Abstract
Phosphatase and tensin homologue (PTEN) regulates cell growth and survival through inhibition of the mammalian target of rapamycin (MTOR) signalling pathway. Germline genetic variation of PTEN is associated with autism, macrocephaly and PTEN hamartoma tumour syndromes. The effect of developmental PTEN somatic mutations on nervous system phenotypes is not well understood, although brain somatic mosaicism of MTOR pathway genes is an emerging cause of cortical dysplasia and epilepsy in the paediatric population.
Here we report two somatic variants of PTEN affecting a single patient presenting with intractable epilepsy and hemimegalencephaly that varied in clinical severity throughout the left cerebral hemisphere. High-throughput sequencing analysis of affected brain tissue identified two somatic variants in PTEN. The first variant was present in multiple cell lineages throughout the entire hemisphere and associated with mild cerebral overgrowth. The second variant was restricted to posterior brain regions and affected the opposite PTEN allele, resulting in a segmental region of more severe malformation, and the only neurons in which it was found by single-nuclei RNA-sequencing had a unique disease-related expression profile.
This study reveals brain mosaicism of PTEN as a disease mechanism of hemimegalencephaly and furthermore demonstrates the varying effects of single- or bi-allelic disruption of PTEN on cortical phenotypes.
Keywords: somatic mosaicism, brain development, hemimegalencephaly (HME), epilepsy
Koboldt et al. reveal brain mosaicism of PTEN as a disease mechanism of hemimegalencephaly, and demonstrate the varying effects of single- or bi-allelic disruption of PTEN on cortical phenotypes.
Introduction
Phosphatase and tensin homologue (PTEN) regulates cell growth and survival as an inhibitory component of the PI3K-AKT3-MTOR signalling pathway.1,2 Genetic variation of PTEN has been associated with a spectrum of disease manifestations depending on genotype and distribution throughout the body. For example, germline heterozygous loss-of-function mutations lead to PTEN hamartoma tumour syndromes, macrocephaly and autism.3 Clinically similar phenotypes to the germline disease are evident in patients harbouring early post-zygotic somatic mosaic mutations disseminated throughout the body.4 ‘Second-hit’ somatic mutations in PTEN result in SOLAMEN syndrome, in which segmental overgrowth affects certain regions of the body, with localized nullizygous disruption of PTEN leading to regions of more severe disease.4,5
PTEN is an important regulator of neurodevelopment and has been implicated through mouse models in cell proliferation, differentiation and migration from early stages of brain development.3 Various congenital structural brain malformations have been observed in patients with pathogenic PTEN germline variants. PTEN somatic mutations are implicated in brain tumour progression; however, the effect of developmental PTEN brain somatic mutations on clinical phenotypes is not well understood. Focal cortical malformations, including hemimegalencephaly, have been linked to brain mosaic mutations in MTOR pathway genes, but these have not yet been explored in the context of PTEN mosaicism.6
Instances of mosaicism provide a unique opportunity to learn about the developmental implications of PTEN disruption in the context of its cell lineage and gene-dosage effects. In this paper, we report a patient with hemimegalencephaly in which the degree of overgrowth and cortical malformation clearly differed throughout the anterior to posterior extent of the left cerebral hemisphere, indicative of variable somatic mosaicism. Two mosaic PTEN mutations with distinct distributions were identified in affected brain tissue, which correlated with imaging and neuropathological findings. We integrated single-cell 3′-RNA sequencing with targeted long-read sequencing to identify affected cell lineages. This analysis revealed the developmental history of each somatic mutation and its contribution to cell types in the brain. Together with additional genomic techniques, we show that PTEN somatic mutations contribute both individually and jointly to aspects of brain overgrowth. Our results implicate brain somatic mutations of PTEN as a mechanism of hemimegalencephaly.
Materials and methods
Human tissues and nucleic acid extraction
We obtained written informed consent to enrol a 1-year-old male patient onto our IRB-approved research study (IRB18-00786, PI: Daniel C. Koboldt) in the Institute for Genomic Medicine at Nationwide Children’s Hospital. Brain tissue was immediately flash-frozen after resection and 30 mg was portioned from each anatomical site for nucleic acid extraction.
Exome and transcriptome sequencing from bulk tissue
DNA isolated from blood and brain tissue was prepared for enhanced exome sequencing using NEBNext® UltraTM II FS DNA Library Prep Kit (New England BioLabs). IDT xGen Exome Research Panel v1.0 enhanced with the xGenCNV Backbone Panel-Tech Access (Integrated DNA Technologies) was used for target enrichment by hybrid capture. Final libraries were sequenced on an Illumina HiSeq 4000 to generate paired-end 151-bp reads. A custom pipeline was used for alignment to human reference genome build GRCh37 and secondary analysis.7
Germline variants were called by GATK v.4.0.5.1 HaplotypeCaller and somatic variants were called by MuTect-2.8,9 Germline variants were filtered based on the following features: GATK quality score (≥30), population frequency (≤1%), depth of sequencing (≥8 reads), variant allele fraction (≥20%), protein-coding region or distance from canonical splice site ≤ 3 bp, damaging prediction scores (CADD ≥ 15; SIFT ≤ 0.05; GERP++ ≥ 2), and relevance of gene–disease association with patient phenotype using OMIM and ClinVar databases. No germline variants were deemed relevant to the patient’s phenotype.
Somatic variants were filtered based on the following features: MuTect-2 = ‘PASS’, GATK quality score (≥30), protein-coding region or distance from canonical splice site ≤ 3 bp, depth of sequencing (≥8 total reads), absence from blood comparator, alternate allele reads (≥5), and variant allele fraction (≥1%). Passing variants were reviewed in Integrated Genomics Viewer to assess for strand bias, read-end bias, or other biases that may suggest sequencing artefacts. Only two somatic variants (PTEN exon 5 and exon 9 variants) met all of these criteria.
RNA isolated from brain tissue was treated with DNase and ribodepletion prior to constructing libraries using Illumina’s TruSeq Stranded Total RNA Kit. Paired-end 151-bp reads were generated on an Illumina HiSeq 4000 and then low-quality reads (q < 10) and adaptor sequences were eliminated from raw reads using bbduk version 37.64. Reads were aligned to the GRCh38.p9 assembly of the human reference genome from NCBI using version 2.6.0c of the aligner STAR.10 Features were counted using featureCounts software against a GFF file from Gencode (v28).11 Comparisons of gene expression among anatomical sites were carried out using DESeq2.12 Gene set enrichment analysis was performed using the fgsea package in R.
ExPASy analysis
PTEN mRNA sequences (reference, exon 5 mutant, and exon 9 mutant) were entered into the ExPASy translate tool using the forward and reverse strands setting to generate translated amino acid sequences for frame 3 and to identify stop codons.13 The reference sequence used for PTEN was uc001kfb.3 (length = 5572) from the UCSC Genome Browser.
Targeted DNA sequencing
The NEBNext® UltraTM II FS Library Prep Kit for Illumina (New England BioLabs) was used to make pre-capture libraries on the Biomek i7 liquid handling automated workstation (Beckman Coulter). The DNA input for each library was 100 ng. DNA was fragmented, end-repaired, and then ligation was performed with xGen Dual Index UMI Adapters at 3.75 µM (Integrated DNA Technologies). The ligation products were purified, without size-selection, with a 0.8× ratio of AMPure XP (Beckman Coulter) and eluted in 20 µl of water. Seven cycles of PCR were performed with 7 µl of ligation product in duplicate reactions for each sample with NEBNext® UltraTM II Q5 Master Mix and universal primers specific for the Illumina adapter sequences, P5 and P7, with a final reaction concentration of 0.2 µM (IDT). The duplicate PCR reactions were combined after cycling, purified with AMPure XP at a 0.9× ratio, and eluted in 22 µl water.
A custom panel targeting 33 genes was designed for the Twist Fast Hybridization kit (Twist Biosciences). The custom set was composed of 2686 probes, creating a 300 kb total panel size. Both expected variants in the PTEN gene were targeted in this panel. Hybridization capture was performed according to the manufacturer protocol, with the following specifications. Two hundred nanograms of each of the pre-capture libraries were combined into two total pools with a final concentration of 1600 ng each. Hybridization was performed in 2 h, followed by 13 cycles of PCR with the NEBNext® UltraTM II Q5 Master Mix. The PCR products were purified with AMPure XP at a 0.9× ratio and eluted in 32 µl water. Sequencing was performed across one lane of a NovaSeq 6000 SP flow cell, resulting in an average of ∼26 million reads passing filter per sample.
Analysis of read counts
Read counts and variant allele frequencies for the PTEN variants were computed from the BAM files by VarScan v2.4.4 (parameters --min-coverage 2 --min-var-freq 0.01 --P-value 0.99) using the mpileup output from SAMtools v1.6 (parameters: -A -B -d 250000 -q 1).14,15
Iso-Seq long-read sequencing from bulk tissue
Total RNA was treated with DNase (Zymo RNA Clean and Concentrator-5) and reverse transcribed using NEBNext Single Cell/Low Input kit for cDNA synthesis following the steps outlined within Iso-Seq Express Template Procedure & Checklist (PN 101–763-800 Version 02, October 2019). The cDNA product was purified and size selected to >1.0 kb using 1.0× ProNex® beads (Promega). Amplification was performed using Takara PrimeSTAR® GXL (2.50 U total) with NEBNext® Single Cell (Cat. #E6421S) and ISO-Seq Express (PN #101-737-500) cDNA primers. PCR products were size selected to >2 kb and 0.5–2 kb using SPRIselectTM paramagnetic beads (Beckman Coulter). Fractionated cDNA was pooled for a final yield of 500 ng at a 1:1 molar ratio of >2 kb and 0.5–2 kb sized cDNA for capture.
Targeted gene enrichment was carried out through cDNA capture using a 31-gene custom panel probe set including the PTEN loci (Supplementary Table 1) following IDT xGen hybridization version 4 kit recommendations (Integrated DNA Technologies). For efficient annealing of custom probes with cDNA template, the hybridization reaction incubated for 16 h. Post-capture PCR was performed using PrimeSTAR® GXL as described earlier. PCR products were purified and size selected >0.5 kb using SPRIselectTM paramagnetic beads with final elution in 12 µl Buffer EB (Qiagen) for SMRTbell library preparation.
A SMRTbell library was prepared using SMRTbell Express Template Prep Kit 2.0 according to PacBio Iso-Seq protocol (PN 101–763-800 Version 02, October 2019). Volumetric calculations were done using SMRT Link Sample Setup for v4 primer and polymerase binding conditions dependent upon initial SMRTbell library molarity. The SMRTbell library was complexed with Sequel I Binding Kit v3.0, purified with 1.0× ProNex® beads and sequenced at 8 pM on-plate loading concentration. The sample was sequenced on a single 1 M SMRT Cell on the PacBio Sequel I platform using v3.0 chemistry and 4-h pre-extension followed by 20-h movie collection.
Iso-Seq long-read sequencing from single cells
Isolated single-cell cDNA from 10x Genomics Chromium Next GEM Single Cell 3′ Reagent Kit v3.1 was subjected to five additional cycles of PCR amplification using NEBNext® UltraTM II Q5 Master Mix (Cat. #M0544L) with recommended NEB NGS PCR thermocycling conditions. Primers used for amplification were 10xG_TSOprimer: 5′-AAGCAGTGGTATCAACGCAGAG-3′ and 10xG_Primer1: 5′-CTACACGACGCTCTTCCGATCT-3′. The resulting PCR product was purified, and size selected using 0.9× SPRIselectTM paramagnetic beads. PTEN loci were enriched by capture following IDT xGen hybridization version 4 kit protocol with slight modifications. A custom 31-gene probe panel was used for hybridization with 500 ng of starting cDNA for a duration of 16 h. Post-capture PCR was performed using NEBNext® UltraTM II Q5 Master Mix with 15 cycles of amplification using 10xG_TSOprimer and 10xG_Primer1. PCR products were purified and size selected >0.5 kb using 0.5× SPRIselectTM paramagnetic beads with final elution being in 12 µl Buffer EB for SMRTbell library preparation.
The SMRTbell library was prepared from 360 ng of capture cDNA using SMRTbell Express Template Prep Kit 2.0 according to PacBio Iso-Seq protocol (PN 101-763-800 Version 02, October 2019). The library was complexed using Sequel II binding kit v2.1 with v4 sequencing primer using volumetric calculations determined by SMRT Link Sample Setup. Final library was loaded at 50 pM on plate concentration and was sequenced on a single 8 M Sequel II SMRT Cell with a 2-h pre-extension followed by a 24-h movie time.
Sequences at each variant position, cell barcodes, and unique molecular index (UMI) sequences were extracted from each read using a custom Python script. Observed cell barcodes were cross-referenced for exact match against a list of all possible 10x Genomics cell barcodes and reads containing invalid barcodes were removed. Duplicate reads containing the exact same UMI sequence were also discarded. Reads were annotated for genotype based on sequence observed at the variant position.
Fluorescence-activated cell sorting
Nuclei were isolated from frozen brain tissue as previously described.16 Briefly, tissue was dissociated by mechanical disruption using a dounce homogenizer. Nuclei were washed and stained with Hoechst 33342 dye, filtered through a 30 µm mesh filter, and resuspended in PBS. Fluorescence-activated cell sorting (FACS) analysis was performed using a BD Influx cell sorter (BD Biosciences). Samples were gated on Hoechst-positive nuclei, debris was excluded using forward and side scatter pulse area parameters (FSC-A and SSC-A), and then aggregates were excluded using pulse width (FSC-W and SSC-W). The purified population of Hoechst-positive nuclei was sorted directly into 10x Genomics reaction buffer.
Single-cell RNA-sequencing
Sorted nuclei from anatomical sites A and C (temporal cortex) were loaded into a 10x Genomics Chromium device for microfluidic-based capture of single nuclei. Reverse transcription, cDNA amplification and library preparation were performed according to the manufacturer protocol for v.3.1 3′-single-cell RNA-sequencing (RNA-seq). Subsequently, libraries were sequenced on an Illumina NovaSeq 6000 instrument to generate paired-end sequencing data. Fastq files were generated using Illumina bcl2fastq software. Data preprocessing, including alignment, filtering, barcode counting and UMI counting, were performed using the 10x Genomics CellRanger software suite following the default parameters. Downstream analysis of gene expression was performed using Seurat v.3 for R.17,18 Briefly, the feature-barcode matrices were log-normalized and top variable genes across single cells were identified using the FindVariableGenes function. Data were scaled to 10000 transcripts per cell. Dimensionality reduction was performed using principal component analysis and then the distance matrix was organized into a K-nearest neighbour graph, partitioned into clusters using Louvain algorithm, and clusters were visualized on a UMAP plot. Top differentially expressed genes for each cluster were computed and cell types were annotated based on expression of known markers. Genotype status from PacBio single-cell sequencing was added to the Seurat object’s metadata slot by joining based on cell barcode. Plots overlaying genotype with cell type were generated using the FeaturePlot function. Differential expression analysis was performed for mutant versus wild-type cells using the FindMarkers function. Gene ontology term enrichment was determined using DAVID Bioinformatics Resources 6.8.
Phospho-S6 immunohistochemistry and image analysis
Fresh frozen sections from brain regions A and C were cryostat sectioned to a thickness of 30 μm. Sections were fixed in 4% paraformaldehyde and stained with neuronal nuclear protein (NeuN; mouse clone MAB377, IgG1; Chemicon; Cat. #MAB377; 1:500) and phospho-S6 protein (Ser240/244; monoclonal rabbit, Cell Signaling Technology; product #5364; 1:1000). Sections were washed and incubated with Donkey anti-Rabbit IgG Alexa Fluor 488 (Thermo Fisher Scientific, Cat. #A-21206; 1:1000) and Donkey anti-Mouse IgG Alexa Fluor 546 (Thermo Fisher Scientific, Cat. #A10036; 1:1000) secondary antibodies.
Images for analysis were acquired from each tissue section (n = 9 per region) using a Zeiss LSM 800 confocal microscope at ×20 magnification with a crop of 0.5× and 2 × 2 stitched tiling. All images were acquired in a single batch with the same calibration settings. One stitched image per section was quantified using ImageJ/FIJI software. Regions of NeuN/phospho-S6 co-staining were manually outlined to acquire mean fluorescence intensity measurements of phospho-S6. Background fluorescence was controlled by subtracting the fluorescence of a region adjacent to each respective measured cell. Outlines were used to quantify neuronal area.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request. The Python script used to analyse single-nuclei long-read sequencing data is available at https://github.com/bedrosian-lab/scGenotyping.
Results
Two somatic PTEN mutations identified in a patient with hemimegalencephaly
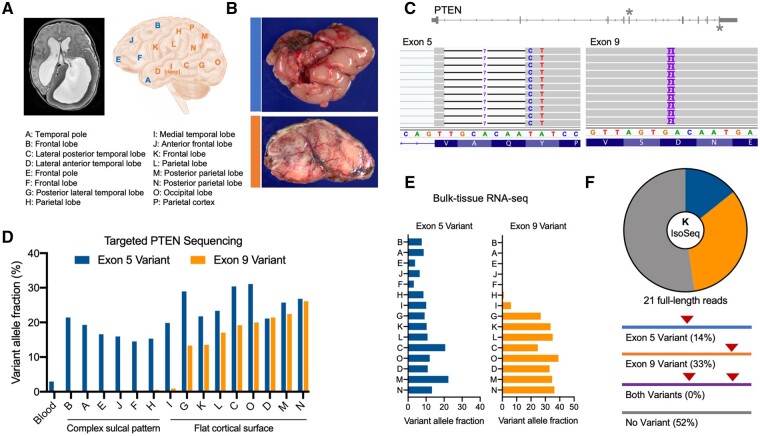
A 1-year-old male undergoing hemispherectomy for left-sided hemimegalencephaly and intractable epilepsy was consented to our research protocol (Supplementary material and Supplementary Fig. 1). Fifteen different anatomical sites were collected for study (Fig. 1A). Notably, there was an anterior-posterior gradient of severity. The anterior cerebrum showed a complex sulcal pattern with small gyri and occasional gyral fusion across sulci. In these areas, the cortical neurons were haphazardly situated and the hexalaminar architecture was ill-defined. By contrast, the posterior cerebrum had a flat, simplified cortical surface (Fig. 1B). At these sites, the cortical ribbon was thicker than expected and neurons were dysmorphic with large nuclei and cell bodies that had irregular processes. In these areas, the expected neuronal heterogeneity of the cortex was reduced and the dysmorphic and non-dysmorphic neurons appeared haphazardly arranged (Supplementary Fig. 1C).
Figure 1.
Two somatic PTEN mutations identified in a patient with hemimegalencephaly patient. (A) MRI and sites of resected brain specimens obtained from a patient undergoing hemispherectomy for intractable epilepsy. See also Supplementary Fig. 1. (B) Gross pathology and SMI-32 immunohistochemistry show sites A and C exhibit either a complex sulcal pattern with abnormal cortical architecture (blue) or flattening and simplification of the cortical surface with enlarged neurons (orange). (C) Exome sequencing of site K reveals two distinct somatic mutations in exons 5 [chr10:89692771 (hg19); chr10:87933013 (hg38)] and 9 [chr10:89725125 (hg19); chr10:87965368 (hg38)] of PTEN. (D) Targeted DNA sequencing of all anatomical sites and blood showing the distribution and variant allele fractions of the two somatic mutations, which correlates with the observed neuropathology. (E) Variant allele fractions of each somatic variant in bulk RNA-seq data. See also Supplementary Fig. 2. (F) Long-read sequencing of PTEN transcripts from site K reveals that the two variants are on opposite alleles.
Exome-sequencing of a resected brain specimen (site K) revealed two distinct variants in PTEN that were not detected at appreciable frequency in the patient’s blood, suggesting that they are somatic mutations rather than germline variation. The first mutation was a complex indel (c.255_262delTGCACAATinsC) at the beginning of exon 5, predicted to cause frameshift and introduce an early stop codon. The second mutation was a 2-base duplication in exon 9 (c.1110_1111dupTG), predicted to cause frameshift and a late stop codon (Fig. 1C and Supplementary Fig. 2A). Mutations that disrupt PTEN can lead to downstream activation of MTOR, which is a known mechanism of developmental brain disorders, including hemimegalencephaly.19 Thus, we performed deep targeted sequencing of PTEN (∼2000× average read depth) in all available brain specimens and blood (Supplementary Fig. 2B). The exon 5 indel was detected in all available brain sites and was detected in blood at 2.95% frequency (55 of 1866 reads), suggesting this variant is also mosaic outside the brain. Indeed, clinical Sanger sequencing performed on buccal cells detected the exon 5 indel at mosaic variant allele fraction (Supplementary Fig. 2C). The exon 9 duplication was detected only in posterior cerebrum, which coincided with the presence of more severe neuropathological findings (Fig. 1D). These results suggest the two variants may collectively contribute to the patient’s overall brain pathology.
To determine the expression level of the two variants in each anatomical site, we performed bulk RNA-seq and quantified the variant allele fractions. Interestingly, we detected the exon 9 duplication at higher frequency compared to the exon 5 indel in RNA-seq data (Fig. 1E). This may be explained by the fact that the exon 5 mutation introduces a premature stop codon, leading to nonsense-mediated decay of transcripts containing the mutation. As expected based on the targeted DNA sequencing results, the exon 9 variant was restricted in expression to posterior cerebrum. The expressed exon 9 variant allele fractions were generally higher than the DNA fractions, which may suggest greater stability of the mutant transcript, but further data would be required to confirm this. To determine whether the two mutations exist on the same allele or opposite alleles, we performed targeted long-read sequencing of PTEN transcripts on the PacBio Sequel II platform (Iso-Seq). Twenty-one reads spanned both variant positions, but no reads contained both mutations, suggesting they are located on opposite alleles (Fig. 1F and Supplementary Fig. 2E).
Tracing the developmental lineage of somatic mutations
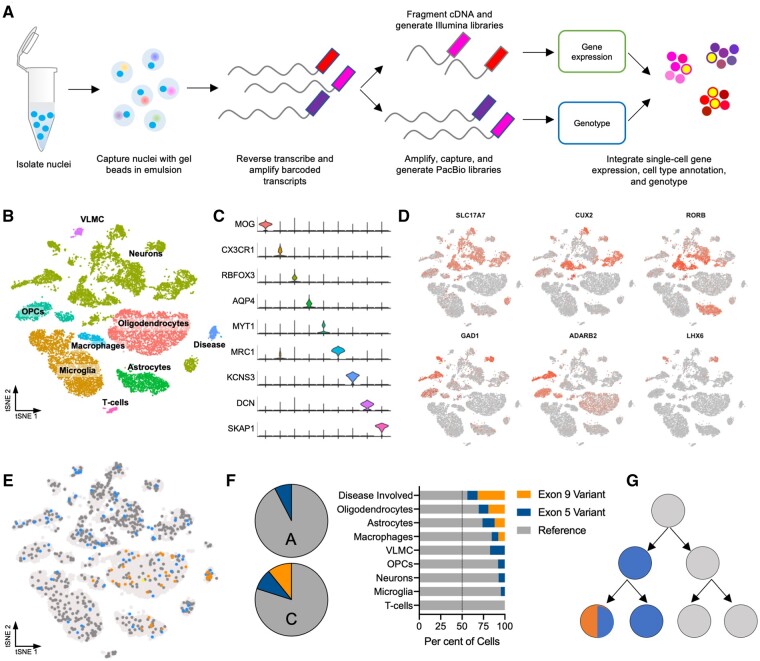
Determining the cell lineages affected by somatic variants is crucial to understand how they contribute to neural circuitry and brain function. We developed an approach to integrate high-throughput single-cell gene expression analysis on the 10x Genomics 3′-RNA-seq platform with targeted PacBio long-read sequencing of barcoded transcripts to determine single-cell genotype at positions across the length of the gene (Fig. 2A). Briefly, we isolated nuclei from archived frozen brain tissue (sites A and C) using FACS and captured ∼18 000 single nuclei following the 10x Genomics protocol (Supplementary Fig. 3A). A portion of the resulting nucleic acids were prepared for 3′-RNA-seq gene expression libraries and the remainder were prepared for targeted long-read sequencing of PTEN transcripts.
Figure 2.
Tracing the developmental lineage of somatic mutations. (A) Experimental schematic for the integration of single-cell 3′-RNA-seq with single-cell targeted long-read sequencing. (B) Cell types identified in brain anatomical sites A and C from single-cell gene expression analysis based on (C) marker gene expression. (D) Markers of cortical neuronal subtypes identified. (E) Overlay of genotype derived from long-read sequencing onto 3′-RNA-seq gene expression data. (F) Variant allele fractions by cell type demonstrate that variants are enriched in disease-involved neurons. (G) Proposed developmental history of two PTEN somatic mutations. OPC = oligodendrocyte precursor cells; VLMC = vascular and leptomeningeal cells.
Approximately 18 000 individual nuclei clustered into nine groups identified based on marker gene expression to represent brain resident cell types (e.g. neurons, astrocytes) as well as immune cells derived from the periphery (e.g. T cells; Fig. 2B, C and Supplementary Fig. 3B and C). A variety of cortical neuronal subtypes were evident among the nuclei (Fig. 2D). One cluster expressed neuronal cell adhesion genes (e.g. CNTN4, NRXN3), signatures of signal transduction (e.g. KCNS3, SCN7A), as well as enrichment of the PI3K-AKT signalling pathway, which we called ‘disease-involved’ neurons (Fig. 2B, C and Supplementary Fig. 3D). Induction of SCN7A in neurons and astrocytes has been associated with seizure activity.20 We overlaid genotype information from the targeted long-read dataset by extracting the mutation status from each long-read at each position and matching the cell barcodes to those in the gene expression dataset. Genotyping 949 cells across all nine cell types revealed that the exon 5 variant was expressed by at least one supporting read in 9.0% of nuclei in section C and 7.6% of nuclei in section A. The exon 9 variant was expressed in 10.8% of nuclei in section C and no nuclei in section A, which is consistent with the results derived from bulk sequencing analysis (Fig. 2E and Supplementary Fig. 3E). The proportion of variant expressing cells may be under-represented by single-nuclei sequencing due to the low level of PacBio coverage (i.e. one read per cell, on average) or the possibility that disease-involved cells may be more difficult to dissociate, capture and sequence due to altered morphology or other properties. Notably, none of the reads expressed both variants, supporting our bulk-tissue PacBio Iso-Seq results showing that the variants exist on opposite alleles, but one oligodendrocyte nucleus from section C separately expressed reads containing the exon 5 variant and reads containing the exon 9 variant, suggesting some cells harbour both variants in trans. There are likely more cells that contain both variants, but sequencing depth (i.e. one long-read per cell, in most cases), as well as the effect of the exon 5 variant causing nonsense mediated decay, limit their detection.
In addition, we analysed the presence of each variant by cell type (Fig. 2F). The exon 5 variant was observed in all cell lineages, including microglia, which originate from the mesoderm during development, in contrast to other CNS cell types, which are ectodermal. This agrees with our finding that the exon 5 variant was indeed present in the patient’s blood at low frequency, demonstrating he is mosaic for this variant outside the CNS. Surprisingly, one cell containing the exon 9 variant was observed in a mesodermal cell type (i.e. macrophages) as well as ectodermal CNS cell types, raising the possibility that this variant is also mosaic at a very low level in multiple germ layers, although it may otherwise be a technical artefact of droplet-based RNA seq. Interestingly, the cluster identified as disease-involved neurons had the highest fraction of mutant cells compared to other cell types (Fig. 2F). Taken together, this analysis suggests a developmental history for the patient’s somatic mutations where they arose sequentially on opposite alleles during germ layer specification in the early embryo (i.e. pre-gastrulation) (Fig. 2G).
Somatic mutations contribute to brain disease
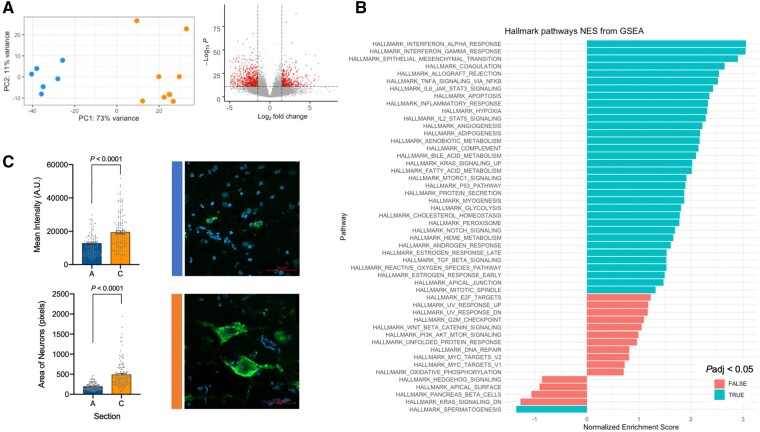
To determine the effects of variant status on gene expression, we compared the transcriptomes of all 15 anatomical sites using bulk RNA-seq data. In a principal component analysis, sites containing both the exon 5 and exon 9 somatic variants clustered separately from sites containing only the exon 5 variant (Fig. 3A). Differentially expressed genes represented pathways including inflammation and MTORC1 signalling, which were enriched in the sites containing both somatic variants (Fig. 3B). To directly test the effect of these somatic variants on MTORC1 signalling, we performed immunohistochemical analysis of phospho-S6, a downstream target of MTOR, in sites containing either the exon 5 variant alone (site A) or both exon 5 and exon 9 variants (site C). Phospho-S6 staining within neurons was significantly enriched in site C, which corresponded to increased neuronal area (Fig. 3E and F).
Figure 3.
Somatic mutations contribute to brain disease. (A) Principal component analysis of bulk RNA-seq data from all 15 brain sites reveals distinct clustering related to genotype (blue: anterior cerebrum/exon 5 variant detected; orange: posterior cerebrum/both variants detected). Volcano plot depicting differentially expressed genes in posterior versus anterior brain samples. (B) Gene set enrichment analysis (GSEA) showing enriched hallmark pathways including MTORC1 signalling. (C) Phospho-S6 immunofluorescence intensity is higher in posterior section C compared to anterior section A, which corresponds to larger neuronal area. Mean ± standard error of the mean. NES = normalized enrichment score.
Discussion
Instances of somatic mosaicism provide a unique opportunity to learn about the developmental implications of PTEN disruption in the context of its cell-lineage and gene-dosage effects. One previous example of a brain-specific somatic mosaic variant in PTEN was reported in a patient with type 2 focal cortical dysplasia.21 Our results implicate brain somatic mutations of PTEN as a disease mechanism of hemimegalencephaly and demonstrate a spectrum of cerebral overgrowth associated with the disruption of one or both alleles. Identifying the affected cell lineages suggests a developmental history for the mutations, arising sequentially in early embryogenesis.
Our results further raise the importance of considering ‘second-hit’ somatic mutations as a moderator of phenotype in cases of germline PTEN disease. Pathogenic germline PTEN variants have been associated with a wide spectrum of disease phenotypes. In particular, variable neurological findings including focal brain malformations have been difficult to explain in the context of germline disease.22-24 One family reported to share the same germline pathogenic PTEN variant exhibited a variable phenotype with three members having macrocephaly and a fourth having hemimegalencephaly.25 This raises the possibility of a ‘second-hit’ somatic mutation affecting only the fourth family member. Similarly, Lhermitte-Duclos disease is a rare hamartomatous overgrowth disorder affecting the cerebellum and associated with germline PTEN mutations. A case series showed that ‘second-hit’ somatic mutations explain the localized cerebellar pathology at least in some cases.26 In our case, double mosaicism of PTEN appears to cause greater MTOR pathway activation, leading to a segmental region of more severe pathology. Direct study of affected brain tissue will be necessary to fully understand the contribution of somatic mutations in moderating PTEN disease phenotypes.
In the future, studying somatic mosaicism in affected tissue at the single-cell level may have advantages in clinical care. In this case, single-cell analyses demonstrated that the patient’s somatic variants were present in multiple germ layers, raising the possibility of increased risk of certain cancers typically associated with PTEN hamartoma tumour syndromes. Single-cell analyses may increase the sensitivity of detecting PTEN mosaicism in the future, allowing for increased surveillance of these patients. Also worth noting is the fact that the patient’s exon 5 variant was observed in buccal cells of ectodermal origin. Buccal cells may be considered as a DNA source for clinical genetic testing of patients with brain disorders in which somatic mosaicism is well established.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Dave Dunaway from the NCH Flow Cytometry Core Facility.
Funding
Funding for this project was provided by the Nationwide Foundation Innovation Fund. T.A.B. is supported by NHGRI (K01 HG011062).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. [DOI] [PubMed] [Google Scholar]
- 2. Wu X, Senechal K, Neshat MS, Whang YE, Sawyers CL.. The PTEN/MMAC1 tumor suppressor phosphatase functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1998;95(26):15587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skelton PD, Stan RV, Luikart BW.. The role of PTEN in neurodevelopment. Mol Neuropsychiatry. 2020;5(Suppl 1):60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nathan N, Keppler-Noreuil KM, Biesecker LG, Moss J, Darling TN.. Mosaic disorders of the PI3K/PTEN/AKT/TSC/mTORC1 signaling pathway. Dermatol Clin. 2017;35(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caux F, Plauchu H, Chibon F, et al. Segmental overgrowth, lipomatosis, arteriovenous malformation and epidermal nevus (SOLAMEN) syndrome is related to mosaic PTEN nullizygosity. Eur J Hum Genet. 2007;15(7):767–73. [DOI] [PubMed] [Google Scholar]
- 6. Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138(Pt 6):1613–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelly BJ, Fitch JR, Hu Y, et al. Churchill: An ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol. 2015;16:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao Y, Smyth GK, Shi W.. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 12. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(Web Server issue):W597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16) :2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lacar B, Linker SB, Jaeger BN, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44(8):941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorter JA, Zurolo E, Iyer A, et al. Induction of sodium channel Na(x) (SCN7A) expression in rat and human hippocampus in temporal lobe epilepsy. Epilepsia. 2010;51(9):1791–800. [DOI] [PubMed] [Google Scholar]
- 21. Schick V, Majores M, Engels G, et al. Activation of Akt independent of PTEN and CTMP tumor-suppressor gene mutations in epilepsy-associated Taylor-type focal cortical dysplasias. Acta Neuropathol. 2006;112(6):715–25. [DOI] [PubMed] [Google Scholar]
- 22. O'Rourke DJ, Twomey E, Lynch SA, King MD.. Cortical dysplasia associated with the PTEN mutation in Bannayan Riley Ruvalcaba syndrome: A rare finding. Clin Dysmorphol. 2012;21(2):91–2. [DOI] [PubMed] [Google Scholar]
- 23. D'Gama AM, Geng Y, Couto JA, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol. 2015;77(4):720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghusayni R, Sachdev M, Gallentine W, Mikati MA, McDonald MT.. Hemimegalencephaly with Bannayan-Riley-Ruvalcaba syndrome. Epileptic Disord. 2018;20(1):30–4. [DOI] [PubMed] [Google Scholar]
- 25. Merks JH, de Vries LS, Zhou XP, et al. PTEN hamartoma tumour syndrome: Variability of an entity. J Med Genet. 2003;40(10):e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou XP, Marsh DJ, Morrison CD, et al. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. Am J Hum Genet. 2003;73(5):1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request. The Python script used to analyse single-nuclei long-read sequencing data is available at https://github.com/bedrosian-lab/scGenotyping.