Abstract
Genomic studies of pediatric acute lymphoblastic leukemia (ALL) have shown remarkable heterogeneity in initial diagnosis, with multiple (sub)clones harboring lesions in relapse-associated genes. However, the clinical relevance of these subclonal alterations remains unclear. We assessed the clinical relevance and prognostic value of subclonal alterations in the relapse-associated genes IKZF1, CREBBP, KRAS, NRAS, PTPN11, TP53, NT5C2, and WHSC1 in 503 ALL cases. Using molecular inversion probe sequencing and breakpoint-spanning polymerase chain reaction analysis we reliably detected alterations with an allele frequency below 1%. We identified 660 genomic alterations in 285 diagnostic samples of which 495 (75%) were subclonal. RAS pathway mutations were common, particularly in minor subclones, and comparisons between RAS hotspot mutations revealed differences in their capacity to drive clonal expansion in ALL. We did not find an association of subclonal alterations with unfavorable outcome. Particularly for IKZF1, an established prognostic marker in ALL, all clonal but none of the subclonal alterations were preserved at relapse. We conclude that, for the genes tested, there is no basis to consider subclonal alterations detected at diagnosis for risk group stratification of ALL treatment.
Introduction
Improvements in the treatment of pediatric acute lymphoblastic leukemia (ALL) have resulted in high overall survival rates, now approaching 90%.1,2 Nevertheless, relapse still remains the most common cause of treatment failure and death in children with ALL, and better recognition of individuals at risk of developing relapse will likely aid further improvements in outcome. Recent studies describing the genomic landscape of relapsed ALL have shown that relapse often originates from a minor (sub)clone at diagnosis, at a cellular fraction often undetectable by routine diagnostic methods.3-5 These minor (sub)clones harbor genomic alterations acquired later during leukemia development, which could potentially contribute to clonal drift, but are unlikely to be essential for initiation of the primary disease. However, selective pressure of the upfront treatment may provide a competitive advantage to subclones that harbor alterations in cancer genes, enabling their selective survival, eventually leading to treatment failure. Both the number and clonal burden of the alterations in these genes are expected to be increased at the time of relapse, compared to initial diagnosis. Indeed, mutations in relapse-associated genes, such as those in the histone acetyltransferase (HAT) domain of the histone methyltransferase CREBBP, can often be traced back to minor subclones in the diagnostic sample.4,6,7
Genomic characterization of relapsed pediatric ALL has revealed multiple alterations that are enriched compared to diagnosis, including activating mutations in RAS pathway genes, HAT domain mutations in CREBBP and deletions or mutations in the B-cell transcription factor IKZF1.6-13 The presence of these aberrations at the time of diagnosis can be of potential prognostic relevance, as has been demonstrated extensively for IKZF1 in many different treatment protocols12,14-19 and can even lead to adjustments in stratification and treatment.14,20 However, it remains unclear whether mutations in relapse-associated genes when present in a minor subclone at initial diagnosis are also clinically relevant.
Subclonal mutations can be identified using deep targeted, next-generation sequencing techniques.21,22 Despite the sensitivity of these techniques, both amplification and sequencing can easily lead to errors that hamper the reliable detection of low-level mosaic mutations. We previously demonstrated that single molecule molecular inversion probes (smMIP), which use unique molecular identifiers to barcode each DNA copy, can correct for sequencing and amplification artefacts, resulting in a reliable detection of low-level mosaic mutations, down to a variant allele frequency of 0.4%.23
In this study we used the smMIP-based sequencing approach to perform deep targeted sequencing of seven relapse-associated genes in a cohort of 503 pediatric ALL samples taken at initial diagnosis, resulting in the detection of 141 clonal and 469 subclonal mutations. In addition, we performed real time quantitative polymerase chain reaction (PCR) to sensitively detect subclonal IKZF1 exon 4-7 deletions (del 4-7), which were found at a similar frequency as full-clonal deletions. Subsequently, we estimated their potential as drivers of clonal expansion and prognostic markers for relapse development.
Methods
In this study we analyzed two cohorts of diagnostic samples from B-cell precursor ALL patients treated according to the Dutch Childhood Oncology Group (DCOG) protocols DCOG-ALL9 (n=131)12,24 and DCOG-ALL10 (n=245) (Online Supplementary Table S1). Both cohorts were representative selections of the total studies12,24 (Online Supplementary Table S2). The median age at diagnosis of the patients in these cohorts was 4 and 5 years, and the median follow-up time, estimated with a reverse Kaplan-Meier method, was 138 and 104 months, respectively.25 Relapse occurred in 18% (24/131) and 11% (27/245) of the patients, while 0.7% (1/131) and 2.8% (7/245) died during the follow-up. DNA was isolated from mononuclear cells obtained from bone marrow or peripheral blood. The median blast percentage of the samples was 92% (Online Supplementary Table S3). To increase the number of patients for the comparisons between relapsed and non-relapsed cases, we used an extended cohort of diagnostic samples from 127 additional ALL patients treated according to the DCOG-ALL9 (n=76) or DCOG-ALL10 (n=51) protocols; this cohort was enriched for patients who had a relapse and also contained 55 patients with T-cell ALL. This latter cohort was not included in the survival analyses. In order to detect mutations preserved in major clones at relapse, we performed Sanger sequencing (73/171) or used previously published Ampliseq-based deep-sequencing data (98/171) to verify alterations observed at diagnosis.26 In accordance with the Declaration of Helsinki, written informed consent was obtained from all patients and/or their legal guardians before enrollment in the study, and the DCOG institutional review board approved the use of excess diagnostic material for this study (OC2017-024).
In order to accurately detect subclonal alterations in diagnostic samples, 166 smMIP were designed in CREBBP, PTPN11, NT5C2, WHSC1, TP53, KRAS and NRAS, seven genes that are frequently mutated in relapsed ALL (Online Supplementary Table S4, Online Supplementary Materials and Methods). IKZF1 and ERG deletion status was assessed using the multiplex ligation-dependent probe amplification assay (MLPA) SALSA P335 ALL-IKZF1 and P327 iAMP-ERG kits, respectively (MRC-Holland, the Netherlands), according to the manufacturer’s instructions and as described before.12,24 Additionally, IKZF1 4-7 deletions were assessed with Sanger sequencing and real-time quantitative PCR, using an IQ SYBR Green supermix (Biorad, USA). For detailed descriptions of the smMIP-based sequencing, IKZF1 deletion detection and data analysis, see the Online Supplementary Materials and Methods (Online Supplementary Figures S1 and S2, Online Supplementary Tables S4-S6).
To test continuous and categorical variables, the nonparametric Wilcoxon signed rank and Fisher exact tests were used, respectively (R packages ggpubr version 0.2 and stats version 3.5.1). Cumulative incidence of relapse (CIR) was estimated by employing a competing-risk model with death as a competing event.27 To assess the statistical difference between CIR, the Gray test28 was applied. To investigate the effect of prognostic factors on relapse, univariate and multivariate Cox proportional hazard regression models were estimated. Competing risk analysis was performed with the R packages cmprsk (version 2.2-7) and survminer (version 0.4.3). Univariate and multivariate Cox models were estimated using R package survival (version 3.1-12). Data were visualized using the R package ggplot2 (version 3.2.1) and cBioPortal MutationMapper.29,30
Results
A total of 503 diagnostic samples from children with ALL (Online Supplementary Table S1) was subjected to targeted deep sequencing of the relapse-associated genes TP53, CREBBP (HAT domain), KRAS, NRAS, PTPN11, NT5C2 and WHSC1 using smMIP, which contain random molecular tags to accurately detect low-level mosaic variants. 23 Each targeted region was covered with an average of 308 unique capture-based consensus reads (Figure 1, Online Supplementary Figure S1A, B), enabling the reliable detection of alterations with allele frequencies even below 1%. A total of 7,836 quality-filtered variants was detected, of which 610 were absent in public and private variant databases and were predicted as pathogenic. The allele frequency of these mutations ranged from 0.03-100% (Figure 2A, Online Supplementary Table S3). The majority of the mutations (473/610; 78%) was found in one of the three RAS pathway genes (KRAS, NRAS, PTPN11), of which 418 (88%) were known hotspot mutations.
In addition to sequencing the seven relapse-associated genes, we performed sensitive screening for IKZF1 deletions, which are strongly associated with the occurrence of relapse. We chose to focus on exon 4-7 deletions, which represent 25% of all IKZF1 deletions, have a similar unfavorable outcome as other IKZF1 deletions,31 and show the strongest clustering of deletion breakpoints, thus enabling their sensitive upfront detection by breakpoint-spanning semi-quantitative PCR.32 Applying this strategy to the 503 diagnostic samples revealed all 22 IKZF1 exon 4-7 deletions previously identified using a standard MLPA method, as well as 28 additional cases carrying deletions that were missed with the MLPA technique. All breakpoints were sequenced to determine their unique breakpoint- spanning sequences (Online Supplementary Table S6). Using a dilution series of a control sample with a full-clonal IKZF1 exon 4-7 deletion, we determined the level of clonality of the deletions, which ranged from 100% down to 0.32% (Figures 1 and 2, Online Supplementary Figure S1C). All but one of the subclonal IKZF1 exon 4-7 deletions had allele frequencies below 10% (Online Supplementary Table S7).
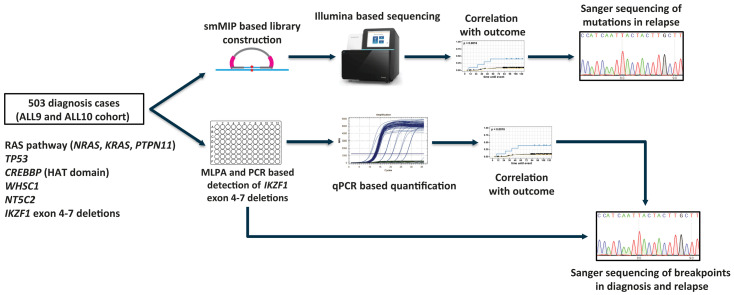
Figure 1.
Schematic representation of the study design. Single-molecule molecular inversion probe-based sequencing approach and real-time quantitative polymerase chain reaction were used in order to detect alterations in known relapse-associated genes in a large cohort of diagnostic samples from patients with acute lymphocytic leukemia. Detected alterations were correlated with outcome and Sanger sequencing was performed on available relapse samples in order to confirm that exactly the same alteration was present in the major clone in relapse. smMIP: single-molecule molecular inversion probe; MLPA: multiplex ligation-dependent probe amplification assay; PCR: polymerase chain reaction; qPCR: real-time quantitative polymerase chain reaction.
Subclonal alterations in relapse-associated genes are common at diagnosis
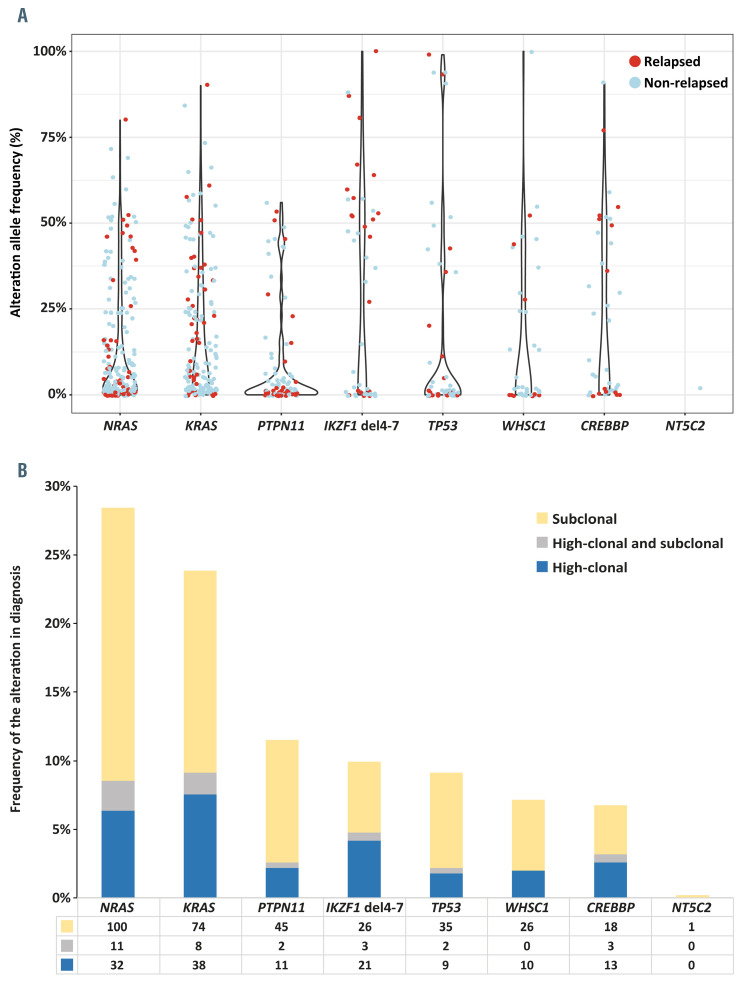
Combining sequence mutations and IKZF1 exon 4-7 deletions, we detected 660 genomic alterations in 285 diagnostic samples, of which 165 (25%) were present in the major fraction of cells (allele frequency ≥25%), which were referred to as high-clonal. The remaining 495 mutations (75%), most of which had an allele frequency <10% were referred to as subclonal (Online Supplementary Figure S2, Online Supplementary Table S7). A total of 147/285 patients carried at least one alteration in a major clone, while 138/285 (48%) patients carried exclusively subclonal alterations. NRAS and KRAS were the most frequently affected genes, showing major clone mutations in 6% and 8% of the cases and subclonal mutations in 20% and 15% of the cases, respectively (Figure 2A, B).
The proportion of subclonal alterations, relative to major clone alterations, was variable among different genes, ranging from 59% for IKZF1 exon 4-7 deletions to 86% for PTPN11 mutations (Figure 2B). Only one thus far unknown (subclonal) NT5C2 mutation (p.Arg507Trp) was identified in a leukemia sample from a patient who did not relapse (Figure 2A, B). Subclonal mutations were relatively common in hyperdiploid ALL (184 cases), particularly for mutations in RAS pathway genes (190/256; 74%), WHSC1 (22/26; 85%) and CREBBP (13/27; 48%) (Online Supplementary Table S8). Major clone WHSC1 mutations were mostly identified in ETV6-RUNX1-positive cases (4/10, 40%).
Potency of RAS pathway genes as drivers of clonal expansion
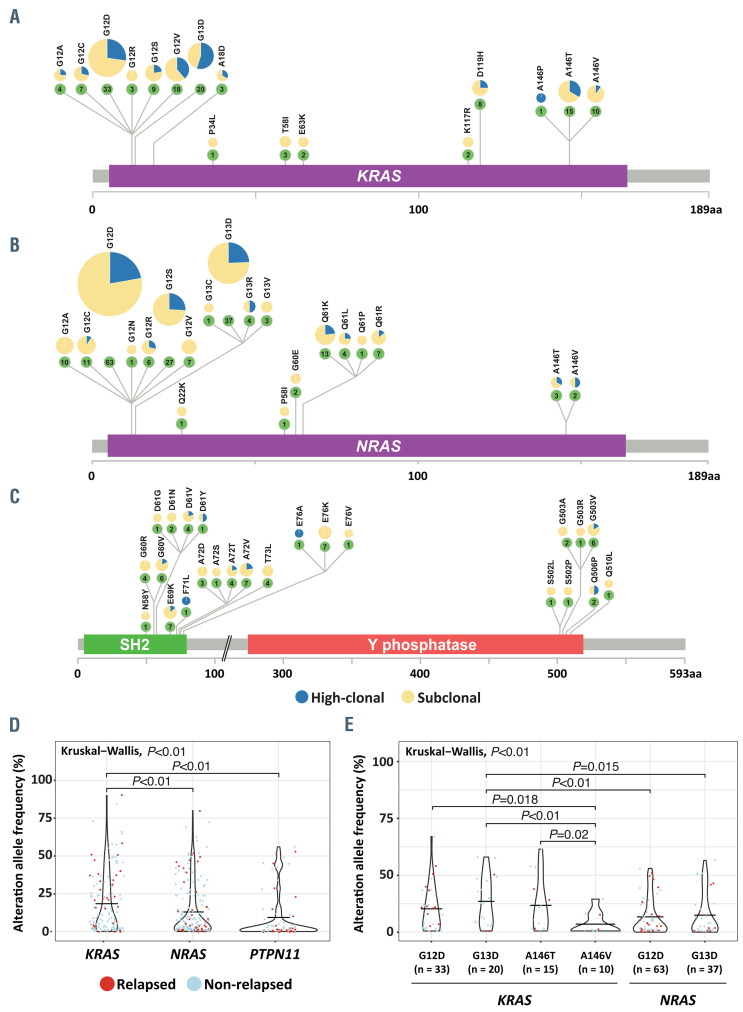
We identified 473 RAS pathway mutations in 225/503 (45%) cases, of which 78% were subclonal (median allele frequency = 3.5%). Over half of the RAS-affected cases were hyperdiploid (>47 chromosomes), in line with previous studies indicating that RAS mutations are associated with hyperdiploidy at diagnosis.10,33 The abundance of these mutations in major and minor clones suggests that these mutations drive clonal expansion during the development of leukemia. Major clone RAS pathway mutations (n=102; all being known hotspots) were found to be mutually exclusive, and 52/102 (51%) of these RAS-mutated cases had at least one additional subclonal mutation in one of the three RAS pathway genes. The mutations mostly affected codons 12 and 13 of KRAS and NRAS (Figure 3AC), and considerable variability in the level of clonality was observed between the different RAS hotspot mutations at the time of diagnosis. For example, NRAS G12A (10 cases), NRAS G12V (7 cases), and PTPN11 E76K (7 cases) were never found to be present in a major clone, whereas 55% (n=11) of the KRAS G13D and 27% (n=9) of the KRAS G12D mutations were found in major clones. With these high numbers of RAS mutations, the variability in clonal burden between hotspot mutations may provide an opportunity to compare the capacity of different hotspots to drive clonal expansion of ALL. In order to test this hypothesis we compared allele frequencies and performed statistical analyses. We found that KRAS hotspot mutations had a significantly higher allele frequency compared to both NRAS and PTPN11 mutations (Wilcoxon signed-rank test, P<0.01) (Figure 3D). When comparing the different hotspot mutations within KRAS, A146V showed the lowest allele frequency, indicating a weaker potential of this hotspot to drive clonal expansion compared to the other KRAS hotspots. Furthermore, the allele frequency of KRAS G13D was significantly higher than that of the NRAS hotspot mutations G13D, G12D and KRAS A146V (Figure 3E). This finding indicates that some RAS hotspot mutations (e.g., KRAS G12D, G13D, A146T) may result in a stronger expansion potential compared to others (e.g., KRAS A146V, NRAS G12D, G13D), and further illustrates the complex heterogeneity of RAS hotspot mutations in their potential to drive clonal expansion.
Figure 2.
Prevalence and distribution of alterations in eight relapse-associated genes. (A) Violin plot showing the variability in mutation allele frequency at diagnosis in the genes studied. The color of the dots indicates whether the mutation was detected in a case without relapse (blue) or with relapse (red). (B) Bar plot showing the frequencies of major clone (high-clonal) and subclonal alterations per case in the genes studied. RAS pathway genes (NRAS, KRAS, PTPN11) were the most frequently mutated. Subclonal mutations (yellow bar) were highly prevalent in all genes tested. A subset of cases had both clonal and subclonal alterations in the same gene (gray bar).
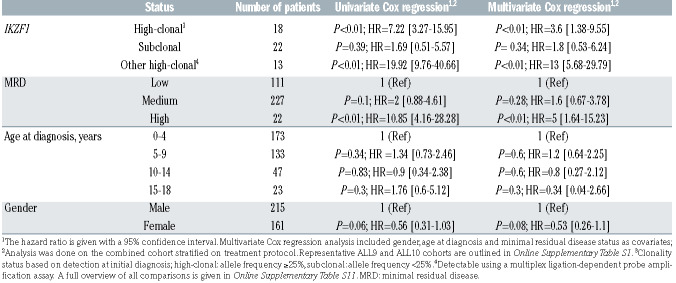
Table 1.
Cox regression analysis in the combined representative ALL9 and ALL10 cohort (n=376).
Relevance of gene alterations to relapse development
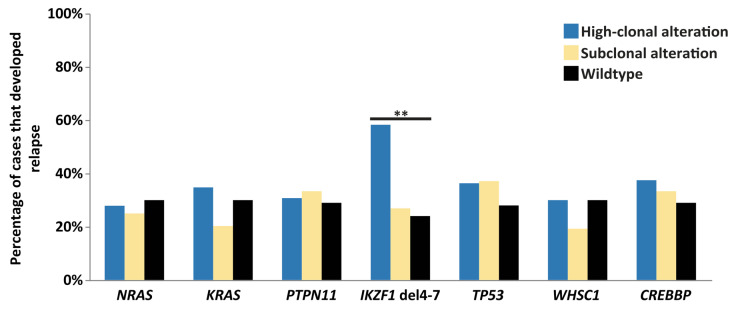
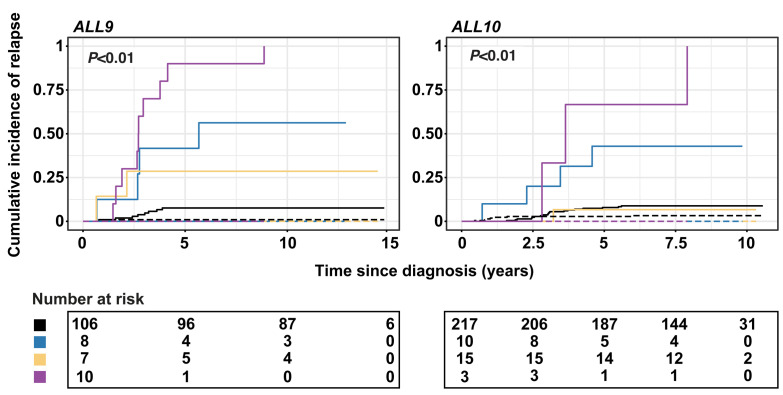
The high number of alterations in these relapse-associated genes at the time of diagnosis triggers the hypothesis that these could be used as prognostic biomarkers for relapse development, even when present at subclonal levels. To test this hypothesis, we first explored whether alterations in each of the eight genes were enriched in diagnostic samples from patients who subsequently relapsed compared to diagnostic samples from patients who did not relapse. In general, subclonal alterations were very common at primary diagnosis in patients who relapsed (60/82; 73%) as well as in patients who did not (165/203; 81%). For high-clonal alterations, we only observed a higher percentage of relapse development in cases with IKZF1 deletions compared to wild-type cases, whereas an association with relapse development was not observed for diagnostic samples with subclonal alterations in any of the genes, including IKZF1 (Figure 4). Furthermore, patients with high-clonal IKZF1 4-7 deletions were more often classified as having high minimal residual disease (MRD; >5×10−4 at day 79 or 84 after start of the treatment) in both representative ALL9 and ALL10 cohorts (Fisher exact test, P<0.01 and P<0.05, respectively), compared to patients without an IKZF1 deletion (Online Supplementary Table S10). The CIR at 5 years was 41.7% (SE 0.04%) and 42.9% (SE 0.03%) in patients with high-clonal IKZF1 4-7 deletions treated according to the ALL9 and ALL10 protocols, respectively (Figure 5). The cause-specific hazard ratio (HRCS) in the two representative cohorts (n=376), estimated with a univariate Cox proportional hazards regression model, revealed an association of high-clonal IKZF1 exon 4-7 deletions with relapse (HR=7.22; 95% CI: 3.27-15.95; P<0.01). In the multivariate Cox model, in which age at diagnosis, gender and MRD status were included, the adjusted HRCS was 3.6 (95% CI: 1.38-9.55; P<0.01) (Table 1, Online Supplementary Table S11). These data are in line with those from earlier studies on these cohorts in which all IKZF1 deletions were included.12,24 However, when we assessed the clinical relevance of subclonal alterations for relapse development in IKZF1, or any of the other genes, Cox regression analysis revealed no significant associations in the combined ALL9 and ALL10 cohorts compared to wild-type cases (Table 1, Online Supplementary Table S11), and the CIR was similar in the two groups (Figure 5). Furthermore, patients with subclonal IKZF1 4-7 deletions did not have significantly different levels of MRD compared to IKZF1 wild-type patients (Online Supplementary Table S10). Since previous studies have shown a lack of association of IKZF1 deletion with relapse in patients who carry a deletion in ERG,34,35 we used MLPA to test whether there was an enrichment of ERG deletions in cases with subclonal IKZF1 exon 4-7 deletions compared to those with clonal IKZF1 exon 4-7, but these deletions were infrequent in both groups (Online Supplementary Table S12).
Tracing of major and minor clone mutations at the time of relapse
To obtain further insight into the clinical relevance of the identified alterations in relapse development, we investigated whether these were preserved in the cases that relapsed. For this analysis, we used all 146 cases that later developed a relapse, of which 82 carried alterations in a major or minor clone in one or more of the genes (Online Supplementary Tables S13 and S14). Overall, we found that for most genes at the time of diagnosis the frequency of subclonal alterations was similar or slightly higher compared to that of the alterations detected in a major clone (Online Supplementary Figure S3A, Online Supplementary Tables S13 and S14).
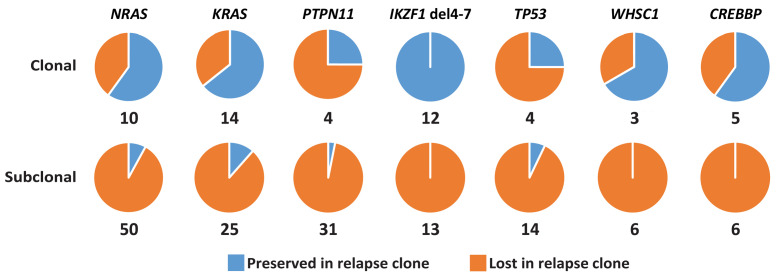
We collected 73 relapse samples from patients who carried these major or minor clone alterations at the time of diagnosis (89%), which enabled us to trace 171 of the 185 sequence mutations, and 25 of the IKZF1 exon 4-7 deletions. We did not assess whether mutations detected at diagnosis were still preserved in minor clones at relapse, since these clones were unlikely to be true relapse drivers. Overall, 56% (22/39) of the tested major clone mutations were found to be preserved in the major clone at relapse, whereas the value for the subclonal mutations was 7% (9/132) (Fisher exact test, P<0.01) (Figure 6, Online Supplementary Figure S3B, Online Supplementary Table S7). For IKZF1 exon 4-7 deletions, the difference was even more striking. Here, the presence of deletions was studied in 19 available relapse samples using breakpoint-spanning PCR, followed by Sanger sequencing to confirm that the breakpoint sequences were identical at diagnosis and relapse (Figure 6, Online Supplementary Figure S3B, Online Supplementary Tables S6 and S7). All major clone IKZF1 exon 4-7 deletions were found to be preserved in the major clone at the time of relapse (n=12), which is in agreement with earlier findings and illustrates their relevance to relapse development in these treatment protocols. 12,24 In contrast, none of the subclonal exon 4-7 deletions in IKZF1 (n=13) was preserved in either the major or a minor clone at relapse. Collectively, the data from the present study indicate that these deletions, when present at initial diagnosis at a subclonal level, do not drive relapse in pediatric ALL.
Figure 3.
Potency of RAS pathway mutations to drive clonal expansion. (A-C) Schematic representation of KRAS (A), NRAS (B) and PTPN11 (C) indicating the prevalence of common hotspot mutations. (D) Violin plot showing allele frequency in hotspot mutations of three investigated RAS pathway genes. The median allele frequency was significantly higher in KRAS, indicating a high potential of KRAS hotspot mutations to drive clonal expansion. (E) Violin plot showing allele frequencies in frequent KRAS and NRAS hotspot mutations. The median allele frequency was significantly higher in KRAS G13D, suggesting their higher potential to drive clonal expansion compared to other RAS hotspot mutations.
Figure 4.
Prevalence of relapse associated genomic alterations at diagnosis. Bar plot showing the percentage of relapses in cases with high-clonal (blue) or subclonal (yellow) mutations in seven relapse-associated genes, and in cases that were wild-type (black) for these genes. Only cases with high-clonal IKZF1 4-7 deletions showed a significantly higher percentage of relapse development compared to wild-type cases (Fisher exact test, P<0.01) (Online Supplementary Table S9).
Figure 5.
Cumulative incidence of relapse for high-clonal and subclonal IKZF1 deletions. The cumulative incidence of relapse (CIR) was estimated using a competing- risk model with death as a competing event. CIR plots are presented for the representative ALL9 (left) and ALL10 (right) cohorts. Lines represent the IKZF1 deletion status and include wild-type (black line), subclonal exon 4-7 deletion (yellow), other high-clonal deletion (purple), and high-clonal exon 4-7 deletion (blue). Straight lines depict relapses and dotted lines death. P-values shown are obtained by employing the Gray test to compare CIR curves. The 5-year CIR was higher in cases with high-clonal IKZF1 deletions, compared to wild-type cases in both representative cohorts.
Discussion
ALL is a heterogeneous disease in which specific genomic alterations show strong associations with relapse risk and outcome. In this study, we assessed the clinical relevance and prognostic value of subclonal alterations in eight genes frequently mutated in relapsed B-cell precursor ALL in a cohort of 503 diagnostic samples. Our data demonstrate that subclonal alterations in these genes are very common at the time of diagnosis, but that these mutations do not provide a basis for risk stratification in pediatric ALL. This finding is particularly relevant for IKZF1 alterations, which are currently used or implemented for treatment stratification in multiple upfront treatment protocols.14,20
Figure 6.
Preservation of clonal and subclonal mutations at the time of relapse. Tracing of major clone (top) and subclonal (bottom) alterations detected in initial diagnosis samples from relapsed patients in the matched relapse samples. The pie charts depict the fractions of preserved (blue) and lost (orange) alterations at the time of relapse.
The selection of these genes was made based on enriched mutation frequencies in relapse found in previous studies. Of all alterations identified in this study, 75% were subclonal at diagnosis, suggesting that these relapseassociated gene mutations accumulate during progression of the leukemia before the initial diagnosis, thereby increasing the clonal complexity. Whereas seven of the genes selected in our study showed this high mutational burden at diagnosis, both in terms of numbers and level of clonality, we identified only a single, not previously reported, subclonal NT5C2 mutation in a non-relapsed case (follow-up time 9.5 years). NT5C2 encodes the cytosolic nucleotidase, which is responsible for inactivating cytotoxic thiopurine monophosphate nucleotides, and activating mutations in this gene are recurrently found in relapsed ALL, mainly T-cell ALL.4,9,36-38 One explanation for the low number of activating NT5C2 mutations at diagnosis is that these mutations decrease cell fitness, and only obtain their selective advantage during treatment with thiopurine.36 If already present at the time of initial diagnosis, these mutations are usually detectable in only a very small subset of cells, far below the detection level of our smMIP analysis.36
Hotspot RAS pathway mutations have been detected in nearly half of the cases, often of the hyperdiploid subtype, and their frequency and clonal burden varied between the different mutations. In our study, we used this variability to compare the potential of different hotspot mutations to drive clonal expansion under physiological conditions. Compared to diagnosis, we observed a less diverse spectrum of KRAS and NRAS hotspot mutations in relapse, with G12D, G12V and G13D together accounting for two-thirds of KRAS and NRAS hotspot mutations found in relapse-fated clones. Studies in other cancers have demonstrated that the prevalence of different RAS pathway mutations varies depending on the type of cancer and tissue of origin, with KRAS mutations G12D, G12V, G13D and G12C being among the most common ones.39,40 Comparison of oncogenic capacities of different RAS hotspots has also been performed using in vitro and in vivo modeling studies, focusing primarily on KRAS. These studies identified KRAS mutations G12D, G12V and G13D as having higher proliferative and transforming potential compared to other common hotspots in various tumors of epithelial origin.39,41,42 Our data indicate that in competition of multiple RAS hotspot mutations, some of these not only confer a proliferative advantage but can also more effectively sustain a treatment-induced selective sweep.4,10
The presence of IKZF1 deletions has been shown to be associated with relapse and survival in multiple clinical ALL studies,12,14-19 and these deletions have been described to play a role in resistance to tyrosine kinase inhibitors and glucocorticoids.43-46 Therefore, with the advance of more sensitive detection techniques, the question of whether subclonal alterations are also associated with relapse is very relevant, both from biological and from clinical perspectives. We here demonstrate that, in contrast to major clone IKZF1 exon 4-7 deletions, cases that carry this deletion only in a subset of the cells do not show an association with relapse. Moreover, whereas all major clone exon 4-7 deletions were preserved in cases that relapsed, none of the relapses from cases with subclonal exon 4-7 deletions at diagnosis carried this deletion. Importantly, the majority of subclonal deletions had allele frequencies below 10% (Online Supplementary Table S7). Therefore, since a threshold to distinguish subclonal from major clonal deletions is difficult, deletions closer to our threshold of 25% should be evaluated with caution. Nevertheless, the difference between major and minor clone IKZF1 4-7 deletions is striking, and the reason behind this remains unclear. Possibly, the functional impact of full-clonal IKZF1 deletions, which arise early during leukemia development, is different from that of deletions that occur in later stages when the leukemia has already expanded. Other deletions in IKZF1 show much less clustering in their breakpoints and, therefore, screening for these subclonal deletions in diagnostic samples is much less efficient. We did not, therefore, directly assess the stability and potential prognostic importance of whole gene and rare intragenic IKZF1 deletions. However, a previous study showed that other IKZF1 deletion subtypes have similar prognostic relevance as exon 4-7 deletions,31 suggesting that subclonal alterations in these other IKZF1 deletions may show the same lack of association.
In summary, we show that subclonal alterations in the relapse-associated genes IKZF1, CREBBP, KRAS, NRAS, PTPN11, TP53, and WHSC1 in pediatric ALL are frequently present at initial diagnosis, often at a subclonal level. At relapse, however, most of these subclonal mutations are lost, suggesting that their selective advantages over wild-type clones during treatment is limited. This finding has direct implications for clinical practice, particularly in the case of IKZF1, where deletion status is used for routine risk stratification. We conclude that, at least for the investigated set of genes, there is no basis for the use of subclonal alterations at initial diagnosis as a prognostic marker.
Supplementary Material
Acknowledgments
We thank Radboud University Medical Center, Department of Human Genetics for bioinformatic support in data analysis.
Funding Statement
Funding: This work was supported by grants from Stichting Kinderen Kankervrij (KIKA 150 to RPK), Stichting Bergh in het Zadel (to RPK), and the China Scholarship Council (CSC201304910347 to JY).
References
- 1.Hunger SP, Lu X, Devidas M, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the Children's Oncology Group. J Clin Oncol. 2012;30(14):1663-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pieters R, de Groot-Kruseman Hd, Velden V, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol. 2016;34(22):2591-2601. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356-361. [DOI] [PubMed] [Google Scholar]
- 4.Ma X, Edmonson M, Yergeau D, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waanders E, Gu Z, Dobson SM, et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discov. 2020;1(1):96-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malinowska-Ozdowy K, Frech C, Schönegger A, et al. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia. 2015;29(8): 1656-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011; 471(7337):235-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irving J, Matheson E, Minto L, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood. 2014;124(23):3420-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzoneva G, Perez-Garcia A, Carpenter Z, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19(3):368-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jerchel IS, Hoogkamer AQ, Ariës IM, et al. RAS pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018; 32(4):931-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuiper RP, Waanders E, van der Velden VH, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24(7):1258-1264. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe JD Wang Y Chan HM, Zet al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45(11):1386-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1- like signature and IKZF1 deletion, but not high CRLF2 expression, in children with Bcell precursor ALL. Blood. 2013;122(15): 2622-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinze L, Moricke A, Zimmermann M, et al. Prognostic impact of IKZF1 deletions in association with vincristine-dexamethasone pulses during maintenance treatment of childhood acute lymphoblastic leukemia on trial ALL-BFM 95. Leukemia. 2017;31(8): 1840-1842. [DOI] [PubMed] [Google Scholar]
- 16.Clappier E, Grardel N, Bakkus M, et al. IKZF1 deletion is an independent prognostic marker in childhood B-cell precursor acute lymphoblastic leukemia, and distinguishes patients benefiting from pulses during maintenance therapy: results of the EORTC Children's Leukemia Group study 58951. Leukemia. 2015;29(11):2154-2161. [DOI] [PubMed] [Google Scholar]
- 17.Ofverholm I, Tran AN, Heyman M, et al. Impact of IKZF1 deletions and PAX5 amplifications in pediatric B-cell precursor ALL treated according to NOPHO protocols. Leukemia. 2013;27(9):1936-1939. [DOI] [PubMed] [Google Scholar]
- 18.Dörge P, Meissner B, Zimmermann M, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98(3):428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asai D, Imamura T, Suenobu S, et al. IKZF1 deletion is associated with a poor outcome in pediatric B-cell precursor acute lymphoblastic leukemia in Japan. Cancer Med. 2013;2(3):412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeoh AEJ, Lu Y, Chin WHN, et al. Intensifying treatment of childhood B-lymphoblastic leukemia with IKZF1 deletion reduces relapse and improves overall survival: results of Malaysia-Singapore ALL 2010 study. J Clin Oncol. 2018;36(26):2726-2735. [DOI] [PubMed] [Google Scholar]
- 21.Absalan F, Ronaghi M. Molecular inversion probe assay. Methods Mol Biol. 2007;396: 315-330. [DOI] [PubMed] [Google Scholar]
- 22.Berglund EC, Lindqvist CM, Hayat S, et al. Accurate detection of subclonal single nucleotide variants in whole genome amplified and pooled cancer samples using HaloPlex target enrichment. BMC Genomics. 2013;14(1):856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Antić Ž, van Reijmersdal SV, et al. Accurate detection of low-level mosaic mutations in pediatric acute lymphoblastic leukemia using single molecule tagging and deep-sequencing. Leuk Lymphoma. 2018; 59(7):1690-1699. [DOI] [PubMed] [Google Scholar]
- 24.Waanders E, van der Velden VH, van der Schoot CE, et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia. 2011;25(2):254-258. [DOI] [PubMed] [Google Scholar]
- 25.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17(4):343-346. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Waanders E, van Reijmersdal SV, et al. Upfront treatment influences the composition of genetic alterations in relapsed pediatric B-cell precursor acute lymphoblastic leukemia. Hemasphere. 2020;4(1):e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putter H, Fiocco M, Geskus RB. Tutorial in biostatistics: competing risks and multi-state models. Stat Med. 2007;26(11):2389-2430. [DOI] [PubMed] [Google Scholar]
- 28.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16(3):1141-1154. [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2(5):401-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boer JM, van der Veer A, Rizopoulos D, et al. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study. Leukemia. 2016;30(1):32-38. [DOI] [PubMed] [Google Scholar]
- 32.Caye A, Beldjord K, Mass-Malo K, et al. Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2013;98(4):597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulsson K, Lilljebjörn H, Biloglav A, et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47(6):672-676. [DOI] [PubMed] [Google Scholar]
- 34.Clappier E, Auclerc MF, Rapion J, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28(1):70-77. [DOI] [PubMed] [Google Scholar]
- 35.Zaliova M, Potuckova E, Hovorkova L, et al. ERG deletions in childhood acute lymphoblastic leukemia with DUX4 rearrangements are mostly polyclonal, prognostically relevant and their detection rate strongly depends on screening method sensitivity. Haematologica. 2019;104(7):1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzoneva G, Dieck CL, Oshima K, et al. Clonal evolution mechanisms in NT5C2 mutant relapsed acute lymphoblastic leukemia. Nature. 2018;553(7689):511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer JA, Wang J, Hogan LE, et al. Relapsespecific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45(3):290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieters R, Huismans DR, Loonen AH, et al. Relation of 5'-nucleotidase and phosphatase activities with immunophenotype, drug resistance and clinical prognosis in childhood leukemia. Leuk Res. 1992; 16(9):873-880. [DOI] [PubMed] [Google Scholar]
- 39.Miller MS, Miller LD. RAS mutations and oncogenesis: not all RAS mutations are created equally. Front Genet. 2012;2:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winters IP, Chiou S-H, Paulk NK, et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun. 2017;8(1):2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stolze B, Reinhart S, Bulllinger L, Fröhling S, Scholl C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep. 2015;5:8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marke R, Havinga J, Cloos J, et al. Tumor suppressor IKZF1 mediates glucocorticoid resistance in B-cell precursor acute lymphoblastic leukemia. Leukemia. 2016; 30(7):1599-1603. [DOI] [PubMed] [Google Scholar]
- 44.Imamura T, Yano M, Asai D, et al. IKZF1 deletion is enriched in pediatric B-cell precursor acute lymphoblastic leukemia patients showing prednisolone resistance. Leukemia. 2016;30(8):1801-1803. [DOI] [PubMed] [Google Scholar]
- 45.Churchman ML, Low J, Qu C, et al. Efficacy of retinoids in IKZF1-mutated BCR-ABL1 acute lymphoblastic leukemia. Cancer Cell. 2015;28(3):343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Veer A, Zaliova M, Mottadelli F, et al. IKZF1 status as a prognostic feature in BCR-ABL1-positive childhood ALL. Blood. 2014;123(11):1691-1698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.