Abstract
A total of 63 clinical isolates of Proteus mirabilis collected over a 19-month period were typed by the Dienes test and ribotyping. Ribotyping was performed using the fully automated RiboPrinter Microbial Characterization System (Qualicon, Wilmington, Del.). Isolates that were indistinguishable by the Dienes test and/or ribotyping were characterized further by pulsed-field gel electrophoresis (PFGE). Most of the isolates represented unique strains as judged by the Dienes test and ribotyping. Forty isolates represented 40 different ribotypes and Dienes types. The remaining 23 isolates were grouped into 13 Dienes types, 12 ribotypes, and 14 PFGE types. The index of discrimination was 0.980 for the Dienes test, 0.979 for ribotyping, and 0.992 for PFGE. Both the Dienes test and ribotyping are useful methods for identifying individual strains of P. mirabilis. The Dienes test is simple, inexpensive, and easy to perform. It can be performed in virtually any laboratory and should be used in the initial epidemiologic characterization of P. mirabilis isolates.
Proteus mirabilis is the clinically most important species within the tribe Protease (1). P. mirabilis accounts for up to 10% of uncomplicated urinary tract infections, is the fifth most common cause of nosocomial urinary tract infections, and may also cause wound infections and sepsis in hospitalized individuals (1, 4, 4a, 8, 11). Importantly, nosocomial strains of P. mirabilis may manifest resistance to several antimicrobial agents including extended-spectrum cephalosporins, fluoroquinolones, and aminoglycosides (4a). Transmission of resistant strains of P. mirabilis in hospitals has been documented by ribotyping and pulsed-field gel electrophoresis (PFGE) (4a).
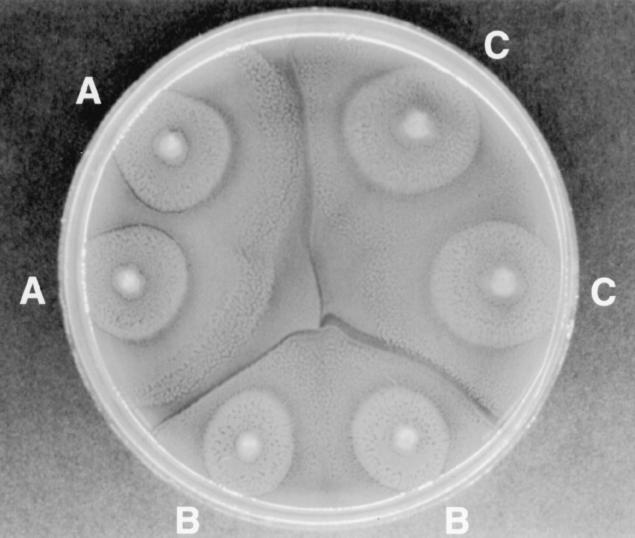
One characteristic feature of P. mirabilis is its ability to swarm over the surface of agar medium. This unique feature was used many years ago to develop a test, the Dienes mutual inhibition test, for identification of unique strains within the species (3). The Dienes test for strain discrimination of P. mirabilis is based on the mutual inhibition of two different strains as they swarm towards one another on an agar surface (3, 12, 13). If the two strains are genetically distinct, a clear line of demarcation will form as the swarming edge of one strain meets the other (Fig. 1). If the two strains are related or identical, there is no mutual inhibition and the swarming edges merge with no visible line of demarcation (Fig. 1). Thus, the test can be used as a simple means of determining whether two or more isolates of P. mirabilis are the same or different (2, 5, 12, 13, 16). The Dienes test has been used as an epidemiologic typing method to detect cross-infection due to P. mirabilis (12–14); however, the discriminatory power of the test as a strain marker has never been compared to modern molecular typing methods such as ribotyping or PFGE.
FIG. 1.
Dienes test showing three different strains, A, B, and C. Note the absence of Dienes lines between identical strains A and A, B and B, and C and C, respectively. Likewise, note the distinct lines of demarcation between strains A, B, and C.
In the present study, a collection of 63 clinical isolates of P. mirabilis was used to evaluate the Dienes test in comparison to ribotyping performed using the automated RiboPrinter Microbial Characterization System (Qualicon, Wilmington, Del.). Isolates that were indistinguishable by the Dienes test and/or ribotyping were further characterized by PFGE. The discriminatory power of the Dienes test and of ribotyping was assessed.
MATERIALS AND METHODS
Bacterial strains.
A collection of 63 clinical isolates of P. mirabilis was studied. This collection consisted of consecutive clinical isolates (one per patient) obtained from specimens of urine, sputum, wounds, blood, and other normally sterile body fluids submitted for culture to the Clinical Microbiology Laboratory of the University of Iowa Hospitals and Clinics (Iowa City, Iowa) between January 1997 and August 1999. All isolates were indole negative and produced typical swarming growth on blood agar. Identification to species level was performed using the Vitek Gram-Negative Identification System (bioMerieux Vitek, Hazelwood, Mo.). All isolates were stored on agar slants at room temperature until used in the study.
Dienes test.
The Dienes test was performed as described by Bale and Hollis (2). Isolates were subcultured from agar slants into sheep blood agar plates (Remel, Lenexa, Kans.) and incubated at 35°C for 24 h. Isolates to be compared were spot inoculated from the original blood agar plate onto the surface of a new blood agar plate. Between three and six different isolates were tested on a single plate (Fig. 1). Inoculated plates were incubated at 35°C in air for 16 to 18 h before reading. The plates were then examined with reflected light for the presence or absence of distinct “Dienes lines” forming between each pair of isolates (Fig. 1). The presence of Dienes lines between two isolates indicated that the isolates were unrelated and thus represented different Dienes types. The lack of Dienes lines indicated that the isolates were indistinguishable and represented the same Dienes type. Each isolate was tested against all other isolates in the collection and by this process was designated Dienes type A, B, C, and so on through type BC (Table 1).
TABLE 1.
Dienes test, ribotype, and PFGE typing data for 63 clinical isolates of P. mirabilis
| Dienes type | No. of isolates | Ribotype (n) | PFGE type (n) |
|---|---|---|---|
| A | 2 | 855.4 | 1 |
| 855.5 | 2 | ||
| C | 2 | 855.7 (2) | 4 (2) |
| E | 2 | 637.8 | 9 |
| 637.9 | 11 | ||
| P | 2 | 863.5 (2) | 10, 11 |
| U | 3 | 855.5 (3) | 12 |
| 13 (2) | |||
| W | 4 | 235.7 | 14 |
| 235.8 | 15 | ||
| 235.9 | 14 | ||
| 235.10 | 15 | ||
| AG | 2 | 855.7 (2) | 4 (2) |
| AH | 1 | 855.7 | 4 |
| AI | 1 | 855.7 | 4 |
| AJ | 1 | 858.2 | 6 |
| AK | 1 | 858.2 | 8 |
| AS | 1 | 851.8 | 3 |
| AT | 1 | 851.8 | 5 |
| 40 different types | 40 | 40 different types |
Ribotyping.
Ribotyping was performed using the RiboPrinter Microbial Characterization System (Qualicon) according to the manufacturer's instructions and as described earlier (6, 10). Isolates were inoculated into tubes containing lysis buffer, placed in a heating block at 80°C for 30 min, and then transferred to the RiboPrinter instrument. Within the RiboPrinter, the remaining steps were entirely automated including cleavage of the DNA using the restriction enzyme EcoRI, size separation using gel electrophoresis, and modified Southern blotting. The DNA fragments were then hybridized with a labeled DNA probe derived from the Escherichia coli rrnB rRNA operon. The bands were detected using a chemiluminescent substrate. An image was captured using a customized charge-coupled device camera and electronically transferred to the systems computer. Each lane of sample data was normalized to a standard marker set. This normalized output was compared with all previously determined samples and reference patterns (6). Similarity coefficients were calculated based upon both band position and relative banding intensity. Isolates were judged to have the same ribotype if the similarity coefficient between their patterns was ≥0.93.
PFGE.
Isolates with the same Dienes type and/or the same ribotype were evaluated further using PFGE performed as described by Pfaller et al. (9). Genomic DNA in agarose was digested with SfiI restriction enzyme, and the resulting fragments were separated by electrophoresis in 1% agarose on the CHEF-DRII (Bio-Rad, Richmond, Calif.) with the following conditions: 200 V for 23 h at switch times ramped from 5 to 40 s. Strains were considered different by PFGE if more than three bands were different (6, 15).
Calculations of discrimination index (DI).
Hunter's generalized formula was used to calculate the DI for the Dienes test, ribotyping, and PFGE (7). The DI is based on the probability that two unrelated strains sampled from the test population will be placed into different groups.
RESULTS AND DISCUSSION
The 63 isolates constituted 53 different Dienes types and 52 different ribotype patterns (Table 1 and Fig. 1 and 2). Forty isolates represented 40 distinctly different strains based upon both Dienes test and ribotype profiles. The remaining 23 isolates included 13 different Dienes types and 12 different ribotypes. Further analysis of these isolates using PFGE identified 14 different PFGE types (Table 1 and Fig. 3). Four clusters of two to four isolates each (10 isolates total) identified by the Dienes test (Dienes types A, E, P, and W) were found to include nine different strains by ribotyping and eight different strains by PFGE. Conversely, the six isolates with ribotype 855.7 and PFGE type 4 represented Dienes types C (two isolates), AG (two isolates), AH (one isolate), and AI (one isolate). One additional cluster of three isolates defined by Dienes type U and ribotype 855.5 contained two isolates with PFGE type 13 and one with PFGE type 12. Finally, two clusters of two isolates each defined by ribotypes 858.2 and 851.8 contained isolates with four different Dienes types and four different PFGE types.
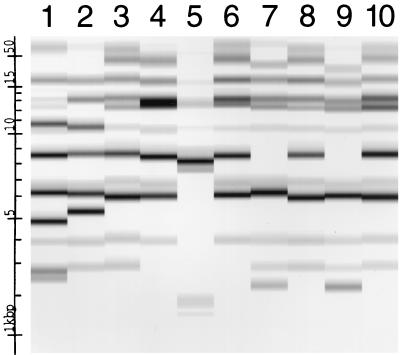
FIG. 2.
Ribotype profiles of 10 different isolates of P. mirabilis. Lane 1, ribotype 851.4; lane 2, ribotype 851.8; lanes 3, 8, and 10, ribotype 855.7; lanes 4 and 6, ribotype 858.2; lane 5, ribotype 859.5; lanes 7 and 9, ribotype 637.8. Molecular size markers in terms of kilobase pairs are shown at the left of the figure.
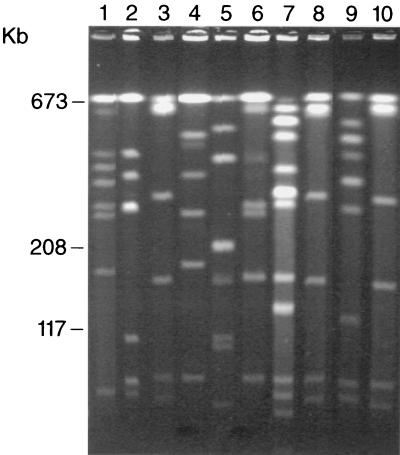
FIG. 3.
PFGE profiles of 10 different isolates of P. mirabilis (same order as in Fig. 2). Lane 1, PFGE type 16; lane 2, PFGE type 3; lanes 3, 8, and 10, PFGE type 4; lane 4, PFGE type 8; lane 5, PFGE type 17; lane 6, PFGE type 6; lane 7, PFGE type 11; lane 9, PFGE type 9.
Both the Dienes test and ribotyping were highly discriminating, with the largest cluster identified by each method constituting only 6.3% (Dienes) to 9.5% (ribotyping) of the isolates (Table 2). The discriminatory power of each method was essentially the same, with DIs of 0.980 and 0.979 for the Dienes test and ribotyping, respectively (Table 2). Although PFGE was performed on only a selected group of isolates, the DI of this method was 0.992 (data not shown) consistent with previous reports documenting the excellent discriminatory power of this method (6).
TABLE 2.
DIs of typing methods used to discriminate among 63 P. mirabilis strains
| Typing method | No. of types | Size (% of strains in largest cluster) | DI |
|---|---|---|---|
| Dienes | 53 | 6.3 | 0.980 |
| Ribotype | 52 | 9.5 | 0.979 |
The goal of this study was to evaluate the usefulness of the Dienes test as an epidemiological typing method for P. mirabilis and to provide the first comparison of this phenotypic typing method with DNA-based typing methods such as ribotyping and PFGE. The discriminatory power of each method was determined with a collection of clinical isolates which were likely to be heterogeneous.
Both the Dienes test and ribotyping typed all isolates, and their discriminatory powers were essentially identical. The discriminatory power of a typing method is defined as its ability to distinguish between unrelated strains and is determined by the number of types defined by the test method and the relative frequencies of those types (7). The DI as described by Hunter (7) provides a means of comparing different typing methods and is based on the probability that two unrelated isolates would be placed into two different typing groups. A DI of >0.90 is desirable if typing results are to be interpreted with confidence (7).
Since most of the strains (84%) had different Dienes types, it is obvious that this simple phenotypic typing method was very discriminatory (DI, 0.980). Among the 63 clinical isolates, only three clusters (two isolates in each cluster) of strains with identical patterns (Dienes type-ribotype-PFGE type) were found. Whether these isolates were epidemiologically linked is unknown, but given the fact that they are indistinguishable by more than one technique and that PFGE is known to be highly discriminatory and capable of identifying linked isolates, these isolates likely represent small clusters of nosocomial transmission.
In contrast with ribotyping and PFGE, the Dienes test is extremely simple to perform and interpret. It takes advantage of easily observed properties of P. mirabilis and requires only an agar plate and an incubator, allowing it to be performed in virtually any laboratory. As shown in the present study, the different Dienes types generally represent strains that also differ at the genetic level. Although several of the small clusters of isolates identified by the Dienes test were shown subsequently to have different DNA profiles, the excellent discriminatory power of this typing method makes it a very practical approach for typing of P. mirabilis. Isolates that are the same Dienes type may require further study using ribotyping or PFGE for optimal strain typing.
In conclusion, the Dienes test is a simple and useful method for epidemiological typing of P. mirabilis. Both ribotyping and PFGE are also highly discriminatory methods; however, the ease of performing the Dienes test coupled with excellent discriminatory power makes it the method of choice for initial epidemiologic characterization of P. mirabilis isolates.
ACKNOWLEDGMENT
We acknowledge the excellent secretarial skills of Kay L. Meyer.
REFERENCES
- 1.Aleksic S, Bockemuhl J. Yersinia and other Enterobacteriaceae. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 483–496. [Google Scholar]
- 2.Bale M J, Hollis R J. Characterization of organisms for epidemiologic purposes: serotyping, pyocin typing, and Dienes test. In: Isenberg H D, editor. Clinical microbiology procedures handbook. Vol. 2. Washington, D.C.: American Society for Microbiology; 1992. pp. 11.14.1–11.14.2. [Google Scholar]
- 3.Dienes L. Reproductive processes in Proteus cultures. Proc Soc Exp Biol Med. 1946;63:265–270. doi: 10.3181/00379727-63-15570. [DOI] [PubMed] [Google Scholar]
- 4.Doern G V, Jones R N, Pfaller M A, Kugler K C, Beach M L The SENTRY Study Group (North America) Bacterial pathogens isolated from patients with skin and soft tissue infections: frequency of occurrence and antimicrobial susceptibility patterns from the SENTRY Antimicrobial Surveillance Program (United States and Canada, 1997) Diagn Microbiol Infect Dis. 1999;34:65–72. doi: 10.1016/s0732-8893(98)00162-x. [DOI] [PubMed] [Google Scholar]
- 4a.Gales, A. C., R. N. Jones, K. A. Gordon, H. S. Sader, W. W. Wilke, M. L. Beach, M. A. Pfaller, G. V. Doern, and The SENTRY Study Group (Latin America). 2000. Activity and spectrum of 22 antimicrobial agents tested against urinary tract infection pathogens in hospitalized patients in Latin America: report from the second year of the SENTRY Antimicrobial Surveillance Program (1998). J. Antimicrob. Chemother., in press. [DOI] [PubMed]
- 5.Hickman F W, Farmer J J., III Differentiation of Proteus mirabilis by bacteriophage typing and the Dienes reaction. J Clin Microbiol. 1976;3:350–358. doi: 10.1128/jcm.3.3.350-358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollis R J, Bruce J L, Fritschel S J, Pfaller M A. Comparative evaluation of an automated ribotyping instrument versus pulsed-field gel electrophoresis for epidemiological investigation of clinical isolates. Diagn Microbiol Infect Dis. 1999;34:263–268. doi: 10.1016/s0732-8893(99)00033-4. [DOI] [PubMed] [Google Scholar]
- 7.Hunter D. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28:1903–1905. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones R N, Kugler K C, Pfaller M A, Winokur P L The SENTRY Surveillance Group, North America. Characteristics of pathogens causing urinary tract infections in hospitals in North America: results from the SENTRY Antimicrobial Surveillance Program, 1997. Diagn Microbiol Infect Dis. 1999;35:55–63. doi: 10.1016/s0732-8893(98)00158-8. [DOI] [PubMed] [Google Scholar]
- 9.Pfaller M A, Hollis R J, Sader H S. Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis. In: Isenberg H D, editor. Clinical microbiology procedures handbook, suppl. 1. Washington, D.C.: American Society for Microbiology; 1994. pp. 10.5.c.1–10.5.c.12. [Google Scholar]
- 10.Pfaller M A, Wendt C, Hollis R J, Wenzel R P, Fritschel S J, Neubar J J, Herwaldt L A. Comparative evaluation of an automated ribotyping system versus pulsed field gel electrophoresis for epidemiological typing of clinical isolates of Escherichia coli and Pseudomonas aeruginosa from patients with recurrent gram-negative bacteremia. Diagn Microbiol Infect Dis. 1996;25:1–8. doi: 10.1016/0732-8893(96)00082-x. [DOI] [PubMed] [Google Scholar]
- 11.Pfaller M A, Jones R N, Doern G V, Kugler K The SENTRY Participants Group. Bacterial pathogens isolated from patients with blood stream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY Antimicrobial Surveillance Program (United States and Canada, 1997) Antimicrob Agents Chemother. 1998;42:1762–1770. doi: 10.1128/aac.42.7.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senior B W. Typing of Proteus strains by proticene production and sensitivity. J Med Microbiol. 1977;10:7–17. doi: 10.1099/00222615-10-1-7. [DOI] [PubMed] [Google Scholar]
- 13.Skirrow M B. The Dienes (mutual inhibition) test in the investigation of Proteus infections. J Med Microbiol. 1969;2:471–477. doi: 10.1099/00222615-2-4-471. [DOI] [PubMed] [Google Scholar]
- 14.Story P. Proteus infection in hospitals. J Pathol Bacteriol. 1954;68:55–62. doi: 10.1002/path.1700680107. [DOI] [PubMed] [Google Scholar]
- 15.Tenover F C, Arbeit R D, Goering R V, Mickelson P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tracy O, Thomson E J. An evaluation of three methods of typing organisms of the genus Proteus. J Clin Pathol. 1972;25:69–72. doi: 10.1136/jcp.25.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]