Abstract
The feasibility of performing a multiplex assay for the detection of human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) RNAs and hepatitis B virus (HBV) DNA is demonstrated. This assay is based (i) on the coamplification of a 142-bp fragment from the gag region of the HIV-1 genome and a 142-bp HIV-1 quantitation standard fragment, a 244-bp fragment from the 5′ noncoding region of the HCV genome, and a 104-bp fragment from the pre-C and C gene regions of the HBV genome, using three sets of specific primers; (ii) on the capacity of these four biotinylated PCR products to hybridize to their specific oligonucleotide probe-coated microspheres; and (iii) on the ability of the flow cytometer to discriminate between distinct fluorescent-microsphere categories. Absence of cross-hybridization between the unrelated oligonucleotide probes and PCR products generated by the multiplex reverse transcription-PCR (RT-PCR) and the highly sensitive detection method allowed us to assess unambiguously the HIV-1 viral load and the infectious status of 35 serologically well-established clinical samples and 20 seronegative blood donor plasma samples tested. The results indicate that multiplex RT-PCR and flow cytometer microsphere-based hybridization assays, when combined, provide a rapid, sensitive, and specific method for the quantitation and detection of the major viral agents of infectious diseases in a single plasma sample.
The development of nucleic acid amplification technologies has found many applications in biomedical research and in diagnostic techniques. The study of infectious diseases is a field that has benefited much from these technologies. Amplification of up to several millionfold of low-copy-number DNA or RNA allows one to make an early and rapid diagnosis of bacterial or viral infections (6, 7, 18, 20). Moreover, techniques for the quantitation of the viral load in plasma have been shown to be of particular interest in monitoring drug therapy and in predicting the disease outcome (12, 15). For several years, attempts have been made to use multiplex PCR or reverse transcription-PCR (RT-PCR) in order to detect several different pathogens or serotypes in coinfected samples after a single reaction (10, 19). In such assays, primer pairs are chosen with respect to their compatibility and ability to simultaneously amplify their specific target sequences. Detection of amplified products is usually done by gel electrophoresis, which discriminates fragments of distinct sizes by ethidium bromide staining or by hybridization using specific DNA probes. Even though multiplex RT-PCR simplifies the procedures, analysis remains tedious and labor-intensive. The development of solid phase hybridization assays greatly improved the practicability of these assays and allowed the detection of different PCR products from one multiplex PCR amplification run (1, 3). Usually, each specific amplified product is detected in a separate well. The amplicon volume becomes a limiting factor. To address this problem, we describe here the use of a flow cytometer and a microsphere-based hybridization assay for the detection of multiplex RT-PCR products in a single tube. This assay relies on the ability of the flow cytometer to resolve multiple microsphere-based assays (5). The red fluorescence intensity (675 nm) emitted by an oligonucleotide probe-coated microsphere population results from the hybridization of its target PCR-amplified sequence with the specific oligonucleotide probe. The simultaneous detection of different target sequences is achieved by mixing several specific oligonucleotide probe-coated microsphere populations, which are differentiated by their level of green fluorescence intensity (525 nm). We demonstrate, using human immunodeficiency virus type 1 (HIV-1), hepatitis C virus (HCV), and hepatitis B virus (HBV) as models, that multiplex RT-PCR and microsphere-based flow cytometric assays constitute an accurate, specific, and rapid method for the simultaneous detection and quantitation of several different viruses in one plasma sample.
MATERIALS AND METHODS
Oligonucleotide primers and probes and thiol-reactive microspheres.
Primers for the amplification of the HIV-1, HCV, and HBV genomes and probes for the specific detection of PCR products are presented in Table 1. These sets of primers were derived from the HIV-1 gag gene (14), the HCV 5′ noncoding region (22, 23), and the pre-C and C regions of the HBV genome (9), respectively. Antisense (SK431) and sense (SK462) primers for HIV-1 and antisense primers for HCV (KY78) and HBV (PB1) were biotinylated at the 5′ ends to allow detection of the amplified products. Oligonucleotide capture probes were modified at the 5′ ends with a thiol linker to allow their covalent coupling to Immunotech-Coulter 3.7-μm-diameter thiol-reactive microspheres. SK102 and CP35 are oligonucleotide probes for HIV-1 detection. They are specific for the detection of amplified HIV-1 RNA and the quantitation standard, respectively. Oligonucleotides KY150 and PB3 allowed detection of amplified HCV RNA and HBV DNA, respectively. Modified 5′-end-biotinylated oligonucleotides complementary to SK102, KY150, and PB3 were synthesized to evaluate the sensitivity of detection.
TABLE 1.
Primers and probes for detection of RNA and DNA of HIV-1, HCV, and HBV viruses
| Virus | Primer or probea | Sequence (5′–3′) | Amplified region and sequence (reference) |
|---|---|---|---|
| HIV-1 | SK431 | TGCTATGTCAGTTCCCCTTGGTTCTCT | gag region, 905–1046 (13) |
| SK462 | AGTTGGAGGACATCAAGCAGCCATGCAAAT | ||
| SK102* | GAGACCATCAATGAGGAAGCTGCAGAATGGGAT | ||
| CP35* | CATAGCACTATAGAACTCTGCAAGCC | ||
| HCV | KY78 | CTCGCAAGCACCCTATCAGGCAGT | 5′ noncoding region, 56–299 (8) |
| KY80 | GCAGAAAGCGTCTAGCCATGGCGT | ||
| KY150* | CATAGTGGTCTGCGGAACCGGTGAGT | ||
| HBV | PB1 | GTTCAAGCCTCCAAGCTGTG | Pre-C and C regions, 1864–1967 (2) |
| PB2 | TCAGAAGGCAAAAAAGAGAGTAACT | ||
| PB3* | CTTTATAAGGATCAATGTCCATGC |
Probes are indicated by asterisks.
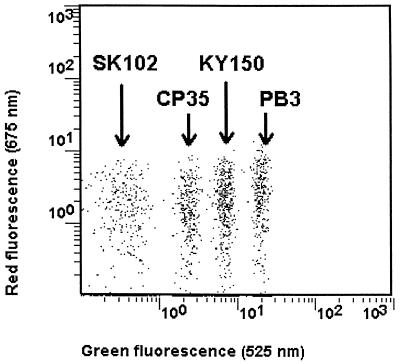
Four distinct intensity levels of green-fluorescent (525 nm) 3.7-μm-diameter thiol-reactive microspheres were used (Fig. 1). SK102, coding for the HIV-1 RNA, was applied to the dimmest green-fluorescent microspheres. CP35, coding for the HIV-1 quantitation standard, was applied to the second-level-intensity microspheres. KY150, coding for the HCV RNA, was applied to the third-level-intensity microspheres, and PB3, coding for the HBV DNA, was applied to the brightest microspheres.
FIG. 1.
Typical dot plots of green fluorescence versus red fluorescence for the mixture of the four oligonucleotide-coated microsphere populations for the simultaneous detection of RT-PCR products from HIV-1, HCV, and HBV nucleic acids.
Clinical samples, preparation, and amplification.
Fifty-five plasma samples were tested. These samples represented five groups: group 1, HCV infection only (n = 13); group 2, HIV-1 infection only (n = 8); group 3, concurrent HIV-1 and HCV infection (n = 12); group 4, concurrent HIV-1 and HBV infection (n = 2); and group 5, seronegative blood donors (n = 20). All plasma samples were kept at −20°C after collection. Extraction of genomic RNA and DNA from these 55 plasma samples was achieved with the AMPLICOR HIV-1 MONITOR Test (reference no. 07 5623 7) according to the recommendations of the manufacturer (Roche). The only difference was in the volume of plasma extracted (100 μl). The extracted RNA and DNA were dissolved in 200 μl of diluent buffer, aliquoted in Safe Lock 0.5-ml Eppendorf tubes for PCR, and stored frozen at −20°C prior to amplification. HIV-1 RT-PCR was done by mixing 25 μl of extract and 25 μl of Working Master Mix from the AMPLICOR HIV-1 MONITOR Test to amplify HIV-1 RNA only. Multiplex RT-PCR was done by mixing 25 μl of extract and 25 μl of Working Master Mix from the AMPLICOR HIV-1 MONITOR Test, to which 5 pmol each of the HCV and HBV primers had been added in order to simultaneously amplify HIV-1 and HCV RNAs and HBV DNA. Amplification was carried out in a single tube in a Biometra Trio Thermoblock with a heated lid. Following 2 min of incubation at 50°C, reverse transcription was allowed to proceed for 30 min at 60°C. After a denaturation step at 95°C, PCR amplification was then carried out with four cycles of 95, 55, and 72°C for 30 s each, followed by 26 cycles of 90, 60, and 72°C for 30 s each. After the last cycle, the temperature was maintained at 72°C while 50 μl of denaturation solution was added to each PCR tube. The RT-PCR-amplified products are referred to as the amplicon.
Flow cytometer microsphere-based hybridization assay.
The simultaneous detection of RT-PCR products was achieved with a flow cytometer microsphere-based hybridization assay. Briefly, 100 μl of a fivefold dilution of denatured amplicon in a hybridization buffer containing 1.2 M NaCl, 0.2 M phosphate (pH 6), 5 mM EDTA, 0.05% Tween 20, 10 mM NaN3, and 5 g of bovine serum albumin per liter was incubated with 10 μl of a microsphere mixture containing 10,000 probe-coated microspheres from each of the four microsphere categories (SK102, CP35, KY150, and PB3) in a Loprodine 0.45-μm-pore-size membrane-bottomed 96-microwell plate (Nunc). Hybridization of RT-PCR products to their cognate probe-coated microspheres was carried out for 90 min with shaking at 37°C. The microplate was then placed on a vacuum manifold to allow five washing cycles with 250 μl of wash solution. Detection of bound biotinylated RT-PCR product to probe-coated microspheres was then done by incubating 100 μl of streptavidin-phycoerythrin cyanin 5 conjugate (Immunotech) for 30 min with shaking at 37°C. Unbound streptavidin-phycoerythrin cyanin 5 conjugate was removed with two more washes, and the microspheres were resuspended in 300 μl of 10 mM phosphate buffer (pH 7) containing 150 mM NaCl, 10 mM NaN3, and 5 g of bovine serum albumin per liter and transferred to a flow cytometer tube. Finally, the microspheres were analyzed on an EPICS-XL MCL flow cytometer (Beckman-Coulter).
Data acquisition and data processing.
Data acquisition was performed at high speed until a minimum of 2,000 microspheres had been acquired. Once each individual microsphere has been analyzed for its green (525-nm) and red (675-nm) fluorescence intensities, software designed in-house computes the data and attributes to each microsphere population a mean red fluorescence intensity value. This value is related directly to the amount of biotinylated RT-PCR product hybridized with the oligonucleotide probe on the microspheres.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the viral sequences are as follows: HCV, D90208; HBV, X02496; and HIV-1, K02083.
RESULTS
Analytical sensitivity of the detection system.
The analytical sensitivity of the flow cytometer microsphere-based hybridization assay for the detection of nucleic acids was studied with biotinylated oligonucleotides complementary to the SK102, KY150, and PB3 oligonucleotide probe-coated microspheres. Serial dilutions of the biotinylated oligonucleotides, specific for HIV-1 and HCV RNAs and HBV DNA, respectively, were assayed by the protocol described in Materials and Methods. The sensitivities found were within a range of 6 to 20 fmol/liter (0.4 × 106 to 1.2 × 106 molecules/100 μl) (data not shown). The small difference observed between these oligonucleotides is not surprising, since it is well known that the hybridization behavior of short oligonucleotides is very dependent on their respective sequences and secondary structures. The microwell plate provided in the AMPLICOR HIV-1 MONITOR Test for HIV-1 RNA RT-PCR product detection gives similar sensitivity (data not shown).
Multiplex RT-PCR combined with microsphere-based hybridization assay.
It is essential to exclude cross-hybridization between oligonucleotide probe-coated microspheres and unrelated RNA amplified by RT-PCR in any assessment of the infectious status of a plasma sample. Using monoamplified RT-PCR plasma samples with a high virus load, we showed that the three amplified fragments for HIV-1, HCV, and HBV hybridized specifically to their cognate oligonucleotide probes. When these amplicons were assayed, using the three HIV-1-, HCV-, and HBV-specific microsphere categories, a high red fluorescence intensity was obtained only with the relevant microsphere category. The signals measured with the unrelated microspheres were lower than the cutoff values calculated from the analysis of 20 seronegative blood donor plasma samples (Table 2). The cutoff values were calculated as the means of the values plus 5 standard deviations; the means and standard deviations, respectively, were 0.75 and 0.34 for SK102 (HIV-1), 0.49 and 0.11 for KY150 (HCV), and 0.44 and 0.13 for PB3 (HBV). Since the CP35 microsphere codes for the HIV-1 quantitation standard, no cutoff value was calculated. A high fluorescence intensity level for this category of microspheres validates both the RT-PCR amplification and the hybridization steps.
TABLE 2.
Cross-hybridization study with monoamplified high-virus-load sample plasma
| Amplified virus | Red fluorescence intensity with the following microsphere categorya:
|
||
|---|---|---|---|
| SK102 (HIV-1) | KY150 (HCV) | PB3 (HBV) | |
| HIV-1 | 923.7 | 0.64 | 0.45 |
| HCV | 0.70 | 564.3 | 0.41 |
| HBV | 0.78 | 0.52 | 1,005.6 |
Cutoff values are as follows: SK102, 2.43; KY150, 1.06; PB3, 1.08.
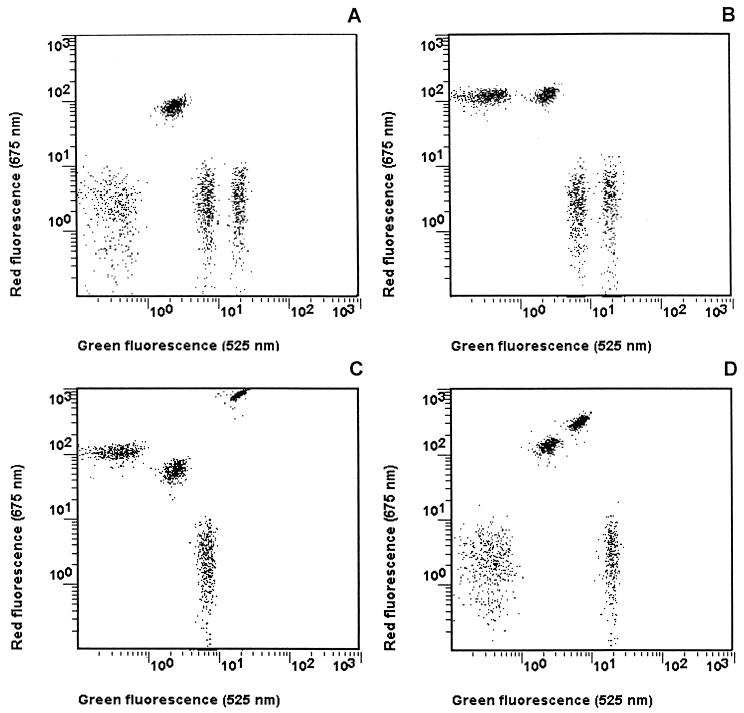
The analysis of the clinical samples from groups 1 to 4 was performed as follow. A specimen was considered positive for a given virus when the signal was higher than the cutoff value for the particular microsphere category. In Table 3 are presented the raw data obtained from these 35 samples. HCV RNA was detected in all of the HCV-positive samples (25 of 25). HIV-1 RNA was detected in all specimens from groups 2 (8 of 8) and 4 (2 of 2) and in 11 of 12 specimens from group 3. The two specimens from group 4 were shown to be positive for HBV. The four dot plots presented in Fig. 2 summarize how, after two simple steps, it is possible to assess unambiguously the viral status of a plasma sample.
TABLE 3.
Detection of HIV-1, HCV, and HBV viremia in plasma samples from 35 seropositive subjects by multiplex RT-PCR and one-tube flow cytometer microsphere-based hybridization assays
| Group | Sample no. | Red fluorescence intensitya with the following microsphere category:
|
|||
|---|---|---|---|---|---|
| SK102 (HIV-1) | CP35 | KY150 (HCV) | PB3 (HBV) | ||
| 3 | 1 | 208.08 | 64.80 | 633.57 | 0.27 |
| 2 | 8.96 | 50.10 | 159.70 | 0.56 | |
| 3 | 182.31 | 43.57 | 483.38 | 0.53 | |
| 4 | 404.24 | 73.60 | 1.16 | 0.49 | |
| 5 | 357.65 | 48.36 | 198.80 | 0.59 | |
| 6 | 0.47 | 30.58 | 502.71 | 0.34 | |
| 7 | 683.50 | 44.62 | 1.67 | 0.43 | |
| 8 | 72.57 | 83.13 | 465.65 | 0.53 | |
| 9 | 219.21 | 86.68 | 526.56 | 0.38 | |
| 10 | 43.95 | 82.65 | 598.81 | 0.30 | |
| 11 | 17.34 | 104.59 | 461.39 | 0.43 | |
| 12 | 846.91 | 63.60 | 390.90 | 0.45 | |
| 2 | 13 | 10.63 | 33.31 | 0.32 | 0.32 |
| 14 | 2.60 | 70.00 | 0.39 | 0.33 | |
| 15 | 59.02 | 111.51 | 0.38 | 0.34 | |
| 16 | 24.64 | 88.03 | 0.48 | 0.32 | |
| 17 | 64.82 | 67.94 | 0.41 | 0.41 | |
| 18 | 35.29 | 94.72 | 0.46 | 0.31 | |
| 19 | 25.57 | 100.72 | 0.42 | 0.45 | |
| 20 | 123.92 | 129.17 | 0.61 | 0.42 | |
| 1 | 21 | 1.06 | 134.87 | 563.56 | 0.39 |
| 22 | 0.92 | 145.09 | 323.19 | 0.35 | |
| 23 | 0.96 | 71.05 | 364.25 | 0.41 | |
| 24 | 0.70 | 138.70 | 1.34 | 0.44 | |
| 25 | 0.65 | 134.99 | 320.43 | 0.39 | |
| 26 | 0.64 | 119.47 | 348.90 | 0.37 | |
| 27 | 0.63 | 122.08 | 454.46 | 0.36 | |
| 28 | 1.30 | 141.40 | 223.29 | 0.36 | |
| 29 | 1.04 | 124.15 | 542.42 | 0.38 | |
| 30 | 0.95 | 119.78 | 430.50 | 0.63 | |
| 31 | 0.73 | 123.04 | 384.46 | 0.26 | |
| 32 | 0.62 | 85.35 | 355.80 | 0.42 | |
| 33 | 0.70 | 126.37 | 564.37 | 0.41 | |
| 4 | 34 | 110.35 | 59.86 | 0.51 | 846.62 |
| 35 | 303.42 | 21.45 | 0.50 | 787.67 | |
Underlined values are those higher than the cutoff value, thus reflecting a positive sample.
FIG. 2.
Typical dot plots of green fluorescence versus red fluorescence for virus-free or seropositive plasma samples. (A) Virus-free plasma sample; (B) HIV-1-seropositive plasma sample; (C) HIV-1- and HBV-seropositive plasma sample; (D) HCV-seropositive plasma sample.
HIV-1 RNA multiplex versus monoplex quantitation.
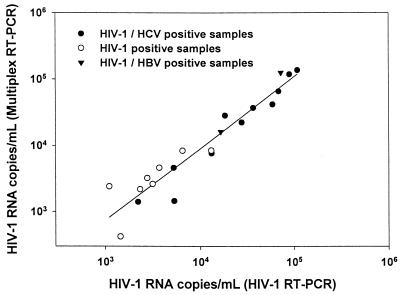
Quantitation of HIV-1 RNA using the flow cytometer microsphere-based hybridization assay can be performed readily, since both microspheres, i.e., those specific for HIV-1 RNA and the quantitation standard, are incubated simultaneously. The number of HIV-1 RNA copies per milliliter was calculated as the ratio of the red fluorescence intensities for HIV-1 RNA (SK102) and the quantitation standard (CP35) from the same assay times a constant value which takes into account the dilutions of the overall extraction procedure and the input number of quantitation standard copies per PCR. Quantitation of the HIV-1 RNA viral load from 21 specimens (8 positive for HIV-1 only, 11 positive for HIV-1 and HCV, and 2 positive for HIV-1 and HBV) was carried out after multiplex or monoplex RT-PCR amplification and microsphere-based hybridization assay. As shown in Fig. 3, the quantitation values for HIV-1 RNA from HIV-1 RT-PCR correlated well with those obtained by multiplex RT-PCR (r = 0.956).
FIG. 3.
Comparison of HIV-1 RNA viral loads (copies per milliliter from 21 specimens after HIV-1 RT-PCR (x axis) and multiplex RT-PCR (y axis). Linear regression was calculated as follows: log (HIV-1 RT-PCR) = 1.07 log (multiplex RT-PCR) − 0.37; r = 0.956.
DISCUSSION
In view of the increasing number of multiple infections in patients and the interest in domains as different as blood banking and disease control, it has become important to find techniques for optimizing the screening of large numbers of samples. The use of the flow cytometer as a tool in microsphere-based fluorimetric assays is a concept that was first described in the 1970s (5). Taking advantage of the ability of the flow cytometer to resolve complex mixtures of microspheres on the basis of size and level of fluorescence, it was shown that this technology was applicable for the simultaneous detection of antibodies specific for different antigens and for the measurement of several soluble antigens in a microsphere-based assay (4, 16). Later, it was reported that this methodology could also be adapted for the detection of PCR-amplified nucleic acid sequences (17, 21). However, none of these studies demonstrated the simultaneous use of multiplex RT-PCR and the microsphere-based hybridization assay. They showed only the detection of a monospecific amplified viral DNA or RNA in a single (21) or multiple (17) microsphere-based hybridization assays.
In this study, we investigated the possibility of combining multiplex RT-PCR amplification and flow cytometer microsphere-based hybridization assays in order to improve screening methods for RT-PCR-based virus detection. As a model, we developed an assay for the simultaneous amplification and detection of HIV-1, HCV, and HBV.
The multiplex RT-PCR amplification of genomic sequences from three different viruses requires the use of three pairs of primers to amplify each viral nucleic acid with a high yield. The primers we used in this study have been described previously (9, 14, 23). They allow the amplification of a 142-bp fragment from the gag region of HIV-1, a 244-bp fragment from the 5′ noncoding region of the HCV genome, and a 104-bp fragment from the pre-C and C gene regions of HBV. The optimization of the multiplex RT-PCR amplification was focused first on the compatibility of these primers during RT-PCR amplification. Only one minor modification of the antisense primer for HBV (9) was necessary. This primer and the KY78 primer for HCV generated a primer-specific by-product during the RT-PCR amplification step that induced unexpectedly high nonspecific binding to the HIV-1 RNA detection microsphere (SK102-coated microsphere) (data not shown). This nonspecific binding was reduced to background noise when the PB1 primer (antisense primer with the 3′ end deleted) was used. Second, primer compatibility for multiplex RT-PCR amplification was demonstrated after analysis of the amplicons obtained from 20 seronegative blood donor plasma samples. Finally, we showed that amplification efficiency was not affected by the simultaneous use of these three pairs of primers. Indeed, the calculated HIV-1 viral load from HIV-1-containing specimens after HIV-1 RT-PCR correlates well with the HIV-1 viral load calculated from the same specimens after the multiplex RT-PCR. These results indicate that the amplification yields of the HIV-1 RNA and the internal standard for the HIV-1 RNA quantitation were similar. More interestingly, we also found also a good correlation for HIV-1 viral load for plasma samples coinfected with HIV-1 and HCV or HBV viruses which were subjected to HIV-1 RT-PCR or multiplex RT-PCR amplification.
Detection of each of the RT-PCR-amplified nucleic acids was achieved in a microsphere-based hybridization assay. Two oligonucleotide capture probes were used for the quantitation of the HIV-1 viral load. SK102 was specific for the detection of the HIV-1 RNA. CP35 was specific for the detection of the quantitation standard, an RNA transcript that is amplified using the same primers as those for the HIV-1 RNA but that contains a specific probe binding region that allows the quantitation standard to be distinguished from the HIV-1 RNA PCR product. Two other specific oligonucleotide capture probes were used for the detection of HBV and HCV. These four oligonucleotide capture probes were coupled to four distinct green-fluorescent microsphere categories. We showed in cross-hybridization experiments that these four oligonucleotide-coated microsphere categories could be used together in the same assay. HIV-1-, HCV-, and HBV-amplified nucleic acid products hybridized specifically to their cognate oligonucleotide capture probe microsphere. No significant hybridization of the RT-PCR products to irrelevant microspheres could be observed. This finding was further confirmed with the analysis of monoinfected plasma samples. None of these samples was found positive for a second viral infection when the fluorescence intensity measured was compared to the cutoff values.
The validation of this approach using two very sensitive methods, i.e., RT-PCR amplification, which increases small quantities of nucleic acid several millionfold, and flow cytometer microsphere-based hybridization assays, which can detect as few as ≤0.4 × 106 biotinylated target molecules per assay, was achieved by investigating a total of 55 plasma samples. Twenty of them, obtained from seronegative blood donors, were analyzed to calculate cutoff values in order to classify samples as positive or negative. The analysis of the remaining 35 serologically characterized clinical plasma samples showed almost perfect correlation with the expected status. Concordance was perfect with the exception of one HIV-1-positive sample in group 3 (sample 6) which was found negative in this study. An aliquot of the same extract amplified only for HIV-1 was also found to be negative for HIV-1 when assayed with the Roche kit. This result indicates that the presence of a high copy number of the HCV genome, demonstrated by the high fluorescence signal measured in the KY150 (HCV) microsphere population, did not hamper amplification of the HIV-1 genome of this sample.
In conclusion, the use of multiplex RT-PCR and a flow cytometer microsphere-based hybridization assay greatly simplified the overall procedures for the detection of several viral pathogens. These data demonstrate that, using these two techniques in combination, it is possible to assess unambiguously and with great sensitivity the infectious status of plasma samples for at least three infectious viruses. One would expect that the simultaneous amplification and detection of more than three nucleic acid sequences could be done using the appropriate primer pairs and oligonucleotide capture probes.
ACKNOWLEDGMENTS
This work was fully supported by Immunotech, a Beckman Coulter company.
We thank particularly J. Fieschi and E. Rouvier for helpful discussions and advice, E. Lagier (Hopital d'Aix en Provence) for providing us with clinical specimens, J. Quintana for providing us with the microspheres, S. Debono and R. Hamelik for writing the data processing software, and H. Rickenberg for critical reading of the manuscript.
REFERENCES
- 1.Bej A K, Mahbubani M H, Miller R, DiCesare J L, Haff L, Atlas R M. Multiplex PCR amplification and immobilized capture probes for detection of bacterial pathogens and indicators in water. Mol Cell Probes. 1990;4:353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- 2.Bichko V, Pushko P, Dreilina D, Pumpen P, Gren E. Subtype ayw variant of hepatitis B virus. DNA primary structure analysis. FEBS Lett. 1985;185:208–212. doi: 10.1016/0014-5793(85)80771-7. [DOI] [PubMed] [Google Scholar]
- 3.DiDomenico N, Link H, Knobel R, Caratsch T, Weschler W, Loewy Z G, Rosenstraus M. Cobas Amplicor™: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin Chem. 1996;42:1915–1923. [PubMed] [Google Scholar]
- 4.Frengen J, Lindmo T, Paus E, Schmid R, Nustad K. Dual analyte assay based on particle types of different size measured by flow cytometry. J Immunol Methods. 1995;178:141–151. doi: 10.1016/0022-1759(94)00252-r. [DOI] [PubMed] [Google Scholar]
- 5.Fulwyler, M. J. 1976. Method for detecting and separating antigens and antibodies in blood or other samples. British patent 15611042.
- 6.Guatelli J C, Gingeras T R, Richman D D. Nucleic acid amplification in vitro: detection of sequences with low copy numbers and application to diagnosis of human immunodeficiency virus type 1 infection. Clin Microbiol Rev. 1989;2:217–226. doi: 10.1128/cmr.2.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart C, Schochetman G, Spira T, Lifson A, Moore J, Galphin J, Sninsky J, Ou C Y. Direct detection of HIV RNA expression in seropositive subjects. Lancet. 1988;ii:596–599. doi: 10.1016/s0140-6736(88)90639-3. [DOI] [PubMed] [Google Scholar]
- 8.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehtovaara P, Uusi-Oukari M, Buchert P, Laaksonen M, Bengtström M, Ranki M. Quantitative PCR for hepatitis B virus with colorimetric detection. PCR Methods Appl. 1993;3:169–175. doi: 10.1101/gr.3.3.169. [DOI] [PubMed] [Google Scholar]
- 10.Mahony J B. Multiplex polymerase chain reaction for the diagnosis of sexually transmitted diseases. Clin Lab Med. 1996;16:61–71. [PubMed] [Google Scholar]
- 11.McHugh T, Miner R C, Logan L H, Stites D P. Simultaneous detection of antibodies to cytomegalovirus and herpes simplex virus by using flow cytometry and microsphere-based fluorescence immunoassay. J Clin Microbiol. 1988;26:1957–1961. doi: 10.1128/jcm.26.10.1957-1961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellors J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 13.Muesing M A, Smith D H, Cabradilla C D, Benton C V, Lasky L A, Capon D J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 14.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schooley R. Correlation between viral load measurements and outcome in clinical trials of antiviral drugs. AIDS. 1995;9(Suppl. 2):S15–S9. [PubMed] [Google Scholar]
- 16.Scillian J J, McHugh T M, Busch M P, Tam M, Fulwyler M J, Chien D Y, Vyas G N. Early detection of antibodies against rDNA-produced HIV proteins with a flow cytometric assay. Blood. 1989;73:2041–2048. [PubMed] [Google Scholar]
- 17.Smith P L, WalkerPeach C R, Fulton R J, DuBois D B. A rapid, sensitive, multiplexed assay for detection of viral nucleic acids using the FlowMetrix System. Clin Chem. 1998;44:2054–2056. [PubMed] [Google Scholar]
- 18.Stoeckl E, Barrett N, Heinz F X, banekovich M, Stingl G, Guggenberger K, Dorner F, Kunz C. Efficiency of the polymerase chain reaction for the detection of human immunodeficiency virus type (HIV-1) DNA in the lymphocytes of infected persons: comparison to antigen-enzyme-linked immunosorbent assay and virus isolation. J Med Virol. 1989;29:249–255. doi: 10.1002/jmv.1890290406. [DOI] [PubMed] [Google Scholar]
- 19.Tenorio A, Echeverria J E, Echeverria I C J M, Tabares E. Detection and typing of human herpes viruses by multiplex polymerase chain reaction. J Virol Methods. 1993;44:261–269. doi: 10.1016/0166-0934(93)90061-u. [DOI] [PubMed] [Google Scholar]
- 20.Vyas G N, Yang G, Murphy E. Transfusion related transmissible diseases: detection by polymerase chain reaction-amplified genes of the microbial agents. Transfus Med Rev. 1994;8:253–266. doi: 10.1016/s0887-7963(94)70117-6. [DOI] [PubMed] [Google Scholar]
- 21.Yang G, Ulrich P P, Aiyer R A, Rawal B D, Vyas G N. Detection of hepatitis B virus in plasma using flow cytometric analyses of polymerase chain reaction-amplified DNA incorporating digoxigenin-11-dUTP. Blood. 1993;81:1083–1088. [PubMed] [Google Scholar]
- 22.Young K K Y, Archer J J, Yokosuka O, Omatat M, Resnick R M. Detection of hepatitis C virus RNA by a combined reverse transcription PCR assay: comparison with nested amplification and antibody testing. J Clin Microbiol. 1995;33:654–657. doi: 10.1128/jcm.33.3.654-657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young K K Y, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]