Abstract
Neoplasms involving plasmacytoid dendritic cells (pDC) include blastic pDC neoplasms (BPDCN) and other pDC proliferations, where pDC are associated with myeloid malignancies: most frequently chronic myelomonocytic leukemia (CMML) but also acute myeloid leukemia (AML), hereafter named pDC-AML. We aimed to determine the reactive or neoplastic origin of pDC in pDC-AML, and their link with the CD34+ blasts, monocytes or conventional DC (cDC) associated in the same sample, by phenotypic and molecular analyses (targeted next-generation sequencing, 70 genes). We compared 15 pDCAML at diagnosis with 21 BPDCN and 11 normal pDC from healthy donors. CD45low CD34+ blasts were found in all cases (10-80% of medullar cells), associated with pDC (4-36%), monocytes in 14 cases (1-10%) and cDC (two cases, 4.8-19%). pDC in pDC-AML harbor a clearly different phenotype from BPDCN: CD4+ CD56– in 100% of cases, most frequently CD303+, CD304+ and CD34+; lower expression of cTCL1 and CD123 with isolated lymphoid markers (CD22/CD7/CD5) in some cases, suggesting a prepDC stage. In all cases, pDC, monocytes and cDC are neoplastic since they harbor the same mutations as CD34+ blasts. RUNX1 is the most commonly mutated gene: detected in all AML with minimal differentiation (M0-AML) but not in the other cases. Despite the low number of cases, the systematic association between M0-AML, RUNX1 mutations and an excess of pDC is puzzling. Further evaluation in a larger cohort is required to confirm RUNX1 mutations in pDC-AML with minimal differentiation and to investigate whether it represents a proliferation of blasts with macrophage and DC progenitor potential.
Introduction
Plasmacytoid dendritic cells (pDC) are hematopoietic cells mainly developed from a myeloid branch including the macrophage DC progenitor (MDP) with monocyte, conventional DC (cDC) and pDC differentiation potential.1–4 Two types of neoplastic counterparts for pDC have been identified: the first is the well-known blastic pDC neoplasm (BPDCN), initially described as CD4+ CD56+ neoplasm;5-10 and the second is defined as mature pDC proliferation (MPDCP) associated with a myeloid neoplasm, frequently chronic myelomonocytic leukemia (CMML), but also myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML), especially with monocytic differentiation.11-17 MPDCP is not formally referenced in the World Health Organization 2017 classification, but mentioned as a differential diagnosis of BPDCN.7,8 As for BPDCN, MPDCP occur predominantly in male patients (75%) with a median age of 69 years18 and frequent lymph nodes or skin lesions. The mature pDC denomination refers to the morphologically mature and CD56– phenotype (as with normal pDC), in the absence of the blastic morphology of BPDCN.8 Flow cytometry or immunohistochemistry are mandatory for BPDCN diagnosis and relatively well defined with CD4+, CD56+, CD123+high, CD303+/-, CD304+/- cells7,19 expressing TCL1 at high levels.20 Conversely, only few cases of MPDCP phenotype have been described.7,14,15 In the same way, the genomic profile of MPDCP is still poorly understood, but a clonal relationship between pDC and the associated neoplasm has been demonstrated with pDC exhibiting leukemic abnormalities such as monosomy 7, trisomy 8, del(5q), CBFB-MYH11 or internal tandem duplication of FLT3 (FLT3-ITD) of blasts in AML16,17,21–23 and the mutational profile of monocytes in CMML.24
We collected AML harboring a heterogeneous phenotypic presentation with a population of immature blasts associated with an excess of pDC, but also monocytes and sometimes cDC. These cases are hereafter referred to as AML with pDC (pDC-AML). The purpose of this study was to better characterize pDC-AML by analyzing the phenotype of immature blasts and pDC and the mutational profiles of each cell population (blasts, pDC, monocytes, cDC) after cell sorting, in order to determine whether they share the same profiles.
Methods
Patient samples
Primary cells are routinely referred to our center for BPDCN suspicion with cytologic and phenotypic arguments in peripheral blood (PB) or bone marrow (BM) (collection DC 2016-27 91). Some cases do not meet the criteria for a BPDCN diagnosis, based on the World Health Organization classification 2017.5,8 Especially, 15 cases were collected based on the association of CD34+ blasts of myeloid origin and pDC (Table 1). All cases were analyzed at diagnosis, except for N35 (day 81 post-induction). The analysis was performed on BM aspiration (n=12) or PB (n=3). BM biopsies were rarely available, preventing from anatomopathology analysis. Fifteen normal BM aspiration performed for peripheral thrombopenia/research of metastatic infiltration, ten PB from healthy donors and 21 previously described cases of BPDCN20 were used as controls after written informed consent. This study was approved by the Besançon Ethics Committee (CPP-Est II, Besançon, France).20
Immunophenotype
Flow cytometry was performed using a FACSCanto II cytometer (BD Biosciences, San Jose, CA, USA) with DIVA 6.2 software (BD Biosciences) after cell labeling with monoclonal antibodies (Online Supplementary Table S1). The mean fluorescence intensity ratio (MFIR) of cTCL1 was obtained by dividing the mean fluorescence intensity (MFI) for cTCL1 by that of the isotype control monoclonal antibody (mAb). Cells were considered positive for cTCL1 expression when MFIR was greater than 2. Isotype control was not used for MFIR of CD123 because of its high expression on pDC; thus MFIR was calculated by dividing the MFI of CD123 on pDC by that on lymphocytes.
Cell sorting
One to 10 million cells were sorted using an ARIA III FACS (Becton Dickinson Biosciences) after cell labeling with mAb (Online Supplementary Table S1) in order to select the populations of interest: T-cells (CD45+high, CD3+), immature blasts (CD34+, CD303–), pDC (CD123+high, CD303+), monocytes (CD14+ or CD64+, CD123+low, CD303–) and cDC (CD34-, CD1c+, CD303–).
Molecular biology
Whole genome amplification was carried out using the REPLI-g® Single Cell kit (Qiagen Hilden, Germany), as recommended by the manufacturer. Next-generation sequencing (NGS) was performed, from a HaloPlexHS Target Enrichment System (Agilent Technologies Inc., Santa Clara, CA, USA) targeting 70 genes (Online Supplementary Table S2), in paired-end, 2x150 cycles on a MiSeq platform (Illumina Inc., San Diego, CA, USA).
Bioinformatics analysis
Raw data were analyzed using in-house bioinformatics pipelines (Online Supplementary Methods) with annotation of the variants via GenerateReportsTM25 yielding variant call files. Finally, several filters were applied to eliminate intronic regions, synonymous mutations and polymorphisms.
Statistical analyses
Statistical analyses were performed using Prism 6.0 software (GraphPad, San Diego, CA, USA). The distribution of MFIR was studied using the D’Agostino-Pearson normality test. Quantitative data were compared using ANOVA. Patient groups were compared using the Mann-Whitney non-parametric test for quantitative variables with a non-Gaussian distribution, the Student t-test for normally distributed variables or the 2 test with Yates’ continuity correction for categorical variables. Correlations between quantitative variables were investigated by linear regression analyses. All statistical tests were two-sided, with a 5% alpha risk. Results are expressed as median (range). For further details see the Online Supplementary Appendix.
Table 1.
Clinical and biological features of the cohort.
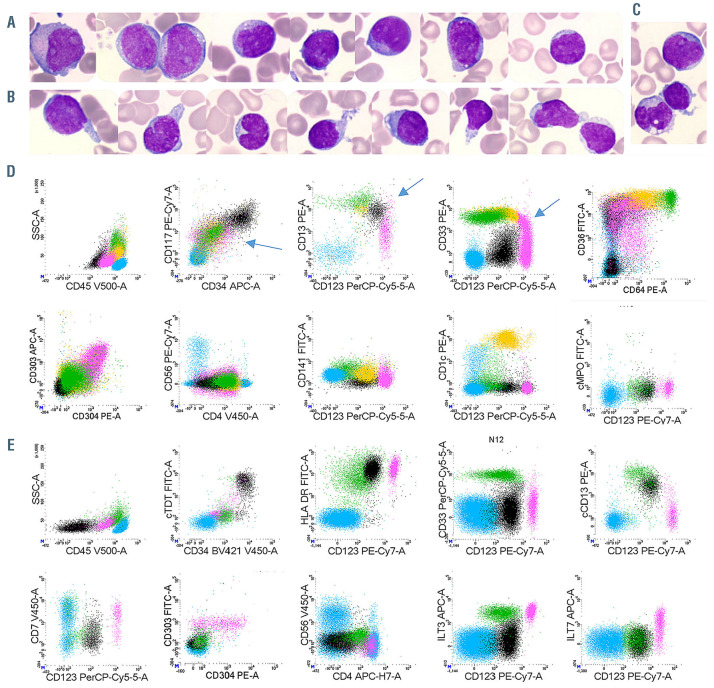
Figure 1.
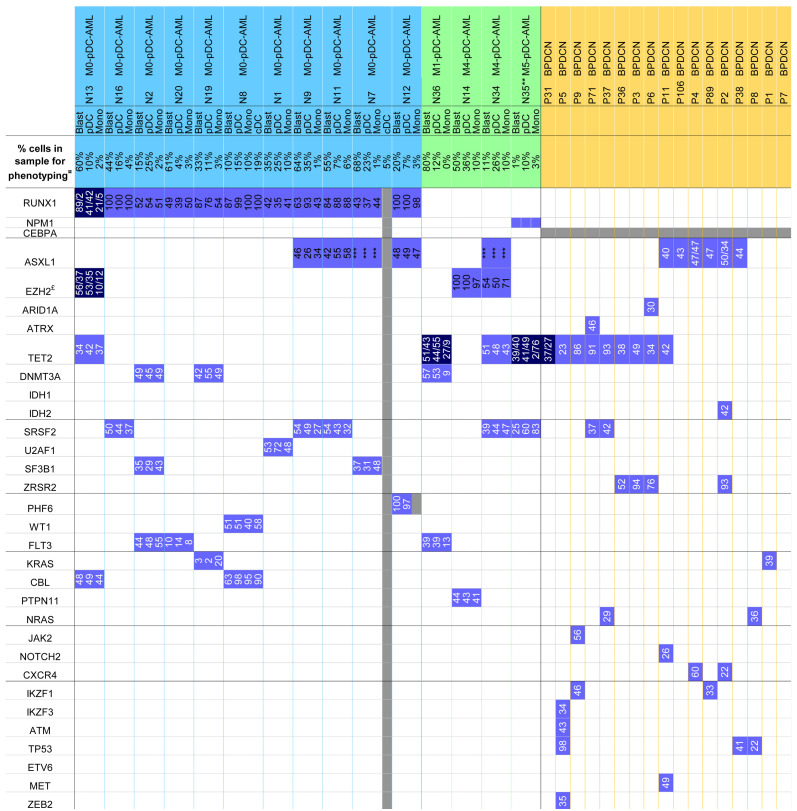
Morphologies and immunophenotypes of populations of interest. (A to C) Representative morphologic aspects of peripheral blood smears from patient N1 (magnification 1,000X). (A) Blast cells are medium sized with a high nuclear cytoplasmic ratio, fine chromatin with proeminent nucleoli. Cytoplasm is basophilic with some rare azurophilic granulations. (B) Plasmacytoid dendritic cells (pDC) are smaller with more mature chromatin. The cytoplasm is less basophilic without granulation but sometimes pseudopodia and small vacuoles under the cytoplasmic membrane. (C) A blast cell (top), a pDC with pseudopodia (center) and a monocyte (bottom). (D to E) Representative immunophenotype after gating on FSC-A vs. FSC-H plus sides catter (SSC) vs. forward scatter (FSC) to select singlets, leucocytes and exclude debris (not shown). Lymphocytes in blue (CD45bright/SSCdim cells); immature blasts in black (CD34+ cells); pDC in pink (CD123bright); monocytes in green (CD123dim, CD33bright, CD64bright); cDC in orange (CD123+, CD33+ CD64dim). (D) Patient N8: acute myeloid leukemia with minimal differentiation (M0-AML) with a continuum of phenotypic acquisition of markers (arrows) from the immature blasts: downregulation of CD13 and CD33 to pDC or upregulation to monocytes and cDC; downregulation of CD117 and CD34 to pDC/monocytes/cDC. (E) Patient N12: M0-AML with Tdt+, HLA-DRbright, CD33-, cCD13+ immature blasts; CD7+ CD4+ CD56– pDC.
Results
Patients
Fifteen patients were included, mainly elderly men with a median age of 70 years (range, 52-87) and a sex ratio of 4:1 (Table 1). Cytological analysis of BM aspiration, associated with phenotypic data, found 11 cases of AML with minimal differentiation (M0-AML in French-American- British [FAB] classification) with a prior history of cytopenia in one of them; one AML without maturation (AML- 1); two acute myelomonocytic leukemia (M4-AML) with a prior history of CMML; and one M5-AML secondary to another myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) analyzed during progression under induction treatment. On BM aspiration, blast cells were observed in all cases, associated with pDC, described as smaller with a more mature chromatin, faint basophilic cytoplasm without granulation, sometimes small vacuoles and short pseudopodia (Figure 1A to C). Lymph node involvement was reported in one patient, and skin lesions in four other patients (26.7%). Pathology analysis of skin lesions found myeloblast infiltration without pDC in patients N2 and N8, and pDC proliferation in patients N9 and N11. No clonal aberration was detected on karyotype for nine cases, while three others had trisomy 13 (N1, N9 and N11), two had chromosome 7 defects (N14, N16), and one had Y loss (N20). Two patients with normal karyotype were rearranged for KMT2A (N8, N35), and a deletion of EZH2 was detected by fluorescence in situ hybridization (FISH) in patient N16.
Immunophenotype identified different subpopulations in patients
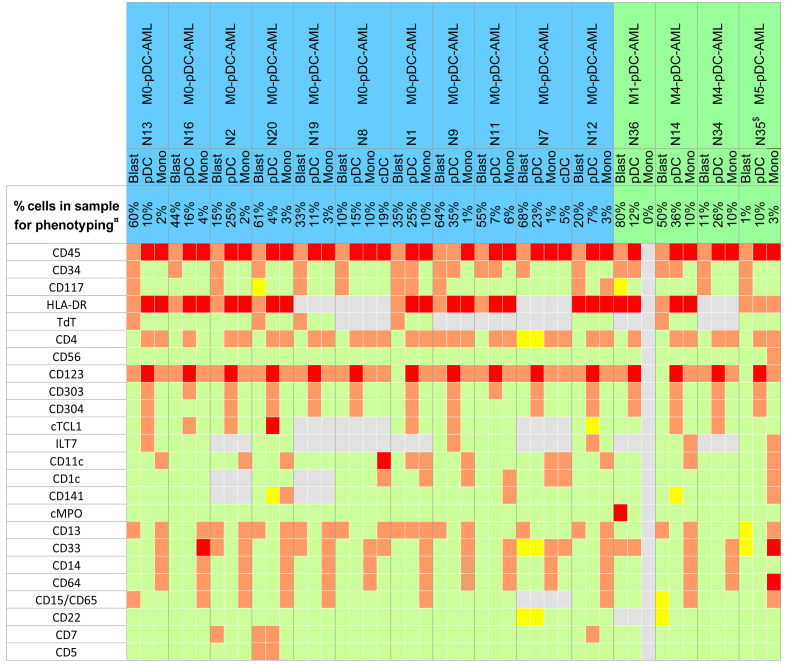
In contrast to BPDCN, cells of interest were heterogeneous, with a significant population of CD45low CD34+ immature myeloid blasts (44%; range, 10-80%) without markers of pDC commitment on the one hand, and an excess of CD4+ CD123+ HLA-DR+ cTCL1+ CD303+ pDC (15%; range, 4-36%) on the other hand. Consequently, these cases more likely fit the description of pDC-AML (or MPDCP associated with AML according to the WHO classification) 8 where pDC were in excess, greater than 4% (median 15%) in our cohort. Indeed, a median of 0.25% of total nucleated cells was detected for the pDC contingent in 15 normal BM aspiration and 0.24% in PB, with similar ranges of 0.02-0.95% and 0.17-0.53%, respectively. For 11 of 15 AML cases (73%), CD45low CD34+ immature blasts were more frequent than pDC, whereas pDC were preponderant in the remaining four cases (range, 10-26%). Monocytes (3%; range 1-10%) were also found in 14 cases, greater in acute myelomonocytic leukemia (M4- AML) than in other cases. Interestingly, cDC were detected in patients N7 and N8 (4.8% and 19% of cells respectively) (Figure 2).
The CD45low immature blasts were CD34+ (15 of 15, 100% of cases), CD117+ (11 of 15; 73%), TdT+ (five of nine; 56%), expressed myeloid markers such as CD13 (11 of 15, 73%) (Figure 1D and E) and/or CD33 (five of 15, 33%); and less frequently CD15/65 (two cases) or myeloperoxidase (case case). Monocyte lineage markers were only expressed on blasts of M4/5-AML patients. T-cell markers were expressed in two cases (CD7 +/- CD5: patients N2 and N20) and the B-cell marker CD22 in two other cases (N7 and N14) which was not sufficient to meet the definition of mixed-phenotype acute leukemia, as CD3/cCD3 or CD19/cCD79a were not expressed (Figure 2). CD123 was expressed in 12 cases of 15 (80%) at low level (MFIR: 15.58 [range, 2.5-63.1], MFI: 2,309 [range 465-8,674]). In two cases, blasts expressed CD4, whereas CD56 was never expressed. The blastic population did not express markers strongly associated with the pDC (cTCL1, CD303 and CD304) or cDC (CD1c, CD11c) lineages.
Figure 2.
Immunophenotypic features of plasmacytoid dendritic cell-acute myeloid leukemia. Positive high on 100% of cells in red (>10x4), positive on 100% of cells in orange (10x2 to 10x4), partially positive in light yellow, negative (<20%) in green, not done in grey. CD15 and CD65 are both labeled by fluorescein isothiocyanate (FITC) in the same tube of our panel. ¤Percentage of cells corresponds to flow cytometry, quantification on the sample used for phenotyping, possibly diluted by peripheral blood. All cases exhibited more than 20% of blasts on bone marrow smears. $Analyses performed on sample obtained after induction of chemotherapy, 70 days after diagnosis. cCD3, CD3, CD19, cCD79a were negative for all cases and all fractions. pDC: plasmacytoid dendritic cells; cDC: conventional DC; Mono: monocytes; AML: acute myeloid leukemia; M0-AML: AML with minimal differentiation.
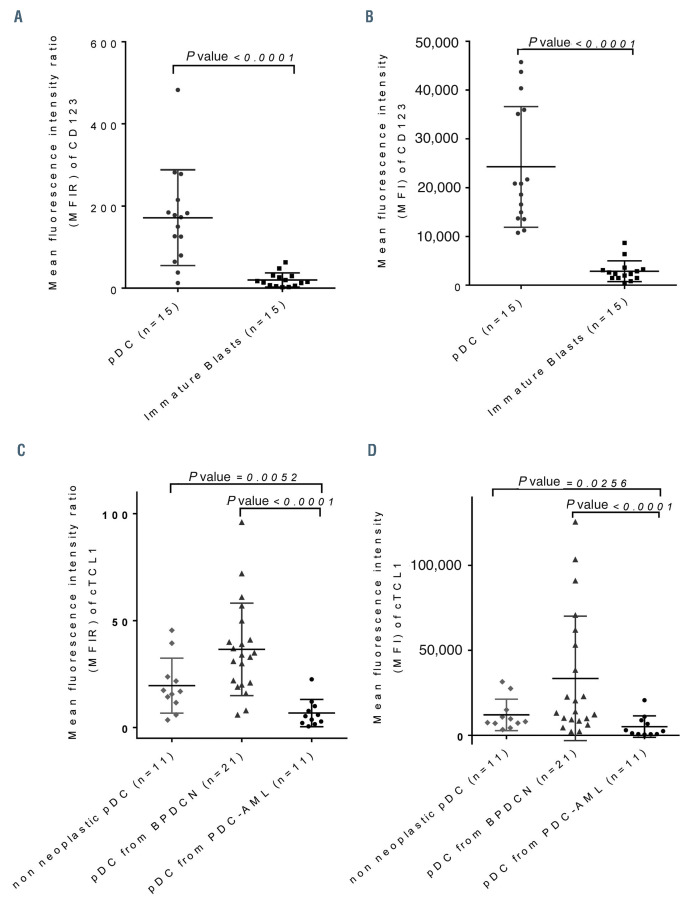
Figure 3.
Expression of CD123 and cTCL1 on plasmacytoid dendritic cells from plasmacytoid dendritic cell-acute myeloid leukemia. (A) Comparison of mean fluorescence intensity ratio (MFIR) of CD123 between plasmacytoid dendritic cells (pDC) and immature CD34+ blasts in pDC-acute myeloid leukemia (pDC-AML). (B) Comparison of MFI of CD123 between pDC and immature CD34+ blasts in pDC-AML. (C) Comparison of MFIR of cTCL1 between pDC from pDC-AML, blastic pDC neoplasms (BPDCN) and non-neoplastic pDC from healthy donors. (D) Comparison of MFI of cTCL1 between pDC from pDC-AML, BPDCN and non-neoplastic pDC from healthy donors. P-values (unpaired Mann-Whitney test) are marked above.
Associated pDC were constantly CD123+high, CD4+/+low and CD303+ (100%), with CD304 expression in most cases (13 of 15, 87%) and none of them expressed CD56. The immaturity marker CD34 was expressed in 33% of cases, whereas TdT in only one case out of nine tested. The CD123 expression level was significantly higher on pDC than on the immature CD34+ blasts (MFIR: 172.9 [range, 12.5-482.6] vs. 15.58 [range, 2.5-63.1] on CD34+ blasts, P<0.0001; MFI: 20,836 [range, 10,755-45,746] vs. 2,309 [range, 465-8,674] on CD34+ blasts, P<0.0001) (Figure 3A and B). cTCL1 was expressed in pDC (nine of 12 cases, 75%) at a statistically lower level (MFIR: 5.3 [range, 0.6-22.6]; MFI: 2,473 [range, 487-20,684]) than in the 21 BPDCN cases (MFIR: 34 [range, 6.0-96.0], P<0.0001; MFI: 13,990 [range, 2,186-125,568], P=0.0006) and even than in normal pDC (MFIR: 16.9 [range, 3.5-45.5], P=0.0052; MFI: 8,106 [range, 3,389-3,1544], P=0.0192) (Figure 3C and D).
Figure 4.
Mutation profile of plasmacytoid dendritic cell-acute myeloid leukemia. Mutations detected by next-generation sequencing with variant allele frequencies (VAF), or Sanger sequencing (especially for ASXL1). Abnormalities in plasmacytoid dendritic cells-acute myeloid leukemia (pDC-AML) and blastic pDC neoplasms (BPDCN) are depicted in: bright blue (monoallelic mutation); dark blue (biallelic mutation); white (absence of mutation); grey (not available). ¤Percentage of cells corresponds to flow cytometry, quantification on the sample used for phenotyping, possibly diluted by peripheral blood (all cases exhibited more than 20% of blasts on bone marrow smears). $Analyses performed on sample obtained after induction of chemotherapy, 70 days after diagnosis. £Fluorescence in situ hybridization (FISH) 7q36 on case N16: loss of EZH2 in 91 of 200 nuclei. ***VAF not available because ASXL1 c.1934dupG;p.Gly646TrpfsX12 was confirmed by Sanger sequencing. Mono: monocytes.
Interestingly, four patients with M0-AML expressed myeloid, B-cell or T-cell markers on both CD34+ blasts and pDC, but with partial and lower expression on pDC than on CD34+ blasts: CD33 and CD22 for N7, CD13 for N1, CD33 for N36, CD5 and CD7 for N20 (Figure 2). A maturation continuum between immature blasts and pDC was observed in some cases (Figure 1D).
cDC were identified by the expression of CD1c and CD11c without CD14 and CD64 expression in patients N7 and N8, and were sorted for molecular studies for patient N8 only.
Mutational profile
The purity of each sorted fraction is depicted in Online Supplementary Table S3. The NGS panel was informative for all cases, with a number of detected mutations ranging from two to six. Fourteen genes were found to be mutated among the 70 explored (Figure 4), corresponding to transcription factors RUNX1 (11 of 15, 73%); epigenetic modifiers ASXL1 (five of 15, 33%), EZH2 (three of 15, 20%), TET2 (four of 15, 27%), DNMT3A (three of 15, 20%); genes involved in splicing SRSF2 (five of 15, 33%), SF3B1 (two of 15, 13%), U2AF1 (one of 15); RAS pathway CBL (three of 15, 19%), KRAS, PTPN11 (one of 15 each); cytokine signaling FLT3 (three of 15, 19%) as well as other genes, such as PFH6 and WT1 (one of 15 each) (Online Supplementary Table S4). Of note, only one case was mutated for NPM1 (patient N35) and none for CEBPA. In BPDCN, 17 out of the 21 of the cohort were studied by NGS, with zero to five mutations per case on 17 genes: TET2 (nine of 17, 53%), ASXL1 (six of 17, 41%), ZRSR2 (four of 17, 24%), TP53 (three of 17, 18%), IKZF1, NRAS, SRSF2, IDH1 (two of 17 each, 12%), ZEB2, MET, ETV6, ATM, IKZF3, CXCR4, NOTCH2, KRAS, JAK2 (one of 17 each) (Figure 4).
Mutations were systematically found in sorted CD34+ immature blasts, pDC, monocyte and cDC of the same sample, and were not detected in the T-cell fraction, thus confirmed to be a non-neoplastic subpopulation (Figure 4). Variant allele frequencies (VAF) were quite similar between cell fractions. However, VAF of the monocyte subpopulation were lower than in blasts and pDC in two cases (N13 and N36), which may indicate that this mutation is subclonal in monocytes (N13) or that there is a mixture of neoplastic and reactive non-neoplastic monocytes (same VAF difference in all detected mutations for N36). Some mutations were subclonal in both blast and monocyte fractions: KRAS for N19, FLT3 for N20 (Figure 4). Overall, the most frequently mutated genes were the transcription factor RUNX1, splicing genes (SRSF2, SF3B1, U2AF1) and epigenetic modifiers DNMT3A and TET2. Interestingly, RUNX1 mutations concerned all M0-AML, while none of the other cases were mutated (M4/5-AML and M1-AML). Consequently, despite the low number of cases, there was a significant association between the M0- AML subtype and RUNX1 mutations (2 with Yates' correction, X2=10.32, 1df, P=0.0013). The majority of RUNX1 mutations detected were frameshift (n=6) or stop gain (n=3), with a biallelic invalidation in patient N13. RUNX1-mutated cases involved mainly males (M/F=8/3), with a median age of 70 years (range, 55-87 years).
Figure 5.
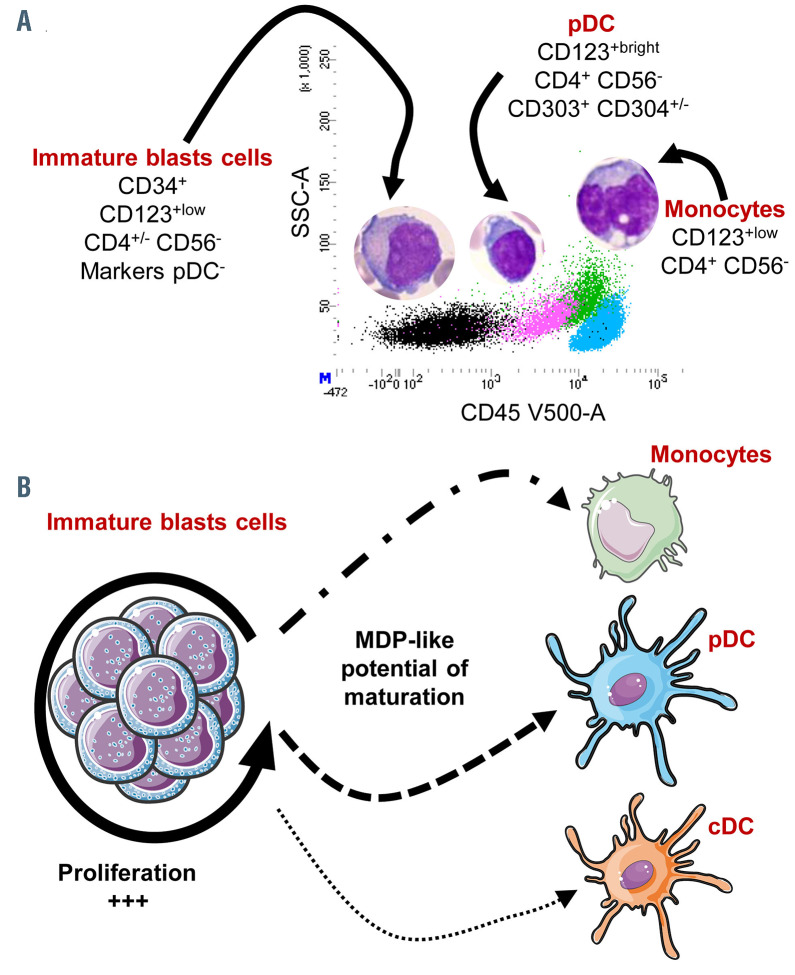
Maturation model in plasmacytoid dendritic cell-acute myeloid leukemia. (A) Representative CD45/SSC dot plot of plasmacytoid dendritic cells-acute myeloid leukemia (pDC-AML), with four populations identified: immature CD34+ blasts in black, pDC in pink, monocytes in green and lymphocyte in blue, with morphologies of these populations depicted above. (B) The maturation model: immature blast cells are mainly proliferative without maturation, but at least part of them conserved MDP (macrophage-DC progenitor)-like potential of maturation leading to variable amounts of clonal pDC, monocytes and conventional DC (cDC).
Discussion
The World Health Organization 2017 classification clearly recognizes BPDCN as a form of pDC neoplasm, while MPDCP is still insufficiently described and probably underdiagnosed8. MPDCP mainly concerns CMML and more rarely MDS or AML. Our study focused on pDC-AML, using a phenotypic and mutation characterization on sorted population of 15 cases. Our gateway for the inclusion of cases is the pDC-like morphology of some blastic cells detected by cytologists in French hospitals. This point precludes us from determining the prevalence of pDC-AML among AML, due to a recruitment bias. We highlight in this study an excess of pDC in a context of AML in order to differentiate them from BPDCN and to precisely dtermine their molecular profile, not well-described so far. The clinical presentation of our cohort was close to that of BPDCN, with elderly patients and a clear male predominance, but skin lesions were less frequent (25% of cases vs. 90% in BPDCN), linked to either pDC (two cases) or myeloblast (two cases) infiltration. The excess of pDC in BM aspiration of pDC-AML was well over the pDC infiltrate in healthy donor BM and PB, considered to be below 1% of total nucleated cells24,26,27 and confirmed by our data. Moreover, pDC are frequently gathered on BM smears, compatible with aggregates and in the only case of BM biopsy, clustered nodules of pDC were present, as reported by pathologists with islands of pDC in MPDCP.13,21,24 In the two cases with prior history of CMML, pDC were not in high excess prior transformation on BM smears. However, in the absence of available sample, flow cytometry assays were not assessed at the CMML stage.
Blasts exhibited an immature phenotype with expression of CD34, CD117 and TdT, which distinguishes these cases from BPDCN. They also frequently express myeloid markers, and the expression of CD123 is definitely weaker than pDC from the same samples or pDC blasts from BPDCN. Of note, strong pDC or cDC lineage markers (CD303, CD304, cTCL1, ILT7 and CD1c, CD141, CD11c) were never expressed on blasts. Focusing on the 11 M0- AML cases, blasts are immature with markers of commitment to the myeloid lineage (CD13, CD33 without MPO) plus expression of lymphoid markers in some cases; this could be a sign of an original progenitor origin or oncogenic dysregulation in pDC-AML compared to other AML.
Only four cases of pDC-AML showed higher infiltration of pDC than blasts: three at diagnosis (N2, N8 and N34) and the fourth at day 81 post-induction (N35). In these 15 cases, the phenotype of pDC is different from pDC in healthy donors and in BPDCN. CD56, is not expressed in our 15 cases, similarly to physiological mature pDC, only CD56+ when stimulated by Flt3L during maturation.28,29 Conversely CD56 is almost systematically expressed in BPDCN5,30,31 except for rare cases.7,32,33 Moreover, the pDC in pDC-AML always expressed CD303 (100%) while 20 to 30% of BPDCN are CD303-6,31 and BPDCN express a lower intensity of CD303 than pDC of pDCAML (data not shown); cTCL-1 expression was substantially lower on pDC of pDC-AML compared to pDC of BPDCN and even pDC of healthy donors (Figure 3B). Finally, CD34 is sometimes expressed on pDC (33% of cases), whereas CD34 is almost never expressed on BPDCN blasts. This phenotype is closer to normal pDC in some respects (CD56–, CD303+, cTCL1+low). pDC lineage maturation is divided in three stages with progressive acquisition of CD303 and CD304 plus downregulation of CD34 and CD117 and gradual loss of CD13, CD33 or CD22 expression34. In our series, the expression of CD34 (six cases) and CD13, CD33 or CD22 (three cases) makes these pDC closer to the intermediate stage of pDC maturation (CD34+, CD303+, CD304+/-, CD123+). Moreover, aberrant markers of myeloid or lymphoid lineage (CD13, CD33, CD11b, CD22, CD7, CD5) on both blasts and pDC in the same patient support the idea of a common origin of these populations, with maturation of CD34+ blasts towards the pDC lineage by downregulation of CD34 and upregulation of CD123, CD303 and CD304. Reinforcing this hypothesis, we clearly show a maturation continuum between blasts and pDC or monocytes (Figure 1D) with CD34 downregulation and upregulation of CD45, pDC markers (CD123, CD304, CD303) on the pDC cells or CD14, CD64 on monocytes, as recently highlighted. 14,15 Another study also described similar cases of leukemia associating immature CD34+ myeloid blasts and CD34+/- CD56– pDC, but considered these cases as an immature subgroup of BPDCN.35 Altogether, a variety of teams described similar cases under different denominations: MPDCP with myeloid neoplasm (MN), pDC proliferations associated with MN, AML/MN with PDC differentiation, AML/MDS-pDC, leukemia of ambiguous lineage, or immature group 1 of BPDCN.14–17,22,23,35,36 There is a need to refine this poorly defined MPDCP, where the “mature” denomination should be omitted because it only refers to the absence of the blastic morphology of BPDCN.8
Our data and others highlight that pDC are present at all stages of maturation in pDC-AML, even CD34+, because they keep their potential for maturation14,15 as already described in pDC-CMML.24 In addition, pDC-AML appear to be quite similar to pPDC-CMML, as clonal monocytes are also frequently detected in our cohort (14 of 15 AML). In this regard, all cases of MPDCP described so far fall within our definition of pDCAML/ CMML/MDS. Interestingly, the expression of CD123 on both blasts and pDC opens up the potential to use new therapies targeting CD123, such as tagraxofusp, IMGN632 or CD123 CAR-T cells.37-40 Crucially, these pDC-AML may respond differently from other AML or BPDCN.
Very few molecular data have been obtained to assert the clonal link between pDC and leukemic cells in CMML and AML.21-23 In our series, using sorted cell populations, we show that the same mutations are shared by blasts, pDC, monocytes and even cDC, thus confirming the neoplastic origin of pDC and their shared clonal origin with blasts, monocytes and cDC (for one tested case). Of note, no unique mutation was identified in pDC but not in the blast compartment, or vice versa, albeit with the limitation that only 70 genes were tested. Thus, no specific mutation arising during the lineage commitment was highlighted. Blasts seem to be broadly blocked in an immature undifferentiated stage in pDC-AML compared to pDC-CMML, and maintain their ability to mature towards pDC, monocytes or even cDC (Figure 5A and B). The immature CD34+ blasts could be a proliferation, at the very least, of a bipotent progenitor with DC (pDC and cDC) and with monocyte potential such as the MDP. The expression of CD22, CD2 and CD5 on pDC and blasts in some cases could also suggest that they derive from a granulocytemonocyte- lymphoid progenitor or from AXL+ SIGLEC6+ DC, recently identified.41
The mutation status obtained does not highlight specific genes for all cases. Many of them are frequently mutated in other myeloid malignancies, involved in epigenetics (ASXL1 and TET2), splicing (SRSF2, SF3B1, U2AF1) or the RAS pathway (CBL, KRAS, PTPN11). This mutation profile is only partially similar to BPDCN, because BPDCN can also be mutated for ETV6, TP53, ZEB2, MET, ATM, IKZF3, JAK, NOTCH2 and CXCR4, plus TET2 with a high frequency (Figure 4). Cases classified as M4/5-AML were always transformations of MDS/MPN and had a similar molecular profile including ASXL1, TET2, SRSF2, CBL or PTPN11 mutations, sometimes with additional mutations responsible for acute transformation (NPM1 mutation for N35). Remarkably, RUNX1 is the most frequently mutated gene in pDC-AML (73% of cases), as already described.17 Moreover, it only concerned M0-AML cases in our study and all cases of M0-AML exhibited this mutation (100%). This point is particularly puzzling considering that this prevalence is markedly different from the described epidemiology of 20-30% of RUNX1 mutations in M0- AML42,43 and knowing that RUNX1 mutation has only been reported once in BPDCN.44 Although, this study only analyzed a small number of cases and the recruitment bias precludes us from determining the frequency of M0-AML and RUNX1 mutations in pDC-AML. Our 11 PDCP-M0- AML cases were 71 years old on average, with a male/female sex ratio of 2.67, consistent with the RUNX1-mutated AML provisional entity.8,42 Unfortunately, given the advanced age of patients with palliative care and low number of cases, prognostic conclusions are impossible. RUNX1 mutations were not detected in the non-neoplastic T-cell fraction, demonstrating that these mutations are somatic. WGA prevented us from definitely obtaining copy number variations, but VAF were higher than 50% for six patients (N8, N9, N11, N12, N16, N19) and a seventh (N13) was double mutated for RUNX1, suggesting secondary alteration of RUNX1, as already described.45
Notably, inhibition of RUNX1 is considered to increase RUNX2 and RUNX3 protein levels, following a complementary compensation mechanism that maintains the entire RUNX family at a constant level.46 Then, RUNX1 invalidation could promote a RUNX2 switch and then a pDC commitment because RUNX2 plays a crucial role in pDC diffentiation.47,48 Further experiments are nevertheless required to confirm this hypothesis.
To conclude, our study identifies a group of pDC-AML requiring a differential diagnosis with BPDCN. They are characterized by an immature myeloid population (CD34+, CD117+/- CD123+low without expression of pDC markers) associated with an excess of pDC (CD123+high, CD4+) that clearly differ from BPDCN neoplastic cells by no expression of CD56, the possible expression of CD34, higher expression of CD303 and lower expression of cTCL1. Molecular data show that these pDC are neoplastic and not reactive, and the mutational landscape of pDCAML appears distinct from BPDCN, notably with frequent RUNX1 mutations. Moreover, a continuous maturation pattern suggests that these pDC as well as monocytes could arise from the immature CD34+ blasts, potentially with MDP properties (Figure 5B). Finally, this study addresses (i) the frequency with which RUNX1-mutated AML are associated with an excess of pDC, (ii) the type of progenitors involved and (iii) its prognostic or therapeutic impact. These questions warrant investigation in an independent and larger cohort of AML.
Supplementary Material
Acknowledgments
The authors would like to thank Hugues Faucheu for helping on the design panel, Véronique Yerly-Motta for helping on the NGS platform and Fiona Ecarnot for English proofreading.
Funding Statement
Funding: This work was supported by Ligue régionale contre le Cancer (CCIRGE-BFC-2016), Fondation ARC (Aides Individuelles DOC20170505805) and Association Laurette Fugain (ALF 2018/08).
References
- 1.Banchereau J, Briere F, Caux C, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18767-811. [DOI] [PubMed] [Google Scholar]
- 2.Jegalian AG, Facchetti F, Jaffe ES. Plasmacytoid dendritic cells: physiologic roles and pathologic states. Adv Anat Pathol. 2009;16(6):392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naik SH, Sathe P, Park H-Y, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8(11):1217-1226. [DOI] [PubMed] [Google Scholar]
- 4.Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007; 8(11):1207-1216. [DOI] [PubMed] [Google Scholar]
- 5.Chaperot L, Bendriss N, Manches O, et al. Identification of a leukemic counterpart of the plasmacytoid dendritic cells. Blood. 2001;97(10):3210-3217. [DOI] [PubMed] [Google Scholar]
- 6.Garnache-Ottou F, Feuillard J, Ferrand C, et al. Extended diagnostic criteria for plasmacytoid dendritic cell leukaemia. Br J Haematol. 2009;145(5):624-636. [DOI] [PubMed] [Google Scholar]
- 7.Facchetti F, Cigognetti M, Fisogni S, Rossi G, Lonardi S, Vermi W. Neoplasms derived from plasmacytoid dendritic cells. Mod Pathol. 2016;29(2):98-111. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th Edition. Lyon, France: International Agency for Research on Cancer; 2017. [Google Scholar]
- 9.Petrella T, Comeau MR, Maynadié M, et al. “Agranular CD4+ CD56+ hematodermic neoplasm” (blastic NK-cell lymphoma) originates from a population of CD56+ precursor cells related to plasmacytoid monocytes. Am J Surg Pathol. 2002;26(7):852-862. [DOI] [PubMed] [Google Scholar]
- 10.Petrella T, Facchetti F. Tumoral aspects of plasmacytoid dendritic cells: what do we know in 2009? Autoimmunity. 2010; 43(3):210-214. [DOI] [PubMed] [Google Scholar]
- 11.Vitte F, Fabiani B, Bénet C, et al. Specific skin lesions in chronic myelomonocytic leukemia: a spectrum of myelomonocytic and dendritic cell proliferations: a study of 42 cases. Am J Surg Pathol. 2012;36(9):1302-1316. [DOI] [PubMed] [Google Scholar]
- 12.Dargent J-L, Delannoy A, Pieron P, Husson B, Debecker C, Petrella T. Cutaneous accumulation of plasmacytoid dendritic cells associated with acute myeloid leukemia: a rare condition distinct from blastic plasmacytoid dendritic cell neoplasm. J Cutan Pathol. 2011;38(11):893-898. [DOI] [PubMed] [Google Scholar]
- 13.Orazi A, Chiu R, O’Malley DP, et al. Chronic myelomonocytic leukemia: the role of bone marrow biopsy immunohistology. Mod Pathol. 2006;19(12):1536-1545. [DOI] [PubMed] [Google Scholar]
- 14.Hamadeh F, Awadallah A, Meyerson HJ, Beck RC. Flow cytometry identifies a spectrum of maturation in myeloid neoplasms having plasmacytoid dendritic cell differentiation. Cytometry B Clin Cytom. 2020; 98(1):43-51. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Wang Y, Chang Y, et al. Myeloid neoplasms with elevated plasmacytoid dendritic cell differentiation reflect the maturation process of dendritic cells. Cytom A. 2020;97(1):61-69. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Feng Y, Deng X, et al. Tumor-forming plasmacytoid dendritic cells in acute myelocytic leukemia: a report of three cases and literature review. Int J Clin Exp Pathol. 2017;10(7):7285-7291. [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao W, Goldberg AD, Famulare C, et al. Acute myeloid leukemia with plasmacytoid dendritic cell differentiation: predominantly Secondary AML, enriched for RUNX1 mutations, frequent cross-lineage antigen expression and poor prognosis. Blood. 2018;132(Suppl 1):S2789. [Google Scholar]
- 18.Bodmer A, Menter T, Juskevicius D, et al. Sharing of a PTPN11 mutation by myelodysplastic bone marrow and a mature plasmacytoid dendritic cell proliferation provides evidence for their common myelomonocytic origin. Virchows Arch. 2017;470(4):469-473. [DOI] [PubMed] [Google Scholar]
- 19.Horny HP, Feller AC, Horst HA, Lennert K. Immunocytology of plasmacytoid T cells: marker analysis indicates a unique phenotype of this enigmatic cell. Hum Pathol. 1987;18(1):28-32. [DOI] [PubMed] [Google Scholar]
- 20.Angelot-Delettre F, Biichle S, Ferrand C, et al. Intracytoplasmic detection of TCL1--but not ILT7-by flow cytometry is useful for blastic plasmacytoid dendritic cell leukemia diagnosis. Cytom A. 2012;81(8):718-724. [DOI] [PubMed] [Google Scholar]
- 21.Vermi W, Facchetti F, Rosati S, et al. Nodal and extranodal tumor-forming accumulation of plasmacytoid monocytes/interferon-producing cells associated with myeloid disorders. Am J Surg Pathol. 2004; 28(5):585-595. [DOI] [PubMed] [Google Scholar]
- 22.Rickmann M, Krauter J, Stamer K, et al. Elevated frequencies of leukemic myeloid and plasmacytoid dendritic cells in acute myeloid leukemia with the FLT3 internal tandem duplication. Ann Hematol. 2011; 90(9):1047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohty M, Jarrossay D, Lafage-Pochitaloff M, et al. Circulating blood dendritic cells from myeloid leukemia patients display quantitative and cytogenetic abnormalities as well as functional impairment. Blood. 2001;98(13):3750-3756. [DOI] [PubMed] [Google Scholar]
- 24.Lucas N, Duchmann M, Rameau P, et al. Biology and prognostic impact of clonal plasmacytoid dendritic cells in chronic myelomonocytic leukemia. Leukemia. 2019;33(10):2466-2480. [DOI] [PubMed] [Google Scholar]
- 25.Viailly P-J, Mareschal S, Bertrand P, et al. GenerateReports: an Ion Torrent plugin summarizing a whole NGS experiment for clinical interpretation. 2015. [Google Scholar]
- 26.Wang W, Khoury JD, Miranda RN, et al. Immunophenotypic characterization of reactive and neoplastic plasmacytoid dendritic cells permits establishment of a 10- color flow cytometric panel for initial workup and residual disease evaluation of blastic plasmacytoid dendritic cell neoplasm. Haematologica. 2021. Apr 1;106(4):1047-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao W, Goldberg AD, Famulare CA, et al. Loss of plasmacytoid dendritic cell differentiation is highly predictive for post-induction measurable residual disease and inferior outcomes in acute myeloid leukemia. Haematologica. 2019;104(7):1378-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comeau MR, Van der Vuurst de Vries A-R, Maliszewski CR, Galibert L. CD123bright plasmacytoid predendritic cells: progenitors undergoing cell fate conversion? J Immunol. 1950 2002;169(1):75-83. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald KPA, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DNJ. Characterization of human blood dendritic cell subsets. Blood. 2002;100(13):4512-4520. [DOI] [PubMed] [Google Scholar]
- 30.Feuillard J, Jacob M-C, Valensi F, et al. Clinical and biologic features of CD4(+)CD56(+) malignancies. Blood. 2002; 99(5):1556-1563. [DOI] [PubMed] [Google Scholar]
- 31.Garnache-Ottou F, Vidal C, Biichlé S, et al. How should we diagnose and treat blastic plasmacytoid dendritic cell neoplasm patients? Blood Adv. 2019;3(24):4238-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Julia F, Dalle S, Duru G, et al. Blastic plasmacytoid dendritic cell neoplasms: clinicoimmunohistochemical correlations in a series of 91 patients. Am J Surg Pathol. 2014;38(5):673-680. [DOI] [PubMed] [Google Scholar]
- 33.Lamar E, Roggy A, Le Calvez G, et al. Blastic plasmacytoid dendritic cell neoplasm: report of a case with atypical cytology and immunophenotype. J Blood Disord. 2015;2(3):1033. [Google Scholar]
- 34.Martín-Martín L, Almeida J, Hernández-Campo PM, Sánchez ML, Lécrevisse Q, Orfao A. Immunophenotypical, morphologic, and functional characterization of maturation- associated plasmacytoid dendritic cell subsets in normal adult human bone marrow. Transfusion. 2009; 49(8):1692-1708. [DOI] [PubMed] [Google Scholar]
- 35.Martín-Martín L, López A, Vidriales B, et al. Classification and clinical behavior of blastic plasmacytoid dendritic cell neoplasms according to their maturation-associated immunophenotypic profile. Oncotarget. 2015;6(22):19204-19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsagarakis NJ, Kentrou NA, Papadimitriou KA, et al. Acute lymphoplasmacytoid dendritic cell (DC2) leukemia: results from the Hellenic Dendritic Cell Leukemia Study Group. Leuk Res. 2010;34(4):438-446. [DOI] [PubMed] [Google Scholar]
- 37.Mani R, Goswami S, Gopalakrishnan B, et al. The interleukin-3 receptor CD123 targeted SL-401 mediates potent cytotoxic activity against CD34+CD123+ cells from acute myeloid leukemia/myelodysplastic syndrome patients and healthy donors. Haematologica. 2018;103(8):1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovtun Y, Jones GE, Adams S, et al. A CD123-targeting antibody-drug conjugate, IMGN632, designed to eradicate AML while sparing normal bone marrow cells. Blood Adv. 2018;2(8):848-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardiros A, Dos Santos C, McDonald T, et al. T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood. 2013;122(18):3138-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pemmaraju N, Lane AA, Sweet KL, et al. Tagraxofusp in blastic plasmacytoid dendritic- cell neoplasm. N Engl J Med. 2019; 380(17):1628-1637. [DOI] [PubMed] [Google Scholar]
- 41.Villani A-C, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017; 356(6335):eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaidzik VI, Teleanu V, Papaemmanuil E, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico- pathologic and genetic features. Leukemia. 2016;30(11):2282. [DOI] [PubMed] [Google Scholar]
- 43.Roumier C, Eclache V, Imbert M, et al. M0 AML, clinical and biologic features of the disease, including AML1 gene mutations: a report of 59 cases by the Groupe Français d’Hématologie Cellulaire (GFHC) and the Groupe Français de Cytogénétique Hématologique (GFCH). Blood. 2003; 101(4):1277-1283. [DOI] [PubMed] [Google Scholar]
- 44.Menezes J, Acquadro F, Wiseman M, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2014;28(4):823-829. [DOI] [PubMed] [Google Scholar]
- 45.Antony-Debré I, Duployez N, Bucci M, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2016; 30(4):999-1002. [DOI] [PubMed] [Google Scholar]
- 46.Morita K, Suzuki K, Maeda S, et al. Genetic regulation of the RUNX transcription factor family has antitumor effects. J Clin Invest. 2017;127(7):2815-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubota S, Tokunaga K, Umezu T, et al. Lineage-specific RUNX2 super-enhancer activates MYC and promotes the development of blastic plasmacytoid dendritic cell neoplasm. Nat Commun. 2019;10(1):1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chopin M, Preston SP, Lun ATL, et al. RUNX2 mediates plasmacytoid dendritic cell egress from the bone marrow and controls viral immunity. Cell Rep. 2016; 15(4):866-878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.