Abstract
Upregulation of the plasma membrane receptor IL1RAP in acute myeloid leukemia (AML) has been reported but its role in the context of the leukemic bone marrow niche is unclear. Here, we studied the signaling events downstream of IL1RAP in relation to leukemogenesis and normal hematopoiesis. High IL1RAP expression was associated with a leukemic GMPlike state, and knockdown of IL1RAP in AML reduced colony-forming capacity. Stimulation with IL1b resulted in the induction of multiple chemokines and an inflammatory secretome via the p38 MAPK and NFkB signaling pathways in IL1RAP-expressing AML cells, but IL1b-induced signaling was dispensable for AML cell proliferation and NFkB-driven survival. IL1RAP was also expressed in stromal cells where IL1b induced expression of inflammatory chemokines and cytokines as well. Intriguingly, the IL1b-induced inflammatory secretome of IL1RAP-expressing AML cells grown on a stromal layer of mesenchymal stem cells affected normal hematopoiesis including hematopoietic stem/progenitor cells while AML cell proliferation was not affected. The addition of Anakinra, an Food and Drug Aministration-approved IL1 receptor antagonist, could reverse this effect. Therefore, blocking the IL1-IL1RAP signaling axis might be a good therapeutic approach to reduce inflammation in the bone marrow niche and thereby promote normal hematopoietic recovery over AML proliferation after chemotherapy.
Introduction
The fate of both acute myeloid leukemia (AML) and normal hematopoietic stem cells (HSC) is critically dependent on interactions with the bone marrow (BM) niche.1-4 Recent data has also suggested that malignant cells can remodel the BM microenvironment into a leukemic BM niche favoring leukemogenesis over normal hematopoiesis.4-6 Plasma membrane (PM) proteins are first in line to respond to signals that arise from the BM niche. One of these PM proteins, interleukin- 1 receptor accessory protein (IL1RAP), is specifically upregulated in stem/progenitor cells from chronic myeloid leukemia (CML) and AML patients but not on normal CD34+ hematopoietic stem/progenitor cells (HSPC).7-10 This has led to several studies that investigated the targetability of IL1RAP as treatment strategy of chronic myeloid leukemia (CML) and AML,9,11-13 but little is known regarding the cell-intrinsic role of IL1RAP in AML stem/progenitor cells. In addition, the canonical IL1RAP signaling axis in AML with respect to the leukemic BM niche has not been studied extensively.
The IL1 family is part of the innate immunity that regulates local inflammatory responses, and its dysregulation may lead to autoinflammatory diseases often caused by excessive IL1b production.14 The IL1 family consists of seven ligands including IL1α, IL1b, IL18, IL33, IL36α, b and g that can bind different IL1 receptors whereby the majority forms a dimer with the IL1RAP co-receptor.15 Activation of the IL1 receptor complex activates the MyD88/IRAK1/IRAK4/TRAF6/TAK1 signaling pathway, which in turn results in activation of NFkB and mitogen-activated protein kinases (MAPK), including p38.16 Many proteins in this pathway are often upregulated in myelodysplastic syndromes (MDS) and AML, which suggests an important role for this pathway in leukemogenesis.17-19 The IL1 signaling route can induce a variety of inflammatory cytokines and chemokines, which has been shown to be an important factor for development and maintenance of MDS.20 In AML, IL1 has been proposed to enhance proliferation and survival. 21-23
Here, we studied the IL1–IL1RAP signaling axis in primary AML patients in the context of the BM niche and revealed that the IL1b-induced secretome impacts on leukemogenesis and most notably on normal hematopoiesis.
Methods
Extensive details on the methods used can be found in the Online Supplementary Appendix.
Primary samples
Neonatal cord blood (CB), mobilized peripheral blood stem cells (PBSC), normal bone marrow (NBM), mesenchymal stromal cells (MSC), MDS, and AML patient material was obtained as described in the Online Supplementary Methods. All healthy individuals and AML patients gave an informed consent in accordance with the Declaration of Helsinki at the University Medical Center Groningen (UMCG) and Martini Hospital Groningen, the Netherlands. All protocols were approved by the Medical Ethical Committee of the UMCG. Details of AML characteristics used in this study can be found in the Online Supplementary Table S1.
Cell (co-)cultures
MSC co-cultures/triple-cultures with CB CD34+, PBSC CD34+ or AML CD34+ cells were performed in Gartner’s medium with the addition of 20 ng/mL granulocyte colony-stimulating factor (G-CSF), N-plate and IL3. Inhibition of the IL1-signaling pathway was established by the addition of 500 ng/mL Anakinra (Swedish Orphan Biovitrum BVBA). In case of triple co-cultures, CB CD34+ cells were transduced with pLKO eGFP to distinguish them from AML cells. Co-cultures were grown at 37°C and 5% CO2 and demi-populated regularly, replacing 50-80% of the volume with fresh or conditioned Gartner’s medium. Suspension cells were used for further analysis.
Colony-forming cell assay
The colony-forming capacity of CB CD34+ cells was evaluated in methylcellulose (1,600 mL) mixed with CM (900 mL) from MSC-AML co-cultures treated for 7 days with or without IL1β and Anakinra.
Lentiviral transfection
For knockdown of IL1RAP, cells were transduced with a pLKO eGFP construct, containing short hairpins against IL1RAP sh1: 5’-TGGCCTTACTCTGATCTGGTATTGGACTA-3’, sh2: 5’-CGGGCATTAATTGATTTCCTACTATATTC-3’,10 or scrambled control 5’-TTCTCCGAACGTGTCACGTT-3’.
Results
IL1RAP is upregulated in acute myeloid leukemia patients and correlates with a leukemic granulocyte-monocyte progenitor cell signature
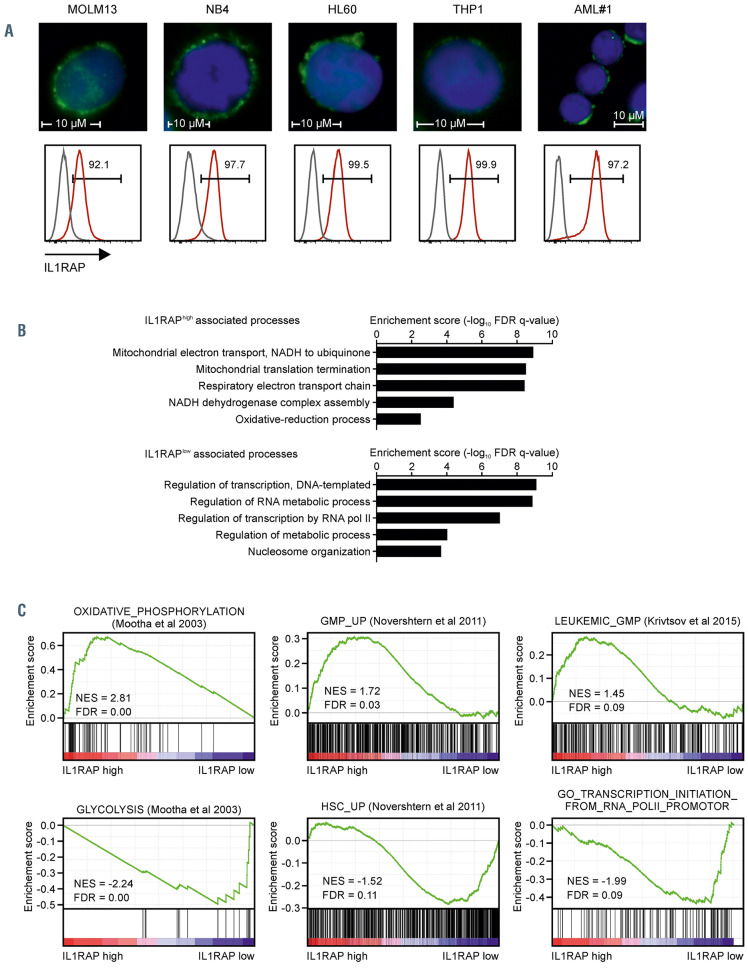
IL1RAP expression was evaluated in NBM CD34+ cells (n=11) and blasts of AML, acute promyelocytic leukemia (APL) and MDS patients (CD34+ or SSClowCD45mid in case of NPM1 mutant AML with CD34 expression <1%). De novo AML patients (n=110), patients that developed AML from an MDS (s-AML) (n=27), APL patients (n=4), and MDS/MDS-excess blasts (MDS-EB) patients (n=13), all showed heterogeneous but on average significantly upregulated expression of IL1RAP (Online Supplementary Figure S1A and B). Immunofluorescent staining showed clear IL1RAP expression on the PM in AML cells representing different genetic subtypes with high IL1RAP expression (Figure 1A).
For 42 AML patients, quantitative proteome data was generated previously7 and we evaluated cellular processes that were enriched in patients with either high or low IL1RAP expression. For 31 of these patients IL1RAP expression was also measured by flow cytometry independently and a significant correlation with our quantitative proteome data was observed (Online Supplementary Figure 1C). Gene ontology (GO) and gene set enrichment analysis (GSEA) was performed on a ranked list based on Pearson correlations of IL1RAP protein expression with the complete quantitative proteome and showed that high expression of IL1RAP was associated with the terms “mitochondrial translation elongation and termination”, “energy production via oxidative phosphorylation” and a “leukemic granulocyte-monocyte progenitor (L-GMP) signature”, whereas AML patients with low IL1RAP expression were associated with “regulation of RNA metabolic processes”, “gene expression” and a glycolysis-enriched HSC-like signature (Figure 1B to C). While IL1A and IL1B were expressed by AML cells at varying levels, no correlations were seen with IL1RAP expression, neither in our quantitative proteome data nor in various published transcriptome datasets (data not shown).24,25 Neither did we find positive correlations of IL1RAP high-expressing AML with RELA, encoding the NFkB transcription factor, MyD88, IRAK1, and TRAF6, all associated with the IL1-IL1RAP downstream pathway (data not shown).
IL1-induced IL1RAP signaling is associated with an inflammatory secretome
Data generated in previous studies comparing gene expression profiles of primary AML and NBM CD34+ cells suggested that the IL1RAP pathway might be actively used in many AML patients since components of the IL1RAP-TAK1 signaling pathway were significantly upregulated in AML CD34+ cells while negative feedback proteins such as IL1R2, IL1RN and MARCH8 were significantly downregulated (Online Supplementary Figure S2A).24 In order to investigate the repertoire of targets downstream of the IL1-IL1RAP axis we performed genomewide transcriptome studies in CD34+ of AML#1 (for details of all AML used in this study, see the Online Supplementary Table S1), THP1, and K562 cells that were stimulated with IL1b for 1 hour (Online Supplementary Table S2). While the IL1RAP receptor can be activated by multiple different cytokines including IL1α, IL1b, IL33, and IL36 depending on its co-receptor IL1R1, IL1RL1 or IL1RL2, and although, e.g., IL33 has also recently been shown to impact on HSC,26 we initially focused on IL1b in our studies. IL1b was chosen since the IL1R1 receptor appeared to be highly expressed in some AML subsets (data not shown) and due to its potential role in inflammation, 27 which we wanted to investigate in more detail.
Figure 1.
High IL1RAP plasma membrane protein expression in acute myeloid leukemia is associated with a leukemic granulocyte-monocyte progenitor signature. (A) Immunofluorescent staining and expression measured by flow cytometry (red histogram) of interleukin-1 receptor accessory protein (IL1RAP) in multiple acute myeloid leukemia (AML) cell lines and one primary AML patient. The grey histogram indicates the unstained control. (B and C). Gene ontology analysis (C) and gene set enrichment analysis (GSEA) (D) on a ranked gene list based on label-free quantitative protein expression in 42 primary AML patient samples.7 Genes were ranked based on Pearson correlation with IL1RAP protein expression. Normalized enrichment score (NES) and false discovery rate (FDR) were used to determine significance. *P<0.05, ***P<0.001.
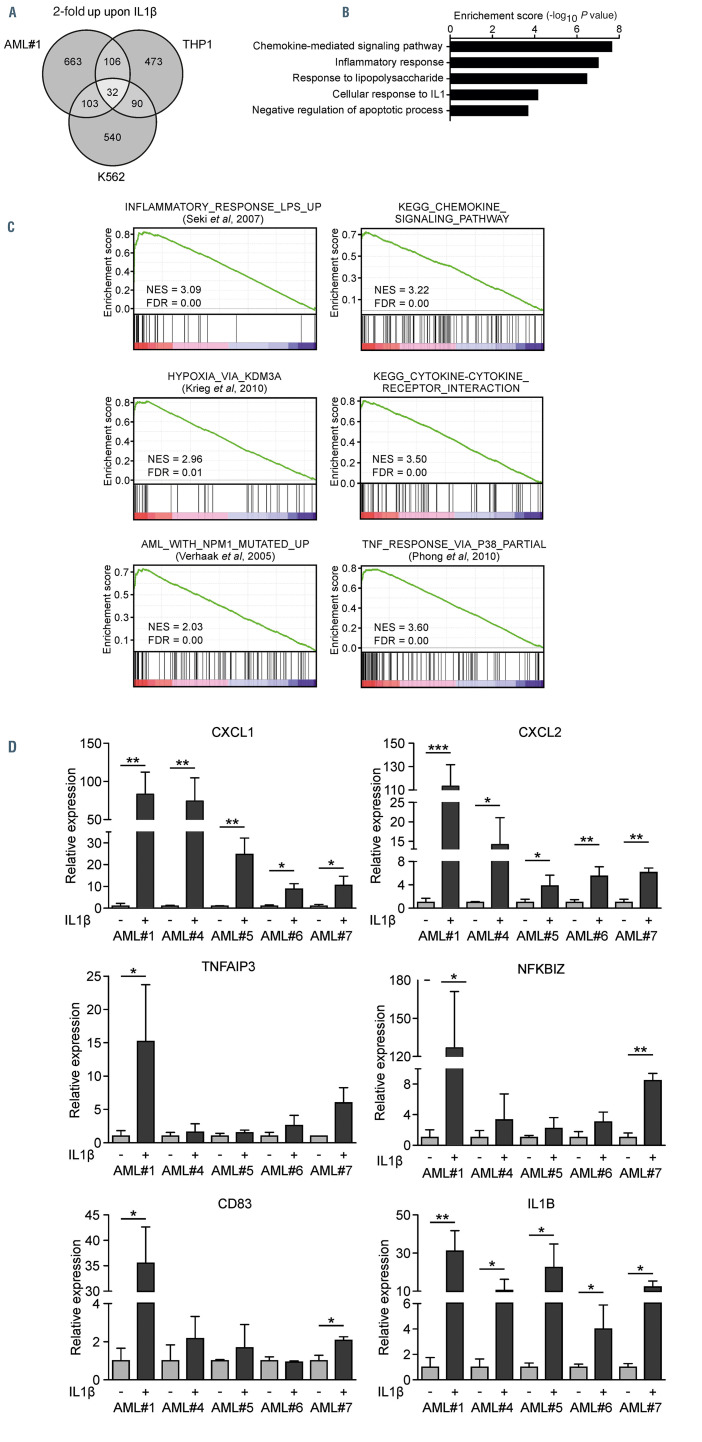
AML#1 and THP1 cells both had high IL1RAP expression whereas K562 showed partial IL1RAP expression (Online Supplementary Figure S2B). We identified 299 genes that were >2-fold upregulated in at least two groups and 32 genes that were >2-fold upregulated in all three groups (Figure 2A). GO analysis on the combination of these genes (331) showed enrichment for genes associated with chemokine signaling, inflammation, response to IL1 and an anti-apoptotic signature (Figure 2B). In addition, GSEA of a ranked gene list of primary AML cells showed significant enrichment in IL1b-stimulated cells for processes associated with “inflammation”, “chemokine signaling”, “TNF signaling via p38”, “hypoxia” and “AML with a NPM1 mutation” (Figure 2C). We confirmed the upregulation of several of the identified genes in an independent set of IL1RAP+ primary AML samples (Figure 2D; Online Supplementary Figure S2C).
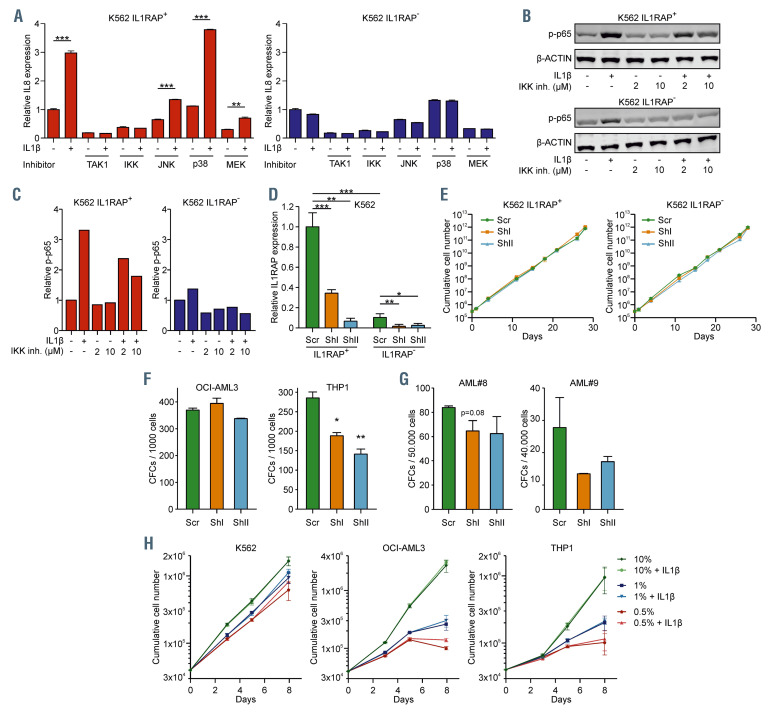
We noted that K562 cells could be divided into a IL1RAP+ and IL1RAP– population. We sorted these populations to elucidate the pathways downstream of IL1RAP (Online Supplementary Figure S3A). Both of them showed similar growth kinetics, clonogenicity in CFC assays, and the IL1RAP expression remained stable over time (Online Supplementary Figure S3B to D). As expected, an upregulation of IL8 upon IL1b stimulation was only observed in IL1RAP+ cells (Figure 3A). The IL1b response was blocked with inhibitors against TAK1 and IKK, whereas inhibition of JNK, p38 and MEK/ERK was less effective (Figure 3A). In accordance, K562 IL1RAP+ cells showed a strong increase in phosphorylation of p65 upon stimulation with IL1b, which could be partially reversed by inhibiting the NFkB pathway with an IKK inhibitor in a dose-dependent manner (Figure 3B to C). These data are in line with previous observations by Bosman et al. who studied the TAK1-NFkB axis.17 Next, we transduced K562 IL1RAP+ cells, K562 IL1RAP– cells (as negative controls), OCIAML3 cells, and THP1 cells with short hairpin RNA (shRNA) against IL1RAP and sorted transduced cells by green fluorescence protein (GFP) positivity (Figure 3D; Online Supplementary Figure S3E and G). Knockdown of IL1RAP did not result in impaired cell proliferation (Figure 3E; Online Supplemental Figure S3H). We observed reduced colony-forming capacity in THP1 cells but not in OCIAML3 cells (Figure 3F). A trend towards reduced CFC capacity upon knockdown of IL1RAP was also observed in primary AML patients #8 and #9 as also observed previously (Figure 3G; Online Supplementary Figure S3I to J).28 Finally, we challenged K562, OCI-AML3 and THP1 cells by serum deprivation and stimulated them with IL1b to determine whether cell viability was controlled by the IL1-IL1RAP axis under stress conditions. Serum starvation- induced loss of viability, however, this was not rescued by addition of IL1b (Figure 3H).
Figure 2.
IL1-induced IL1RAP signaling is associated with an inflammatory secretome. (A) Transcriptome analysis of genes 2-fold upregulated in one primary acute myeloid leukemia (AML) patient (AML#1) and two AML cell lines upon stimulation with IL1b. (B) Gene ontology analysis on 331 genes that were 2-fold upregulated in at least two out of three groups (THP1, K562 and AML#1). (C) Gene set enrichment analysis analysis on a ranked gene list of AML#1. Genes were ranked from upregulated to downregulated upon stimulation with IL1b. Normalized enrichment score (NES) and false discovery rate (FDR) were used to determine significance. (D) Quantitative real-time polymerase chain reaction analysis of five primary AML patient samples ± IL1b stimulation. Bars indicate mean ± standard deviation of biological triplicates. Statistical analysis was performed by a one-tailed Student’s t-test. *P<0.05; ** P<0.01; ***P<0.001.
Synergism of the IL1-IL1RAP signaling with other active signaling pathways in acute myeloid leukemia
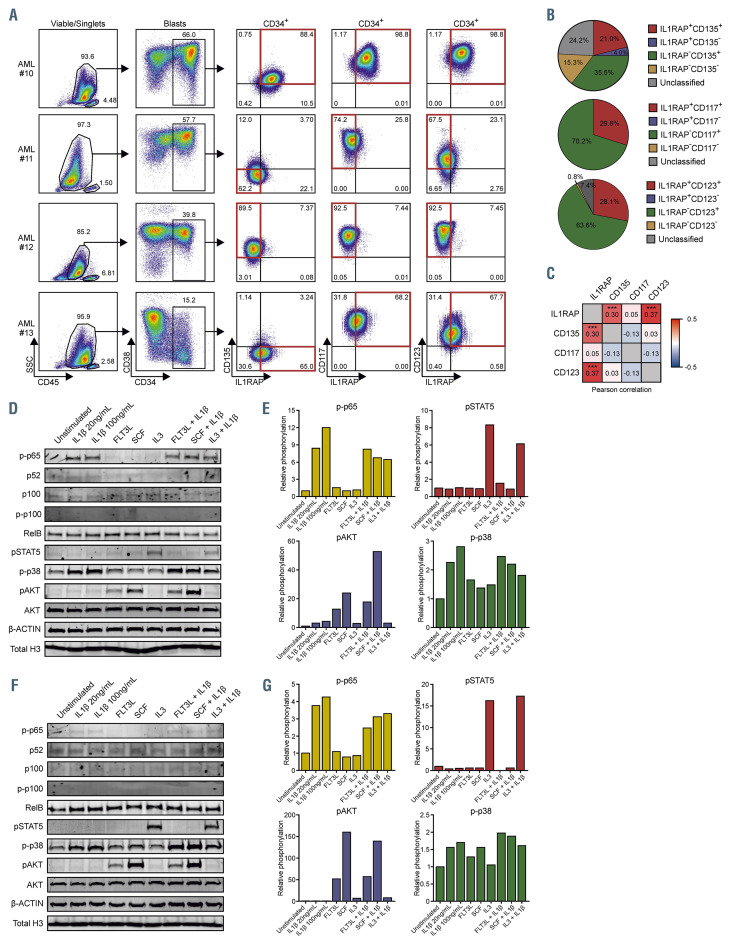
IL1RAP was recently described to be directly associated with FLT3 (CD135) and c-kit (CD117) receptors.28 We analyzed co-expression of IL1RAP with signaling receptors including CD135, CD117 and IL3 receptor (CD123) in immature AML stem progenitor cells (CD34+ or SSClowCD117+ in case of CD34 expression <1%) by flow cytometry in a cohort of 124 primary AML patients of which four representative examples are shown (Figure 4A). Subsequently, patients were defined as single positive, double positive, or double negative when at least 50% of the cells resided within either one of the gates (Figure 4A and B). These analyses revealed that 21% of the patients were CD135+IL1RAP+, while 4% expressed IL1RAP without CD135 (Figure 4B); 29.8% and 28.1% of the patients were IL1RAP+CD117+ and IL1RAP+CD123+, respectively, and we did not identify patients that expressed IL1RAP without any detectable CD117 or CD123 (Figure 4B). Expression of IL1RAP, as quantified by flow cytometry (mean fluorescense intensity [MFI]), correlated significantly with CD123 expression, and to a lesser degree with CD135, but no significant correlations were found with CD117 (Figure 4C; Online Supplementary Figure 4A). These observations indicate that IL1RAP signaling can co-occur in cells that are also hardwired for FLT3 ligand (FLT3L), stem cell factor (SCF) and/or IL3-induced signal transduction, and in fact might influence those pathways as well. In addition, Muto et al. showed that MDS HSPC switch from canonical to non-canonical NFkB signaling in response to inflammatory signals like IL1b, which might also occur in AML cells.29 In order to investigate both hypotheses, we isolated CD34+ cells from AML patients #10-13 and stimulated them with IL1b, FLT3L, SCF, IL3, or a combination thereof. Activation of signal transduction pathways was determined by western blotting for phosphorylated p65 (p-p65), phosphorylated p100 (p-p100), p52/p100, RelB, phosphorylated STAT5 (pSTAT5), phosphorylated p38 (p-p38), and phosphorylated AKT (pAKT), (Figure 4D to G; Online Supplementary Figure S4B to E). Figure 4D to E illustrates AML patient #10 that expressed high levels of IL1RAP, FLT3, CD117 and CD123 within the CD34+ blast compartment. Stimulation with IL1b resulted in downstream activation of the canonical NFkB-(p65) and p38 pathways, whereas we observed limited activation of non-canonical NFkB (p52/p100, RelB, and p-p100), IL3 activated the STAT5 signaling pathway, FLT3L and SCF both activated the PI3K-AKT signaling pathway. Co-stimulation of IL1b with SCF resulted in a slightly increased downstream activation of the PI3K-AKT signaling, although no additive effects were seen on pSTAT5 or p-p38 (Figure 4D to E). In AML#13, which expressed IL1RAP, CD123 and CD117 and low levels of CD135, we observed that IL1b led to activation of p-p38 and a moderate activation of only canonical NFkB, IL3 induced pSTAT5, but again the IL3 signaling was not further potentiated by co-stimulation with IL1b (Online Supplementary Figure 4B to C). The third example (AML#11) showed increased p-p65 levels upon IL1b stimulation whereas the non-canonical NFkB was active at baseline but not further enhanced upon IL1b stimulation as read out by p52/p100 levels (Figure 4F to G). AML#11 responded to FLT3L, SCF and IL3 stimulation, but co-stimulation with IL1b did not further enhance activation of any of these signaling pathways (Figure 4F and G). The fourth patient sample (AML#12) had limited IL1RAP expression and showed no activation of neither canonical nor non-canonical NFkB upon stimulation with IL1b, no additional activation of the p-p38 signal that was already highly activated at baseline, and no effects of co-stimulation with IL1b were seen on IL3-induced pSTAT5 (Online Supplemental Figure S4D to E). Further evaluation of the non-canonical NFκB pathway showed that some primary AML patients already have high baseline non-canonical NFκB activity compared to THP1 cells. We did not observe differences in baseline non-canonical NFkB activity in K562 IL1RAP+ and K562 IL1RAP- cells (Online Supplementary Figure S4F and G). Neither did we observe synergistic effects of IL1b with FLT3L or IL3 in THP1 cells on downstream phosphorylation of ERK (pERK), cJUN (pcJUN) and AKT (pAKT) (Online Supplementary Figure S4H, I, K and L). Addition of the anti-IL1RAP monoclonal antibody (α-IL1RAP MAb), that could partly rescue IL1- induced upregulation of IL8 and CXCL1, did not alter phosphorylation levels of AKT and cJUN upon stimulation with IL1b in combination with FLT3L or IL3 (Online Supplementary Figure S4J to L). In summary, both the canonical NFkB and p38 pathway can be activated by IL1b in primary AML patients whereby downstream canonical NFkB signaling might be correlated to IL1RAP expression levels. We did not observe synergism of IL1b with other signaling molecules including FLT3L, SCF and IL3 on downstream phosphorylation of p65, p38, STAT5, AKT, ERK and cJUN.
Figure 3.
IL1-IL1RAP mediated activation of the NFkB signaling does not rescue proliferation under stress conditions, but IL1RAP knockdown results in reduced colony-forming capacity. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis in K562 IL1RAP+ and IL1RAP– cells treated with TAK1, NFkB, JNK, p38 and MEK inhibitors and subsequently stimulated with IL1b. Bars indicate mean ± standard deviation (SD) of technical triplicates. (B) Western blot of K562 IL1RAP+ and IL1RAP- treated with or without IL1b and/or IKK inhibitor (IKK inh). (C) Quantification of western blot in panel C, p-p65 was normalized to b-ACTIN. (D) Interleukin-1 receptor accessory protein (IL1RAP) mRNA levels measured by qPCR in K562 IL1RAP+ and IL1RAP- cells transduced with short hairpin (shRNA) including a non-targeting control (scr) and shRNA targeting IL1RAP (shI and shII). Bars indicate mean ± SD of technical triplicates. (E) Growth curves of K562 IL1RAP+ and IL1RAP- cells (n=3) ± knockdown of IL1RAP. (F) Colony-forming cell (CFC) output of OCI-AML3 and THP1 ± knockdown of IL1RAP. Bars indicate mean ± SD of technical duplicates. (G) CFC output of acute myeloid leukemia (AML) patient 8 (AML#8) and AML#9 ± knockdown of IL1RAP. Bars indicate mean ± SD of technical duplicates. (H) Growth curves after serum depletion of K562, OCI-AML3 and THP1 cells (n=3) ± IL1b. Statistical analysis in all panels was performed using a Student’s t-test. *P<0.05; **P<0.01; ***P<0.001.
Figure 4.
Synergism of the IL1-IL1RAP signaling with other active signaling pathways in acute myeloid leukemia. (A) Gating strategy of co-expressing Interleukin- 1 receptor accessory protein (IL1RAP), CD135, CD123 and CD117 measured by flow cytometry within four representative primary acute myeloid leukemia (AML) patients. (B) Pie-chart showing co-expression of IL1RAP with CD135, CD123 and CD117 in blasts of 124 primary acute myeloid leukemia (AML) patients. At least 50% of the total amount of cells in a specific group was used as a cutoff to include a patient in a certain group; otherwise, patients were called “unclassified”. (C) Pearson correlation of IL1RAP with CD135, CD123 and CD117 based on mean fluorescense intensity (MFI) (n=124). (D) Western blot of primary patient CD34+ blasts positive for IL1RAP, CD135, CD117 and CD123. Cells were stimulated with IL1b, FLT3L, stem cell factor (SCF), IL3 or a combination of these cytokines. (E) Quantification of western blot in panel D. p-p65 was normalized to total H3, pSTAT5 and p-p38 were normalized to b-ACTIN and pAKT was normalized to AKT. (F) Western blot of primary patient CD34+ blasts with low IL1RAP and CD135 expressing but positive for CD117 and CD123. Cells were stimulated with IL1b, FLT3L, SCF, IL3 or a combination of these cytokines. (G) Quantification of western blot in panel (F). p-p65 was normalized to total H3, pSTAT5 and p-p38 were normalized to b-ACTIN and pAKT was normalized to AKT. ***P<0.001.
Figure 5.
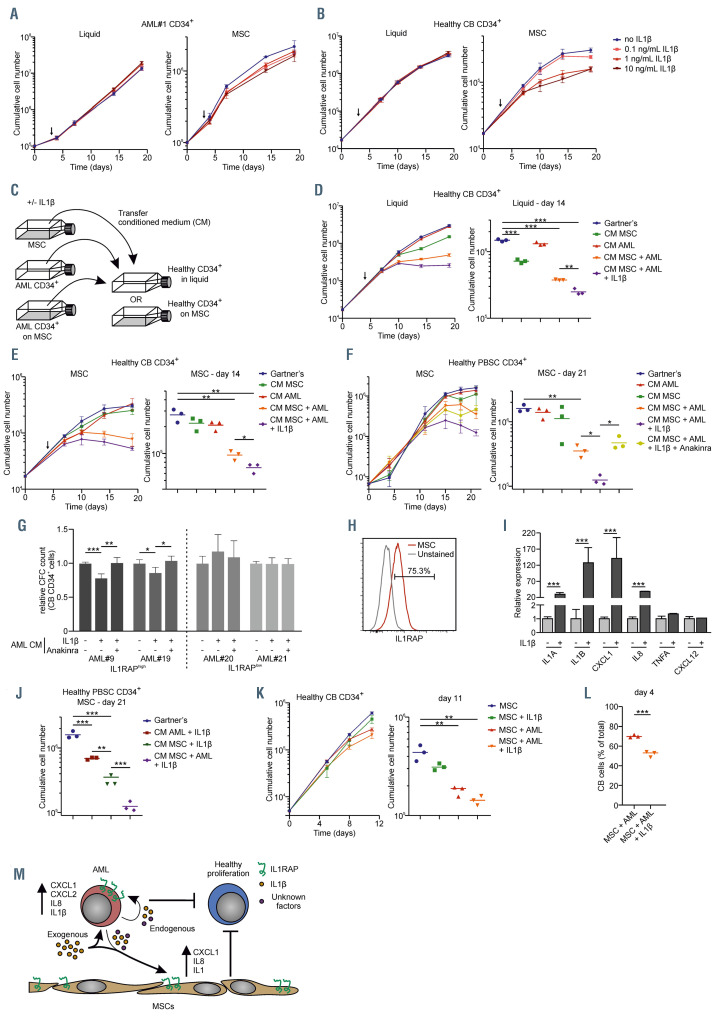
The IL1-IL1RAP signaling pathway affects normal hematopoiesis but not acute myeloid leukemia cell growth in the context of human mesenchymal stromal cells. (A) Growth curve of acute myeloid leukemia (AML) patient 1 (AML#1) CD34+ cells in liquid culture (left) and on a stromal layer of mesenchymal stromal cells (MSC) (right) ±IL1b in different concentrations. The arrow indicates from whereon IL1b was added. (B) Growth curve of CB CD34+ cells in liquid culture (left) and on a stromal layer of MSC (right) ±IL1b in different concentrations. The arrow indicates from whereon IL1b was added. (C) Schematic overview of experimental setup of co-cultures and conditioned medium (CM) transferring. (D and E) Growth curve (left) and cumulative cell number on day 14 (right) of CB CD34+cells in liquid (D) and on a stromal layer of MSC (E) with the addition of CM from a MSC culture (CM MSC), AML#1 CD34+ liquid culture (CM AML), AML#1 CD34+ MSC co-culture (CM MSC + AML), and AML#1 CD34+ MSC co-culture with the addition of 10 ng/mL IL1b (CM MSC + AML + IL1b). CM was added at day 4 (arrow) and every following demi-population. (F) Growth curve (left) and cumulative cell number on day 21 (right) of CD34+ peripheral blood stem cells (PBSC) grown on MSC with the CM from AML#18 (co-)cultures. Treatment conditions were similar to the once described in the legend of Figure 5D and E, adding one condition including AML CD34+ co-culture with the addition of 10 ng/mL IL1b and 500 ng/mL Anakinra (CM MSC + AML + IL1b + Anakinra). CM was added at day 0 and at every following demi-population. (G) Colony-forming cell (CFC) assay of cord blood (CB) CD34+ treated with CM of IL1RAPhigh AML (AML#9 and AML#19) and IL1RAPlow AML (AML#20 and AML#21), which were cultured for 7 days on a MSC stromal-layer in the presence or absence of IL1b and Anakinra, before CM was harvested. Data of two biological duplicates are shown relative to the untreated condition. (H) Interleukin-1 receptor accessory protein (IL1RAP) expression on MSC measured by flow cytometry. (I) Quantitative real-time polymerase chain reaction of MSC stimulated with and without IL1b. Statistical analysis was performed using a Student’s t-test. (J) Cumulative cell number on day 21 of CD34+ PBSC grown on MSC (experimental setup identical to panel F) including conditions AML + IL1b and MSC + IL1b. Gartner’s and CM MSC + AML + IL1b (identical to panel F) has been added for direct comparison. (K) Growth curve (left) and cumulative cell number on day 11 (right) of CB CD34+ cells in triple co-culture with MSC and AML#16 CD34+ cells ±IL1b (L) Percentage of CB cells in triple co-culture with MSC and AML#22 ±IL1b at day 4. (M) Schematic model how AML cells might impact on normal hematopoiesis in the bone marrow niche, in part via the IL1-IL1RAP axis. Statistical analysis in all panels was performed using a Student’s t-test.* P<0.05; **P<0.01; ***P<0.001.
The IL1-IL1RAP signaling axis reduces proliferation of normal hematopoietic cells but does not affect acute myeloid leukemia cell growth
We showed that IL1RAP is often upregulated in AML cells and that the receptor is functional, inducing an inflammatory gene expression signature. However, we found no evidence for a cell-intrinsic role for the IL1- IL1RAP axis in controlling cell proliferation or survival under stress conditions. Therefore, we wondered whether the inflammatory secretome induced via the IL1-IL1RAP signaling route plays a role in inducing an inflammatory BM niche that favors AML cell proliferation over normal hematopoiesis. Therefore, IL1RAP-expressing AML CD34+ cells were grown in liquid culture conditions or on MSC, in the absence or presence of IL1b. The addition of IL1b had limited impact on the proliferative capacity of primary AML CD34+ cells, neither when grown in liquid culture conditions nor when grown in co-culture with MSC (Figure 5A). Similarly, IL1b did not affect the proliferation of CB CD34+ cells when grown in liquid culture conditions, however, a marked, dose-dependent, reduction in proliferation was noted when cultured on MSC (Figure 5B). From the MSC/AML cultures, the conditioned medium (CM) was harvested and transferred to CBderived or PB-derived CD34+ cell cultures to determine the effects on proliferation and differentiation (Figure 5C). CM was harvested every time cultures were demi-populated by taking a third of the volume and transferring it to CB- or PB-derived CD34+ cell cultures in a 1:1 ratio (v/v) (see the Online Supplementary Methods for more details)
The addition of the CM of AML#1 grown in liquid culture conditions did not affect CB CD34+ growth, neither in liquid culture conditions nor when grown on MSC (Figure 5D-E). Possibly, this relates to the high levels of TGFb, BMP and angiopoietins that are known to be secreted by stromal cells. These factors negatively impact on the cell cycle and can preserve stemness, which would coincide with the observation that CD34+ cell populations were better maintained under stromal coculture conditions (Online Supplementary Figure 5C and D). CM harvested from MSC, only marginally reduced CB cell growth in liquid culture and not when grown on MSC (Figure 5D to E). Strikingly, the CM harvested from AML#1 grown on MSC negatively impacted on normal CB proliferation, both in liquid cultures and on MSC, which was even further aggravated when the AML cultures were treated with IL1b (Figure 5 D and E; Online Supplementary Figure S5A and B). The percentage of DAPI-positive CB cells was slightly increased upon treatment with CM from AML cells, compared to the addition of Gartner’s medium (Online Supplementary Figure 5B). Similar results on cell proliferation and survival were obtained using adult PB CD34+ cells that were cultured in presence of CM from a different AML (AML#18; Figure 5F; Online Supplementary Figure 5E). Importantly, the negative phenotype could be partially rescued by addition of Anakinra, an FDAapproved inhibitor of the IL1 receptor, and consequently, the IL1RAP pathway (Figure 5F and Online Supplementary Figure S5F to H). The CB CD34+ percentage, but not absolute CD34+ cell counts, were increased 3 days after addition of CM of AML co-cultures (Online Supplementary Figure S5C and D). We observed no clear differences in the CD34+CD38+/CD34+CD38– distribution after addition of CM to CB cells grown in liquid culture or on MSC, respectively (data not shown). For a more functional HSPC analysis, we performed a CFC assay with CB CD34+ cells supplemented with CM of AML-MSC co-cultures. Two high-expressing IL1RAP (AML#9 and #19) and two low-expressing IL1RAP (AML#20 and #21) AMLs were grown for 7 days on a stromal layer of MSCs with or without IL1b/Anakinra (Online Supplementary Figure S5I). IL8 was strongly upregulated by IL1b in the IL1RAP+ AML cells (AML#19), but not in the IL1RAP– AML cells (AML #20), and Anakinra abrogated this effect (Online Supplementary Figure S5F). At day 7, CM was harvested and added to the methylcellulose mixture in a 1:2 v/v ratio. Only the addition of CM from AMLs with a high IL1RAP expression resulted in a significant reduction of colony-forming potential of CB CD34+ cells, which was completely reversed by the addition of Anakinra (Figure 5G; Online Supplementary Figure 5J).
Likely, the stromal cells play an important role in mediating the negative effects of IL1b on the proliferation of healthy CD34+ HSPCs. Surprisingly, MSCs expressed high levels of IL1RAP and were responsive to IL1b comparable to what was seen for IL1RAP+ AML cells (Figure 5H-I). Both the CM of AML cells as well as MSCs treated with IL1b affected cell proliferation of healthy HSPCs, which was further enhanced when CM from AML + MSC + IL1b was used (Figure 5J, Online Supplementary Figure 5H). This would argue that both the stromal cells as well as AML cells participate in generating an inflammatory environment. In addition, two triple co-cultures were performed with MSC, AML CD34+ cells of two IL1RAP+ AML patients (AML#16 and #22), and CB CD34+ cells (Figure 5J to K; Online Supplementary Figure S5K to L). We observed reduced CB proliferation in the presence of AML#16, which was further reduced by the addition of IL1b (Figure 5J). CB in co-culture with AML#22 without IL1b did not result in reduced CB proliferation, but this was the case when cultured in the presence of IL1b (Online Supplementary Figure S5K). We observed a slight increase of Annexin V+ cells at day 16, which might not account fully for the reduced normal hematopoiesis. These data propose a model in which interplay between AML cells and MSC results in an inflammatory secretome that impairs normal hematopoiesis (Figure 5K). This negative phenotype of normal hematopoiesis is partly IL1-induced and the addition of exogenous IL1b can aggravate this observed phenotype.
Discussion
The fate of normal and leukemic stem cells critically depends on signals arising from the BM niche.30-32 Many of them initiate signal transduction in target cells by binding to PM receptors. The identification and functional analyses of such AML-specific PM proteins will help our understanding of leukemia initiation, progression and maintenance. Here, we studied IL1RAP, which was associated with a L-GMP like signature suggesting that cells from IL1RAP+ AML patients differ significantly in their origin, metabolic state, and cell cycle state, compared IL1RAP–/low patients.33,34 High IL1RAP expression in normal karyotype AML patients showed worsened overall survival suggesting that indeed these are different AML subtypes.35 Also at the subclonal level, IL1RAP expression is associated with different biology, as an IL1RAP+ NRASmutated subclone differed significantly from an IL1RAPWT1- mutated subclone, while both subclones contained similar founder mutations.7 The IL1RAP+ NRAS-mutated subclone was strongly enriched for L-GMP and inflammatory gene signatures. The connection between dysregulation of the Ras-pathway and IL1 signaling was previously investigated in non-small cell lung cancer in the context of GATA2 dependency.36 Combined, these data indicate that IL1RAP expression can be used to distinguish distinct biological characteristics of leukemic clones.
A recent study indicated that IL1RAP can also co-dimerize with CD117 and CD135 receptors, which resulted in an amplification of survival and proliferation signaling in AML.28 In our dataset, FLT3 and CD123, but not CD117 expression, correlated well with IL1RAP expression. Stimulation of these receptors with FLT3L, IL3 and SCF showed heterogeneous downstream activation between different AML but no clear synergism was observed when FLT3, CD123 and CD117 were co-stimulated with IL1b. While the hypothesis that IL1RAP receptors can interact with other receptors and thereby affect signal transduction is certainly intriguing, further studies are required to resolve these issues.
We showed that the IL1RAP pathway was functional in primary AML CD34+ cells and could be activated by IL1b. Various pathways were activated downstream, including canonical NFkB signaling. Muto et al. showed that MDS HSPC could switch to non-canonical NFkB signaling in response to inflammation, resulting in a competitive advantage over normal HSPC.29 In general, we observed rather low levels of proteins involved in active non-canonical NFkB signaling, which was not further increased upon stimulation with IL1b suggesting that this switch might be less relevant for AML stem cells. Previously, studies showed reduced clonogenicity in AML cell lines as well as reduced engraftment capacity upon knockdown or inhibition of IL1RAP using antagonistic antibodies.28,35 We also found a reduction in CFC frequency upon knockdown of IL1RAP in THP1 cells or primary IL1RAP+ AML cells, but not in K562 or OCI-AML3 cells. It was also shown that IL1RAP-targeting antibodies resulted in reduced cell proliferation, however, we did not observe any difference in cell proliferation upon knockdown of IL1RAP.28,35 Possibly, a reduction in expression is dissimilar to an antibody-mediated block in signaling. Importantly, we did observe an IL1b–induced activation of canonical NFkB signaling in IL1RAP-expressing AML, but this did not result in NFkBdriven survival under stress conditions such as serum deprivation.
Intriguingly, the IL1b-induced inflammatory secretome of AML cells grown on MSC affected normal hematopoietic proliferation and HSPC clonogenicity, while AML cells were much less affected. This observation was specific for IL1RAP-expressing AML cells that were cultured on stromal cells as CM of AML cells and MSC alone did not affect normal hematopoiesis. We observed only a mild increase in DAPI percentage or Annexin V positivity in CB CD34+ cells, suggesting that an increase in apoptosis does not fully explain the loss of cell growth. Recently, Waclawiczek et al. showed that transcriptionally remodeled MSC, due to the presence of AML cells, resulted in suppression of HSPC but did not affect their viability.37 Along the same lines, Miraki- Moud et al. suggested that AML cells do not impair the survival of normal HSC but do inhibit their differentiation, from which HSC can recover once removed from the leukemic environment.38 Single-cell sequencing studies of the mouse BM provided a very detailed description of the cellular heterogeneity within the BM niche, which was remodeled upon stress or MLL-AF9 leukemia engraftment.39,40 This remodeling affected function and maturation of BM stromal cells resulting in the loss of signaling molecules known to be essential for normal hematopoiesis.39 Similarly, studies showed BM remodeling via exosome secretion, TGF-b, Notch and inflammatory signals.41,42 Together, these findings suggest that leukemic cells can impact on normal hematopoiesis in multiple ways. Likely, there are more proteins secreted by AML and/or MSC that also influence normal hematopoietic proliferation, as Anakinra could not fully rescue the negative phenotype. For example, Carter and colleagues showed that IL1b can result in a Cox2-dependent secretion of prostaglandin E2 (PGE2) by MSC, which ultimately resulted in b-catenin-mediated augmented chemotherapy resistance in AML cells.43 Besides inflammatory factors, GATA2 was also upregulated ~2.6-fold in AML cells upon IL1β stimulation. p38-dependent GATA2 activation has been associated with poor overall survival and increased transcriptional activation of IL1b and CXCL2.44,45 We hypothesize that this p38-dependent activation of GATA2 is part of a positive feedback loop in IL1RAP-expressing AML that results in an inflammatory niche.
The formation of an inflammatory niche possibly plays a role in the early stages of leukemia development as well. It has been shown that hematopoietic clones harboring a pre-leukemia mutation in Tet methylcytosine dioxygenase 2 (TET2) can outgrow non-mutated clones after inflammatory stress, which in turn might be aggravated by the fact that TET2 knockout mice show increased levels of inflammatory proteins including IL1b, IL-6 and chemokines including Cxcl1-3 and Pf4.46,47 Upon aging, the BM niche changes and becomes more senescent and as a result, via a senescent-associated secretory phenotype (SASP), more inflammatory.48-50 Although the data are limited, it is enticing to consider that an inflammatory BM niche might accelerate clonal expansion and that the IL1- IL1RAP signaling axis plays an important role already in early stages of leukemic initiation.
Overall, our study contributes to the understanding of the role that plasma membrane receptors play in the leukemic BM niche. Such insights might aid further development of therapies aimed at specifically targeting factors that are essential for leukemogenesis. Inhibition of the IL1-IL1RAP signaling axis might be a good therapeutic approach to reduce inflammation in the BM niche and thereby promote normal hematopoietic recovery over AML proliferation after chemotherapy.
Supplementary Material
Funding Statement
Funding: This work was supported by a grant from the European Research Council (ERC-2011-StG 281474-huLSCtargeting) awarded to JJS. The UMCG/MPDI program is acknowledged for the scholarship awarded to SS.
References
- 1.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baryawno N, Przybylski D, Kowalczyk MS, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915-1932.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizo A, Vellenga E, G. dH, Schuringa JJ. Signaling pathways in self-renewing hematopoietic and leukemic stem cells: do all stem cells need a niche? Hum Mol Genet. 2006;15 Spec No 2:R210-R219. [DOI] [PubMed] [Google Scholar]
- 4.Goulard M, Dosquet C, Bonnet D. Role of the microenvironment in myeloid malignancies. Cell Mol Life Sci. 2018;75(8):1377-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duarte D, Hawkins ED, Akinduro O, et al. Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell. 2018;22(1):64-77.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schepers K, Campbell TB, Passegue E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16(3):254-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer B, Prick J, Pruis MG, et al. Prospective isolation and characterization of genetically and functionally distinct AML subclones. Cancer Cell. 2018;34(4): 674-689.e8. [DOI] [PubMed] [Google Scholar]
- 8.Bonardi F, Fusetti F, Deelen P, van GD, Vellenga E, Schuringa JJ. A Proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Mol Cell Proteomics. 2013;12(3):626-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaras M, Johnels P, Hansen N, et al. Isolation and killing of candidate chronic myeloid leukemia stem cells by antibody targeting of IL-1 receptor accessory protein. Proc Natl Acad Sci U S A. 2010;107(37):16280-16285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barreyro L, Will B, Bartholdy B, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120(6):1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Askmyr M, Agerstam H, Hansen N, et al. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 2013;121(18):3709-3713. [DOI] [PubMed] [Google Scholar]
- 12.Agerstam H, Hansen N, von Palffy S, et al. IL1RAP antibodies block IL-1-induced expansion of candidate CML stem cells and mediate cell killing in xenograft models. Blood. 2016;128(23):2683-2693. [DOI] [PubMed] [Google Scholar]
- 13.Agerstam H, Karlsson C, Hansen N, et al. Antibodies targeting human IL1RAP (IL1R3) show therapeutic effects in xenograft models of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2015; 112(34):10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010; 10(2):89-102. [DOI] [PubMed] [Google Scholar]
- 15.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39(6):1003-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281(1):8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosman MC, Schepers H, Jaques J, et al. The TAK1-NF-kappaB axis as therapeutic target for AML. Blood. 2014;124(20):3130-3140. [DOI] [PubMed] [Google Scholar]
- 18.Smith MA, Choudhary GS, Pellagatti A, et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat Cell Biol. 2019;21(5):640-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasparini C, Celeghini C, Monasta L, Zauli G. NF-kappaB pathways in hematological malignancies. Cell Mol Life Sci. 2014; 71(11):2083-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li AJ, Calvi LM. The microenvironment in myelodysplastic syndromes: niche-mediated disease initiation and progression. Exp Hematol. 2017;55:3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang T, Haman A, Goncalves O, et al. Interleukin 1 enhances growth factordependent proliferation of the clonogenic cells in acute myeloblastic leukemia and of normal human primitive hemopoietic precursors. J Exp Med. 1988;168(2):463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delwel R, van Buitenen C, Salem M, et al. Interleukin-1 stimulates proliferation of acute myeloblastic leukemia cells by induction of granulocyte-macrophage colonystimulating factor release. Blood. 1989; 74(2):586-593. [PubMed] [Google Scholar]
- 23.Bradbury D, Bowen G, Kozlowski R, Reilly I, Russell N. Endogenous interleukin-1 can regulate the autonomous growth of the blast cells of acute myeloblastic leukemia by inducing autocrine secretion of GMCSF. Leukemia. 1990;4(1):44-47. [PubMed] [Google Scholar]
- 24.de Jonge HJ, Woolthuis CM, Vos AZ, et al. Gene expression profiling in the leukemic stem cell-enriched CD34(+) fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia. 2011;25(12):1825-1833. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013; 368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capitano ML, Griesenauer B, Guo B, Cooper S, Paczesny S, Broxmeyer HE. The IL-33 Receptor/ST2 acts as a positive regulator of functional mouse bone marrow hematopoietic stem and progenitor cells. Blood Cells Mol Dis. 2020;84:102435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281(1):8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell K, Barreyro L, Todorova TI, et al. IL1RAP potentiates multiple oncogenic signaling pathways in AML. J Exp Med. 2018;215(6):1709-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muto T, Walker CS, Choi K, et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat Immunol. 2020; 21(5): 535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schepers K, Campbell TB, Passegue E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16(3):254-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nat Rev Immunol. 2017;17(9):573-590. [DOI] [PubMed] [Google Scholar]
- 32.Mendez-Ferrer S, Bonnet D, Steensma DP, et al. Bone marrow niches in haematological malignancies. Nat Rev Cancer. 2020; 20(5):285-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goardon N, Marchi E, Atzberger A, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19(1):138-152. [DOI] [PubMed] [Google Scholar]
- 34.Ye M, Zhang H, Yang H, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell. 2015;17(5):611-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barreyro L, Will B, Bartholdy B, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120(6):1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar MS, Hancock DC, Molina-Arcas M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven nonsmall cell lung cancer. Cell. 2012; 149(3):642-655. [DOI] [PubMed] [Google Scholar]
- 37.Waclawiczek A, Hamilton A, Rouault-Pierre K, et al. Mesenchymal niche remodeling impairs hematopoiesis via stanniocalcin 1 in acute myeloid leukemia. J Clin Invest. 2020;130(6):3038-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miraki-Moud F, Anjos-Afonso F, Hodby KA, et al. Acute myeloid leukemia does not deplete normal hematopoietic stem cells but induces cytopenias by impeding their differentiation. Proc Natl Acad Sci U S A. 2013;110(33):13576-13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baryawno N, Przybylski D, Kowalczyk MS, et al. A cellular taxonomy of the bone marrow stroma in Hhmeostasis and leukemia. Cell. 2019;177(7):1915-1932.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tikhonova AN, Dolgalev I, Hu H, et al. The bone marrow microenvironment at singlecell resolution. Nature. 2019;569(7755):222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar B, Garcia M, Weng L, et al. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia. 2018;32(3):575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schepers K, Pietras EM, Reynaud D, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a selfreinforcing leukemic niche. Cell Stem Cell. 2013;13(3):285-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter BZ, Mak PY, Wang X, et al. An ARCregulated IL1beta/Cox-2/PGE2/beta- Catenin/ARC circuit controls leukemiamicroenvironment interactions and confers drug resistance in AML. Cancer Res. 2019; 79(6):1165-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vicente C, Vazquez I, Conchillo A, et al. Overexpression of GATA2 predicts an adverse prognosis for patients with acute myeloid leukemia and it is associated with distinct molecular abnormalities. Leukemia. 2012;26(3):550-554. [DOI] [PubMed] [Google Scholar]
- 45.Katsumura KR, Ong IM, DeVilbiss AW, Sanalkumar R, Bresnick EH. GATA factordependent positive-feedback circuit in acute myeloid leukemia cells. Cell Rep. 2016;16(9):2428-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai Z, Kotzin JJ, Ramdas B, et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 2018;23(6):833-849.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gnani D, Crippa S, Della Volpe L, et al. An early-senescence state in aged mesenchymal stromal cells contributes to hematopoietic stem and progenitor cell clonogenic impairment through the activation of a proinflammatory program. Aging Cell. 2019; 18(3):e12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farr JN, Xu M, Weivoda MM, et al. Targeting cellular senescence prevents agerelated bone loss in mice. Nat Med. 2017; 23(9):1072-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014; 20(8):833-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.