of the GUVs over time and observed no spontaneous ILV formation either with or without the inclusion of dhCer in the lipid mixture (Figures 5A, 5B, 5B′, 5D, and 5D′). Interestingly, when we added human DEGS1 recombinant enzyme to the GUVs, we were able to observe the formation of ILVs in the GUVs that contained dhCer within 5 min (Figures 5A, 5C, 5C′, 5E, and 5E′). ILV formation required ATP and occurred at a higher efficiency at 37°C than 25°C, suggesting that it was indeed promoted by DEGS1 enzyme activity (Figure S5). When we added the DEGS1 inhibitor fenretinide to the system, the formation of ILVs by DEGS1 was blocked (Figures 5F, 5J, and 5J′); the ethanol solvent control had no such inhibitory effect (Figures 5F, 5H, and 5H′). Therefore, the activity of DEGS1 was sufficient to drive the formation of ILV in vitro by converting dhCer to Cer. These data further support that the catalytic activity of DEGS1 promotes membrane invagination for the initiation of exosome biogenesis.
Figure 5.
DEGS1 drives the formation of ILVs in the giant unilamellar vesicles (GUVs) in vitro
(A) Table summary of the percentages of GUVs that formed ILVs within 5 min with or without dhCer and recombinant DEGS1 protein.
(B–E) Representative images of GUVs in (A).
(F) Table summary of the percentages of GUVs that showed ILV formation within 5 min with and without DEGS1 protein in the presence of ethanol (EtOH) solvent or 0.2 μM fenretinide, a DEGS1 inhibitor.
(G–J)Representative images of GUVs in (F). Scale bar: 5 μm.
See also Figure S5.
Ifc promotes exosome secretion by blocking autophagy-mediated degradation of MVEs
We and others have reported autophagy induction as a common outcome of DEGS1/ifc inhibition not only in Drosophila but also in mammalian cell lines and animal models (Barbarroja et al., 2015; Holland et al., 2007; Jung et al., 2017; Kraveka et al., 2007). In addition, we previously identified the endosomal localization of Ifc (Jung et al., 2017) and herein showed the regulation of DEGS1/Ifc in exosome secretion. Therefore, we hypothesized that DEGS1/Ifc functions as the pivot of MVE fate between autophagic degradation versus fusion with the plasma membrane for exosome release.
To evaluate the interrelationship between autophagy and exosome pathways, we inhibited autophagy by feeding the Drosophila larvae 3-methyladenine (3-MA) and chloroquine to block the formation of autophagosome and prevent autophagosomes-lysosome fusion, respectively (Jung et al., 2017). Treatment of larvae with 3-MA and chloroquine both resulted in ref(2)P accumulation, showing that autophagy was inhibited (Figure S6). Inhibiting autophagy by 3-MA and chloroquine both increased the numbers of GFP-CD63+ vesicles released in the control background (Figures 6A–6D). This result suggests that GFP-CD63 puncta in MVEs underwent autophagy-mediated degradation in ifc-KO, which helps explain why we found an insignificant decrease in ILV numbers in MVEs but a more profound decrease in released GFP-CD63.
Figure 6.
Ifc promotes exosome secretion by blocking autophagy-mediated degradation of MVEs
(A–C) Representative confocal images of Drosophila eye imaginal discs with dorsally expressed GFP-CD63 treated with ddH2O (A-A‴), 3-methyladenine (3-MA, B-B‴), and chloroquine (C-C‴) showing merged images with DAPI in blue (A,B,C), mCherry in red (A′,B′,C′), anti-CD63 staining in white (A″,B″,C″), and enlarged images of the ROI (white box) with linearly enhanced brightness and contrast to show GFP-CD63 puncta in the MF (A‴,B‴,C‴).
(D) Quantification of GFP-CD63 puncta.
(E–G) Representative confocal images of Drosophila eye imaginal discs with dorsally expressed GFP-CD63 and ifc(WT)-mCherry treated with ddH2O (E-E‴), 3-methyladenine (3-MA, F-F‴), and chloroquine (G-G‴) showing merged images with DAPI in blue (E,F,G), Ifc(WT)-mCherry in red (E′,F′,G′), anti-CD63 staining in white (E″,F″,G″), and enlarged images of the ROI with linearly enhanced brightness and contrast to show GFP-CD63 puncta in the MF (E‴,F‴,G‴).
(H) Quantification of GFP-CD63 puncta.
Scale bars: 10 μm. The bar charts show the mean ± SEM of at least three independent experiments. ∗∗∗p < 0.001; ns: Not significant (Student's t-test).
See also Figure S6.
Consistent with the results shown in Figure 3, overexpression of ifc(WT)-mCherry increased the number of exosomes (Figures 6A and 6E). Interestingly, feeding 3-MA or chloroquine to larvae overexpressing ifc did not further increase the number of GFP-CD63 exosomes in the ventral morphogenetic furrow (Figures 6E–6H). The lack of an additive effect from treating ifc-overexpressing larvae with autophagy inhibitors suggests that Ifc promotes the production of exosomes by inhibiting autophagy.
Human DEGS1 is present in the endosomal system
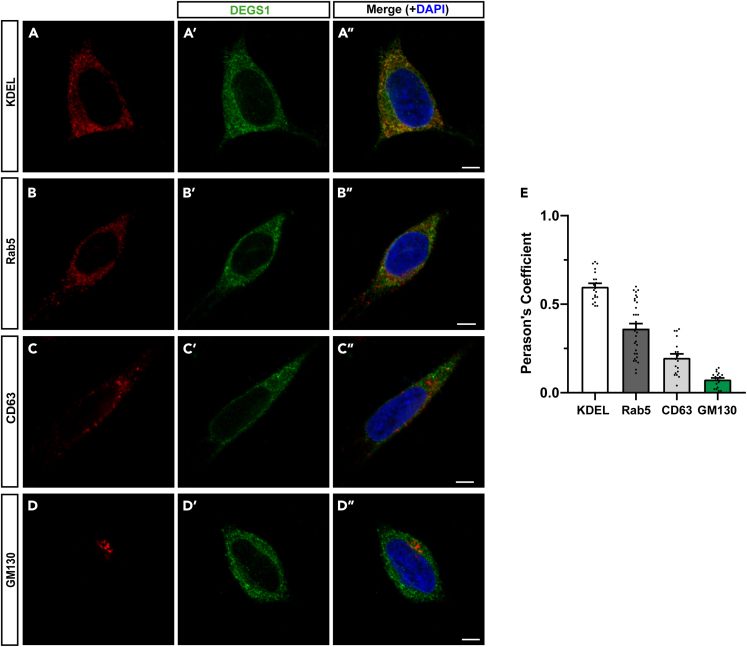
The human ortholog of Drosophila is DEGS1, which is conventionally believed to locate in the ER. We examined the subcellular localization of DEGS1 by immunostaining in human neuroblastoma SH-SY5Y cells. We found that DEGS1 had the most significant colocalization with the ER marker KDEL with a Pearson's coefficient of r = 0.599, consistent with conventional knowledge. However, we also identified obvious colocalization of DEGS1 with the endosomal protein Rab5 (r = 0.362), and to lesser extents with the Golgi protein GM130 (r = 0.074) and CD63 (r = 0.197), which locates on intracellular vesicles (Figure 7). Therefore, while DEGS1 predominantly resides in the ER to produce Cer, its presence in the endosomal compartment suggests it may function locally to control ILV formation and MVE fate to regulate exosome production and release.
Figure 7.
The subcellular localization of human DEGS1 in SH-SY5Y cells
(A–D)Representative confocal images of SH-SY5Y cells immunostained with antibodies against specific subcellular compartments including anti-KDEL for the ER (A), anti-Rab5 for the endosomes (B), anti-CD63 for vesicles (C), and anti-GM130 for the Golgi apparatus (D). The cells were co-stained with anti-DEGS1 antibodies (A′-D′) and merged with DAPI (A″-D″).
(E) Pearson's coefficient of DEGS1 and the markers of subcellular compartments. Scale bars: 5 μm. The bar chart shows the mean ± SEM of at least three independent experiments.
Discussion
In the present study, we developed an in vivo system for the analysis of exosome production using Drosophila eye imaginal discs with the dorsal expression of GFP-CD63. The validity of the system was supported by two additional exosome markers, Flo2 and TSG101, as well as size measurement by nanoparticle tracking analysis. We showed in vivo and in vitro that the release of exosomes from the dorsal half of Drosophila eye imaginal discs to the ventral morphogenetic furrow correlated with not only the level but also the activity of the dihydroceramide desaturase, ifc. Our genetic analysis and GUV assay support a clear role for Ifc in the early stage of ILV formation in promoting membrane invagination. In addition, ifc also affects the degradation of MVEs via autophagy. Overexpression of ifc inhibits autophagy to preserve MVEs thus increasing exosome secretion. Together, our data show that DEGS1/Ifc promotes membrane invagination of ILV formation during exosome biogenesis and prevents the autophagic degradation of MVEs, thus promoting exosome formation and release.
Between the ESCRT-dependent and Cer-mediated mechanisms of exosome biogenesis, it is generally believed that the ESCRT-dependent mechanism is involved in the selective sorting of cargos, whereas the Cer-dependent mechanisms are important to control the initial membrane budding (Larios et al., 2020; Teis et al., 2010; Trajkovic et al., 2008). Here, we found that overexpression of ifc(WT)-mCherry could rescue the exosome numbers in the heterozygous mutant of vps25 but not in that of hrs, suggesting that ifc does not regulate cargo loading. However, Kajimoto et al. (2013) show that the continuous Cer catabolism produces sphingosine 1-phosphate that is required for cargo sorting into ILVs in human cells. Exosome membranes share similar lipid composition with lipid rafts, both highly enriched for cholesterol and sphingolipids (Hebbar et al., 2008). Lipid rafts are rigid membrane microdomains that serve as a platform for protein-protein interaction and are essential for protein activation and signal transduction (Lingwood and Simons, 2010). Cer-rich domains have been recognized to affect raft properties by competing and displacing cholesterol (Megha and London, 2004) and promoting the formation of highly ordered gel phases (Bieberich, 2018). The Cer-rich membrane domains may not only serve as a permissive environment to promote membrane invagination for ILV formation but may also behave as a platform to facilitate the recruitment of ESCRT components. Although DEGS1 has been widely believed to reside in the ER, its presence outside of the ER has been reported previously (Beauchamp et al., 2009; Murakami et al., 2015). We have reported that Ifc predominantly resides in the endosomes in Drosophila photoreceptors (Jung et al., 2017). Here, we showed that human DEGS1 also had an obvious presence in the endosomal system, suggesting an on-site conversion of dhCer into Cer for the dynamic regulation of membrane property. How the levels of Cer vs. dhCer on local membranes influence biological processes such as ILV formation need to be further investigated.
Several previous studies have shown that inhibition of autophagosome-lysosome degradation enhances exosome secretion (Danzer et al., 2012; Fader et al., 2008; Iguchi et al., 2016). Interestingly, inhibition of autophagy by bafilomycin A1 (BafA1) not only increases the secretion of CD63+ exosomes but also increases the detection of autophagic proteins p62 and LC3 in exosomes (Minakaki et al., 2018). However, autophagy proteins ATG12–ATG3 have been shown to interact with the ESCRT-associated protein Alix to promote late endosome to lysosome trafficking as well as exosome biogenesis (Murrow et al., 2015). In addition, knockout of ATG5 but not ATG7 significantly reduces the number of exosomes, suggesting that not all of the autophagy core components participate in the regulation of exosome release (Guo et al., 2017). In addition, exosomes purified from cultured cortical neurons treated with BafA1 show a ∼2-fold increase in certain sphingolipid species, including Cer and dhCer, indicating BafA1 affects the lipid composition of exosomes (Miranda et al., 2018). Leidal et al. (2020) report that LC3 and LC3-conjugation machinery recruit specific cargos to pack into LC3-positive EVs for secretion, a process that does not require the ESCRT complex but is dependent on the activity of neutral sphingomyelinase 2, an enzyme that hydrolyzes sphingomyelin to produce Cer. In the present study, we showed that exosome production increased with the inhibition of autophagy by either 3-MA or chloroquine. However, combining autophagy inhibition and ifc overexpression did not further increase exosome production, implying that ifc overexpression inhibits autophagy in opposite to ifc-KO which induces autophagy. These findings suggest that the discrepancy we found in the level of decrease in ILV numbers/density and that in secreted GFP-CD63 puncta in ifc-KO may be due to differences in autophagy-mediated degradation of MVEs. Membrane Cer/dhCer may play an important role in controlling the fate of MVEs between autophagic degradation and secretion of exosomes; further study is warranted to elucidate the mechanism in detail.
While the effects of Cer have been extensively studied in various contexts, our findings draw attention to dhCer, which is a long-ignored sphingolipid species. Although dhCer is structurally different from Cer only in the lack of the 4,5-double bond, the two sphingolipid species have widely different biophysical properties and bioactivities. Several previous studies have hinted at a role for dhCer in the nervous system. Jung et al. (2017) show that eye-specific ifc-KO results in activity-dependent neurodegeneration and demonstrated that ifc can function cell non-autonomously. Inherited DEGS1 deficiencies lead to hypomyelination in the central and peripheral nervous systems (Dolgin et al., 2019; Karsai et al., 2019; Pant et al., 2019). Because exosomes are important for myelination and demyelination, whether altered exosome production is the underlying cause of defects seen in patients with inherited DEGS1 deficiencies warrants further investigation.
Limitations of the study
There are some limitations to the study. First, we used GFP-CD63 as a surrogate for exosomes for the in vivo imaging experiments, and the regulation of exosome populations that do not contain CD63 was not directly analyzed. Second, we were not able to perform western blot analysis of the secreted exosomes because we could not collect a sufficient amount of samples owing to the small size of Drosophila eye imaginal discs. Without assessing the presence of exosome markers and the absence of non-exosome contaminants, we could not completely rule out the possible presence of extracellular vesicles other than exosomes in the in vitro analyses. Third, for the in vivo observation of exosomes, most experiments were conducted using the eye imaginal discs, which is a developing tissue in Drosophila. Although we show that human DEGS1 is also present in the endosomal system, further investigation is needed to elucidate whether DEGS1 functions to control exosome formation and release in other tissues, life stages, and animals.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-CD63 | Developmental Studies Hybridoma Bank (DSHB) | Cat# H5C6;RRID: AB_528158 |
| Anti-RFP | Rockland | Cat# 600-401-379; RRID: AB_2209751 |
| Anti-HA | Cell Signaling Technology | Cat# 3724;RRID: AB_1549585 |
| Anti-mCherry | GeneTex | Cat# GTX630189; RRID: AB_2888193 |
| Anti-Actin | Millipore | Cat# MAB1501; RRID: AB_2223041 |
| Anti-mouse-HRP | Jackson ImmunoResearch Laboratories, Inc. | Cat# 115-005-003; RRID: AB_2338447 |
| Anti-DEGS1 | Abcam | Cat# ab167169 |
| Anti-KDEL | Santa Cruz Biotechnology | Cat# sc-58774; RRID: AB_784161 |
| Anti-Rab5 | Santa Cruz Biotechnology | Cat# sc-46692; RRID: AB_628191 |
| Anti-GM130 | Abcam | Cat# ab169276 |
| Anti-Rabbit IgG-Alexa Fluor 488 | Jackson ImmunoResearch Laboratories, Inc. | Cat# 111-545-003; RRID: AB_2338046 |
| Anti-mouse IgG-Cy3 | Jackson ImmunoResearch Laboratories, Inc. | Cat# 115-165-003; RRID: AB_2338680 |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | ThermoFisher | Cat# D1306; |
| Fenretinide | Tocris Bioscience | 1396 |
| 3-Methyladenine | Sigma | 5142234 |
| Chloroquine | Sigma | C6628-25G |
| Sucrose | Sigma | S7903 |
| NADPH | Sigma | 1010782400 |
| ATP | Sigma | A2383 |
| Recombinant human DEGS1 protein | Abcam | ab160100 |
| Experimental models: Cell lines | ||
| Human: SH-SY5Y | ATCC | CRL-2266 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: ey-FLP; cl, FRT40A/CyO | Bloomington Drosophila Stock Center | RRID: BDSC_5622 |
| D. melanogaster: FRT40A/CyO | Bloomington Drosophila Stock Center | RRID: BDSC_5615 |
| D. melanogaster: DE-GAL4/TM3sb | Bloomington Drosophila Stock Center | RRID: BDSC_29650 |
| D. melanogaster: UAS-mCherry-CAAX | Bloomington Drosophila Stock Center | RRID: BDSC_59021 |
| D. melanogaster: UAS-hTSG101-HA | Bloomington Drosophila Stock Center | RRID: BDSC_77977 |
| D. melanogaster: hrsD28 | Bloomington Drosophila Stock Center | RRID: BDSC_54574 |
| D. melanogaster: vps25A3 | Bloomington Drosophila Stock Center | RRID: BDSC_39633 |
| D. melanogaster: ifc-KO, FRT40A | Jung et al. (2017) | N/A |
| D. melanogaster: UAS-ifc(WT)-mCherry | Jung et al. (2017) | N/A |
| D. melanogaster: UAS-EGFP-CD63 | Gift from Dr. Suzanne Eaton (Panakova et al., 2005) | N/A |
| D. melanogaster: UAS-ifc(C3mut)-mCherry | This study | N/A |
| D. melanogaster: UAS-Flo2-RFP | Bischoff et al. (2013) | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ RRID: SCR_002798 |
| Fiji (ImageJ) | National Institutes of Health (NIH) | https://imagej.net/software/fiji/ RRID: SCR_002285 |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| RRID:SCR_010279 | ||
| Power Director | CyberLink Corporation | 19.3.2724.0 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shu-Yi Huang (shuyih@ntuh.gov.tw)
Materials availability
Drosophila strains generated in the study are available upon request from the Lead Contact with a material transfer agreement.
Experimental model and subject details
Experiments were performed in compliance with the guidelines of the Environmental Health and Safety Center of National Taiwan University College of Medicine and National Taiwan University Hospital. Experiments involving gene recombination were approved by the Biosafety Committees of National Taiwan University College of Medicine and National Taiwan University Hospital.
Drosophila strains and genetics
Drosophila stocks were maintained at 25°C on standard medium and genetic crosses were performed following standard fly husbandry procedures. Male and Female flies were equally represented in all experiments. Information on individual fly strains can be found on FlyBase (flybase.org) unless otherwise noted in the above Key Resources Table. To circumvent the larval lethality of systemic ifc-KO, the ifc-KO eye imaginal discs used in the study were whole-eye clones where ifc was knocked out by the FLP-FRT system (Stowers and Schwarz, 1999). We used ey-FLP to express flippase recombinase in the eye discs, which renders the post-mitotic recombination of the FRT-containing ifc-KO chromosome against the opposite FRT-containing chromosome in the heterozygous ifc-KO cell, resulting in the homozygous ifc-KO clone and its sister clone. In our system, the opposite chromosome contained cl, a recessive cell lethal mutation. Therefore, after the FLP-FRT recombination, the sister clone will be eliminated due to homozygous cl, thus creating ifc-KO whole-eye clones.
Method details
Immunohistochemistry and confocal microscopy
For immunofluorescence staining, larval eye imaginal discs from 3rd instar larvae were dissected, fixed, washed, and immunostained with primary and secondary antibodies as previously described (Jung et al., 2017). Human SH-SY5Y cells were fixed in 4% paraformaldehyde for 10 minutes, blocked with 1% BSA at 25°C for 1hr, incubated with primary antibodies at 4°C overnight, and then incubated with secondary antibodies at 25°C for 1 hr. The antibodies used were listed in the above Key Resources Table. All samples were mounted in Vectashield (Vector Laboratories) and analyzed on a Carl Zeiss LSM880 confocal microscope with a 100x/1.46 oil objective at a pixel size of 0.08 μm x 0.08 μm x 0.39 μm using the LAS AF software. Imaging data were processed and quantified using Photoshop (CS6, Adobe), Illustrator (CS6, Adobe), and Fiji (National Institutes of Health; Schindelin et al., 2012).
Nanoparticle tracking analysis
Fifty to 100 eye imaginal discs were dissected from 3rd instar larvae and incubated in sterile-filtered Schneider medium (Gibco 21720024) with low-speed agitation (7 rpm) at room temperature for 2.5 h. The supernatant was then separated by low-speed centrifugation at 3,000 x g for 15 min to remove large debris, followed by another centrifugation at 12,000 x g for 15 min. The samples were then passed through a Smart SEC single column (System Biosciences, USA) for EV isolation. The size distribution was determined using NanoSight NS300 (Malvern Panalytical Ltd, United Kingdom) under constant flow (flow rate = 70) at 25°C. The recordings were analyzed by NTA 3.4 software Build 3.4.003 with the detection threshold set to 6.
Transmission electron microscopy and image analysis of MVE morphology
Exosome samples were prepared as described for nanoparticle tracking analysis. Transmission electron microscopy was performed as previously described (Jung et al., 2017). To characterize the MVE morphology, the fields of imaging for the characterization within the eye of each genotype were selected randomly, and the image files were blinded for the analyses of both the area of MVE and the number of ILV. The area and number of MVE were quantified using Photoshop.
Timelapse live imaging of exosome secretion
For live imaging of exosome secretion by spinning disc confocal microscopy, timelapse images were acquired using a Zeiss spinning disc microscope equipped with a Yokogawa CSU-X1 scanner unit and a high-resolution CCD (Phtometrics PRISM 95B). Images were acquired through a 40X objective with an exposure time of 200 ms every 5 minutes for 80 min. For live imaging of exosome secretion by light sheet microscopy, the Gaussian intensity distribution of the laser was projected to an annular ring pattern on the customized aluminum coating mask (thickness of 1,500 angstroms). The masked laser ring image was then projected to a set of galvanometer scanners (6215H, Cambridge Technology) which were composed of a pair of achromatic lenses (Thorlabs, AC254-100-A, Achromat, Ø1”, 400–750 nm), aligned in a 4f arrangement. After passing through the scanning mirror set, the ring pattern was magnified through a relay lens (Thorlabs, AC254-250-A and AC254-350-A Ø1” Achromat, 400–750 nm) and conjugated to the back focal plane of the excitation objective (Nikon CFI Plan Fluorite Objective, 0.30 NA, 3.5 mm WD). The annular pattern was projected to the rear focal plane of the excitation objective and forms a self-reconstructive Bessel beam by optical interference. The signal was detected by a 25X and an sCMOS camera (Hamamatsu, Orca Flash 4.0 v3 sCMOS). Thirty frames were acquired at each time point, with an exposure time of 30 ms for an area of 300 μm x 300 μm. The images for each time point were converted into 4D time series by Imaris 9.1.1. (Bitplane AG).
Site-directed mutagenesis of UAS-ifc mutants
To generate point mutations in the catalytic domain of UAS-ifc transgene, the pUASt-ifc was used as a backbone and site-directed mutagenesis was done with QuikChange Kit (Agilent 200523) following the manufacturer’s instruction. All constructs were verified by sequencing. Transgenic flies were generated by embryo injection according to standard protocols of integrase-mediated insertion (Bischof et al., 2007) followed by a germline transmission screen. Primer sequences for ifc(C3mut) were: (FWD) GCCAACGAGGCCGCCGACTTTCCG; (REV) GGCGGCCTCGTTGGCGTAGCCCAC.
Western blotting
The protein extracts were prepared with dissected fly tissues that were homogenized for 10 min at 4°C in 1x RIPA buffer containing protease inhibitors (ThermoFisher A32955), followed by centrifugation at 16,000 x g for 15min at 4°C to remove the debris. The supernatant of the homogenates was mixed with 4X sample buffer (20% glycerol, 4% SDS, 100 mM Tris pH 6.8, 0.002% Bromophenol blue), boiled for 10 min, separated by SDS-PAGE, and transferred in a wet-transfer tank to PVDF membranes (Millipore, IPVH00010) following manufacturer’s instructions (Bio-Rad). PVDF membranes were incubated with 5% non-fat milk or 5% BSA in TBST [10 mM Tris (pH 8.0), 150 mM NaCl, 0.1% Tween 20] for 1 h at room temperature and then incubated with primary antibodies at 4°C overnight. Membranes were then washed three times with TBST for 10 min each before incubating with secondary antibodies in TBST for 1 h at room temperature, washed with TBST three times, and developed with ECL reagents (Millipore, WBKLS0500). The signals were captured by the BioSpectrumTM 600 Imaging System (UVP Ltd), and the relative intensities of bands were quantified by densitometry using the Fiji software (National Institutes of Health, Schindelin et al., 2012).
Sphingolipidomics
Sphingolipid extraction was performed as described (Jung et al., 2017). Briefly, 500 3rd instar larvae were snap-frozen in liquid nitrogen and homogenized by grinding.
Giant unilamellar vesicle (GUV) assay
GUV was prepared by the electroformation method (Lee et al., 2018). Briefly, the lipid mixture of Dioleoylphosphatidylcholine (DOPC) and cholesterol was dissolved in chloroform at 1:0.67 molar ratio to the final concentration of 15mM. For lipid mixture with dhCer, the lipid mixture included DOPC:dhCer:cholesterol at a 1:1:0.67 molar ratio to the total concentration of 15mM. The lipid mixture was then deposited into a chamber made with a rubber O-ring on an electrically conductive ITO glass. The setup was vacuum dried at 50°C for 1 h, and the solvent was evaporated thereby producing a thin lipid film. The chamber was then re-filled with 2ml of 400mM sucrose. Depending on the experimental conditions, 0.2 μM of the DEGS1 inhibitor fenretinide might be added to the sucrose solution. After sealing the chamber with another piece of ITO glass, the setup was supplied with a sinusoidal voltage waveform of 3.5 V amplitude at 10 Hz frequency and kept at 65°C overnight. The unilamellar vesicles were cooled down and resuspended in 200 μl of 400mM glucose, and confocal images were acquired using Zeiss Cell Observer SD to show the morphology of GUV at time = 0 – 30 min. The recombinant DEGS1 protein at the concentration of 9.62 nM and co-factors (NADPH 2 mM, MgCl2 2.4 mM, NaCl 50 mM, and sucrose 50 mM) were then supplemented to the GUV/sucrose mixture, with or without ATP at 4 mM. The post-treatment images were acquired after 5 min and 30 min of incubation at 25°C or 37°C.
Quantification and statistical analysis
Quantitative data were analyzed using a two-tailed unpaired Student’s t-test or one-way ANOVA with Tukey’s multiple comparison test. Data analysis was performed using GraphPad Prism 5. All data in bar graphs are shown as mean ± SEM. The quantified GFP-CD63 puncta in the experimental groups were normalized to the average of the controls which was set as 1. P values of less than 0.05 were regarded as statistically significant. All quantifications included at least three experimental replicates.
Acknowledgments
The authors are grateful to Drs. Guang-Chao Chen, Robin Hiesinger, Jennifer Jin, Guang Lin, Ya-Wen Liu, Yi Henry Sun, Chao-Wen Wang, Chi-Kuang Yao, and Mr. Fei-Yang Tzou for their comments on the manuscript. The authors sincerely thank Dr. Ya-Wen Liu and Ms. Yu-Chen Chang for helping us establish the GUV system. We thank the staff of the Imaging Core at the First Core Lab of National Taiwan University College of Medicine, the Second, Third, and Sixth Core Labs of the Department of Medical Research, National Taiwan University Hospital, and the Electron Microscopy Laboratory, Tzu Chi University, for technical support. We also thank Brain Research Center (BRC) from National Tsing Hua University for instrument support. BRC is founded by Deep Cultivation Grant from Ministry of Science and Technology and Ministry of Education, R.O.C. We thank WellGenetics, Inc. for the embryo injection service. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This work was supported by the Ministry of Science and Technology of Taiwan grants 107-2635-B-002-001, 109-2311-B-002-017- to S.-Y. Huang and 108-2311-B-002-011-MY3 to C.-C.-C.C.; National Taiwan University Hospital grant 109-21 and 109-N4554 to S.-Y. Huang; and National Taiwan University grant 109L104308 to C.-C. Chan.
Author contributions
S.-Y.H. and C.-C.C. conceived the study. C.-Y.W., J.-G.J., W.-S.L., and C.-W.L. carried out the fly experiments. J.-G.J. developed the in vivo exosome release assay. C.-Y.W. and H.-C.H. carried out the EM analysis. C.-Y.W., L.-A.C., and A.-S.C. designed and performed live imaging. C.-Y.W. designed and carried out the GUV experiments. P.-H.C., W.-S.L., and S.-Y.H. did the immunostaining of the mammalian cells. C.-Y.W., W.-S.L., and S.-Y.H. prepared the figures. S.-Y.H. prepared the original draft. S.-Y.H. and C.-C.C. reviewed and edited the manuscript with comments from all other authors. S.-Y.H. and C.-C.C. obtained the funding for the project. S.-Y.H. and C.-C.C. supervised the project.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103437.
Contributor Information
Chih-Chiang Chan, Email: chancc1@ntu.edu.tw.
Shu-Yi Huang, Email: shuyih@ntuh.gov.tw.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adell M.A., Vogel G.F., Pakdel M., Muller M., Lindner H., Hess M.W., Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J.Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L., Seow Y., Schapira A.H., Gardiner C., Sargent I.L., Wood M.J., Cooper J.M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011;42:360–367. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarroja N., Rodriguez-Cuenca S., Nygren H., Camargo A., Pirraco A., Relat J., Cuadrado I., Pellegrinelli V., Medina-Gomez G., Lopez-Pedrera C., et al. Increased dihydroceramide/ceramide ratio mediated by defective expression of degs1 impairs adipocyte differentiation and function. Diabetes. 2015;64:1180–1192. doi: 10.2337/db14-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp E., Tekpli X., Marteil G., Lagadic-Gossmann D., Legrand P., Rioux V. N-Myristoylation targets dihydroceramide Δ4-desaturase 1 to mitochondria: partial involvement in the apoptotic effect of myristic acid. Biochimie. 2009;91:1411–1419. doi: 10.1016/j.biochi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Bieberich E. Sphingolipids and lipid rafts: novel concepts and methods of analysis. Chem. Phys. Lipids. 2018;216:114–131. doi: 10.1016/j.chemphyslip.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M., Gradilla A.C., Seijo I., Andres G., Rodriguez-Navas C., Gonzalez-Mendez L., Guerrero I. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat.Cell Biol. 2013;15:1269–1281. doi: 10.1038/ncb2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski J.S., Jankowski H., Bond D.R., Mccague S.B., Munro B.R., Predebon M.J., Scarlett C.J., Skelding K.A., Weidenhofer J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018;17:211. doi: 10.1186/s12944-018-0854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer K.M., Kranich L.R., Ruf W.P., Cagsal-Getkin O., Winslow A.R., Zhu L., Vanderburg C.R., Mclean P.J. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin V., Straussberg R., Xu R., Mileva I., Yogev Y., Khoury R., Konen O., Barhum Y., Zvulunov A., Mao C., Birk O.S. DEGS1 variant causes neurological disorder. Eur. J. Hum.Genet. 2019;27:1668–1676. doi: 10.1038/s41431-019-0444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Sanchez D., Furlan M., Colombo M.I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Gross J.C., Chaudhary V., Bartscherer K., Boutros M. Active Wnt proteins are secreted on exosomes. Nat.Cell Biol. 2012;14:1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- Guo H., Chitiprolu M., Roncevic L., Javalet C., Hemming F.J., Trung M.T., Meng L., Latreille E., Tanese De Souza C., Mcculloch D., et al. Atg5 Disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev.Cell. 2017;43:716–730 e7. doi: 10.1016/j.devcel.2017.11.018. [DOI] [PubMed] [Google Scholar]
- Hebbar S., Lee E., Manna M., Steinert S., Kumar G.S., Wenk M., Wohland T., Kraut R. A fluorescent sphingolipid binding domain peptide probe interacts with sphingolipids and cholesterol-dependent raft domains. J. Lipid Res. 2008;49:1077–1089. doi: 10.1194/jlr.M700543-JLR200. [DOI] [PubMed] [Google Scholar]
- Holland W.L., Brozinick J.T., Wang L.P., Hawkins E.D., Sargent K.M., Liu Y., Narra K., Hoehn K.L., Knotts T.A., Siesky A., et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5:167–179. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Iguchi Y., Eid L., Parent M., Soucy G., Bareil C., Riku Y., Kawai K., Takagi S., Yoshida M., Katsuno M., et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain. 2016;139:3187–3201. doi: 10.1093/brain/aww237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W.H., Liu C.C., Yu Y.L., Chang Y.C., Lien W.Y., Chao H.C., Huang S.Y., Kuo C.H., Ho H.C., Chan C.C. Lipophagy prevents activity-dependent neurodegeneration due to dihydroceramide accumulation in vivo. EMBO Rep. 2017;18:1150–1165. doi: 10.15252/embr.201643480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto T., Okada T., Miya S., Zhang L., Nakamura S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013;4:2712. doi: 10.1038/ncomms3712. [DOI] [PubMed] [Google Scholar]
- Karsai G., Kraft F., Haag N., Korenke G.C., Hanisch B., Othman A., Suriyanarayanan S., Steiner R., Knopp C., Mull M., et al. DEGS1-associated aberrant sphingolipid metabolism impairs nervous system function in humans. J. Clin.Invest. 2019;129:1229–1239. doi: 10.1172/JCI124159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Kramer-Albers E.M., Bretz N., Tenzer S., Winterstein C., Mobius W., Berger H., Nave K.A., Schild H., Trotter J. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin. Appl. 2007;1:1446–1461. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- Kraveka J.M., Li L., Szulc Z.M., Bielawski J., Ogretmen B., Hannun Y.A., Obeid L.M., Bielawska A. Involvement of dihydroceramide desaturase in cell cycle progression in human neuroblastoma cells. J. Biol. Chem. 2007;282:16718–16728. doi: 10.1074/jbc.M700647200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios J., Mercier V., Roux A., Gruenberg J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J.Cell Biol. 2020;219 doi: 10.1083/jcb.201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Chiang Y.L., Liu J.T., Chen Y.X., Lee C.H., Chen Y.L., Hwang I.S. Emerging roles of air gases in lipid bilayers. Small. 2018;14 doi: 10.1002/smll.201802133. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Kim J.R., Hu Y., Khan R., Kim S.J., Bharadwaj K.G., Davidson M.M., Choi C.S., Shin K.O., Lee Y.M., et al. Cardiomyocyte specific deficiency of serine palmitoyltransferase subunit 2 reduces ceramide but leads to cardiac dysfunction. J. Biol. Chem. 2012;287:18429–18439. doi: 10.1074/jbc.M111.296947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal A.M., Huang H.H., Marsh T., Solvik T., Zhang D., Ye J., Kai F., Goldsmith J., Liu J.Y., Huang Y.H., et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat.Cell Biol. 2020;22:187–199. doi: 10.1038/s41556-019-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tang X., Taylor K.G., Dupre D.B., Yappert M.C. Conformational characterization of ceramides by nuclear magnetic resonance spectroscopy. Biophys. J. 2002;82:2067–2080. doi: 10.1016/S0006-3495(02)75554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lloyd T.E., Atkinson R., Wu M.N., Zhou Y., Pennetta G., Bellen H.J. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- Megha, London E. Ceramide selectively displaces cholesterol from ordered lipid domains (rafts): implications for lipid raft structure and function. J. Biol. Chem. 2004;279:9997–10004. doi: 10.1074/jbc.M309992200. [DOI] [PubMed] [Google Scholar]
- Minakaki G., Menges S., Kittel A., Emmanouilidou E., Schaeffner I., Barkovits K., Bergmann A., Rockenstein E., Adame A., Marxreiter F., et al. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy. 2018;14:98–119. doi: 10.1080/15548627.2017.1395992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Shimamoto T., Nagano H., Tsuruno M., Okuhara H., Hatanaka H., Tojo H., Kodama Y., Funato K. Producing human ceramide-NS by metabolic engineering using yeast Saccharomyces cerevisiae. Sci. Rep. 2015;17:16319. doi: 10.1038/srep16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A.M., Lasiecka Z.M., Xu Y., Neufeld J., Shahriar S., Simoes S., Chan R.B., Oliveira T.G., Small S.A., Di Paolo G. Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat. Commun. 2018;9:291. doi: 10.1038/s41467-017-02533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C.M., Halder G. Characterization of a dorsal-eye Gal4 line in Drosophila. Genesis. 2010;48:3–7. doi: 10.1002/dvg.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L., Malhotra R., Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat.Cell Biol. 2015;17:300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panakova D., Sprong H., Marois E., Thiele C., Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Pant D.C., Dorboz I., Schluter A., Fourcade S., Launay N., Joya J., Aguilera-Albesa S., Yoldi M.E., Casasnovas C., Willis M.J., et al. Loss of the sphingolipid desaturase DEGS1 causes hypomyelinating leukodystrophy. J. Clin.Invest. 2019;129:1240–1256. doi: 10.1172/JCI123959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielska M., Szulc Z.M., Kurowska E., Hogan E.L., Bielawski J., Bielawska A., Bhat N.R. Cytokine-induced release of ceramide-enriched exosomes as a mediator of cell death signaling in an oligodendroglioma cell line. J. Lipid Res. 2016;57:2028–2039. doi: 10.1194/jlr.M070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehler A.M., Xiang W., Spitzer P., May V.E., Meixner H., Rockenstein E., Chutna O., Outeiro T.F., Winkler J., Masliah E., Klucken J. Autophagy modulates SNCA/alpha-synuclein release, thereby generating a hostile microenvironment. Autophagy. 2014;10:2171–2192. doi: 10.4161/auto.36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J.Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers R.S., Schwarz T.L. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Saksena S., Judson B.L., Emr S.D. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P., Franke S., Zahringer U., Sperling P., Heinz E. Identification and characterization of a sphingolipid delta 4-desaturase family. J. Biol. Chem. 2002;277:25512–25518. doi: 10.1074/jbc.M202947200. [DOI] [PubMed] [Google Scholar]
- Thery C., Boussac M., Veron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brugger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Vaccari T., Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev.Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Vallabhaneni K.C., Penfornis P., Dhule S., Guillonneau F., Adams K.V., Mo Y.Y., Xu R., Liu Y., Watabe K., Vemuri M.C., Pochampally R. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget. 2015;6:4953–4967. doi: 10.18632/oncotarget.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Baixauli F., Mittelbrunn M., Fernandez-Delgado I., Torralba D., Moreno-Gonzalo O., Baldanta S., Enrich C., Guerra S., Sanchez-Madrid F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016;7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Hurley J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.