With great interest we read the article by Diaz and colleagues1 providing further evidence of a neurodevelopmental disorder caused by bi-allelic variants disrupting the function of YIF1B, by reporting a second patient cohort and a mouse model. We had earlier reported six individuals from five unrelated families, harbouring bi-allelic protein truncating mutations in YIF1B, presenting with a progressive encephalopathy with various degrees of movement disorders, microcephaly and epilepsy.2
We here describe eight additional individuals from eight independent families harbouring protein altering YIF1B variants, including four individuals with homozygous or compound heterozygous missense variants (Fig. 1A–D and Supplementary Figs 1–6). We provide functional evidence that these missense variants impact on YIF1B function, and compare the clinical phenotype between these new and all previously reported cases to further delineate the mutational landscape and clinical phenotype associated with this new disease entity, which Online Mendelian Inheritance in Man (OMIM) recently named ‘Kaya-Barakat-Masson syndrome’ (KABAMAS, OMIM #619125).
Figure 1.
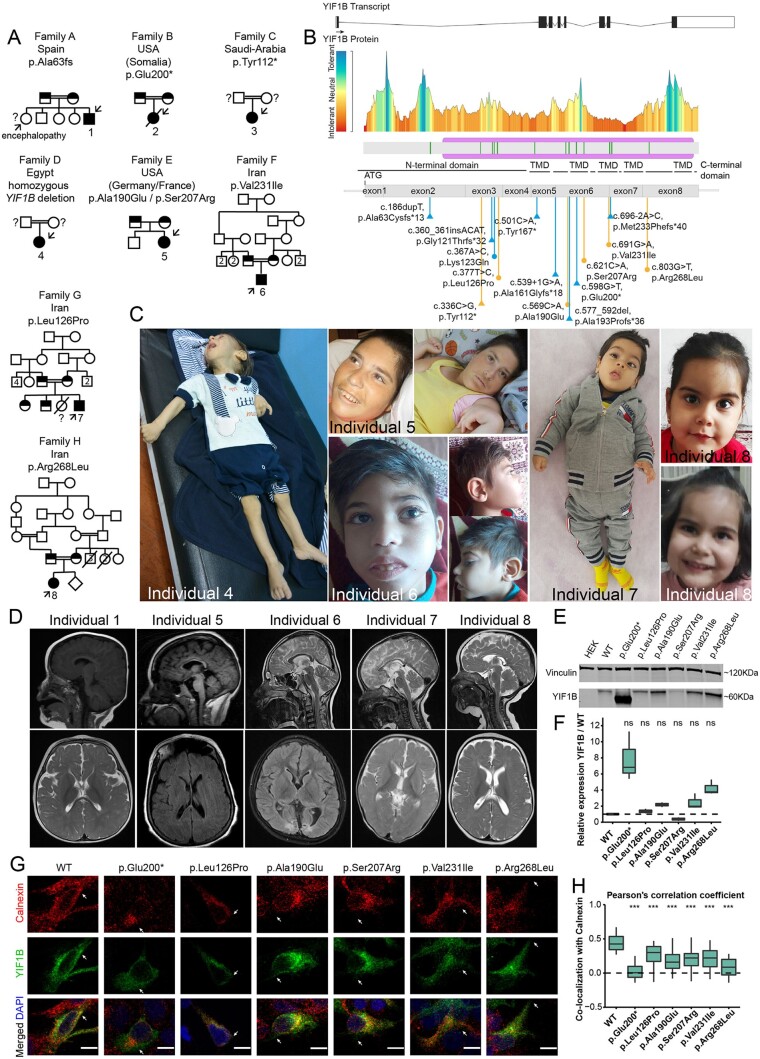
Pedigrees of families, clinical hallmarks and functional investigations of identified YIF1B variants. (A) Family pedigrees of ascertained families. Affected individuals with homozygous YIF1B variants and healthy parents with confirmed heterozygous YIF1B variants are indicated in filled and half filed symbols, respectively. Presumed carrier parents that were not available for segregation analysis are indicated with open circles or squares and a question mark. Affected individuals with confirmed genotype are indicated with an arrow and numbered. Not-tested affected siblings presenting with similar phenotypes are indicated with a question mark. Consanguineous parents are indicated with a double connection line. Males are squares, females are circles; deceased individuals are marked with a diagonal line. (B) Drawing of the YIF1B transcript and YIF1B protein, including the tolerance landscape as determined by MetaDome analysis, which displays regional tolerance or intolerance to missense or synonymous variation. All currently known YIF1B variants from Almuhaizea et al.2 and Diaz et al.1 (blue) and those reported herein (orange) encountered in individuals with Kaya-Barakat-Masson syndrome are indicated. Missense and truncating variants are indicated with circles and triangles, respectively. (C) Images of Individuals 4–7 and 8 at age of 1 year, 27 years, 7.5 years, 11 months and 4.5 years, respectively. No gross dysmorphic features were observed. Note the neurological posture in Individuals 4, 5 and 7. (D) T1 and T2-weighted brain MRI images of Individuals 1 and 5–8 in sagittal and axial planes. Note the various degrees of cerebral atrophy, cerebellar hypoplasia, thin corpus callosum and white matter abnormalities. (E) Immunoblotting detecting proteins of wild-type and YIF1B variants upon transient transfection for 48 h in HEK293 cells. Endogenous vinculin served as a normalization control. Full length blots are provided in Supplementary Fig. 11. (F) Western blot quantification was performed using biological triplicates, normalized to vinculin for each sample and to the wild-type YIF1B control. Box plots represent interquartile range (IQR); line is median; and whiskers extend to 1.5× IQR, dots are outliers. Wilcoxon signed-rank test; ns = not significant. Full-length blots used for quantification are provided in Supplementary Fig. 11. (G) Representative images of immunofluorescence of HEK293 cells, transfected for 24 h with wild-type or mutant eGFP-YIF1B expression plasmids, and co-stained for the ER marker calnexin (red) and DAPI (blue). Scale bar = 10 µm. (H) Quantification of calnexin-YIF1B co-localization (n = 30 cells for each variant) using Pearson’s correlation coefficient (range: −1 negative correlation, 1 max correlation). Box plots represent IQR; line is median; and whiskers extend to 1.5× IQR. ***P < 0.001, Kruskal-Wallis test with post hoc Dunn’s test.
Individual 1 is currently a 5-year-old male, the fifth child of consanguineous parents from Spain, born after an uneventful pregnancy and uncomplicated delivery. Developmental delay was noticed early on. He developed a severe encephalopathy, is non-verbal, has severe motor impairment with poor head control, axial hypotonia, peripheral hypertonia and upper extremity dystonia. He is unable to sit independently. He had a febrile seizure at 12 months and developed epileptic seizures at 21 months, initially treated with levetiracetam followed by valproate. EEG showed bilateral focal fronto-temporal activity and MRI showed cortical atrophy and thin corpus callosum. Hypotelorism and flat occiput were noticed. Whole exome sequencing (WES) identified a homozygous truncating variant in YIF1B (c.186dupT, p.Ala63fs), heterozygous in the unaffected parents. His 23-year-old sister was not investigated, but has a similar encephalopathy although without epilepsy.
Individual 2 is a female born to consanguineous Somali parents, who required hospitalization because of feeding problems at age of 2 months. She displayed severe global developmental delay, with no developmental milestones, and epileptic seizures. EEG showed frequent multifocal epileptiform discharges and at times evidence of burst suppression. Seizure control was obtained with phenobarbital and levetiracetam. MRI imaging at 10 months showed reduced cerebral white matter volume with atrophic prominence of ventricles and cerebellar hypoplasia. She became ventilation dependent and deceased at the age of 15 months. WES identified a homozygous truncating YIF1B variant (c.598G>T, p.Glu200*), heterozygous in the unaffected parents.
Interestingly, both p.Ala63fs and p.Glu200* are recurrent variants, identified in five and three independent families, respectively.1,2 The p.Ala63fs variant was previously found in four Arab families, and given historic migrations of Arabs to Spain this might suggest a founder mutation. Similar, all families harbouring p.Glu200* are from Somali descent, likely indicating a founder mechanism.
Individual 3 is a female born to consanguineous parents from Saudi Arabia who showed lack of developmental milestones, congenital microcephaly, severe failure to thrive, spastic quadriparesis, axial hypotonia and hypoventilation with pons atrophy, cerebellar and corpus callosum hypoplasia and white matter alterations. WES identified a homozygous truncating YIF1B variant (c.336C>G, p.Tyr112*).
Individual 4 is a 1-year-old male born to consanguineous parents from Egypt, which showed severe developmental delay starting at the age of 2 months with spasticity and dystonia, with progressive psychomotor deterioration and feeding difficulties. Whole genome sequencing identified a homozygous chr19:38796532–38812925 (GRCh37/hg19) deletion that included the YIF1B promoter, YIF1B exon 1–7, and exon 1 of KCNK6, a potassium channel without a known OMIM phenotype.
All but one previously reported family1,2 presented with protein truncating variants. Interestingly, we identified four additional families with missense variants that resulted in a similar clinical phenotype.
Individual 5 is a 27-year-old female, born at full term to non-consanguineous parents of French and German descent. Because of developmental delay, she received medical attention at ∼7 months of age. She was able to sit independently at age of ∼2–3 years, but lost this capability. She never walked independently. Speech was limited to few words and lost upon regression. She currently communicates through noises and facial expression. She is currently managed for medically refractory generalized epilepsy, with EEG at age of 26 years noting tonic seizures, and multifocal and diffuse discharges. MRI at age of 25 years showed generalized cerebral and cerebellar volume loss with severe thinning of the corpus callosum. Dysmorphic features include a long face, widely-spaced teeth, a history of gingival hyperplasia, high arched palate, and bitemporal hirsutism. WES identified compound heterozygous missense variants in YIF1B (c.569C>A, p.Ala190Glu and c.621C>A, p.Ser207Arg), which were both absent in the unaffected brother.
Individual 6 is a currently 7.5-year-old male born to consanguineous parents from Iran that presented in early infancy with developmental delay and hypotonia. Epileptic spasms, axial dystonia and limb spasticity subsequently developed (Supplementary Video 1). His best motor achievements included independent sitting and pencil grasp, but no speech development or eye contact. WES identified a homozygous missense variant in YIF1B (c.691G>A, p.Val231Ile), heterozygous in the unaffected parents.
Individual 7 is an 11-month-old male born to consanguineous parents from Iran. He first came to medical attention at age of 4 years because of severe global developmental delay. He failed to develop any milestones and developed axial dystonia and limb spasticity but no epilepsy. Brain MRI showed a thin corpus callosum. WES identified a homozygous missense variant in YIF1B (c.377T>C, p.Leu126Pro), heterozygous in the unaffected parents and siblings.
Finally, Individual 8 is currently 4.5 years old, a female born to consanguineous parents from Iran that presented with global developmental delay, microcephaly and hypotonia, developing into limb spasticity, dystonia, dyskinesia and oculomotor apraxia, without epilepsy. Brain MRI showed global atrophy and a thin corpus callosum. She could sit independently but was unable to stand and spoke only a few words. Metabolic screening was unremarkable. WES identified a homozygous missense variant in YIF1B (c.803G>T, p.Arg268Leu).
Interestingly, all YIF1B missense variants encountered localized in or close to the transmembrane domains (previously shown to be required for YIF1B function3) and were absent from gnomAD, except for p.Val231Ile, which was found once in heterozygous state (minor allele frequency 0.00040). All changed highly conserved residues, had a CADD score >22, and in silico analysis predicted pathogenicity (Fig. 1B, Supplementary Figs 7–9, Supplementary Table 1 and Supplementary material).
As primary cells of affected individuals were unavailable, we introduced the encountered missense variants by site-directed mutagenesis4,5 in a YIF1B expression plasmid, and first tested protein expression of these mutants upon transient transfection in HEK cells. Missense variants assessed did not result in significantly reduced YIF1B proteins levels (Fig. 1E and F). To investigate subcellular localization of wild-type and mutant YIF1B, we performed co-staining for YIF1B and the endoplasmic reticulum (ER) marker calnexin (Fig. 1G). Whereas wild-type YIF1B showed a high co-localization with the ER, as previously found,6 YIF1B variants showed significantly reduced co-localization (Fig. 1H). Previously, YIF1B was found to interact with RAB6A7 and TAPL.3 Upon co-transfection of RAB6A and YIF1B, we found significantly reduced co-localization between both proteins for all YIF1B variants, except p.Leu126Pro and p.Ser207Arg (Supplementary Fig. 10A and D). In contrast, all tested YIF1B missense variants showed reduced co-localization with TAPL (Supplementary Fig. 10B and E). YIF1B overexpression diminishes co-localization of TAPL and lysosomal markers.3 In agreement, whereas overexpression of wild-type YIF1B resulted in low co-localization correlation between TAPL and the lysosomal marker LAMP2, overexpression of mutant YIF1B failed to diminish this co-localization (Supplementary Fig. 10C and F). Also, co-localization correlation between YIF1B and LAMP2 was higher for mutant YIF1B. Together this indicates that the assessed missense YIF1B variants show mislocalization, reduced co-localization with known interactors and reduced functionality compared to wild-type YIF1B.
Including the eight individuals described herein, 24 individuals from 19 families have currently been identified, harbouring bi-allelic truncating/whole gene deletion (n = 18), or missense variants (n = 6) in YIF1B (Table 1 and Supplementary Table 2). All individuals presented early in life with a progressive encephalopathy with global developmental delay and cognitive impairment, after uneventful perinatal development. Almost all had feeding problems, axial hypotonia and limb spasticity, with seizures (varying from myoclonic jerks, to generalized tonic clonic seizures and infantile spasms) in around two-thirds of the cases. Around half of the individuals showed signs of dystonia, dyskinesia or microcephaly. Whereas hypoventilation was relatively frequent in the cohort described by Diaz et al.,1 in total this was only found in five individuals and seems to correlate to brainstem atrophy. Two-thirds of the individuals have brain imaging abnormalities, including white matter alterations, cerebral atrophy, corpus callosum hypoplasia and cerebellar hypoplasia. Interestingly, limited developmental milestones, such as head control, independent sitting, and limited speech were only observed in individuals harbouring missense variants, reaching statistical significant differences between the truncating and missense group after Bonferroni correction (P = 0.0001783, P = 0.001694, P = 0.001694, respectively). This might suggest that the encountered missense variants harbour some residual YIF1B activity in vivo, in agreement with our in vitro functional investigations. Other clinical features were not significantly different between the truncating and missense cohort (Table 1 and Supplementary Table 2).
Table 1.
Overview of core clinical phenotypes of 24 individuals harbouring bi-allelic variants in YIF1B
| Summary | Total (%) | Truncating (%) | Missense (%) | Fisher's exact test |
|||
|---|---|---|---|---|---|---|---|
| OR | P | Low CI | High CI | ||||
| Truncating mutation | 18/24 (75) | – | – | – | – | – | – |
| Missense mutation | 6/24 (25) | – | – | – | – | – | – |
| Female | 13/24 (54.2) | 10/18 (56.3) | 3/6 (50) | 1.238 | 1.000 | 0.128 | 11.999 |
| Deceased | 5/24 (20.8) | 5/18 (27.8) | 0/6 (0) | Infinity | 0.280 | 0.305 | Infinity |
| Growth parameters | |||||||
| Microcephaly | 15/23 (65.2) | 11/17 (64.7) | 4/6 (66.7) | 0.920 | 1.000 | 0.065 | 8.949 |
| Level of functioning | |||||||
| Global developmental delay | 23/23 (100) | 17/17 (100) | 6/6 (100) | 0.000 | 1.000 | 0.000 | Infinity |
| Head control | 5/23 (30.4) | 0/17 (0) | 5/6 (83.3) | 0.000 | 0.000 | 0.000 | 0.189 |
| Sitting | 4/23 (17.4) | 0/17 (0) | 4/6 (66.7) | 0.000 | 0.002 | 0.000 | 0.351 |
| Standing | 2/23 (8.7) | 0/17 (0) | 2/6 (33.3) | 0.000 | 0.059 | 0.000 | 1.733 |
| Walking | 2/22 (9.1) | 0/16 (0) | 2/6 (33.3) | 0.000 | 0.065 | 0.000 | 1.844 |
| Cognitive impairment | 23/23 (100) | 17/17 (100) | 6/6 (100) | 0.000 | 1.000 | 0.000 | Infinity |
| Speech development | 4/23 (17.4) | 0/17 (0) | 4/6 (66.7) | 0.000 | 0.002 | 0.000 | 0.351 |
| Progressive speech loss | 3/4 (75) | n/a (n/a | 3/4 (75) | ||||
| Hypoventilation | 5/18 (22.7) | 5/16 (31.3) | 0/2 (0) | Infinity | 1.000 | 0.069 | Infinity |
| Feeding problems/swallowing difficulties | 19/23 (82.6) | 16/17 (94.1) | 3/6 (50) | 13.390 | 0.040 | 0.796 | 883.373 |
| Autistic behaviour | 4/13 (30.8) | 2/8 (25) | 2/5 (40) | 0.529 | 1.000 | 0.025 | 10.812 |
| Neurology | |||||||
| Seizures | 14/22 (63.6) | 10/16 (62.5) | 4/6 (66.7) | 0.840 | 1.000 | 0.059 | 8.247 |
| Spasticity | 21/22 (95.5) | 15/16 (93.8) | 6/6 (100) | 0.000 | 1.000 | 0.000 | 103.833 |
| Hypotonia | 19/23 (82.6) | 15/17 (88.2) | 4/6 (66.7) | 3.497 | 0.271 | 0.197 | 63.558 |
| Dystonia | 11/23 (47.8) | 8/17 (47.1) | 3/6 (50) | 0.893 | 1.000 | 0.091 | 8.729 |
| Dyskinesia or tremor | 9/20 (45) | 7/14 (50) | 2/6 (33.3) | 1.932 | 0.642 | 0.195 | 28.137 |
| Eyes and ears | |||||||
| Ear/hearing loss | 0/16 (0) | 0/12 (0) | 0/4 (0) | 0.000 | 1.000 | 0.000 | Infinity |
| Strabismus | 10/18 (55.6) | 5/12 (41.7) | 5/6 (83.3) | 0.159 | 0.152 | 0.003 | 2.093 |
| Optic atrophy | 2/20 (10) | 2/14 (14.3) | 0/6 (0) | Infinity | 1.000 | 0.078 | Infinity |
| Retinal involvement | 0/18 (0) | 0/12 (0) | 0/6 (0) | 0.000 | 1.000 | 0.000 | Infinity |
| Cataract | 0/18 (0) | 0/12 (0) | 0/6 (0) | 0.000 | 1.000 | 0.000 | Infinity |
| Nystagmus | 5/19 (26.3) | 4/13 (30.8) | 1/6 (16.7) | 2.137 | 1.000 | 0.147 | 130.886 |
| Brain imaging | |||||||
| White matter/myelinization abnormalities | 7/24 (29.3) | 6/18 (33.3) | 1/6 (16.7) | 2.416 | 0.629 | 0.196 | 137.539 |
| Cerebellar hypoplasia | 8/23 (34.8) | 7/17 (41.2) | 1/6 (16.7) | 3.333 | 0.369 | 0.275 | 188.570 |
| Corpus callosum hypoplasia | 12/22 (54.5) | 9/16 (56.3) | 3/6 (50) | 1.271 | 1.000 | 0.128 | 12.729 |
| Cerebral atrophy/parenchymal volume loss | 10/24 (41.7) | 8/18 (50) | 2/6 (33.3) | 1.570 | 1.000 | 0.170 | 21.636 |
| Pons/brain stem atrophy | 5/22 (18.2) | 5/16 (31.3) | 0/6 (0) | Infinity | 0.266 | 0.352 | Infinity |
CI = confidence interval; n/a = not applicable; n/d = not determined; OR = odds ratio. P-values in bold indicate significance.
Together, our previous work2 and this study, and that of Diaz et al.1 defines a previously unrecognized neurodevelopmental disorder, presenting with severe to profound neurodevelopmental delay, cognitive impairment, neurological sequelae, seizures and microcephaly. Long term follow-up of individuals with YIF1B variants will help to delineate this new disease entity further.
Data availability
All data are available from the corresponding author upon reasonable request, except for primary patient sequencing data, which cannot be made available due to consent regulations.
Supplementary Material
Acknowledgements
We would like to thank the individuals described herein and their families for participating in this study. We would like to thank all members of the Barakat lab for helpful discussion. We thank Rupert Abele (Frankfurt) for providing the TAPL plasmid, and Gerben Schaaf (Rotterdam) and Grazia M.S. Mancini (Rotterdam) for providing the LAMP2 and Calnexin antibody, respectively.
Funding
R.D. is supported by a China Scholarship Council (CSC) PhD Fellowship (201906300026) for her PhD studies at the Erasmus Medical Center, Rotterdam, the Netherlands. B.P. was funded by PI19/01155 and B2017/BMD-3721. The work by K.J.C.L. and S.T.A. was supported by the King Abdullah University of Science and Technology (KAUST) through the baseline fund and the Award No. FCC/1/1976–25 and REI/1/4446–01 from the Office of Sponsored Research (OSR). N.K.’s laboratory is supported by KFSHRC seed grants (RAC2120022), King Salman Center for Disability Research (KSCDR#2180 004) and King Abdulaziz City for Science and Technology (KACST#14-MED2007-20). T.S.B.’s laboratory is supported by the Netherlands Organization for Scientific Research (ZonMw Veni, Grant 91617021), an Erasmus MC Fellowship 2017 and Erasmus MC Human Disease Model Award 2018.
Competing interests
A.B. is an employee of GeneDx, Inc. T.R.D.S, P.B. and A.B.A. are employees of CENTOGENE GmbH. All other authors declare no conflict of interest.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Diaz J, Gérard X, Emerit M-B, et al. YIF1B mutations cause a post-natal neurodevelopmental syndrome associated with Golgi and primary cilium alterations. Brain. 2020;143(10):2911–2928. [DOI] [PubMed] [Google Scholar]
- 2. AlMuhaizea M, AlMass R, AlHargan A, et al. Truncating mutations in YIF1B cause a progressive encephalopathy with various degrees of mixed movement disorder, microcephaly, and epilepsy. Acta Neuropathol. 2020;139(4):791–794. [DOI] [PubMed] [Google Scholar]
- 3. Graab P, Bock C, Weiss K, et al. Lysosomal targeting of the ABC transporter TAPL is determined by membrane-localized charged residues. J Biol Chem. 2019;294(18):7308–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanderson LE, Lanko K, Alsagob M, et al. Bi-allelic variants in HOPS complex subunit VPS41 cause cerebellar ataxia and abnormal membrane trafficking. Brain. 2021;144(3):769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weerts MJA, Lanko K, Guzmán-Vega FJ. et al. Delineating the molecular and phenotypic spectrum of the SETD1B-related syndrome. Genet Med. Published online 3 August 2021. doi:10.1038/s41436-021-01246-2 [DOI] [PMC free article] [PubMed]
- 6. Alterio J, Masson J, Diaz J, et al. Yif1B is involved in the anterograde traffic pathway and the Golgi architecture. Traffic. 2015;16(9):978–993. [DOI] [PubMed] [Google Scholar]
- 7. Al Awabdh S, Miserey-Lenkei S, Bouceba T, et al. A new vesicular scaffolding complex mediates the G-protein-coupled 5-HT1A receptor targeting to neuronal dendrites. J Neurosci. 2012;32(41):14227–14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request, except for primary patient sequencing data, which cannot be made available due to consent regulations.